New DNA Methylation Signals for Malignant Pleural Mesothelioma Risk Assessment
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
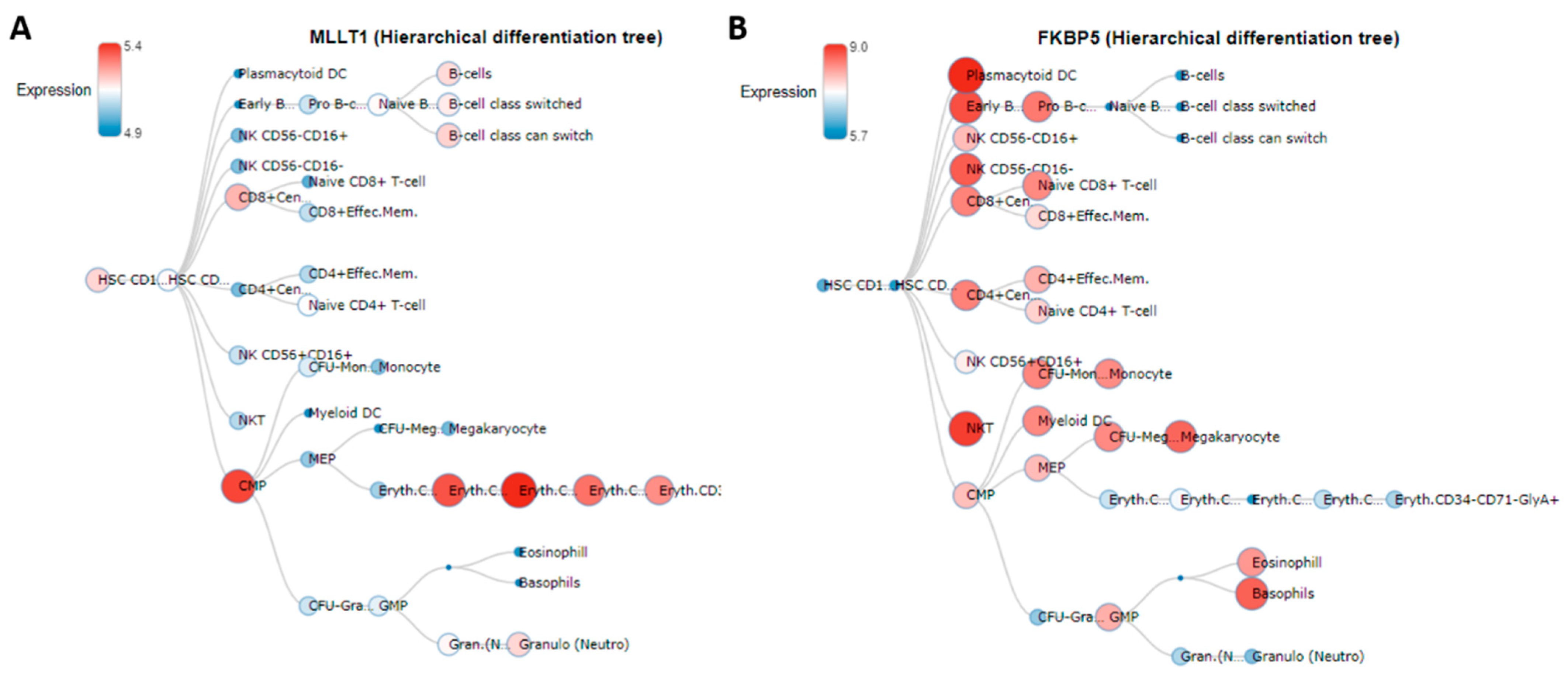
2.1. Epigenome-Wide Association Study (EWAS)
2.2. Receiver Operation Characteristics (ROC) for Case-Control Discrimination
2.3. Interaction Analysis
2.4. Validation and Replication
3. Discussion
Limitation of the Study
4. Material and Methods
4.1. Study Population
4.2. Exposure Assessment
4.3. Blood DNAm Analysis and Beta-Value Extraction
4.4. Batch Effect, Population Stratification and White Blood Cells Estimations
4.5. Statistical Analyses
4.5.1. Epigenome-Wide Association Study
4.5.2. Statistical Power
4.5.3. Interaction Analysis
4.6. Validation and Replication
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sekido, Y. Molecular pathogenesis of malignant mesothelioma. Carcinogenesis 2013, 34, 1413–1419. [Google Scholar] [CrossRef]
- Rossini, M.; Rizzo, P.; Bononi, I.; Clementz, A.; Ferrari, R.; Martini, F.; Tognon, M.G. New Perspectives on Diagnosis and Therapy of Malignant Pleural Mesothelioma. Front. Oncol. 2018, 8, 91. [Google Scholar] [CrossRef] [PubMed]
- Furuya, S.; Chimed-Ochir, O.; Takahashi, K.; David, A.; Takala, J. Global Asbestos Disaster. Int. J. Environ. Res. Public Health 2018, 15, 1000. [Google Scholar] [CrossRef] [PubMed]
- Straif, K.; Benbrahim-Tallaa, L.; Baan, R.; Grosse, Y.; Secretan, B.; El Ghissassi, F.; Bouvard, V.; Guha, N.; Freeman, C.; Galichet, L.; et al. A review of human carcinogens—Part C: Metals, arsenic, dusts, and fibres. Lancet Oncol. 2009, 10, 453–454. [Google Scholar] [CrossRef]
- Jaurand, M.C. Mechanisms of fiber-induced genotoxicity. Environ. Health Perspect. 1997, 105, 1073–1084. [Google Scholar]
- Kelsey, K.T.; Yano, E.; Liber, H.L.; Little, J.B. The in vitro genetic effects of fibrous erionite and crocidolite asbestos. Br. J. Cancer 1986, 54, 107–114. [Google Scholar] [CrossRef]
- Iwatsubo, Y.; Pairon, J.C.; Boutin, C.; Ménard, O.; Massin, N.; Caillaud, D.; Orlowski, E.; Galateau-Salle, F.; Bignon, J.; Brochard, P. Pleural mesothelioma: Dose-response relation at low levels of asbestos exposure in a French population-based case-control study. Am. J. Epidemiol. 1998, 148, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Howel, D.; Arblaster, L.; Swinburne, L.; Schweiger, M.; Renvoize, E.; Hatton, P. Routes of asbestos exposure and the development of mesothelioma in an English region. Occup. Environ. Med. 1997, 54, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Kanherkar, R.R.; Bhatia-Dey, N.; Csoka, A.B. Epigenetics across the human lifespan. Front. Cell Dev. Biol. 2014, 2, 49. [Google Scholar] [CrossRef] [PubMed]
- Ferrante, D.; Bertolotti, M.; Todesco, A.; Mirabelli, D.; Terracini, B.; Magnani, C. Cancer mortality and incidence of mesothelioma in a cohort of wives of asbestos workers in Casale Monferrato, Italy. Environ. Health Perspect. 2007, 115, 1401–1405. [Google Scholar] [CrossRef]
- Guarrera, S.; Viberti, C.; Cugliari, G.; Allione, A.; Casalone, E.; Betti, M.; Ferrante, D.; Aspesi, A.; Casadio, C.; Grosso, F.; et al. Peripheral Blood DNA Methylation as Potential Biomarker of Malignant Pleural Mesothelioma in Asbestos-Exposed Subjects. J. Thorac. Oncol. 2019, 14, 527–539. [Google Scholar] [CrossRef]
- Matullo, G.; Guarrera, S.; Betti, M.; Fiorito, G.; Ferrante, D.; Voglino, F.; Cadby, G.; Di Gaetano, C.; Rosa, F.; Russo, A.; et al. Genetic variants associated with increased risk of malignant pleural mesothelioma: A genome-wide association study. PLoS ONE 2013, 8, e61253. [Google Scholar] [CrossRef]
- Cugliari, G.; Catalano, C.; Guarrera, S.; Allione, A.; Casalone, E.; Russo, A.; Grosso, F.; Ferrante, D.; Viberti, C.; Aspesi, A.; et al. DNA Methylation of FKBP5 as Predictor of Overall Survival in Malignant Pleural Mesothelioma. Cancers 2020, 12, 3470. [Google Scholar] [CrossRef]
- Betti, M.; Aspesi, A.; Sculco, M.; Matullo, G.; Magnani, C.; Dianzani, I. Genetic predisposition for malignant mesothelioma: A concise review. Mutat. Res. 2019, 781, 1–10. [Google Scholar] [CrossRef]
- Guo, G.; Chmielecki, J.; Goparaju, C.; Heguy, A.; Dolgalev, I.; Carbone, M.; Seepo, S.; Meyerson, M.; Pass, H.I. Whole-exome sequencing reveals frequent genetic alterations in BAP1, NF2, CDKN2A, and CUL1 in malignant pleural mesothelioma. Cancer Res. 2015, 75, 264–269. [Google Scholar] [CrossRef]
- Fasanelli, F.; Baglietto, L.; Ponzi, E.; Guida, F.; Campanella, G.; Johansson, M.; Grankvist, K.; Johansson, M.; Assumma, M.B.; Naccarati, A.; et al. Hypomethylation of smoking-related genes is associated with future lung cancer in four prospective cohorts. Nat. Commun. 2015, 15, 10192. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar]
- Bononi, A.; Napolitano, A.; Pass, H.I.; Yang, H.; Carbone, M. Latest developments in our understanding of the pathogenesis of mesothelioma and the design of targeted therapies. Expert. Rev. Respir. Med. 2015, 9, 633–654. [Google Scholar] [CrossRef] [PubMed]
- Vandermeers, F.; Neelature, S.; Costa, C.; Hubaux, R.; Cosse, J.P.; Willems, L. The role of epigenetics in malignant pleural mesothelioma. Lung Cancer 2013, 81, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tang, N.; Rishi, A.K.; Pass, H.I.; Wali, A. Methylation profile landscape in mesothelioma: Possible implications in early detection, disease progression, and therapeutic options. Methods Mol. Biol. 2015, 1238, 235–247. [Google Scholar] [PubMed]
- Goto, Y.; Shinjo, K.; Kondo, Y.; Shen, L.; Toyota, M.; Suzuki, H.; Gao, W.; An, N.; Fujii, M.; Murakami, H.; et al. Epigenetic profiles distinguish malignant pleural mesothelioma from lung adenocarcinoma. Cancer Res. 2009, 69, 9073–9082. [Google Scholar] [CrossRef] [PubMed]
- Christensen, B.C.; Houseman, E.A.; Poage, G.M.; Godleski, J.J.; Bueno, R.; Sugarbaker, D.J.; Wiencke, J.K.; Nelson, H.H.; Marsit, C.J.; Kelsey, K.T. Integrated profiling reveals a global correlation between epigenetic and genetic alterations in mesothelioma. Cancer Res. 2010, 70, 5686–5694. [Google Scholar] [CrossRef]
- Fischer, J.R.; Ohnmacht, U.; Rieger, N.; Zemaitis, M.; Stoffregen, C.; Kostrzewa, M.; Buchholz, E.; Manegold, C.; Lahm, H. Promoter methylation of RASSF1A, RARbeta and DAPK predict poor prognosis of patients with malignant mesothelioma. Lung Cancer 2006, 54, 109–116. [Google Scholar] [CrossRef]
- Matosin, N.; Halldorsdottir, T.; Binder, E.B. Understanding the molecular mechanisms underpinning gene by environment interactions in psychiatric disorders: The FKBP5 model. Biol. Psychiatry 2018, 83, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.B.; Hong, Y.; Dhe-Paganon, S.; Sup Yoon, H. FKBP family proteins: Immunophilins with versatile biological functions. Neurosignals 2008, 16, 318–325. [Google Scholar] [CrossRef]
- Li, L.; Lou, Z.; Wang, L. The role of FKBP5 in cancer aetiology and chemoresistance. Br. J. Cancer 2011, 104, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Romano, S.; D’Angelillo, A.; Romano, M.F. Pleiotropic roles in cancer biology for multifaceted proteins FKBPs. Biochim. Biophys. Acta 2015, 1850, 2061–2068. [Google Scholar] [CrossRef]
- Staibano, S.; Mascolo, M.; Ilardi, G.; Siano, M.; De Rosa, G. Immunohistochemical analysis of FKBP51 in human cancers. Curr. Opin. Pharmacol. 2011, 11, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Zannas, A.S.; Jia, M.; Hafner, K.; Baumert, J.; Wiechmann, T.; Pape, J.C.; Arloth, J.; Ködel, M.; Martinelli, S.; Roitman, M.; et al. Epigenetic upregulation of FKBP5 by aging and stress contributes to NF-kappaB-driven inflammation and cardiovascular risk. Proc. Natl. Acad. Sci. USA 2019, 116, 11370–11379. [Google Scholar] [CrossRef]
- Yoshikawa, Y.; Sato, A. Biallelic germline and somatic mutations in malignant mesothelioma: Multiple mutations in transcription regulators including mSWI/SNF genes. Int. J. Cancer 2015, 136, 560–571. [Google Scholar] [CrossRef]
- Zhou, J.; Ng, Y.; Chng, W.J. ENL: Structure, function, and roles in hematopoiesis and acute myeloid leukemia. Cell Mol. Life Sci. 2018, 75, 3931–3941. [Google Scholar] [CrossRef] [PubMed]
- Canino, C.; Luo, Y.; Marcato, P.; Blandino, G.; Pass, H.I.; Cioce, M. STAT3-NFkB/DDIT3/CEBPbeta axis modulates ALDH1A3 expression in chemoresistant cell subpopulations. Oncotarget 2015, 6, 12637–12653. [Google Scholar] [CrossRef]
- Bagger, F.O.; Kinalis, S.; Rapin, N. BloodSpot: A database of healthy and malignant haematopoiesis updated with purified and single cell mRNA sequencing profiles. Nucleic Acids Res. 2019, 47, 881–885. [Google Scholar] [CrossRef] [PubMed]
- Schins, R.P.; Donaldson, K. Nuclear Factor Kappa-B Activation by Particles and Fibers. Inhal. Toxicol. 2000, 12, 317–326. [Google Scholar] [CrossRef]
- Tan, Z.; Lu, W.; Li, X.; Yang, G.; Guo, J.; Yu, H.; Li, Z.; Guan, F. Altered N-Glycan expression profile in epithelial-to-mesenchymal transition of NMuMG cells revealed by an integrated strategy using mass spectrometry and glycogene and lectin microarray analysis. J. Proteome Res. 2014, 13, 2783–2795. [Google Scholar] [CrossRef] [PubMed]
- Kale, V.P.; Hengst, J.A.; Desai, D.H.; Dick, T.E.; Choe, K.N.; Colledge, A.L.; Takahashi, Y.; Sung, S.S.; Amin, S.G.; Yun, J.K. A novel selective multikinase inhibitor of ROCK and MRCK effectively blocks cancer cell migration and invasion. Cancer Lett. 2014, 354, 299–310. [Google Scholar] [CrossRef]
- Tanrikulu, A.C.; Abakay, A.; Komek, H.; Abakay, O. Prognostic value of the lymphocyte-to-monocyte ratio and other inflammatory markers in malignant pleural mesothelioma. Environ. Health Prev. Med. 2016, 21, 304–311. [Google Scholar] [CrossRef]
- Zhao, Z.S.; Manser, E. PAK and other Rho-associated kinases—Effectors with surprisingly diverse mechanisms of regulation. Biochem. J. 2005, 386 Pt 2, 201–214. [Google Scholar] [CrossRef]
- Nishimura, Y.; Kumagai-Takei, N.; Matsuzaki, H.; Lee, S.; Maeda, M.; Kishimoto, T.; Fukuoka, K.; Nakano, T.; Otsuki, T. Functional alteration of natural killer cells and cytotoxic T lymphocytes upon asbestos exposure and in malignant mesothelioma patients. Biomed. Res. Int. 2015, 2015, 238431. [Google Scholar] [CrossRef]
- Maeda, M.; Nishimura, Y.; Kumagai, N.; Hayashi, H.; Hatayama, T.; Katoh, M.; Miyahara, N.; Yamamoto, S.; Hirastuka, J.; Otsuki, T. Dysregulation of the immune system caused by silica and asbestos. J. Immunotoxicol. 2010, 7, 268–278. [Google Scholar] [CrossRef]
- Miura, Y.; Nishimura, Y.; Katsuyama, H.; Maeda, M.; Hayashi, H.; Dong, M.; Hyodoh, F.; Tomita, M.; Matsuo, Y.; Uesaka, A.; et al. Involvement of IL-10 and Bcl-2 in resistance against an asbestos-induced apoptosis of T cells. Apoptosis 2006, 11, 1825–1835. [Google Scholar] [CrossRef]
- Cristaudo, A.; Bonotti, A.; Guglielmi, G.; Fallahi, P.; Foddis, R. Serum mesothelin and other biomarkers: What have we learned in the last decade? J. Thorac. Dis. 2018, 10, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Andujar, P.; Wang, J.; Descatha, A.; Galateau-Sallé, F.; Abd-Alsamad, I.; Billon-Galland, M.A.; Blons, H.; Clin, B.; Danel, C.; Housset, B.; et al. p16INK4A inactivation mechanisms in non-small-cell lung cancer patients occupationally exposed to asbestos. Lung Cancer 2010, 67, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Dianzani, I.; Gibello, L.; Biava, A.; Giordano, M.; Bertolotti, M.; Betti, M.; Ferrante, D.; Guarrera, S.; Betta, G.P.; Mirabelli, D.; et al. Polymorphisms in DNA repair genes as risk factors for asbestos-related malignant mesothelioma in a general population study. Mutat. Res. 2006, 599, 124–134. [Google Scholar] [CrossRef]
- Betti, M.; Ferrante, D.; Padoan, M.; Guarrera, S.; Giordano, M.; Aspesi, A.; Mirabelli, D.; Casadio, C.; Ardissone, F.; Ruffini, E.; et al. XRCC1 and ERCC1 variants modify malignant mesothelioma risk: A case-control study. Mutat. Res. 2011, 708, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Betti, M.; Casalone, E.; Ferrante, D.; Aspesi, A.; Morleo, G.; Biasi, A.; Sculco, M.; Mancuso, G.; Guarrera, S.; Righi, L.; et al. Germline mutations in DNA repair genes predispose asbestos-exposed patients to malignant pleural mesothelioma. Cancer Lett. 2017, 405, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Ferrante, D.; Mirabelli, D.; Tunesi, S.; Terracini, B.; Magnani, C. Pleural mesothelioma and occupational and non-occupational asbestos exposure: A case-control study with quantitative risk assessment. Occup. Environ. Med. 2016, 73, 147–153. [Google Scholar] [CrossRef]
- Du, P.; Zhang, X.; Huang, C.C.; Jafari, N.; Kibbe, W.A.; Hou, L.; Lin, S.M. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010, 11, 587. [Google Scholar] [CrossRef]
- Lehne, B.; Drong, A.W.; Loh, M.; Zhang, W.; Scott, W.R.; Tan, S.T.; Afzal, U.; Scott, J.; Jarvelin, M.R.; Elliott, P.; et al. A coherent approach for analysis of the Illumina HumanMethylation450 BeadChip improves data quality and performance in epigenome-wide association studies. Genome Biol. 2015, 16, 37. [Google Scholar] [CrossRef]
- Campanella, G.; Polidoro, S.; Di Gaetano, C.; Fiorito, F.; Guarrera, S.; Krogh, V.; Palli, D.; Panico, S.; Sacerdote, C.; Tumino, R.; et al. Epigenetic signatures of internal migration in Italy. Int. J. Epidemiol. 2015, 44, 1442–1449. [Google Scholar] [CrossRef]
- Di Gaetano, C.; Voglino, F.; Guarrera, S.; Fiorito, G.; Rosa, F.; Di Blasio, A.M.; Manzini, P.; Dianzani, I.; Betti, M.; Cusi, D.; et al. An overview of the genetic structure within the Italian population from genome-wide data. PLoS ONE 2012, 7, e43759. [Google Scholar] [CrossRef] [PubMed]
- Houseman, E.A.; Accomando, W.P.; Koestler, D.C.; Christensen, B.C.; Marsit, C.J.; Nelson, H.H.; Wiencke, J.K.; Kelsey, K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform. 2012, 13, 86. [Google Scholar] [CrossRef] [PubMed]
- Ehrich, M.; Nelson, M.R.; Stanssens, P.; Zabeau, M.; Liloglou, T.; Xinarianos, G.; Cantor, C.R.; Field, J.K.; van den Boom, D. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc. Natl. Acad. Sci. USA 2005, 102, 15785–15790. [Google Scholar] [CrossRef] [PubMed]
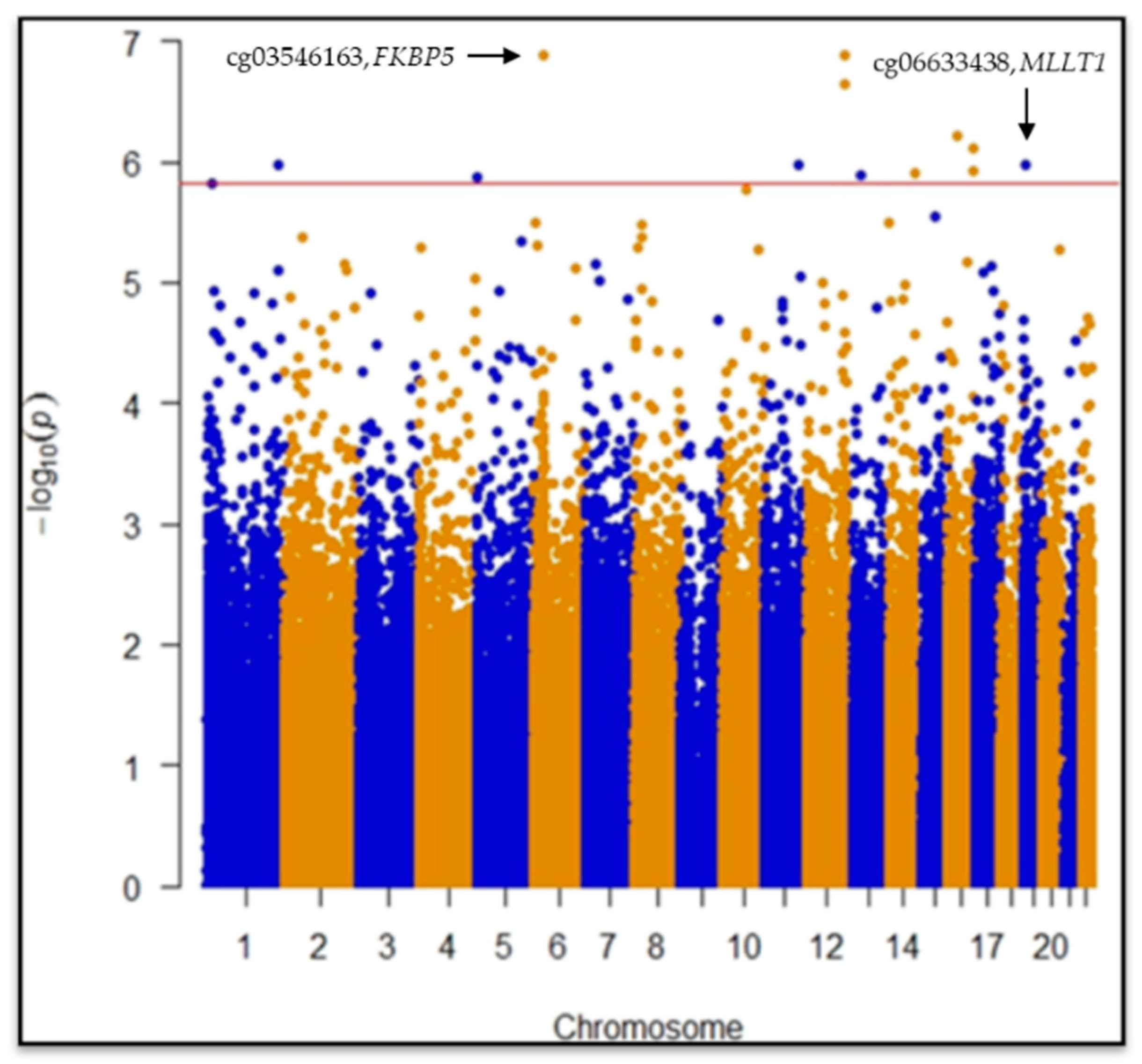
| Probe ID | Chr | Map Position | Gene Symbol | Ucsc Refgene Group | Snp Probe | Effect Size | Standard Error | p Value | Fdr | Significance |
|---|---|---|---|---|---|---|---|---|---|---|
| cg02869235 | 12 | 124726864 | rs73223527 | 0.058 | 0.011 | 1.3 × 10−7 | 0.028 | *§ | ||
| cg03546163 | 6 | 35654363 | FKBP5 | 5′UTR | −0.089 | 0.016 | 1.3 × 10−7 | 0.028 | *§₼ | |
| cg02353048 | 12 | 124718401 | 0.033 | 0.006 | 2.2 × 10−7 | 0.032 | *§ | |||
| cg06633438 | 19 | 6272158 | MLLT1 | Body | 0.069 | 0.014 | 1.0 × 10−6 | 0.049 | *§₼ | |
| cg18860329 | 13 | 43354421 | C13orf30 | TSS1500 | 0.050 | 0.010 | 1.3 × 10−6 | 0.049 | *§ | |
| cg19782190 | 14 | 103487004 | CDC42BPB | Body | 0.043 | 0.009 | 1.2 × 10−6 | 0.049 | *§ | |
| cg06834916 | 5 | 95610 | 0.037 | 0.008 | 1.4 × 10−6 | 0.049 | *§ | |||
| cg09479650 | 16 | 85578516 | rs4843449 | 0.037 | 0.007 | 1.2 × 10−6 | 0.049 | *§ | ||
| cg26680989 | 16 | 85560739 | rs80332660 | 0.036 | 0.007 | 7.6 × 10−7 | 0.049 | *§ | ||
| cg25409554 | 1 | 234871422 | 0.034 | 0.007 | 1.1 × 10−6 | 0.049 | *§ | |||
| cg01201399 | 16 | 30793389 | ZNF629 | Body | 0.030 | 0.006 | 6.1 × 10−7 | 0.049 | *§ | |
| cg17283266 | 11 | 111717611 | ALG9 | Body | −0.030 | 0.006 | 1.1 × 10−6 | 0.049 | *§ |
| Model | AUC | DeLong’s Test |
|---|---|---|
| BM (asbestos exposure, age, gender and WBCs) | 0.75 | Reference |
| BM + cg03546163 (FKBP5) | 0.89 | 2.1 × 10−7 |
| BM + cg06633438 (MLLT1) | 0.89 | 6.3 × 10−8 |
| DNAm | Asbestos Exposure | OR | Std. Error | 95% CI | p Value |
|---|---|---|---|---|---|
| cg03546163 (FKBP5) | |||||
| Hypo | Low | 2.79 | 1.51 | 1.26|6.33 | 0.013 |
| Hyper | High | 7.21 | 1.54 | 3.17|17.27 | 4.6 × 10−6 |
| Hypo | High | 20.84 | 1.59 | 8.71|53.96 | 5.5 × 10−11 |
| cg06633438 (MLLT1) | |||||
| Hyper | Low | 1.29 | 1.63 | 0.70|3.81 | 0.258 |
| Hypo | High | 7.27 | 1.55 | 3.17|17.65 | 5.3 × 10−6 |
| Hyper | High | 11.71 | 1.57 | 4.97|29.64 | 5.9 × 10−8 |
| Variable | Controls (Male 100, Female 37) | |||||
|---|---|---|---|---|---|---|
| Min | 1st Q | Median | Mean | 3rd Q | Max | |
| Age | 41.60 | 57.41 | 65.65 | 64.59 | 72.63 | 90.94 |
| Asbestos exposure | −2.71 | −0.97 | −0.48 | −0.44 | 0.09 | 1.73 |
| Monocytes | 0.00 | 0.05 | 0.06 | 0.07 | 0.08 | 0.26 |
| Granulocytes | 0.36 | 0.54 | 0.60 | 0.62 | 0.68 | 0.99 |
| Natural Killer | 0.00 | 0.04 | 0.07 | 0.08 | 0.11 | 0.29 |
| B cells | 0.00 | 0.07 | 0.09 | 0.09 | 0.11 | 0.19 |
| CD4+ T | 0.00 | 0.10 | 0.14 | 0.14 | 0.19 | 0.35 |
| CD8+ T | 0.00 | 0.03 | 0.06 | 0.07 | 0.10 | 0.23 |
| Variable | Cases (Male 113, Female 50) | |||||
|---|---|---|---|---|---|---|
| Min | 1st Q | Median | Mean | 3rd Q | Max | |
| Age | 33.90 | 61.19 | 68.68 | 67.59 | 75.17 | 90.80 |
| Asbestos exposure | −2.71 | −0.21 | 0.39 | 0.37 | 0.98 | 2.94 |
| Monocytes | 0.00 | 0.05 | 0.07 | 0.08 | 0.10 | 0.20 |
| Granulocytes | 0.37 | 0.67 | 0.74 | 0.74 | 0.81 | 1.03 |
| Natural Killer | 0.00 | 0.02 | 0.05 | 0.06 | 0.08 | 0.23 |
| B cells | 0.00 | 0.05 | 0.06 | 0.06 | 0.08 | 0.16 |
| CD4+ T | 0.00 | 0.03 | 0.07 | 0.08 | 0.11 | 0.22 |
| CD8+ T | 0.00 | 0.00 | 0.02 | 0.03 | 0.04 | 0.22 |
| Smoking Habits | Cases (163) | Controls (137) | ||
|---|---|---|---|---|
| n | % | n | % | |
| Current smokers | 29 | 17.79 | 30 | 21.90 |
| Former smokers | 54 | 33.13 | 60 | 43.80 |
| Never smokers | 75 | 46.01 | 47 | 34.31 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cugliari, G.; Allione, A.; Russo, A.; Catalano, C.; Casalone, E.; Guarrera, S.; Grosso, F.; Ferrante, D.; Sculco, M.; La Vecchia, M.; et al. New DNA Methylation Signals for Malignant Pleural Mesothelioma Risk Assessment. Cancers 2021, 13, 2636. https://doi.org/10.3390/cancers13112636
Cugliari G, Allione A, Russo A, Catalano C, Casalone E, Guarrera S, Grosso F, Ferrante D, Sculco M, La Vecchia M, et al. New DNA Methylation Signals for Malignant Pleural Mesothelioma Risk Assessment. Cancers. 2021; 13(11):2636. https://doi.org/10.3390/cancers13112636
Chicago/Turabian StyleCugliari, Giovanni, Alessandra Allione, Alessia Russo, Chiara Catalano, Elisabetta Casalone, Simonetta Guarrera, Federica Grosso, Daniela Ferrante, Marika Sculco, Marta La Vecchia, and et al. 2021. "New DNA Methylation Signals for Malignant Pleural Mesothelioma Risk Assessment" Cancers 13, no. 11: 2636. https://doi.org/10.3390/cancers13112636
APA StyleCugliari, G., Allione, A., Russo, A., Catalano, C., Casalone, E., Guarrera, S., Grosso, F., Ferrante, D., Sculco, M., La Vecchia, M., Pirazzini, C., Libener, R., Mirabelli, D., Magnani, C., Dianzani, I., & Matullo, G. (2021). New DNA Methylation Signals for Malignant Pleural Mesothelioma Risk Assessment. Cancers, 13(11), 2636. https://doi.org/10.3390/cancers13112636









