Comprehensive Plasma Metabolomic Profile of Patients with Advanced Neuroendocrine Tumors (NETs). Diagnostic and Biological Relevance
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
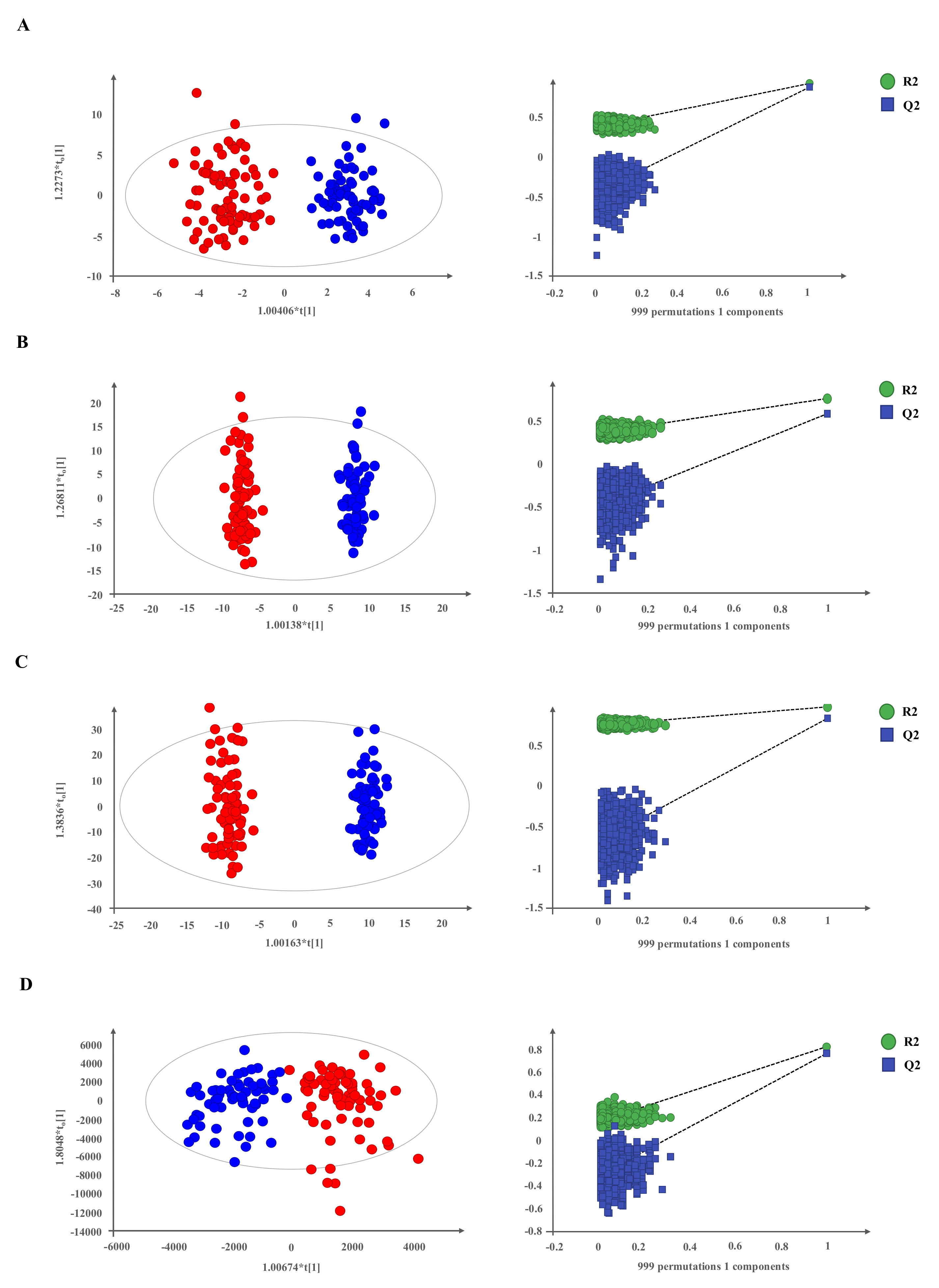
2.1. Metabolomic Profiling in Plasma of Patients with Neuroendocrine Tumors
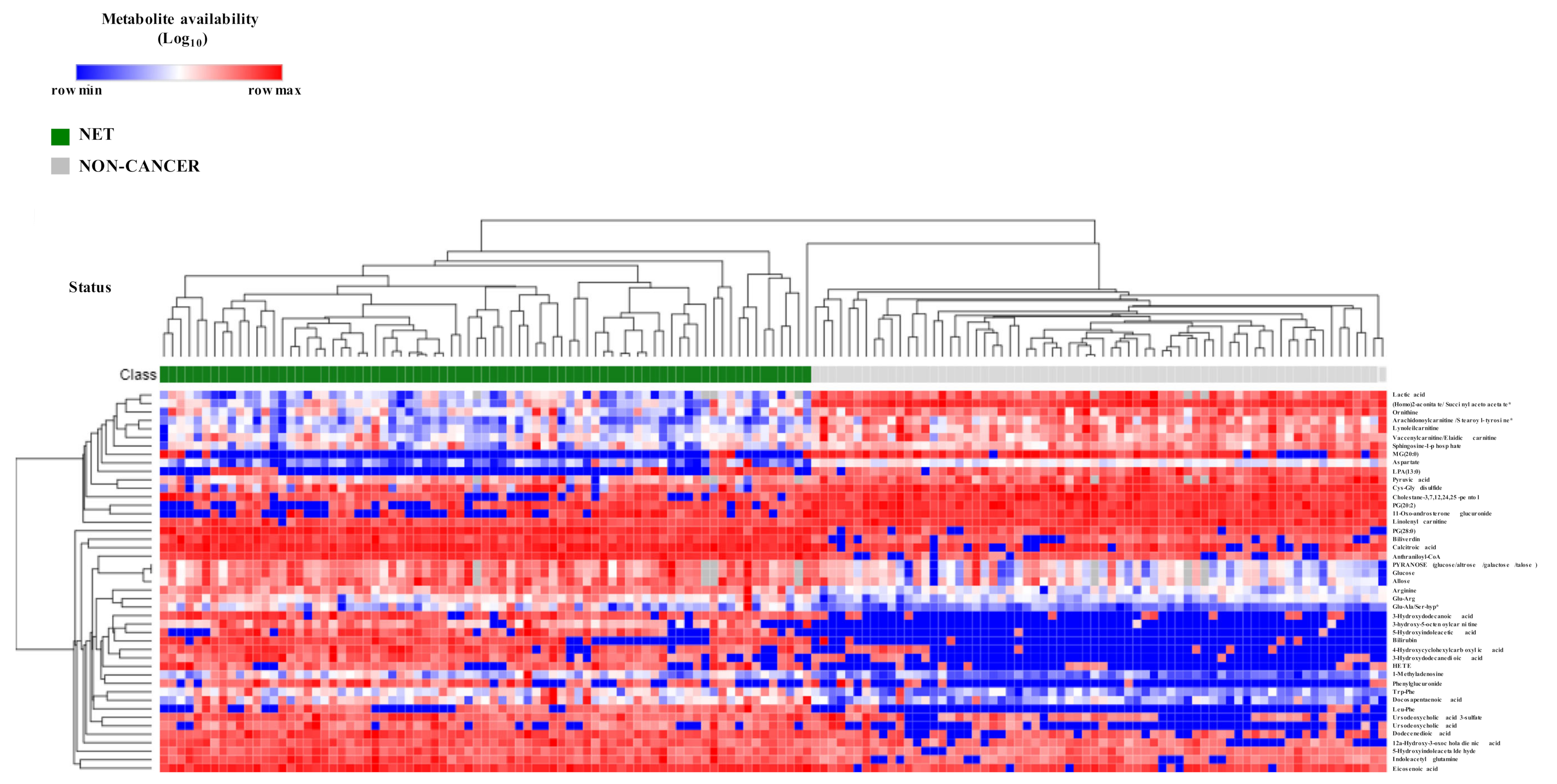
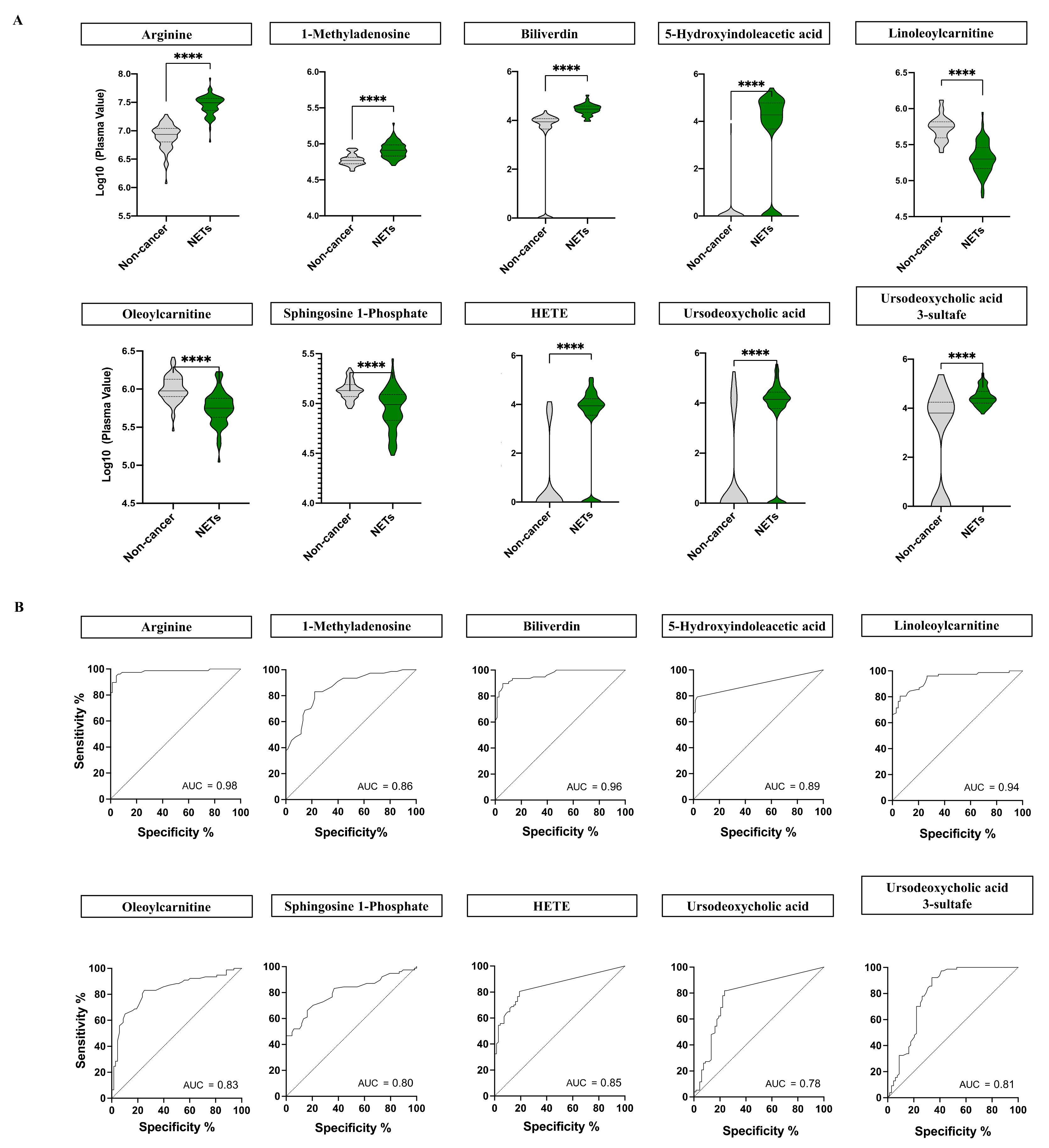
2.2. NETs Show a Particular Signature of Metabolites with Diagnostic Potential
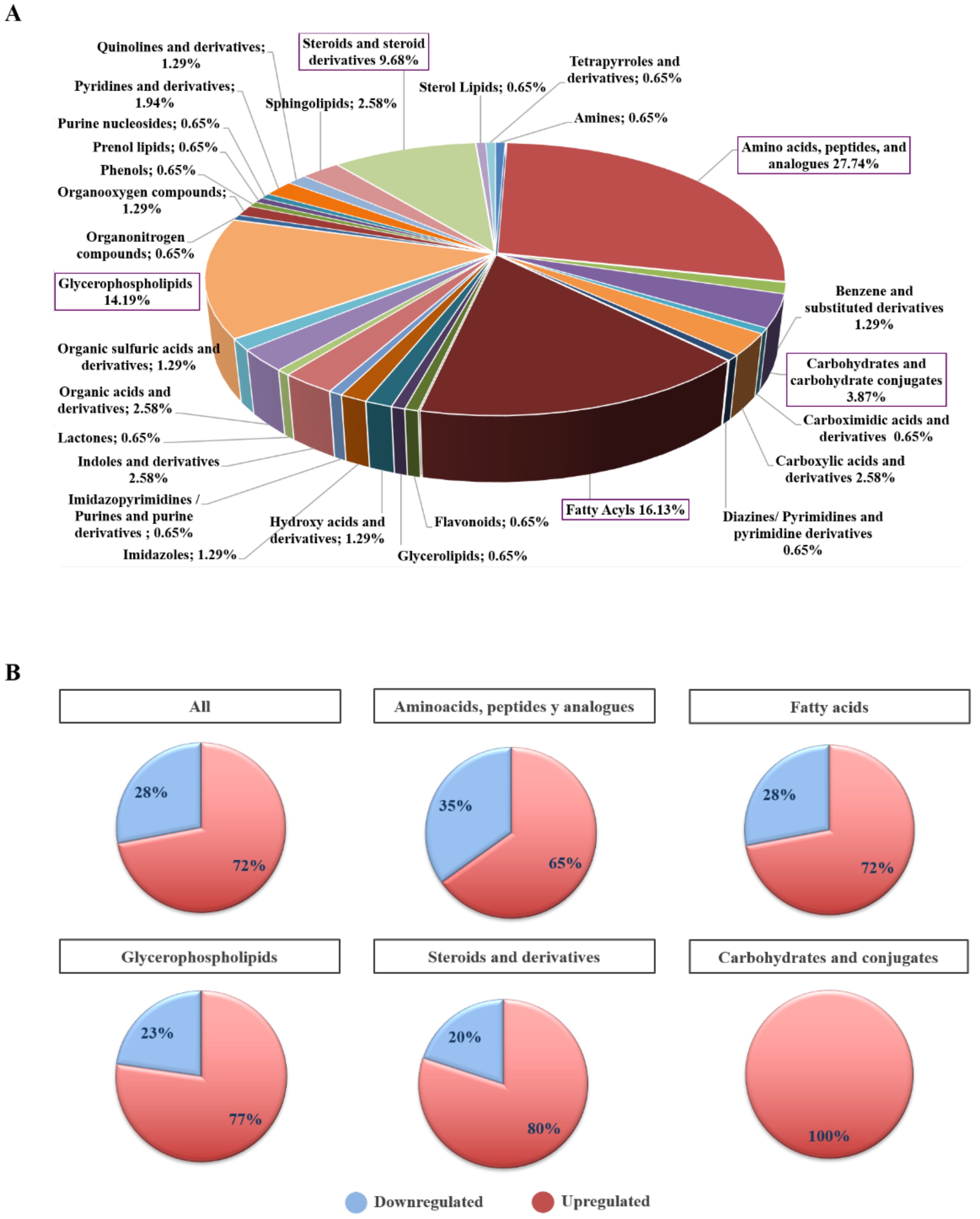
2.3. Biochemical and Functional Nature of Identified Metabolites in NET Patients
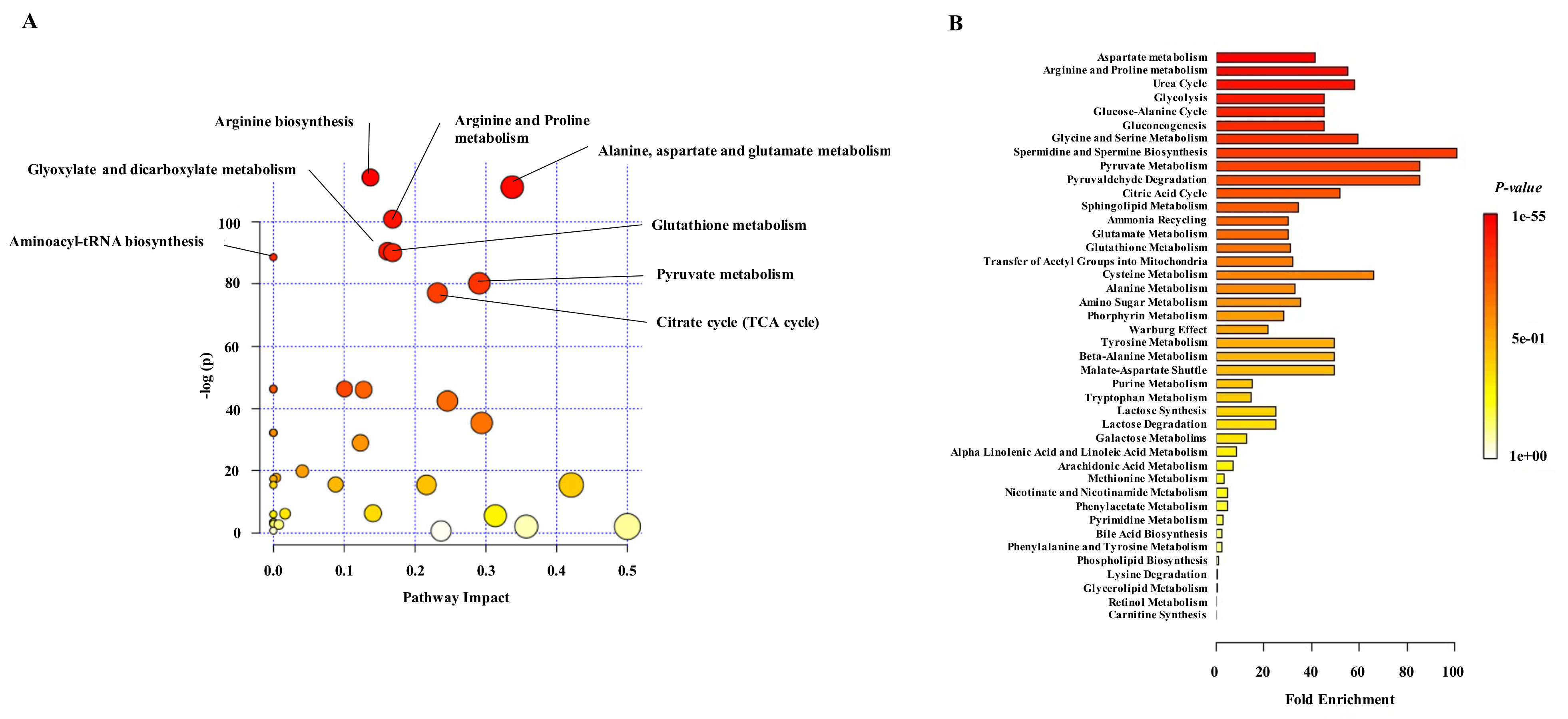
2.4. Differential Metabolites in NET Patients Are Related with Molecular Pathways Associated with Cancer
3. Discussion
4. Material and Methods
4.1. Study Population
4.2. Multiplatform Metabolic Fingerprinting
4.2.1. Data Processing
4.2.2. Statistical Analysis
4.2.3. Annotation and Compound Identification
4.2.4. Targeted Analysis
4.3. Metabolite Classification and Pathway Analysis
4.4. Clinical and Molecular Data Analysis
4.5. Heatmap and Hierarchical Clustering
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cairns, R.A.; Harris, I.S.; Mak, T.W. Regulation of Cancer Cell Metabolism. Nat. Rev. Cancer 2011, 11, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Suhre, K.; Shin, S.-Y.; Petersen, A.-K.; Mohney, R.P.; Meredith, D.; Wägele, B.; Altmaier, E.; CARDIoGRAM; Deloukas, P.; Erdmann, J.; et al. Human Metabolic Individuality in Biomedical and Pharmaceutical Research. Nature 2011, 477, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, L.B.; Gui, D.Y.; Vander Heiden, M.G. Altered Metabolite Levels in Cancer: Implications for Tumour Biology and Cancer Therapy. Nat. Rev. Cancer 2016, 16, 680–693. [Google Scholar] [CrossRef] [PubMed]
- Newgard, C.B. Metabolomics and Metabolic Diseases: Where Do We Stand? Cell Metab. 2017, 25, 43–56. [Google Scholar] [CrossRef]
- Oakman, C.; Tenori, L.; Claudino, W.M.; Cappadona, S.; Nepi, S.; Battaglia, A.; Bernini, P.; Zafarana, E.; Saccenti, E.; Fornier, M.; et al. Identification of a Serum-Detectable Metabolomic Fingerprint Potentially Correlated with the Presence of Micrometastatic Disease in Early Breast Cancer Patients at Varying Risks of Disease Relapse by Traditional Prognostic Methods. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2011, 22, 1295–1301. [Google Scholar] [CrossRef]
- Martín-Blázquez, A.; Díaz, C.; González-Flores, E.; Franco-Rivas, D.; Jiménez-Luna, C.; Melguizo, C.; Prados, J.; Genilloud, O.; Vicente, F.; Caba, O.; et al. Untargeted LC-HRMS-Based Metabolomics to Identify Novel Biomarkers of Metastatic Colorectal Cancer. Sci. Rep. 2019, 9, 20198. [Google Scholar] [CrossRef]
- Bertini, I.; Cacciatore, S.; Jensen, B.V.; Schou, J.V.; Johansen, J.S.; Kruhøffer, M.; Luchinat, C.; Nielsen, D.L.; Turano, P. Metabolomic NMR Fingerprinting to Identify and Predict Survival of Patients with Metastatic Colorectal Cancer. Cancer Res. 2012, 72, 356–364. [Google Scholar] [CrossRef]
- Deja, S.; Porebska, I.; Kowal, A.; Zabek, A.; Barg, W.; Pawelczyk, K.; Stanimirova, I.; Daszykowski, M.; Korzeniewska, A.; Jankowska, R.; et al. Metabolomics Provide New Insights on Lung Cancer Staging and Discrimination from Chronic Obstructive Pulmonary Disease. J. Pharm. Biomed. Anal. 2014, 100, 369–380. [Google Scholar] [CrossRef]
- González-Flores, E.; Serrano, R.; Sevilla, I.; Viúdez, A.; Barriuso, J.; Benavent, M.; Capdevila, J.; Jimenez-Fonseca, P.; López, C.; Garcia-Carbonero, R. SEOM Clinical Guidelines for the Diagnosis and Treatment of Gastroenteropancreatic and Bronchial Neuroendocrine Neoplasms (NENs) (2018). Clin. Transl. Oncol. 2019, 21, 55–63. [Google Scholar] [CrossRef]
- Hofland, J.; Kaltsas, G.; de Herder, W.W. Advances in the Diagnosis and Management of Well-Differentiated Neuroendocrine Neoplasms. Endocr. Rev. 2020, 41, 371–403. [Google Scholar] [CrossRef]
- Dasari, A.; Shen, C.; Halperin, D.; Zhao, B.; Zhou, S.; Xu, Y.; Shih, T.; Yao, J.C. Trends in the Incidence, Prevalence, and Survival Outcomes in Patients With Neuroendocrine Tumors in the United States. JAMA Oncol. 2017, 3, 1335–1342. [Google Scholar] [CrossRef]
- Nuñez-Valdovinos, B.; Carmona-Bayonas, A.; Jimenez-Fonseca, P.; Capdevila, J.; Castaño-Pascual, Á.; Benavent, M.; Barrio, J.J.P.; Teule, A.; Alonso, V.; Custodio, A.; et al. Neuroendocrine Tumor Heterogeneity Adds Uncertainty to the World Health Organization 2010 Classification: Real-World Data from the Spanish Tumor Registry (R-GETNE). Oncologist 2018, 23, 422–432. [Google Scholar] [CrossRef]
- Stevenson, M.; Lines, K.E.; Thakker, R.V. Molecular Genetic Studies of Pancreatic Neuroendocrine Tumors: New Therapeutic Approaches. Endocrinol. Metab. Clin. N. Am. 2018, 47, 525–548. [Google Scholar] [CrossRef]
- Brandi, M.L.; Gagel, R.F.; Angeli, A.; Bilezikian, J.P.; Beck-Peccoz, P.; Bordi, C.; Conte-Devolx, B.; Falchetti, A.; Gheri, R.G.; Libroia, A.; et al. Guidelines for Diagnosis and Therapy of MEN Type 1 and Type 2. J. Clin. Endocrinol. Metab. 2001, 86, 5658–5671. [Google Scholar] [CrossRef]
- Thakker, R.V. Multiple Endocrine Neoplasia Type 1 (MEN1) and Type 4 (MEN4). Mol. Cell. Endocrinol. 2014, 386, 2–15. [Google Scholar] [CrossRef]
- Rednam, S.P.; Erez, A.; Druker, H.; Janeway, K.A.; Kamihara, J.; Kohlmann, W.K.; Nathanson, K.L.; States, L.J.; Tomlinson, G.E.; Villani, A.; et al. Von Hippel-Lindau and Hereditary Pheochromocytoma/Paraganglioma Syndromes: Clinical Features, Genetics, and Surveillance Recommendations in Childhood. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, e68–e75. [Google Scholar] [CrossRef]
- Jett, K.; Friedman, J.M. Clinical and Genetic Aspects of Neurofibromatosis 1. Genet. Med. Off. J. Am. Coll. Med. Genet. 2010, 12, 1–11. [Google Scholar] [CrossRef]
- Larson, A.M.; Hedgire, S.S.; Deshpande, V.; Stemmer-Rachamimov, A.O.; Harisinghani, M.G.; Ferrone, C.R.; Shah, U.; Thiele, E.A. Pancreatic Neuroendocrine Tumors in Patients with Tuberous Sclerosis Complex. Clin. Genet. 2012, 82, 558–563. [Google Scholar] [CrossRef]
- Scarpa, A.; Chang, D.K.; Nones, K.; Corbo, V.; Patch, A.-M.; Bailey, P.; Lawlor, R.T.; Johns, A.L.; Miller, D.K.; Mafficini, A.; et al. Whole-Genome Landscape of Pancreatic Neuroendocrine Tumours. Nature 2017, 543, 65–71. [Google Scholar] [CrossRef]
- Jiao, Y.; Shi, C.; Edil, B.H.; de Wilde, R.F.; Klimstra, D.S.; Maitra, A.; Schulick, R.D.; Tang, L.H.; Wolfgang, C.L.; Choti, M.A.; et al. DAXX/ATRX, MEN1, and MTOR Pathway Genes Are Frequently Altered in Pancreatic Neuroendocrine Tumors. Science 2011, 331, 1199–1203. [Google Scholar] [CrossRef]
- Karpathakis, A.; Dibra, H.; Pipinikas, C.; Feber, A.; Morris, T.; Francis, J.; Oukrif, D.; Mandair, D.; Pericleous, M.; Mohmaduvesh, M.; et al. Prognostic Impact of Novel Molecular Subtypes of Small Intestinal Neuroendocrine Tumor. Clin. Cancer Res. 2016, 22, 250–258. [Google Scholar] [CrossRef]
- Kinross, J.M.; Drymousis, P.; Jiménez, B.; Frilling, A. Metabonomic Profiling: A Novel Approach in Neuroendocrine Neoplasias. Surgery 2013, 154, 1185–1192; discussion 1192–1193. [Google Scholar] [CrossRef]
- Imperiale, A.; Poncet, G.; Addeo, P.; Ruhland, E.; Roche, C.; Battini, S.; Cicek, A.E.; Chenard, M.P.; Hervieu, V.; Goichot, B.; et al. Metabolomics of Small Intestine Neuroendocrine Tumors and Related Hepatic Metastases. Metabolites 2019, 9, 300. [Google Scholar] [CrossRef]
- Fahy, E.; Subramaniam, S.; Murphy, R.C.; Nishijima, M.; Raetz, C.R.H.; Shimizu, T.; Spener, F.; van Meer, G.; Wakelam, M.J.O.; Dennis, E.A. Update of the LIPID MAPS Comprehensive Classification System for Lipids. J. Lipid Res. 2009, 50, S9–S14. [Google Scholar] [CrossRef]
- López-López, Á.; Godzien, J.; Soldevilla, B.; Gradillas, A.; López-Gonzálvez, Á.; Lens-Pardo, A.; La Salvia, A.; Del Carmen Riesco-Martínez, M.; García-Carbonero, R.; Barbas, C. Oxidized Lipids in the Metabolic Profiling of Neuroendocrine Tumors - Analytical Challenges and Biological Implications. J. Chromatogr. A 2020, 1625, 461233. [Google Scholar] [CrossRef]
- Sansone, A.; Lauretta, R.; Vottari, S.; Chiefari, A.; Barnabei, A.; Romanelli, F.; Appetecchia, M. Specific and Non-Specific Biomarkers in Neuroendocrine Gastroenteropancreatic Tumors. Cancers 2019, 11, 1113. [Google Scholar] [CrossRef]
- Costello, L.C.; Franklin, R.B. The Intermediary Metabolism of the Prostate: A Key to Understanding the Pathogenesis and Progression of Prostate Malignancy. Oncology 2000, 59, 269–282. [Google Scholar] [CrossRef]
- Anderson, N.M.; Mucka, P.; Kern, J.G.; Feng, H. The Emerging Role and Targetability of the TCA Cycle in Cancer Metabolism. Protein Cell 2018, 9, 216–237. [Google Scholar] [CrossRef]
- Jiang, L.; Boufersaoui, A.; Yang, C.; Ko, B.; Rakheja, D.; Guevara, G.; Hu, Z.; DeBerardinis, R.J. Quantitative Metabolic Flux Analysis Reveals an Unconventional Pathway of Fatty Acid Synthesis in Cancer Cells Deficient for the Mitochondrial Citrate Transport Protein. Metab. Eng. 2017, 43, 198–207. [Google Scholar] [CrossRef]
- Reuter, S.E.; Evans, A.M. Carnitine and Acylcarnitines. Clin. Pharmacokinet. 2012, 51, 553–572. [Google Scholar] [CrossRef]
- Gao, B.; Lue, H.-W.; Podolak, J.; Fan, S.; Zhang, Y.; Serawat, A.; Alumkal, J.J.; Fiehn, O.; Thomas, G.V. Multi-Omics Analyses Detail Metabolic Reprogramming in Lipids, Carnitines, and Use of Glycolytic Intermediates between Prostate Small Cell Neuroendocrine Carcinoma and Prostate Adenocarcinoma. Metabolites 2019, 9, 82. [Google Scholar] [CrossRef] [PubMed]
- Powell, W.S.; Rokach, J. Biosynthesis, Biological Effects, and Receptors of Hydroxyeicosatetraenoic Acids (HETEs) and Oxoeicosatetraenoic Acids (Oxo-ETEs) Derived from Arachidonic Acid. Biochim. Biophys. Acta 2015, 1851, 340–355. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Song, B.-L.; Xu, C. Cholesterol Metabolism in Cancer: Mechanisms and Therapeutic Opportunities. Nat. Metab. 2020, 2, 132–141. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, S.; Uccella, S.; Finzi, G.; Albarello, L.; Sessa, F.; Capella, C. Localization of Vascular Endothelial Growth Factor and Its Receptors in Digestive Endocrine Tumors: Correlation with Microvessel Density and Clinicopathologic Features. Hum. Pathol. 2003, 34, 18–27. [Google Scholar] [CrossRef]
- Keshet, R.; Erez, A. Arginine and the Metabolic Regulation of Nitric Oxide Synthesis in Cancer. Dis. Model. Mech. 2018, 11. [Google Scholar] [CrossRef]
- Xu, Y. Targeting Lysophosphatidic Acid in Cancer: The Issues in Moving from Bench to Bedside. Cancers 2019, 11, 1523. [Google Scholar] [CrossRef]
- Panagiotopoulos, A.A.; Kalyvianaki, K.; Castanas, E.; Kampa, M. Eicosanoids in Prostate Cancer. Cancer Metastasis Rev. 2018, 37, 237–243. [Google Scholar] [CrossRef]
- Chau, L.-Y. Heme Oxygenase-1: Emerging Target of Cancer Therapy. J. Biomed. Sci. 2015, 22, 22. [Google Scholar] [CrossRef]
- Lamberti, G.; Brighi, N.; Maggio, I.; Manuzzi, L.; Peterle, C.; Ambrosini, V.; Ricci, C.; Casadei, R.; Campana, D. The Role of MTOR in Neuroendocrine Tumors: Future Cornerstone of a Winning Strategy? Int. J. Mol. Sci. 2018, 19, 747. [Google Scholar] [CrossRef]
- Yao, J.C.; Shah, M.H.; Ito, T.; Bohas, C.L.; Wolin, E.M.; Van Cutsem, E.; Hobday, T.J.; Okusaka, T.; Capdevila, J.; de Vries, E.G.E.; et al. Everolimus for Advanced Pancreatic Neuroendocrine Tumors. N. Engl. J. Med. 2011, 364, 514–523. [Google Scholar] [CrossRef]
- Yao, J.C.; Fazio, N.; Singh, S.; Buzzoni, R.; Carnaghi, C.; Wolin, E.; Tomasek, J.; Raderer, M.; Lahner, H.; Voi, M.; et al. Everolimus for the Treatment of Advanced, Non-Functional Neuroendocrine Tumours of the Lung or Gastrointestinal Tract (RADIANT-4): A Randomised, Placebo-Controlled, Phase 3 Study. Lancet Lond. Engl. 2016, 387, 968–977. [Google Scholar] [CrossRef]
- Zabala-Letona, A.; Arruabarrena-Aristorena, A.; Martín-Martín, N.; Fernandez-Ruiz, S.; Sutherland, J.D.; Clasquin, M.; Tomas-Cortazar, J.; Jimenez, J.; Torres, I.; Quang, P.; et al. MTORC1-Dependent AMD1 Regulation Sustains Polyamine Metabolism in Prostate Cancer. Nature 2017, 547, 109–113. [Google Scholar] [CrossRef]
- Gerner, E.W.; Meyskens, F.L. Polyamines and Cancer: Old Molecules, New Understanding. Nat. Rev. Cancer 2004, 4, 781–792. [Google Scholar] [CrossRef]
- Chalishazar, M.D.; Wait, S.J.; Huang, F.; Ireland, A.S.; Mukhopadhyay, A.; Lee, Y.; Schuman, S.S.; Guthrie, M.R.; Berrett, K.C.; Vahrenkamp, J.M.; et al. MYC-Driven Small-Cell Lung Cancer Is Metabolically Distinct and Vulnerable to Arginine Depletion. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 5107–5121. [Google Scholar] [CrossRef]
- Kim, K.; Yeo, S.-G.; Yoo, B.C. Identification of Hypoxanthine and Phosphoenolpyruvic Acid as Serum Markers of Chemoradiotherapy Response in Locally Advanced Rectal Cancer. Cancer Res. Treat. Off. J. Korean Cancer Assoc. 2015, 47, 78–89. [Google Scholar] [CrossRef]
- Naz, S.; Garcia, A.; Rusak, M.; Barbas, C. Method Development and Validation for Rat Serum Fingerprinting with CE-MS: Application to Ventilator-Induced-Lung-Injury Study. Anal. Bioanal. Chem. 2013, 405, 4849–4858. [Google Scholar] [CrossRef]
- Godzien, J.; Kalaska, B.; Adamska-Patruno, E.; Siroka, J.; Ciborowski, M.; Kretowski, A.; Barbas, C. Oxidized Glycerophosphatidylcholines in Diabetes through Non-Targeted Metabolomics: Their Annotation and Biological Meaning. J. Chromatogr. B Analyt. Technol. Biomed. Life. Sci. 2019, 1120, 62–70. [Google Scholar] [CrossRef]
- Dudzik, D.; Zorawski, M.; Skotnicki, M.; Zarzycki, W.; Kozlowska, G.; Bibik-Malinowska, K.; Vallejo, M.; García, A.; Barbas, C.; Ramos, M.P. Metabolic Fingerprint of Gestational Diabetes Mellitus. J. Proteomics 2014, 103, 57–71. [Google Scholar] [CrossRef]
- Wheelock, Å.M.; Wheelock, C.E. Trials and Tribulations of ’omics Data Analysis: Assessing Quality of SIMCA-Based Multivariate Models Using Examples from Pulmonary Medicine. Mol. Biosyst. 2013, 9, 2589–2596. [Google Scholar] [CrossRef]
- Gil de la Fuente, A.; Godzien, J.; Fernández López, M.; Rupérez, F.J.; Barbas, C.; Otero, A. Knowledge-Based Metabolite Annotation Tool: CEU Mass Mediator. J. Pharm. Biomed. Anal. 2018, 154, 138–149. [Google Scholar] [CrossRef]
- Mamani-Huanca, M.; Gradillas, A.; Gil de la Fuente, A.; López-Gonzálvez, Á.; Barbas, C. Unveiling the Fragmentation Mechanisms of Modified Amino Acids as the Key for Their Targeted Identification. Anal. Chem. 2020, 92, 4848–4857. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinforma. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. Data, Information, Knowledge and Principle: Back to Metabolism in KEGG. Nucleic Acids Res. 2014, 42, D199–D205. [Google Scholar] [CrossRef] [PubMed]
- Jewison, T.; Su, Y.; Disfany, F.M.; Liang, Y.; Knox, C.; Maciejewski, A.; Poelzer, J.; Huynh, J.; Zhou, Y.; Arndt, D.; et al. SMPDB 2.0: Big Improvements to the Small Molecule Pathway Database. Nucleic Acids Res. 2014, 42, D478–D484. [Google Scholar] [CrossRef]
- Habibzadeh, F.; Habibzadeh, F.; Habibzadeh, P.; Yadollahie, M. On Determining the Most Appropriate Test Cut-off Value: The Case of Tests with Continuous Results. Biochem. Med. 2016, 26, 297–307. [Google Scholar] [CrossRef]
| Compound | Formula | Mass (Da) | RT (Min) | % Change | p-Value | p(corr) | VIP | Mass Error (ppm) | Analytical Platform | Identification Source | Confidence Level | HMDB Code |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amines | ||||||||||||
| Triethylamine | C6H15N | 101.1204 | 10.28 | −53 | 0.0009 | 0 | CE-MS | DB | 3 | HMDB0032539 | ||
| Amino acids, peptides and analogues | ||||||||||||
| Arginine | C6H14N4O2 | 174.1131 | 9.98 | 243 | 4.58 × 10−35 | −0.80 | 2.33 | 15 | CE-MS LC-MS(+) | DB | 2 | HMDB0000517 |
| Arg-Val | C11H23N5O3 | 273.1822 | 9.95 | 66 | 0.0004 | 8 | CE-MS | DB | 3 | HMDB0028722 | ||
| Aspartate | C4H7NO4 | 133.0375 | 13.80 | −32 | 4.98 × 10−13 | 2 | CE-MS | DB | 2 | HMDB0000191 | ||
| Cys-Gly | C5H10N2O3S | 178.0412 | 12.19 | −29 | 8.00 × 10−9 | 0 | CE-MS | DB | 3 | HMDB0000078 | ||
| Cys-Gly disulfide | C8H15N3O5S2 | 297.0455 | 12.19 | −41 | 1.39 × 10−8 | 0.49 | 1.63 | 1 | CE-MS | DB | 3 | HMDB0000709 |
| Cysteineglutathione disulfide | C13H22N4O8S2 | 426.0912 | 13.95 | −37 | 7.40 × 10−6 | 8 | CE-MS | DB | 3 | HMDB0000656 | ||
| Dimethyl-Arginine (symmetric) | C8H18N4O2 | 202.1428 | 10.67 | 25 | 0.0006 | 1 | CE-MS | DB | 2 | HMDB0003334 | ||
| GalactosylhydroxyLys | C12H24N2O8 | 324.1558 | 11.49 | 42 | 0.0014 | 8 | CE-MS | DB | 3 | HMDB0000600 | ||
| gamma-Glu-orn * | C10H19N3O5 | 261.1335 | 11.30 | 34 | 0.0103 | 4 | CE-MS | DB | 3 | HMDB0002248 | ||
| Glu-Ala * | C8H14N2O5 | 218.0904 | 14.40 | 135 | 4.38 × 10−14 | 1 | CE-MS | DB | 3 | HMDB0006248 | ||
| Glu-Arg | C11H21N5O5 | 303.1554 | 11.49 | 87 | 6.28 × 10−14 | −0.52 | 1.55 | 4 | CE-MS | DB | 3 | HMDB0028813 |
| Glu-Asp | C9H14N2O7 | 262.0801 | 3.78 | 54 | 0.0036 | 7 | LC-MS(−) | MSMS | 2 | HMDB0030419 | ||
| Glu-hyp | C10H16N2O6 | 260.0993 | 12.98 | 37 | 5.12 × 10−7 | 6 | CE-MS | DB | 3 | HMDB0011161 | ||
| Glu-Lys/Ɛ-Glu-Lys | C11H21N3O5 | 275.1481 | 12.32 | 67 | 1.20 × 10−9 | 2 | CE-MS | DB | 3 | HMDB0029154 | ||
| Glu-Lys/Ɛ-Glu-Lys | C11H21N3O5 | 275.1481 | 11.37 | 58 | 1.55 × 10−7 | 2 | CE-MS | DB | 3 | HMDB0029155 | ||
| Glutamine | C5H10N2O3 | 146.0691 | 1.34 | 69 | 0.0006 | 1 | LC-MS(+) LC-MS(−) | MSMS | 2 | HMDB0000641 | ||
| Glu-Val | C10H18N2O5 | 246.1217 | 14.68 | 48 | 0.0009 | 1 | CE-MS LC-MS(+) | DB | 3 | HMDB0028832 | ||
| Glycine | C2H5NO2 | 75.0320 | 9.75 | 32 | 0.0202 | - | GC-MS | Fiehn | 2 | HMDB0000123 | ||
| Gly-Pro | C7H12N2O3 | 172.0831 | 15.89 | −20 | 0.0275 | 10 | CE-MS | DB | 3 | HMDB0000721 | ||
| Homocitrulline | C7H15N3O3 | 189.1118 | 13.52 | 44 | 0.0027 | 3 | CE-MS | DB | 3 | HMDB0000679 | ||
| Iminodiacetic acid | C4H7NO4 | 133.0375 | 12.87 | −37 | 0.0158 | - | GC-MS CE-MS | Fiehn | 2 | HMDB0011753 | ||
| Indoleacetyl glutamine | C15H17N3O4 | 303.1219 | 1.70 | 203 | 1.43 × 10−6 | −0.58 | 2.05 | 2 | LC-MS(−) LC-MS(+) | DB | 3 | HMDB0013240 |
| Leu-hyp | C11H20N2O4 | 244.1423 | 2.00 | 87 | 0.0018 | 2 | LC-MS(−) | DB | 3 | HMDB0028867 | ||
| Leu-Phe | C15H22N2O3 | 278.1630 | 2.14 | Presented in cancer group | 3.54 × 10−5 | −0.60 | 2.45 | 0 | LC-MS(−) | DB | 3 | HMDB0013243 |
| Lys-Asp * | C10H19N3O5 | 261.1335 | 11.30 | 34 | 0.0103 | 4 | CE-MS | DB | 3 | HMDB0028947 | ||
| Methionine S-oxide | C5H11NO3S | 165.0478 | 14.06 | 110 | 3.30 × 10−9 | 11 | CE-MS | DB | 2 | HMDB0002005 | ||
| N2-Methyl-lysine | C7H16N2O2 | 160.1208 | 10.67 | −77 | 6.97 × 10−9 | 2 | CE-MS | DB | 2 | HMDB0002038 | ||
| N2-Methylproline | C6H11NO2 | 129.0791 | 14.59 | −43 | 0.0031 | 1 | CE-MS | DB | 2 | HMDB0059649 | ||
| N6-Acetyl-hydroxy-lysine * | C8H16N2O4 | 204.1103 | 12.78 | −55 | 0.0002 | 3 | CE-MS | DB | 3 | HMDB0033891 | ||
| N-acetyl-lysine | C8H16N2O3 | 188.1155 | 13.70 | 20 | 0.0002 | 3 | CE-MS | DB | 2 | HMDB0000206 | ||
| Ornithine | C5H12N2O2 | 132.0906 | 9.66 | −39 | 5.58 × 10−19 | 0.65 | 1.65 | 6 | CE-MS | DB | 2 | HMDB0000214 |
| Phenylalanine | C9H11NO2 | 165.0770 | 13.88 | −58 | 0.0050 | 12 | CE-MS GC-MS | DB | 2 | HMDB0000159 | ||
| Pipecolic acid | C6H11NO2 | 129.0790 | 10.14 | 49 | 0.0307 | - | GC-MS | Fiehn | 2 | HMDB0000716 | ||
| Pyroglutamine | C5H8N2O2 | 128.0583 | 11.48 | 68 | 1.41 × 10−5 | 2 | CE-MS | DB | 3 | HMDB0062558 | ||
| Ser-Ala * | C6H12N2O4 | 176.0789 | 13.07 | −38 | 1.37 × 10−8 | 4 | CE-MS | DB | 3 | HMDB0029032 | ||
| Ser-hyp * | C8H14N2O5 | 218.0904 | 14.40 | 135 | 4.38 × 10−14 | 1 | CE-MS | DB | 3 | HMDB0029040 | ||
| Ser-Val * | C8H16N2O4 | 204.1103 | 12.78 | −55 | 0.0002 | 3 | CE-MS | DB | 3 | HMDB0029052 | ||
| Stearoyl-tyrosine * | C27H45NO4 | 447.3349 | 5.15 | −66 | 9.67 × 10−17 | 0.75 | 3.74 | 1 | LC-MS(+) | DB | 3 | HMDB0062343 |
| Suberylglycine | C10H17NO5 | 231.1106 | 1.19 | Presented in cancer group | 0.0001 | −0.73 | 3.82 | 1 | LC-MS(−) | DB | 3 | HMDB0000953 |
| Thr-Ala | C7H14N2O4 | 190.0954 | 13.83 | 90 | 5.09 × 10−11 | 0 | CE-MS | DB | 3 | HMDB0029054 | ||
| Thr-Gly * | C6H12N2O4 | 176.0789 | 13.07 | −38 | 1.37 × 10−8 | 4 | CE-MS | DB | 3 | HMDB0029061 | ||
| Trp-Phe | C20H21N3O3 | 351.1582 | 2.32 | 74 | 3.73 × 10−11 | −0.63 | 1.56 | 1 | LC-MS(−) | DB | 3 | HMDB0029090 |
| Val-Leu | C11H22N2O3 | 230.1616 | 12.83 | 51 | 0.0012 | 6 | CE-MS | DB | 3 | HMDB0029131 | ||
| Benzene and substituted derivatives | ||||||||||||
| Mandelic acid | C8H8O3 | 152.0473 | 2.28 | 335 | 0.0037 | 3 | LC-MS(−) | DB | 3 | HMDB0000703 | ||
| 3-phenylprop-2-en-1-yloxysulfonic acid | C9H10O4S | 214.0299 | 2.77 | 118 | 0.0005 | 2 | LC-MS(−) | DB | 3 | HMDB0135284 | ||
| Carbohydrates and carbohydrate conjugates | ||||||||||||
| Allose | C6H12O6 | 180.0633 | 17.10 | 139 | 1.32 × 10−14 | 0.62 | 1.44 | - | GC-MS | Fiehn | 2 | HMDB0001151 |
| Glucose | C6H12O6 | 180.0633 | 17.45 | 174 | 1.98 × 10−15 | 0.62 | 1.64 | - | GC-MS | Fiehn | 2 | HMDB0000122 |
| Glycerol | C3H8O3 | 92.0473 | 9.28 | 45 | 0.0000 | - | GC-MS | Fiehn | 2 | HMDB0000131 | ||
| Mannitol | C6H14O6 | 182.0790 | 17.59 | 196 | 0.0017 | - | GC-MS | Fiehn | 2 | HMDB0000765 | ||
| Phenylglucuronide | C12H14O7 | 270.0739 | 0.93 | 308 | 0.0052 | −0.52 | 2.37 | 2 | LC-MS(−) | DB | 3 | HMDB0060014 |
| PYRANOSE (glucose/altrose/galactose/talose) | C6H12O6 | 180.0633 | 17.24 | 194 | 1.61 × 10−16 | 0.63 | 1.71 | - | GC-MS | Fiehn | 2 | |
| Carboximidic acids and derivatives | ||||||||||||
| Acetylspermidine | C9H21N3O | 187.1671 | 9.15 | 38 | 6.27 × 10−6 | 7 | CE-MS | DB | 3 | HMDB0001276 | ||
| Carboxylic acids and derivatives | ||||||||||||
| 1-Aminocyclohexanecarboxylic acid | C7H13NO2 | 143.0944 | 13.89 | −32 | 0.0366 | 1 | CE-MS | DB | 3 | HMDB0038249 | ||
| di-Hydroxymelatonin * | C13H16N2O4 | 264.1110 | 1.34 | 46 | 0.0268 | 2 | LC-MS(−) | DB | 3 | HMDB0061136 | ||
| Edetic Acid | C10H16N2O8 | 292.0906 | 0.24 | 35 | 0.0001 | 1 | LC-MS(+) | DB | 3 | HMDB0015109 | ||
| Isocitric acid Citric acid | C6H8O7 | 192.0270 | 0.23 | 64 | 5.83 × 10−5 | 0 | LC-MS(−) | MSMS | 2 | HMDB0000193HMDB0000094 | ||
| Diazines/Pyrimidines and pyrimidine derivatives | ||||||||||||
| 5,6-Dihydrothymine | C5H8N2O2 | 128.0570 | 12.22 | 56 | 0.0169 | 12 | CE-MS | DB | 3 | HMDB0000079 | ||
| Fatty acyls | ||||||||||||
| 3-carboxy-4-methyl-5-propyl-2-furanpropanoic acid (CMPF) | C12H16O5 | 240.0998 | 3.78 | 69 | 0.004 | 2 | LC-MS(+) | MSMS | 2 | HMDB0061112 | ||
| 8-amino-7-oxo-nonanoic acid * | C9H17NO3 | 187.1208 | 2.06 | 418 | 0.0220 | 2 | LC-MS(−) | DB | 3 | |||
| Arachidonic acid | C20H32O2 | 304.2402 | 7.13 | 62 | 1.59 × 10−5 | 1 | LC-MS(−) | MSMS | 2 | HMDB0001043 | ||
| beta-Phenylalanoyl- CoA * | C30H45N8O17P3S | 914.1836 | 2.83 | 73 | 7.07 × 10−3 | 1 | LC-MS(−) | DB | 3 | |||
| beta-Phenylalanoyl-CoA * | C30H45N8O17P3S | 914.1836 | 3.62 | 65 | 8.78 × 10−3 | 0 | LC-MS(−) | DB | 3 | |||
| DG(31:0) | C34H66O5 | 554.4910 | 8.18 | −37 | 0.0008 | 0 | LC-MS(+) | DB | 3 | HMDB0093505 | ||
| Docosapentaenoic acid | C22H34O2 | 330.2558 | 7.25 | 75 | 6.87 × 10−9 | −0.55 | 1.51 | 1 | LC-MS(−) | DB | 3 | HMDB0006528 |
| Dodecenedioic acid | C12H20O4 | 228.1362 | 3.50 | 191 | 9.87× 10−8 | −0.60 | 2.24 | 2 | LC-MS(−) | DB | 3 | HMDB0000933 |
| Eicosapentaenoic acid | C20H30O2 | 302.2246 | 6.77 | 65 | 3.88 × 10−4 | 2 | LC-MS(−) | DB | 3 | HMDB0001999 | ||
| Eicosatrienoic acid | C20H34O2 | 306.2558 | 7.39 | 82 | 1.85 × 10−10 | 1 | LC-MS(−) | DB | 3 | HMDB0010378 | ||
| Eicosenoic acid | C20H38O2 | 310.2871 | 8.34 | 79 | 2.75 × 10−10 | −0.51 | 1.50 | 1 | LC-MS(−) | DB | 3 | HMDB0002231 |
| Glucosylgalactosylhydroxylysine | C18H34N2O13 | 486.2093 | 12.38 | 46 | 1.73 × 10−5 | 7 | CE-MS | DB | 3 | HMDB0000585 | ||
| HETE | C20H32O3 | 320.2351 | 5.92 | Presented in cancer group | 1.24 × 10−5 | −0.63 | 2.64 | 0 | LC-MS(−) | MSMS | 2 | HMDB0060101 |
| MG(18:2) | C21H38O4 | 354.2770 | 6.81 | 116 | 0.0007 | 2 | LC-MS(+) | MSMS | 2 | HMDB0011538 | ||
| MG(20:0) | C23H46O4 | 386.3396 | 7.79 | −86 | 1.30 × 10−12 | 5 | LC-MS(+) | DB | 3 | HMDB0072859 | ||
| N-palmitoyl glutamic acid * | C21H39NO5 | 385.2828 | 4.13 | 62 | 0.004 | 0 | LC-MS(+) | DB | 3 | |||
| Oleic acid | C18H34O2 | 282.2559 | 20.47 | 67 | 0.0013 | - | GC-MS LC-MS(+) | Fiehn | 2 | HMDB0000207 | ||
| Vaccenic acid | C18H34O2 | 282.2559 | 20.56 | 23 | 0.0158 | - | GC-MS | Fiehn | 2 | HMDB0041480 | ||
| 3-hydroxy-5-octenoylcarnitine | C15H27NO5 | 301.1889 | 4.23 | Presented in cancer group | 3.57 × 10−2 | −0.72 | 3.96 | 3 | LC-MS(−) | DB | 3 | |
| 3-Hydroxy-5-tetradecenoylcarnitine * | C21H39NO5 | 385.2828 | 4.13 | 62 | 0.004 | 0 | LC-MS(+) | DB | 3 | HMDB0013330 | ||
| 9-Decenoylcarnitine | C17H31NO4 | 313.2232 | 12.99 | −15 | 0.0325 | 7 | CE-MS | DB | 3 | HMDB0013205 | ||
| Arachidonoylcarnitine * | C27H45NO4 | 447.3349 | 5.15 | −66 | 9.67 × 10−17 | 0.75 | 3.74 | 1 | LC-MS(+) | DB | 3 | HMDB0062343 |
| α-Linolenyl carnitine | C25H43NO4 | 421.3192 | 4.91 | −41 | 1.79 × 10−8 | 0.53 | 2.26 | 0 | LC-MS(+) | DB | 3 | HMDB0006319 |
| Linoleyl carnitine | C25H45NO4 | 423.3349 | 5.14 | −59 | 4.03 × 10−16 | 0.73 | 3.40 | 0 | LC-MS(+) | DB | 3 | HMDB0006469 |
| Oleoylcarnitine Elaidic carnitine | C25H47NO4 | 425.3504 | 5.43 | −41 | 1.41 × 10−8 | 0.51 | 2.40 | 1 | LC-MS(+) | MSMS | 2 | HMDB0006351 HMDB0006464 |
| Flavonoids | ||||||||||||
| Anthraniloyl-CoA | C28H41N8O17P3S | 886.1523 | 0.23 | 186 | 3.47 × 10−21 | −0.69 | 2.39 | 3 | LC-MS(−) | DB | 3 | |
| Glycerolipids | ||||||||||||
| 11-Oxo-androsterone glucuronide | C25H36O9 | 480.2359 | 3.43 | −45 | 8.06 × 10−6 | 0.53 | 2.06 | 1 | LC-MS(−) | DB | 3 | HMDB0010338 |
| Glycerophospholipids | ||||||||||||
| LPC(16:0)-OH | C24H50NO8P | 511.3274 | 4.12 | 25 | 0.008 | 0 | LC-MS(+) LC-MS(−) | MSMS | 2 | |||
| LPC(16:0)-OH | C24H50NO8P | 511.3274 | 4.21 | 37 | 0.0005 | 0 | LC-MS(+) LC-MS(−) | MSMS | 2 | |||
| LPC(18:0)-OH | C26H54NO8P | 539.3587 | 4.71 | 27 | 0.0100 | 0 | LC-MS(+) LC-MS(−) | MSMS | 2 | |||
| LPC(18:2)-OH | C26H50NO8P | 535.3274 | 4.41 | Presented in cancer group | 2.19 × 10−6 | 0 | LC-MS(+) LC-MS(−) | MSMS | 2 | |||
| LPA(13:0) | C16H33O7P | 368.1963 | 5.23 | −57 | 4.95 × 10−3 | 0.56 | 2.29 | 7 | LC-MS(−) | DB | 3 | HMDB0114760 |
| LPC(22:1) | C30H60NO7P | 577.4107 | 6.99 | 66 | 2.14 × 10−5 | 2 | LC-MS(−) | MSMS | 2 | HMDB0010399 | ||
| LPE(16:0) | C21H44NO7P | 453.2855 | 5.65 | 25 | 0.008 | 0 | LC-MS(+) | MSMS | 2 | HMDB0011473 | ||
| LPE(20:5) | C25H42NO7P | 499.2699 | 5.08 | 179 | 2.80 × 10−6 | 0 | LC-MS(+) | MSMS | 2 | HMDB0011489 | ||
| LPE(20:5) | C25H42NO7P | 499.2699 | 5.16 | 55 | 0.001 | 0 | LC-MS(+) LC-MS(−) | MSMS | 2 | HMDB0011489 | ||
| LPE(22:6) | C27H44NO7P | 525.2855 | 5.37 | 28 | 0.001 | 0 | LC-MS(+) | MSMS | 2 | HMDB0011526 | ||
| LPE(22:6) | C27H44NO7P | 525.2855 | 5.45 | 39 | 0.0005 | 0 | LC-MS(+) | MSMS | 2 | HMDB0011496 | ||
| LPE(P-16:0) | C21H44NO6P | 437.2906 | 5.83 | 36 | 0.002 | 0 | LC-MS(+) | DB | 3 | HMDB0011152 | ||
| LPI(16:1) | C25H47O12P | 570.2805 | 5.51 | 97 | 2.63 × 10−3 | 1 | LC-MS(−) | MSMS | 2 | |||
| LPS(18:0) | C24H48NO9P | 525.3066 | 6.71 | 104 | 9.62 × 10−6 | 2 | LC-MS(−) | MSMS | 2 | |||
| PC(32:0) | C40H80NO8P | 733.5622 | 10.47 | 51 | 3.91 × 10−7 | 2 | LC-MS(+) | DB | 3 | HMDB0007871 | ||
| PC(38:2) | C46H88NO8P | 813.6247 | 11.94 | 39 | 0.0004 | 0 | LC-MS(+) | MSMS | 2 | HMDB0007987 | ||
| PC(38:5) | C46H82NO8P | 807.5778 | 9.82 | −28 | 5.05 × 10−5 | 2 | LC-MS(+) | MSMS | 2 | HMDB0008156 | ||
| PE(34:2) PE(O-34:3) | C39H74NO7P | 699.5203 | 10.25 | −33 | 0.001 | 1 | LC-MS(+) | MSMS | 2 | HMDB0011343 | ||
| PE(38:6) | C43H74NO8P | 763.5151 | 9.31 | −31 | 0.0087 | 1 | LC-MS(+) | MSMS | 2 | HMDB0009294 | ||
| PG(20:2) | C26H49O9P | 536.3114 | 7.41 | −58 | 3.44 × 10−11 | 0.59 | 2.33 | 4 | LC-MS(−) | DB | 3 | |
| PG(28:0) | C34H67O10P | 666.4471 | 7.64 | 115 | 8.69 × 10−15 | −0.61 | 2.75 | 5 | LC-MS(+) | DB | 3 | HMDB0116681 |
| PS(39:5) | C45H78NO10P | 823.5363 | 10.46 | 25 | 1.60 × 10−7 | 5 | LC-MS(+) | DB | 3 | |||
| Hydroxy acids and derivatives | ||||||||||||
| 3-Hydroxydodecanedioic acid | C12H22O5 | 246.1467 | 2.79 | Presented in cancer group | 2.42 × 10−6 | −0.68 | 2.97 | 0 | LC-MS(−) | MSMS | 2 | HMDB0000413 |
| 3-Hydroxydodecanoic acid | C12H24O3 | 216.1725 | 4.62 | Presented in cancer group | 2.32 × 10−11 | −0.72 | 2.79 | 2 | LC-MS(−) | MSMS | 2 | HMDB0000387 |
| Imidazoles | ||||||||||||
| Methylimidazole | C4H6N2 | 82.0538 | 11.48 | 64 | 6.41 × 10−6 | 9 | CE-MS | DB | 3 | |||
| Urocanate Nicotinamide N-oxide | C6H6N2O2 | 138.0435 | 11.02 | 24 | 0.001 | 4 | CE-MS | DB | 3 | HMDB0034174 HMDB0002730 | ||
| Imidazopyrimidines/Purines and purine derivatives | ||||||||||||
| Hypoxanthine | C5H4N4O | 136.0402 | 14.19 | −34 | 8.20 × 10−6 | 13 | CE-MS | DB | 2 | HMDB0000157 | ||
| Indoles and derivatives | ||||||||||||
| 3-Indoleacetic acid | C10H9NO2 | 175.0633 | 17.94 | 131 | 2.62 × 10−7 | - | GC-MS | Fiehn | 2 | HMDB0000197 | ||
| 5-Hydroxyindole | C8H7NO | 133.0527 | 0.76 | 37 | 3.82 × 10−2 | 1 | LC-MS(−) | DB | 3 | HMDB0059805 | ||
| 5-Hydroxyindoleacetaldehyde | C10H9NO2 | 175.0633 | 2.51 | 150 | 2.63 × 10−5 | −0.51 | 2.55 | 1 | LC-MS(+) | MSMS | 2 | HMDB0004073 |
| 5-Hydroxyindoleacetic acid | C10H9NO3 | 191.0582 | 0.80 | Presented in cancer group | 3.28 × 10−8 | 0 | LC-MS(+) | MSMS | 2 | HMDB0000763 | ||
| Lactones | ||||||||||||
| N-(4-Coumaroyl)-homoserine lactone | C13H13NO4 | 247.0845 | 1.34 | 55 | 0.0006 | 2 | LC-MS(+) | DB | 3 | |||
| Organic acids and derivatives | ||||||||||||
| (Homo)2-aconitate* | C8H10O6 | 202.0477 | 0.26 | −75 | 7.87 × 10−59 | 0.84 | 2.98 | 11 | LC-MS(−) | DB | 3 | |
| Lactic acid | C3H6O3 | 90.0317 | 6.06 | −77 | 4.72 × 10−36 | −0.92 | 2.58 | - | GC-MS | Fiehn | 2 | HMDB0001311 |
| Pyruvic acid | C3H4O3 | 88.0160 | 5.89 | −58 | 1.06 × 10−14 | −0.69 | 1.78 | - | GC-MS | Fiehn | 2 | HMDB0000243 |
| Succinylacetoacetate * | C8H10O6 | 202.0477 | 0.26 | −75 | 7.87 × 10−59 | 0.84 | 2.98 | 11 | LC-MS(−) | DB | 3 | HMDB0240258 |
| Organic sulfuric acids and derivatives | ||||||||||||
| Indoxylsulfuric acid | C8H7NO4S | 213.0095 | 1.00 | 42 | 3.47 × 10−2 | 1 | LC-MS(−) | MSMS | 2 | HMDB0000682 | ||
| p-Phenolsulfonic acid | C6H6O4S | 173.9986 | 0.65 | 174 | 3.45 × 10−3 | 1 | LC-MS(−) | MSMS | 2 | HMDB0060015 | ||
| Organonitrogen compounds | ||||||||||||
| Phosphocholine | C5H14NO4P | 183.0660 | 5.37 | 32 | 0.003 | 1 | LC-MS(+) | MSMS | 2 | |||
| Organooxygen compounds | ||||||||||||
| 4-Hydroxycyclohexylcarboxylic acid | C7H12O3 | 144.0786 | 0.76 | 789 | 5.08 × 10−10 | −0.71 | 2.99 | 1 | LC-MS(−) | DB | 3 | HMDB0001988 |
| Acetyl-N-formyl-5-methoxykynurenamine (AFMK) * | C13H16N2O4 | 264.1110 | 1.34 | 46 | 0.0268 | 2 | LC-MS(−) | DB | 3 | HMDB0004259 | ||
| Phenols | ||||||||||||
| 4-Methylcatechol | C7H8O2 | 124.0524 | 1.23 | −43 | 1.68 × 10−2 | 2 | LC-MS(−) | DB | 3 | HMDB0000873 | ||
| Prenol lipids | ||||||||||||
| Retinol | C20H30O | 286.2296 | 7.04 | 34 | 0.0003 | 2 | LC-MS(+) | MSMS | 2 | HMDB0000305 | ||
| Purine nucleosides | ||||||||||||
| 1-Methyladenosine | C11H15N5O4 | 281.1132 | 12.49 | 41 | 3.68 × 10−16 | −0.58 | 1.20 | 3 | CE-MS | DB | 2 | HMDB0003331 |
| Pyridines and derivatives | ||||||||||||
| Norcotinine | C9H10N2O | 162.0790 | 11.48 | 34 | 0.0039 | 2 | CE-MS | DB | 3 | HMDB0001297 | ||
| Piperideine | C5H9N | 83.0739 | 13.77 | 28 | 0.0239 | 5 | CE-MS | DB | 3 | |||
| Serotonine | C10H12N2O | 176.0954 | 11.40 | 228 | 0.0007 | 3 | CE-MS | DB | 2 | HMDB0001046 | ||
| Quinolines and derivatives | ||||||||||||
| 8-Hydroxycarteolol | C16H24N2O4 | 308.1732 | 13.90 | −31 | 0.0246 | 1 | CE-MS | DB | 3 | HMDB0060990 | ||
| Quinoline | C9H7N | 129.0578 | 2.51 | 99 | 0.0008 | 1 | LC-MS(+) | DB | 3 | HMDB0033731 | ||
| Sphingolipids | ||||||||||||
| Cer(35:0) | C35H69NO3 | 551.5277 | 11.39 | 58 | 4.24 × 10−8 | 3 | LC-MS(−) | DB | 3 | |||
| Cer(36:1) | C36H71NO3 | 565.5434 | 11.74 | 42 | 0.0003 | 1 | LC-MS(+) | DB | 3 | HMDB0004950 | ||
| SM(36:0) | C41H85N2O6P | 732.6145 | 10.14 | 41 | 0.0014 | 1 | LC-MS(+) | MSMS | 2 | HMDB0012087 | ||
| Sphingosine-1-phosphate | C18H38NO5P | 379.2488 | 5.02 | −30 | 2.17 × 10−8 | 0.50 | 2.06 | 0 | LC-MS(+) | MSMS | 2 | HMDB0000277 |
| Steroids and steroid derivatives | ||||||||||||
| 12a-Hydroxy-3-oxocholadienic acid | C24H34O4 | 386.2457 | 4.27 | 144 | 3.66 × 10−6 | −0.54 | 2.12 | 0 | LC-MS(−) | MSMS | 2 | HMDB0000385 |
| Biliverdin | C33H34N4O6 | 582.2478 | 4.73 | 300 | 1.47 × 10−16 | −0.72 | 2.62 | 0 | LC-MS(−) | DB | 3 | HMDB0001008 |
| Hydroxy-3-oxo-4-cholestenoate | C27H42O4 | 430.3083 | 5.53 | 54 | 2.78 × 10−6 | LC-MS(+) LC-MS(−) | MSMS | 2 | ||||
| Calcitroic acid | C23H34O4 | 374.2457 | 7.39 | 84 | 6.25 × 10−11 | −0.54 | 1.76 | 6 | LC-MS(−) | DB | 3 | HMDB0006472 |
| Chenodeoxycholic acid 3-glucuronide * | C30H48O10 | 568.3248 | 4.33 | 94 | 2.10 × 10−3 | 1 | LC-MS(−) | DB | 3 | |||
| Cholestane-3,7,12,24,25-pentol | C27H48O5 | 452.3501 | 7.77 | −56 | 3.81 × 10−10 | 0.56 | 2.03 | 2 | LC-MS(−) | DB | 3 | HMDB0000483 |
| Cholestane-3,7,12,25-tetrol-3-glucuronide | C33H56O10 | 612.3873 | 4.71 | 97 | 4.89 × 10−5 | 0 | LC-MS(−) | DB | 3 | HMDB0010355 | ||
| Cortisone acetate | C23H30O6 | 402.2042 | 3.86 | 63 | 5.89 × 10−4 | 9 | LC-MS(−) | DB | 3 | HMDB0015459 | ||
| Dehydroepiandrosterone 3-glucuronide Dehydroisoandrosterone 3-glucuronide | C25H36O8 | 464.2410 | 3.91 | −51 | 1.01 × 10−5 | 2 | LC-MS(−) | DB | 3 | HMDB0010348 HMDB0010327 | ||
| Deoxycholic acid 3-glucuronide * | C30H48O10 | 568.3248 | 4.33 | 94 | 2.10 × 10−3 | 1 | LC-MS(−) | DB | 3 | HMDB0002596 | ||
| ecdysone 25-O-D-glucopyranoside | C33H54O11 | 626.3666 | 4.41 | 88 | 1.02 × 10−4 | 1 | LC-MS(−) | DB | 3 | |||
| Pregnanediol | C21H36O2 | 320.2715 | 6.35 | −51 | 4.21 × 10−5 | 0 | LC-MS(−) | DB | 3 | HMDB0004025 | ||
| Pregnanolone sulfate | C21H34O5S | 398.2127 | 3.64 | 55 | 5.27 × 10−3 | 0 | LC-MS(−) | DB | 3 | HMDB0240591 | ||
| Ursodeoxycholic acid | C24H40O4 | 392.2927 | 4.34 | 188 | 3.93 × 10−2 | −0.51 | 2.35 | 1 | LC-MS(−) | MSMS | 2 | HMDB0000946 |
| Ursodeoxycholic acid 3-sulfate | C24H40O7S | 472.2494 | 3.75 | 130 | 4.38 × 10−3 | −0.54 | 2.36 | 1 | LC-MS(−) | DB | 3 | HMDB0002642 |
| Sterol lipids | ||||||||||||
| 24-Hydroxygeminivitamin D3 | C32H54O5 | 518.3971 | 6.79 | −37 | 2.82 × 10−7 | 1 | LC-MS(+) | DB | 3 | |||
| Tetrapyrroles and derivatives | ||||||||||||
| Bilirubin | C33H36N4O6 | 584.2635 | 3.85 | 674 | 3.79 × 10−8 | −0.68 | 2.63 | 4 | LC-MS(−) LC-MS(+) | MSMS | 2 | HMDB0000054 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soldevilla, B.; López-López, A.; Lens-Pardo, A.; Carretero-Puche, C.; Lopez-Gonzalvez, A.; La Salvia, A.; Gil-Calderon, B.; Riesco-Martinez, M.C.; Espinosa-Olarte, P.; Sarmentero, J.; et al. Comprehensive Plasma Metabolomic Profile of Patients with Advanced Neuroendocrine Tumors (NETs). Diagnostic and Biological Relevance. Cancers 2021, 13, 2634. https://doi.org/10.3390/cancers13112634
Soldevilla B, López-López A, Lens-Pardo A, Carretero-Puche C, Lopez-Gonzalvez A, La Salvia A, Gil-Calderon B, Riesco-Martinez MC, Espinosa-Olarte P, Sarmentero J, et al. Comprehensive Plasma Metabolomic Profile of Patients with Advanced Neuroendocrine Tumors (NETs). Diagnostic and Biological Relevance. Cancers. 2021; 13(11):2634. https://doi.org/10.3390/cancers13112634
Chicago/Turabian StyleSoldevilla, Beatriz, Angeles López-López, Alberto Lens-Pardo, Carlos Carretero-Puche, Angeles Lopez-Gonzalvez, Anna La Salvia, Beatriz Gil-Calderon, Maria C. Riesco-Martinez, Paula Espinosa-Olarte, Jacinto Sarmentero, and et al. 2021. "Comprehensive Plasma Metabolomic Profile of Patients with Advanced Neuroendocrine Tumors (NETs). Diagnostic and Biological Relevance" Cancers 13, no. 11: 2634. https://doi.org/10.3390/cancers13112634
APA StyleSoldevilla, B., López-López, A., Lens-Pardo, A., Carretero-Puche, C., Lopez-Gonzalvez, A., La Salvia, A., Gil-Calderon, B., Riesco-Martinez, M. C., Espinosa-Olarte, P., Sarmentero, J., Rubio-Cuesta, B., Rincón, R., Barbas, C., & Garcia-Carbonero, R. (2021). Comprehensive Plasma Metabolomic Profile of Patients with Advanced Neuroendocrine Tumors (NETs). Diagnostic and Biological Relevance. Cancers, 13(11), 2634. https://doi.org/10.3390/cancers13112634






