International Multi-Site Initiative to Develop an MRI-Inclusive Nomogram for Side-Specific Prediction of Extraprostatic Extension of Prostate Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Statistical Considerations
2.2. Benchmark Comparisons
3. Results
3.1. Study Population
3.2. Inter-Site Variabilities
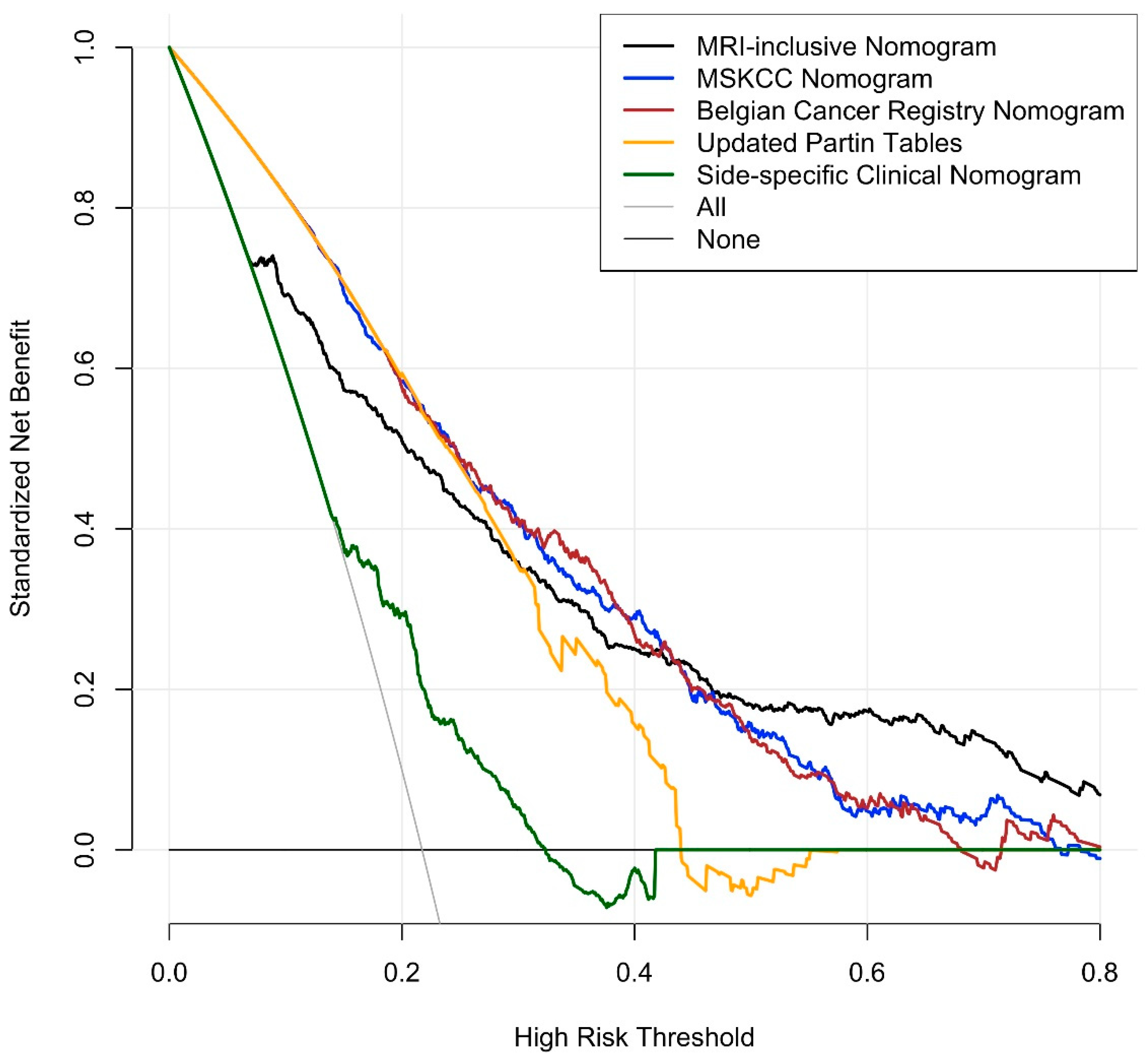
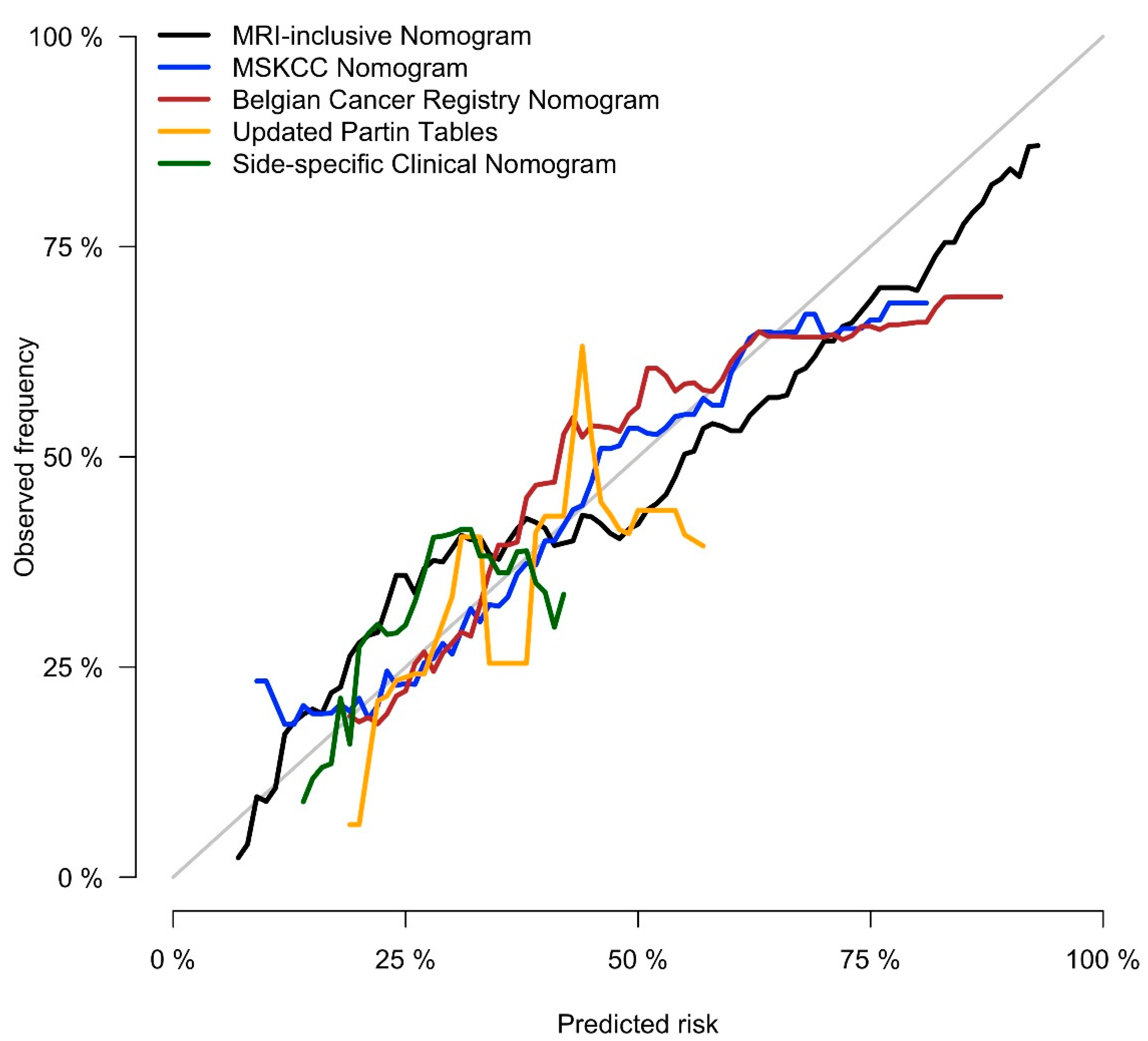
3.3. Nomogram and Benchmarks
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Woo, S.; Suh, C.H.; Eastham, J.A.; Zelefsky, M.J.; Morris, M.J.; Abida, W.; Scher, H.I.; Sidlow, R.; Becker, A.S.; Wibmer, A.G.; et al. Comparison of Magnetic Resonance Imaging-stratified Clinical Pathways and Systematic Transrectal Ultrasound-guided Biopsy Pathway for the Detection of Clinically Significant Prostate Cancer: A Systematic Review and Meta-analysis of Randomized Controlled Trials. Eur. Urol. Oncol. 2019, 2, 605–616. [Google Scholar] [CrossRef]
- Pagniez, M.A.; Kasivisvanathan, V.; Puech, P.; Drumez, E.; Villers, A.; Olivier, J. Predictive Factors of Missed Clinically Significant Prostate Cancers in Men with Negative Magnetic Resonance Imaging: A Systematic Review and Meta-Analysis. J. Urol. 2020, 204, 24–32. [Google Scholar] [CrossRef]
- Falagario, U.G.; Ratnani, P.; Lantz, A.; Jambor, I.; Dovey, Z.; Verma, A.; Treacy, P.-J.; Sobotka, S.; Martini, A.; Bashorun, H.; et al. Staging Accuracy of Multiparametric Magnetic Resonance Imaging in Caucasian and African American Men Undergoing Radical Prostatectomy. J. Urol. 2020, 204, 82–90. [Google Scholar] [CrossRef]
- De Rooij, M.; Hamoen, E.H.; Witjes, J.A.; Barentsz, J.O.; Rovers, M.M. Accuracy of Magnetic Resonance Imaging for Local Staging of Prostate Cancer: A Diagnostic Meta-analysis. Eur. Urol. 2016, 70, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.A.; Rodin, D.M.; Harisinghani, M.; Dahl, D.M. Impact of preoperative endorectal MRI stage classification on neurovascular bundle sparing aggressiveness and the radical prostatectomy positive margin rate. Urol. Oncol. Semin. Orig. Investig. 2009, 27, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Hoogenes, J.; Wright, I.; Matsumoto, E.D.; Shayegan, B. Utility of preoperative 3 Tesla pelvic phased-array multiparametric magnetic resonance imaging in prediction of extracapsular extension and seminal vesicle invasion of prostate cancer and its impact on surgical margin status: Experience at a Canadian academic tertiary care centre. Can. Urol. Assoc. J. 2017, 11, E174–E178. [Google Scholar] [CrossRef] [PubMed][Green Version]
- McClure, T.D.; Margolis, D.J.A.; Reiter, R.E.; Sayre, J.W.; Thomas, M.A.; Nagarajan, R.; Gulati, M.; Raman, S.S. Use of MR Imaging to Determine Preservation of the Neurovascular Bundles at Robotic-assisted Laparoscopic Prostatectomy. Radiology 2012, 262, 874–883. [Google Scholar] [CrossRef] [PubMed]
- Rud, E.; Baco, E.; Klotz, D.; Rennesund, K.; Svindland, A.; Berge, V.; Lundeby, E.; Wessel, N.; Hoff, J.-R.; Berg, R.E.; et al. Does Preoperative Magnetic Resonance Imaging Reduce the Rate of Positive Surgical Margins at Radical Prostatectomy in a Randomised Clinical Trial? Eur. Urol. 2015, 68, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Kozikowski, M.; Malewski, W.; Michalak, W.; Dobruch, J. Clinical utility of MRI in the decision-making process before radical prostatectomy: Systematic review and meta-analysis. PLoS ONE 2019, 14, e0210194. [Google Scholar] [CrossRef]
- Rayn, K.N.; Bloom, J.B.; Gold, S.A.; Hale, G.R.; Baiocco, J.A.; Mehralivand, S.; Czarniecki, M.; Sabarwal, V.K.; Valera, V.; Wood, B.J.; et al. Added Value of Multiparametric Magnetic Resonance Imaging to Clinical Nomograms for Predicting Adverse Pathology in Prostate Cancer. J. Urol. 2018, 200, 1041–1047. [Google Scholar] [CrossRef]
- Martini, A.; Gupta, A.; Lewis, S.C.; Cumarasamy, S.; Haines, K.G.; Briganti, A.; Montorsi, F.; Tewari, A.K. Development and internal validation of a side-specific, multiparametric magnetic resonance imaging-based nomogram for the prediction of extracapsular extension of prostate cancer. BJU Int. 2018, 122, 1025–1033. [Google Scholar] [CrossRef]
- Morlacco, A.; Sharma, V.; Viers, B.R.; Rangel, L.J.; Carlson, R.E.; Froemming, A.T.; Karnes, R.J. The Incremental Role of Magnetic Resonance Imaging for Prostate Cancer Staging before Radical Prostatectomy. Eur. Urol. 2017, 71, 701–704. [Google Scholar] [CrossRef]
- Feng, T.S.; Sharif-Afshar, A.R.; Wu, J.; Li, Q.; Luthringer, D.; Saouaf, R.; Kim, H.L. Multiparametric MRI Improves Accuracy of Clinical Nomograms for Predicting Extracapsular Extension of Prostate Cancer. Urology 2015, 86, 332–337. [Google Scholar] [CrossRef]
- Zapała, P.; Dybowski, B.; Bres-Niewada, E.; Lorenc, T.; Powała, A.; Lewandowski, Z.; Gołębiowski, M.; Radziszewski, P. Predicting side-specific prostate cancer extracapsular extension: A simple decision rule of PSA, biopsy, and MRI parameters. Int. Urol. Nephrol. 2019, 51, 1545–1552. [Google Scholar] [CrossRef]
- Nyarangi-Dix, J.; Wiesenfarth, M.; Bonekamp, D.; Hitthaler, B.; Schütz, V.; Dieffenbacher, S.; Mueller-Wolf, M.; Roth, W.; Stenzinger, A.; Duensing, S.; et al. Combined Clinical Parameters and Multiparametric Magnetic Resonance Imaging for the Prediction of Extraprostatic Disease—A Risk Model for Patient-tailored Risk Stratification When Planning Radical Prostatectomy. Eur. Urol. Focus 2020, 6, 1205–1212. [Google Scholar] [CrossRef]
- Lebacle, C.; Roudot-Thoraval, F.; Moktefi, A.; Bouanane, M.; De La Taille, A.; Salomon, L. Integration of MRI to clinical nomogram for predicting pathological stage before radical prostatectomy. World J. Urol. 2016, 35, 1409–1415. [Google Scholar] [CrossRef]
- Chen, Y.; Yu, W.; Fan, Y.; Zhou, L.; Yang, Y.; Wang, H.; Jiang, Y.; Wang, X.; Wu, S.; Jin, J. Development and comparison of a Chinese nomogram adding multi-parametric MRI information for predicting extracapsular extension of prostate cancer. Oncotarget 2016, 8, 22095–22103. [Google Scholar] [CrossRef][Green Version]
- Rosenkrantz, A.B.; Ginocchio, L.A.; Cornfeld, D.; Froemming, A.T.; Gupta, R.T.; Turkbey, B.; Westphalen, A.C.; Babb, J.; Margolis, D.J. Interobserver Reproducibility of the PI-RADS Version 2 Lexicon: A Multicenter Study of Six Experienced Prostate Radiologists. Radiology 2016, 280, 793–804. [Google Scholar] [CrossRef]
- Weaver, J.K.; Kim, E.H.; Vetter, J.M.; Shetty, A.; Grubb, R.L.; Strope, S.A.; Andriole, G.L. Prostate Magnetic Resonance Imaging Provides Limited Incremental Value Over the Memorial Sloan Kettering Cancer Center Preradical Prostatectomy Nomogram. Urology 2018, 113, 119–128. [Google Scholar] [CrossRef]
- Zanelli, E.; Giannarini, G.; Cereser, L.; Zuiani, C.; Como, G.; Pizzolitto, S.; Crestani, A.; Valotto, C.; Ficarra, V.; Girometti, R. Head-to-head comparison between multiparametric MRI, the partin tables, memorial sloan kettering cancer center nomogram, and CAPRA score in predicting extraprostatic cancer in patients undergoing radical prostatectomy. J. Magn. Reson. Imaging 2019, 50, 1604–1613. [Google Scholar] [CrossRef]
- Jansen, B.H.; Nieuwenhuijzen, J.A.; Oprea-Lager, D.E.; Yska, M.J.; Lont, A.P.; van Moorselaar, R.J.; Vis, A.N. Adding multiparametric MRI to the MSKCC and Partin nomograms for primary prostate cancer: Improving local tumor staging? Urol. Oncol. Semin. Orig. Investig. 2019, 37, 181.e1–181.e6. [Google Scholar] [CrossRef]
- Weinreb, J.C.; Barentsz, J.O.; Choyke, P.L.; Cornud, F.; Haider, M.A.; Macura, K.J.; Margolis, D.J.A.; Schnall, M.D.; Shtern, F.; Tempany, C.M.; et al. PI-RADS Prostate Imaging—Reporting and Data System: 2015, Version 2. Eur. Urol. 2016, 69, 16–40. [Google Scholar] [CrossRef]
- Barentsz, J.O.; Richenberg, J.; Clements, R.; Choyke, P.; Verma, S.; Villeirs, G.; Rouviere, O.; Logager, V.; Fütterer, J.J. ESUR prostate MR guidelines 2012. Eur. Radiol. 2012, 22, 746–757. [Google Scholar] [CrossRef]
- Van Buuren, S.; Groothuis-Oudshoorn, K. mice: Multivariate Imputation by Chained Equations in R. J. Stat. Softw. 2011, 45, 1–67. [Google Scholar] [CrossRef]
- Vickers, A.J.; Elkin, E.B. Decision Curve Analysis: A Novel Method for Evaluating Prediction Models. Med. Decis. Mak. 2006, 26, 565–574. [Google Scholar] [CrossRef]
- Memorial_Sloan_Kettering_Cancer_Center. Prediction Tools/Prostate Cancer Nomograms/Pre-Radical Prostatectomy. Available online: https://www.mskcc.org/nomograms/prostate/pre_op (accessed on 26 September 2019).
- Eifler, J.B.; Feng, Z.; Lin, B.M.; Partin, M.T.; Humphreys, E.B.; Han, M.; Epstein, J.I.; Walsh, P.C.; Trock, B.J.; Partin, A.W. An updated prostate cancer staging nomogram (Partin tables) based on cases from 2006 to 2011. BJU Int. 2013, 111, 22–29. [Google Scholar] [CrossRef]
- Tosco, L.; De Coster, G.; Roumeguère, T.; Everaerts, W.; Quackels, T.; Dekuyper, P.; Van Cleynenbreugel, B.; Van Damme, N.; Van Eycken, E.; Ameye, F.; et al. Development and External Validation of Nomograms To Predict Adverse Pathological Characteristics After Robotic Prostatectomy: Results of a Prospective, Multi-institutional, Nationwide series. Eur. Urol. Oncol. 2018, 1, 338–345. [Google Scholar] [CrossRef]
- Steuber, T.; Graefen, M.; Haese, A.; Erbersdobler, A.; Chun, F.K.-H.; Schlom, T.; Perrotte, P.; Huland, H.; Karakiewicz, P.I. Validation of a Nomogram for Prediction of Side Specific Extracapsular Extension at Radical Prostatectomy. J. Urol. 2006, 175, 939–944. [Google Scholar] [CrossRef]
- Vickers, A.J.; Van Calster, B.; Steyerberg, E.W. A simple, step-by-step guide to interpreting decision curve analysis. Diagn. Progn. Res. 2019, 3, 1–8. [Google Scholar] [CrossRef]
- Vickers, A.J.; Cronin, A.M. Everything You Always Wanted to Know About Evaluating Prediction Models (But Were Too Afraid to Ask). Urology 2010, 76, 1298–1301. [Google Scholar] [CrossRef]
- Zorn, K.C.; Gallina, A.; Hutterer, G.C.; Walz, J.; Shalhav, A.L.; Zagaja, G.P.; Valiquette, L.; Gofrit, O.N.; Orvieto, M.A.; Taxy, J.B.; et al. External Validation of a Nomogram for Prediction of Side-Specific Extracapsular Extension at Robotic Radical Prostatectomy. J. Endourol. 2007, 21, 1345–1352. [Google Scholar] [CrossRef]
- Fütterer, J.J.; Heijmink, S.W.T.P.J.; Scheenen, T.W.J.; Jager, G.J.; De Kaa, C.A.H.; Witjes, J.A.; Barentsz, J.O. Prostate Cancer: Local Staging at 3-T Endorectal MR Imaging—Early Experience. Radiology 2006, 238, 184–191. [Google Scholar] [CrossRef]
- Akin, O.; Riedl, C.C.; Ishill, N.M.; Moskowitz, C.S.; Zhang, J.; Hricak, H. Interactive dedicated training curriculum improves accuracy in the interpretation of MR imaging of prostate cancer. Eur. Radiol. 2010, 20, 995–1002. [Google Scholar] [CrossRef][Green Version]
- Wibmer, A.; Vargas, H.A.; Donahue, T.F.; Zheng, J.; Moskowitz, C.; Eastham, J.; Sala, E.; Hricak, H. Diagnosis of Extracapsular Extension of Prostate Cancer on Prostate MRI: Impact of Second-Opinion Readings by Subspecialized Genitourinary Oncologic Radiologists. Am. J. Roentgenol. 2015, 205, W73–W78. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, G.; Zhao, L.; Mao, L.; Li, X.; Yan, W.; Xiao, Y.; Lei, J.; Sun, H.; Jin, Z. Radiomics Based on Multiparametric Magnetic Resonance Imaging to Predict Extraprostatic Extension of Prostate Cancer. Front. Oncol. 2020, 10, 940. [Google Scholar] [CrossRef]
- Losnegård, A.; Reisæter, L.A.R.; Halvorsen, O.J.; Jurek, J.; Assmus, J.; Arnes, J.B.; Honoré, A.; Monssen, J.A.; Andersen, E.; Haldorsen, I.S.; et al. Magnetic resonance radiomics for prediction of extraprostatic extension in non-favorable intermediate- and high-risk prostate cancer patients. Acta Radiol. 2020, 61, 1570–1579. [Google Scholar] [CrossRef]
- Cuocolo, R.; Stanzione, A.; Faletti, R.; Gatti, M.; Calleris, G.; Fornari, A.; Gentile, F.; Motta, A.; Dell’Aversana, S.; Creta, M.; et al. MRI index lesion radiomics and machine learning for detection of extraprostatic extension of disease: A multicenter study. Eur. Radiol. 2021, 1–9. [Google Scholar] [CrossRef]
- Stanzione, A.; Cuocolo, R.; Cocozza, S.; Romeo, V.; Persico, F.; Fusco, F.; Longo, N.; Brunetti, A.; Imbriaco, M. Detection of Extraprostatic Extension of Cancer on Biparametric MRI Combining Texture Analysis and Machine Learning: Preliminary Results. Acad. Radiol. 2019, 26, 1338–1344. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, H.; Ahmad, A.E.; Chandrasekar, T.; Klotz, L.; Emberton, M.; Haider, M.A.; Taneja, S.S.; Arora, K.; Fleshner, N.; Finelli, A.; et al. Comparison of Magnetic Resonance Imaging and Transrectal Ultrasound Informed Prostate Biopsy for Prostate Cancer Diagnosis in Biopsy Naïve Men: A Systematic Review and Meta-Analysis. J. Urol. 2020, 203, 1085–1093. [Google Scholar] [CrossRef]
- Popita, C.; Popiţa, A.-R.; Andrei, A.; Rusu, A.; Petruţ, B.; Kacso, G.; Bungărdean, C.; Bolog, N.; Coman, I. Local staging of prostate cancer with multiparametric-MRI: Accuracy and inter-reader agreement. Med. Pharm. Rep. 2020, 93, 150–161. [Google Scholar] [CrossRef]
- Matsuoka, Y.; Ishioka, J.; Tanaka, H.; Kimura, T.; Yoshida, S.; Saito, K.; Fujii, Y.; Kihara, K. Impact of the Prostate Imaging Reporting and Data System, Version 2, on MRI Diagnosis for Extracapsular Extension of Prostate Cancer. Am. J. Roentgenol. 2017, 209, W76–W84. [Google Scholar] [CrossRef]
- Kuroiwa, K.; Shiraishi, T.; Ogawa, O.; Usami, M.; Hirao, Y.; Naito, S. Clinicopathological Research Group for Localized Prostate Cancer Investigators Discrepancy Between Local and Central Pathological Review of Radical Prostatectomy Specimens. J. Urol. 2010, 183, 952–957. [Google Scholar] [CrossRef] [PubMed]
- Ghadjar, P.; Hayoz, S.; Genitsch, V.; Zwahlen, D.R.; Hölscher, T.; Gut, P.; Guckenberger, M.; Hildebrandt, G.; Müller, A.-C.; Putora, P.M.; et al. Importance and outcome relevance of central pathology review in prostatectomy specimens: Data from the SAKK 09/10 randomized trial on prostate cancer. BJU Int. 2017, 120, E45–E51. [Google Scholar] [CrossRef]
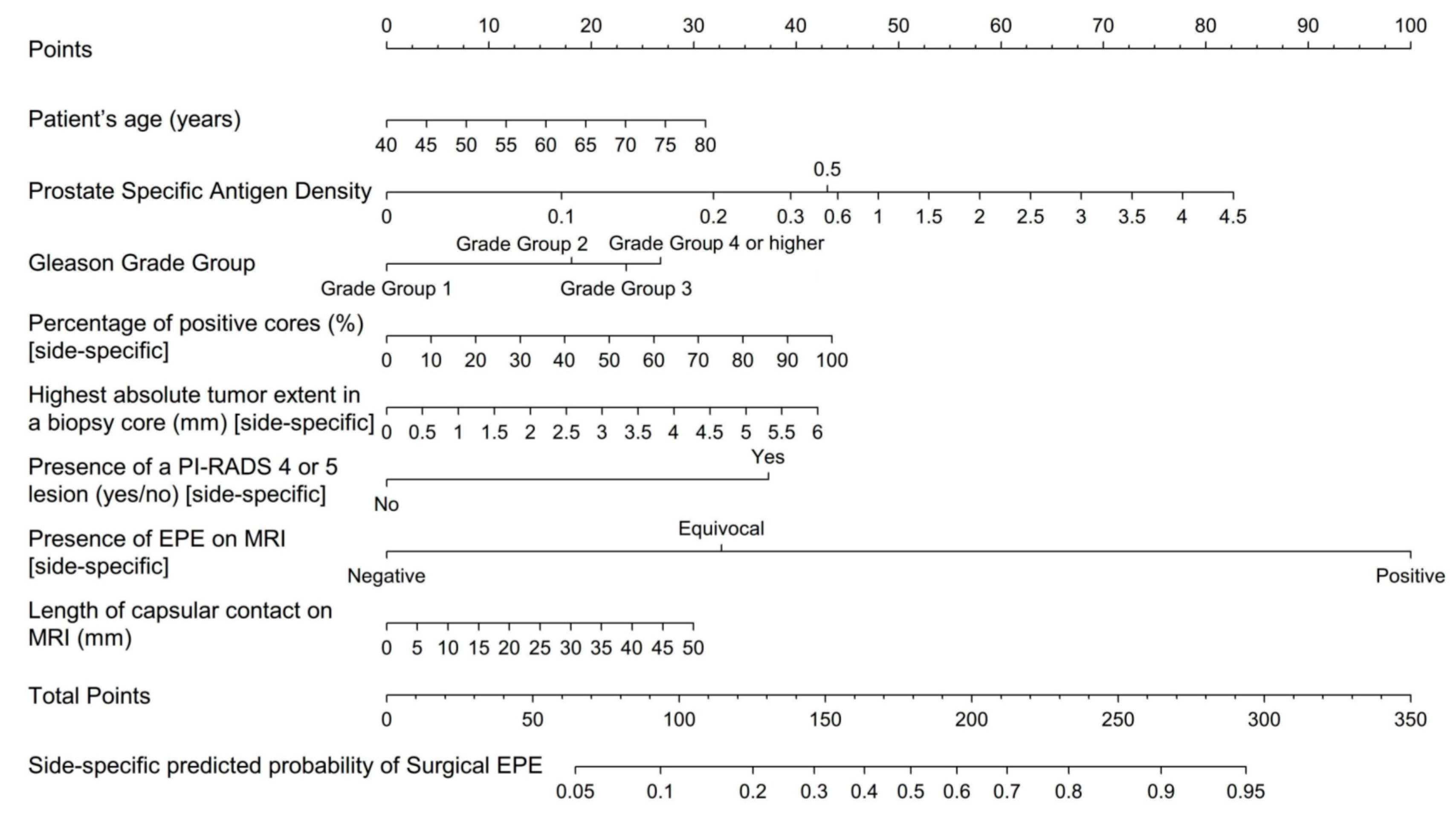
| Authors | Country | No. | Whole Gland vs. Side-Specific | MRI Variables | Benchmark Models/ Benchmark Clinical Data | Main Finding |
|---|---|---|---|---|---|---|
| Rayn et al. [10] | USA | 532 | Whole gland | NIH suspicion score EPE: present vs. absent Largest lesion diameter | MSKCC nomogram Partin tables | MRI in addition to clinical nomograms increases predictive ability. |
| Martini et al. [11] | USA | 561 | Side-specific | EPE: absent vs. present | PSA, Bx Gleason grade group, % cancer in Bx cores | MRI-inclusive model for the side-specific prediction of EPE. |
| Morlacco et al. [12] | USA | 501 | Whole gland | EPE: absent vs. present | Partin tables CAPRA score | MRI-inclusive models outperform clinical-based models alone. |
| Feng et al. [13] | USA | 112 | Whole gland | EPE: absent vs. present | MSKCC nomogram Partin tables | MRI improved accuracy of existing clinical nomograms. |
| Zapala et al. [14] | Poland | 88 | Side-specific | Likert score (1–5) EPE: present vs. absent Largest lesion diameter | PSA, cT, number and % positive Bx cores, % cancer in Bx cores, Bx Gleason score | Lesion diameter ≥ 15 mm on MRI is an independent predictor of EPE. |
| Nyarangi-Dix et al. [15] | Germany | 264 | Side-specific | EPE: ESUR score (1–5) Prostate volume Capsular contact length | MSKCC nomogram Nomogram by Steuber et al. | Combining MRI and clinical parameters outperformed clinical nomograms. |
| Lebacle et al. [16] | France | 1743 | Whole gland | EPE: present vs. absent | PSA, Gleason score, prostate weight, cT | MRI-inclusive model is more accurate than clinical and biopsy data alone. |
| Chen et al. [17] | China | 706 | Side-specific | EPE risk score (1–5) | Age, cT, PSA, Bx Gleason grade groups, % positive Bx cores, % cancer in bx cores | MRI-inclusive model is more accurate than clinical and biopsy data alone. |
| Weaver et al. [19] | USA | 236 | Whole gland | PI-RADS score EPE: present vs. absent | MSKCC nomogram | A combined model (MRI + MSKCC) provides no additional benefit over the MSKCC nomogram alone. |
| Jansen et al. [20] | Netherlands | 430 | Whole gland | EPE: present vs. absent | MSKCC nomogram Partin tables | The addition of MRI to the MSKCC and Partin nomograms did not increase diagnostic accuracy. |
| Zanelli et al. [21] | Italy | 73 | Whole gland | PI-RADS score EPE: ESUR score (1–5) | MSKCC nomogram CAPRA score | Combination of MRI + clinical models outperforms clinical models for two radiologists, but not for a third. |
| Parameter | Overall | Institution | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | p-Value # | |||
| Number of Patients | 840 | 100 | 82 | 98 | 96 | 100 | 44 | 100 | 100 | 20 | 100 | ||
| Clinical Data | Age (years) | 64.0 [59.0, 68.0] | 60.5 [55.0, 66.0] | 66.0 [61.0, 69.0] | 59.0 [53.3, 64.0] | 66.0 [62.0, 71.0] | 65.0 [62.0, 67.3] | 65.5 [58.8, 73.3] | 65.0 [60.0, 68.0] | 63.5 [60.0, 66.0] | 65.0 [63.0, 68.0] | 63.0 [59.0, 68.0] | <0.001 |
| PSA (ng/mL) Missing: 0.1% | 7.1 [5.2, 10.7] | 6.1 [4.5, 8.3] | 7.9 [5.7, 10.1] | 5.4 [4.3, 7.6] | 9.0 [6.5, 14.2] | 11.8 [7.3, 18.0] | 8.3 [5.2, 13.9] | 6.0 [5.0, 9.0] | 6.4 [5.2, 8.8] | 7.3 [6.1, 9.6] | 7.2 [5.4, 10.4] | <0.001 | |
| PSA Density (ng/mL2) Missing: 0.2% | 0.2 [0.1, 0.3] | 0.2 [0.1, 0.3] | 0.1 [0.1, 0.3] | 0.2 [0.1, 0.3] | 0.2 [0.1, 0.3] | 0.3 [0.2, 0.4] | 0.2 [0.1, 0.3] | 0.1 [0.1, 0.2] | 0.2 [0.1, 0.2] | 0.2 [0.1, 0.3] | 0.2 [0.1, 0.3] | <0.001 | |
| Systematic Biopsy Data | Positive Biopsy Cores (%) Missing: 3.2% | 25.0 [12.5, 41.7] | 35.9 [21.4, 59.8] | 16.7 [8.3, 41.7] | 41.7 [25.0, 54.6] | 30.0 [18.3, 50.0] | 20.0 [10.0, 40.0] | 16.7 [9.9, 35.7] | 25.0 [8.3, 41.7] | 25.0 [10.0, 40.0] | 33.3 [31.7, 46.7] | 16.7 [8.3, 33.3] | <0.001 |
| Highest Gleason Grade Group | |||||||||||||
| 1 | 216 (25.7) | 18 (18.0) | 17 (20.7) | 26 (26.5) | 22 (22.9) | 36 (36.0) | 8 (18.2) | 26 (26.0) | 33 (33.0) | 0 | 30 (30.0) | <0.001 | |
| 2 | 293 (34.9) | 45 (45.0) | 35 (42.7) | 44 (44.9) | 20 (20.8) | 29 (29.0) | 11 (25.0) | 40 (40.0) | 33 (33.0) | 0 | 36 (36.0) | ||
| 3 | 97 (11.5) | 12 (12.0) | 4 (4.9) | 19 (19.4) | 6 (6.2) | 6 (6.0) | 8 (18.2) | 11 (11.0) | 14 (14.0) | 0 | 17 (17.0) | ||
| 4 or higher | 163 (19.4) | 23 (23.0) | 21 (25.6) | 9 (9.2) | 43 (44.8) | 16 (16.0) | 13 (29.5) | 14 (14.0) | 9 (9.0) | 3 (15.0) | 12 (12.0) | ||
| Cancer only on targeted biopsy | 71 (8.5) | 2 (2.0) | 5 (6.1) | 0 | 5 (5.2) | 13 (13.0) | 4 (9.1) | 9 (9.0) | 11 (11.0) | 17 (85.0) | 5 (5.0) | <0.001 | |
| Maximum tumor extent (mm) Missing: 11.9% | 4.0 [1.5, 8.0] | 6.0 [3.0, 9.0] | 4.0 [1.0, 12.0] | Missing * | 5.0 [2.0, 8.0] | 3.0 [2.0, 8.0] | 1.0 [0.0, 5.0] | 5.0 [3.0, 9.0] | 4.0 [2.0, 7.0] | 1.6 [1.6, 1.80] | 5.0 [2.0, 7.0] | <0.001 | |
| Data | Highest PI-RADS score | ||||||||||||
| 1 | 9 (1.1) | 0 | 0 | 0 | 0 | 0 | 0 | 5 (5.0) | 0 | 0 | 4 (4.0) | <0.001 | |
| 2 | 31 (3.7) | 10 (10.0) | 7 (8.5) | 4 (4.1) | 0 | 1 (1.0) | 0 | 2 (2.0) | 6 (6.0) | 0 | 1 (1.0) | ||
| 3 | 83 (9.9) | 11 (11.0) | 14 (17.1) | 9 (9.2) | 4 (4.2) | 9 (9.0) | 6 (13.6) | 11 (11.0) | 6 (6.0) | 2 (10.0) | 11 (11.0) | ||
| 4 | 339 (40.4) | 31 (31.0) | 26 (31.7) | 55 (56.1) | 41 (42.7) | 34 (34.0) | 17 (38.6) | 38 (38.0) | 45 (45.0) | 13 (65.0) | 39 (39.0) | ||
| 5 | 378 (45.0) | 48 (48.0) | 35 (42.7) | 30 (30.6) | 51 (51.1) | 56 (56.0) | 21 (47.7) | 44 (44.0) | 43 (43.0) | 5 (25.0) | 45 (45.0) | ||
| Cases with PI-RADSv2 ≥ 4 | 717 (85.4) | 79 (79.0) | 61 (74.4) | 85 (86.7) | 92 (95.8) | 90 (90.0) | 38 (86.4) | 82 (82.0) | 88 (88.0) | 18 (90.0) | 84 (84.0) | 0.005 | |
| Maximum Lesion Diameter (cm) | 1.5 [1.1, 2.0] | 1.6 [1.2, 2.2] | 1.4 [1.0, 2.1] | 1.3 [1.0, 1.6] | 1.6 [1.1, 2.4] | 1.6 [1.2, 2.3] | 1.5 [1.2, 2.2] | 1.3 [1.0, 1.9] | 1.6 [1.2, 2.0] | 1.2 [1.0, 1.5] | 1.4 [1.0, 1.8] | <0.001 | |
| Length of Capsular Contact (mm) | 10.0 [4.0, 17.0] | 12.0 [4.0, 23.3] | 13.0 [8.0, 20.0] | 8.0 [5.0, 12.0] | 16.0 [8.8, 25.0] | 7.0 [2.0, 13.5] | 13.0 [7.8, 22.3] | 8.0 [0.0, 14.0] | 15.0 [11.0, 20.0] | 10.5 [7.8, 14.3] | 0.0 [0.0, 12.0] | <0.001 | |
| Presence of ECE | |||||||||||||
| Negative | 487 (58.0) | 47 (47.0) | 50 (61.0) | 77 (78.6) | 33 (34.4) | 55 (55.0) | 11 (25.0) | 71 (71.0) | 63 (63.0) | 5 (25.0) | 75 (75.0) | <0.001 | |
| Equivocal | 284 (33.8) | 42 (42.0) | 30 (36.6) | 15 (15.3) | 58 (60.4) | 36 (36.0) | 26 (59.1) | 18 (18.0) | 26 (26.0) | 12 (60.0) | 21 (21.0) | ||
| Positive | 69 (8.2) | 11 (11.0) | 2 (2.5) | 6 (6.1) | 5 (5.2) | 9 (9.0) | 7 (15.9) | 11 (11.0) | 11 (11.0) | 3 (15.0) | 4 (4.0) | ||
| ECE on prostatectomy specimen (standard of reference) | 320 (38.1) | 42 (42.0) | 24 (29.3) | 28 (28.6) | 34 (35.4) | 35 (35.0) | 21 (47.7) | 44 (44.0) | 43 (43.0) | 6 (30.0) | 43 (43.0) | 0.136 | |
| Statistical Model | Area under the Receiver Operator Characteristics Curve (95% Confidence Intervals) |
|---|---|
| MRI-inclusive Nomogram | 0.828 (0.805, 0.852) |
| MSKCC Pre-Radical Prostatectomy Nomogram [26] | 0.675 (0.638, 0.712) * |
| Belgian Cancer Registry Nomogram [28] | 0.679 (0.641, 0.716) * |
| Updated Partin Tables [27] | 0.601 (0.563, 0.640) * |
| Side-Specific Clinical Nomogram [29] | 0.650 (0.619, 681) * |
| Nomogram Model | Mean Area under the Receiver Operator Characteristics Curve (Range) | |
|---|---|---|
| With Imputation * | Without Imputation * | |
| MRI-inclusive Nomogram | 0.821 (0.762, 0.880) | 0.799 (0.738, 0.857) |
| MSKCC Pre-Radical Prostatectomy Nomogram [26] | 0.678 (0.605, 0.725) | 0.684 (0.587, 0.806) |
| Belgian Cancer Registry Nomogram [28] | 0.681 (0.599, 0.731) | 0.684 (0.599, 0.777) |
| Updated Partin Tables [27] | 0.600 (0.533, 0.678) | 0.607 (0.536, 0.708) |
| Side-Specific Clinical Nomogram [29] | 0.652 (0.585, 0.727) | 0.626 (0.535, 0.727) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wibmer, A.G.; Kattan, M.W.; Alessandrino, F.; Baur, A.D.J.; Boesen, L.; Franco, F.B.; Bonekamp, D.; Campa, R.; Cash, H.; Catalá, V.; et al. International Multi-Site Initiative to Develop an MRI-Inclusive Nomogram for Side-Specific Prediction of Extraprostatic Extension of Prostate Cancer. Cancers 2021, 13, 2627. https://doi.org/10.3390/cancers13112627
Wibmer AG, Kattan MW, Alessandrino F, Baur ADJ, Boesen L, Franco FB, Bonekamp D, Campa R, Cash H, Catalá V, et al. International Multi-Site Initiative to Develop an MRI-Inclusive Nomogram for Side-Specific Prediction of Extraprostatic Extension of Prostate Cancer. Cancers. 2021; 13(11):2627. https://doi.org/10.3390/cancers13112627
Chicago/Turabian StyleWibmer, Andreas G., Michael W. Kattan, Francesco Alessandrino, Alexander D. J. Baur, Lars Boesen, Felipe Boschini Franco, David Bonekamp, Riccardo Campa, Hannes Cash, Violeta Catalá, and et al. 2021. "International Multi-Site Initiative to Develop an MRI-Inclusive Nomogram for Side-Specific Prediction of Extraprostatic Extension of Prostate Cancer" Cancers 13, no. 11: 2627. https://doi.org/10.3390/cancers13112627
APA StyleWibmer, A. G., Kattan, M. W., Alessandrino, F., Baur, A. D. J., Boesen, L., Franco, F. B., Bonekamp, D., Campa, R., Cash, H., Catalá, V., Crouzet, S., Dinnoo, S., Eastham, J., Fennessy, F. M., Ghabili, K., Hohenfellner, M., Levi, A. W., Ji, X., Løgager, V., ... Shukla-Dave, A. (2021). International Multi-Site Initiative to Develop an MRI-Inclusive Nomogram for Side-Specific Prediction of Extraprostatic Extension of Prostate Cancer. Cancers, 13(11), 2627. https://doi.org/10.3390/cancers13112627







