Bone Marrow Mesenchymal Stromal Cells in Multiple Myeloma: Their Role as Active Contributors to Myeloma Progression
Abstract
Simple Summary
Abstract
1. Multiple Myeloma and the Bone Marrow Microenvironment
2. Characterization of Mesenchymal Stromal Cells in Multiple Myeloma
2.1. Mesenchymal Stromal Cells (MSCs)
2.2. Physiological and Pathological Roles of MSCs in the BM
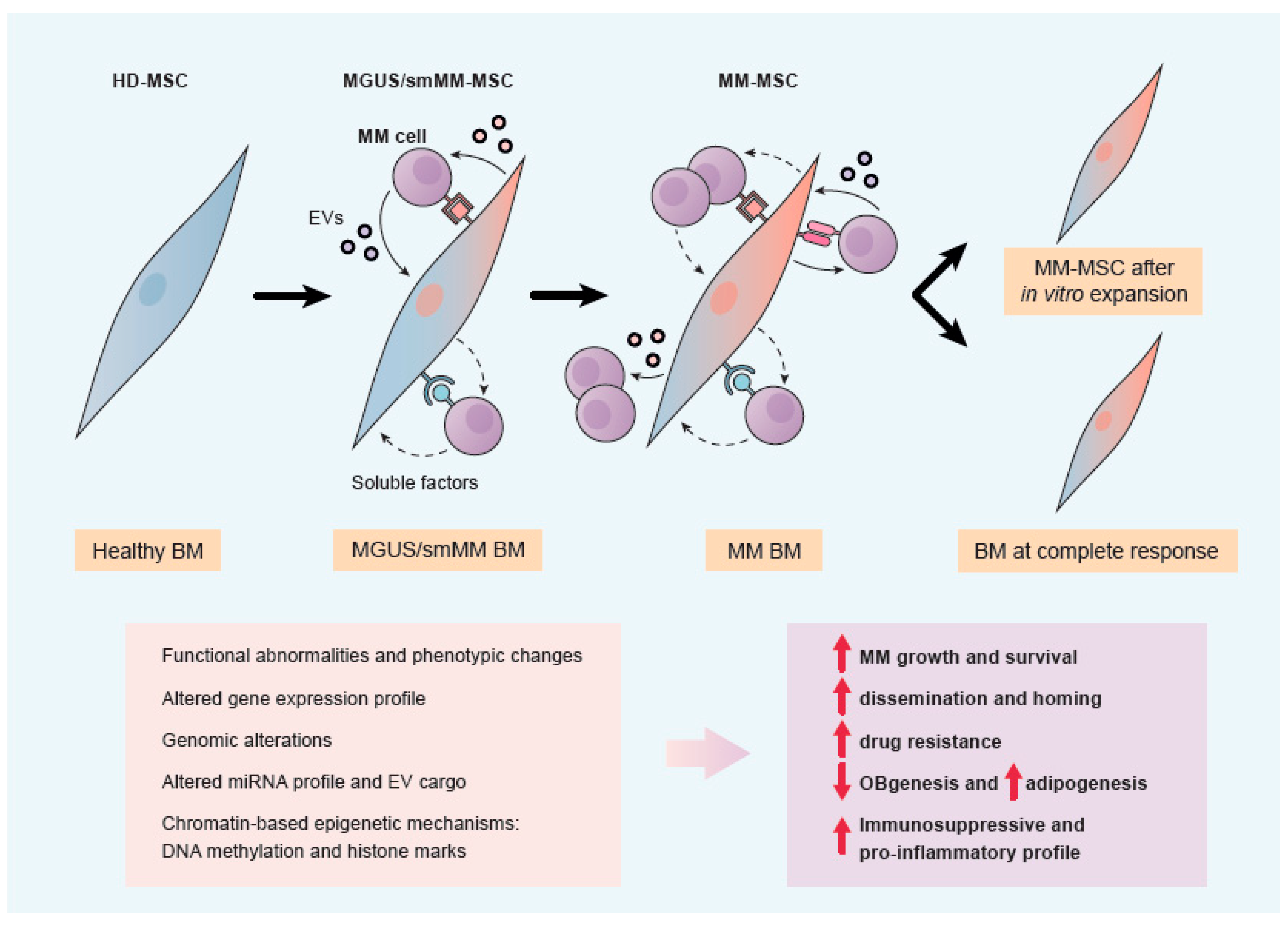
2.3. Comparison of MSCs from Healthy Donors (HD-MSCs) and Myeloma Patients MM-MSCs)
2.3.1. Studies from MSCs after Expansion vs. Fresh MSCs
2.3.2. Studies of MSCs in Monoculture vs. MSCs in Co-Culture with MM Cells
2.3.3. Studies of MSCs in 2D vs. 3D In Vitro Platforms
2.4. Transition from HD-MSCs to MM-MSCs
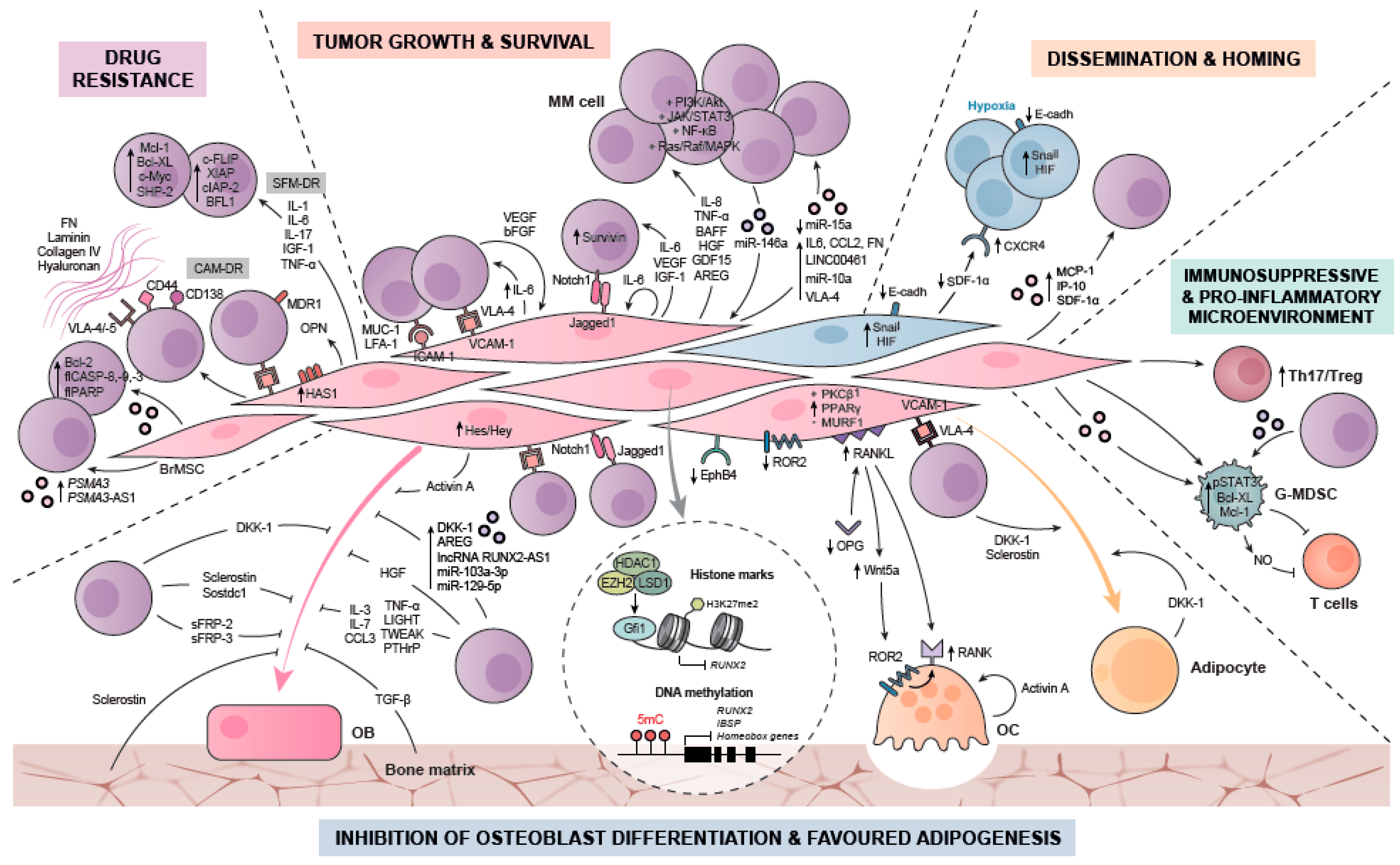
3. Biological Roles of MSCs in MM pathology
3.1. Contribution of MM-MSCs to Tumor Growth and Survival
3.2. Contribution of MM-MSCs to Drug Resistance
3.3. Contribution of MM-MSCs to Dissemination and Homing
3.4. Contribution of MM-MSCs to Myeloma Bone Disease (MBD): Suppressed Osteoblast and Favoured Adipocyte Differentiation
3.4.1. Mechanisms of Osteoblast (OB) Suppression
Soluble Factors (Including Inhibitors of Wnt and BMP Signaling; Cytokines, Chemokines, PTHrP)
Adhesion Interactions of Myeloma Cells with MSCs or Pre-OBs
Deregulated Expression of Surface Molecules such as EphB4 and FZD5/Ror2 on Osteoprogenitor Cells and OBs
Extracellular Vesicles from Myeloma Cells and MicroRNAs Mediating Osteogenic Suppression
Long-Term Inhibition of OB Differentiation by Epigenetic Modifications
3.4.2. Mechanisms of Osteoclast Activation
3.4.3. Mechanisms of Adipocyte Formation
3.5. Contribution to a Pro-Inflammatory and Immunosuppressive Microenvironment
4. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- van de Donk, N.W.C.J.; Pawlyn, C.; Yong, K.L. Multiple Myeloma. Lancet 2021, 397, 410–427. [Google Scholar] [CrossRef]
- Kim, E.B.; Yee, A.J.; Raje, N. Treatment of Smoldering Multiple Myeloma: Ready for Prime Time? Cancers 2020, 12, 1223. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Hideshima, T.; Mitsiades, C.; Tonon, G.; Richardson, P.G.; Anderson, K.C. Understanding Multiple Myeloma Pathogenesis in the Bone Marrow to Identify New Therapeutic Targets. Nat. Rev. Cancer 2007, 7, 585–598. [Google Scholar] [CrossRef] [PubMed]
- Manier, S.; Sacco, A.; Leleu, X.; Ghobrial, I.M.; Roccaro, A.M. Bone Marrow Microenvironment in Multiple Myeloma Progression. J. Biomed. Biotechnol. 2012, 2012, 157496. [Google Scholar] [CrossRef]
- Kawano, Y.; Moschetta, M.; Manier, S.; Glavey, S.; Görgün, G.T.; Roccaro, A.M.; Anderson, K.C.; Ghobrial, I.M. Targeting the Bone Marrow Microenvironment in Multiple Myeloma. Immunol. Rev. 2015, 263, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, G.; Munshi, N.C. Pathogenesis beyond the Cancer Clone(s) in Multiple Myeloma. Blood 2015, 125, 3049–3058. [Google Scholar] [CrossRef]
- Podar, K.; Chauhan, D.; Anderson, K. Bone Marrow Microenvironment and the Identification of New Targets for Myeloma Therapy. Leukemia 2009, 23, 10–24. [Google Scholar] [CrossRef]
- Korde, N.; Kristinsson, S.Y.; Landgren, O. Monoclonal Gammopathy of Undetermined Significance (MGUS) and Smoldering Multiple Myeloma (SMM): Novel Biological Insights and Development of Early Treatment Strategies. Blood 2011, 117, 5573–5581. [Google Scholar] [CrossRef]
- Meads, M.B.; Gatenby, R.A.; Dalton, W.S. Environment-Mediated Drug Resistance: A Major Contributor to Minimal Residual Disease. Nat. Rev. Cancer 2009, 9, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Friedenstein, A.J.; Petrakova, K.V.; Kurolesova, A.I.; Frolova, G.P. Heterotopic of Bone Marrow. Analysis of Precursor Cells for Osteogenic and Hematopoietic Tissues. Transplantation 1968, 6, 230–247. [Google Scholar] [CrossRef] [PubMed]
- Rastegar, F.; Shenaq, D.; Huang, J.; Zhang, W.; Zhang, B.-Q.; He, B.-C.; Chen, L.; Zuo, G.-W.; Luo, Q.; Shi, Q.; et al. Mesenchymal Stem Cells: Molecular Characteristics and Clinical Applications. World J. Stem Cells 2010, 2, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Salem, H.K.; Thiemermann, C. Mesenchymal Stromal Cells: Current Understanding and Clinical Status. Stem Cells 2010, 28, 585–596. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal Criteria for Defining Multipotent Mesenchymal Stromal Cells. The International Society for Cellular Therapy Position Statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Squillaro, T.; Peluso, G.; Galderisi, U. Clinical Trials with Mesenchymal Stem Cells: An Update. Cell Transpl. 2016, 25, 829–848. [Google Scholar] [CrossRef]
- Viswanathan, S.; Shi, Y.; Galipeau, J.; Krampera, M.; Leblanc, K.; Martin, I.; Nolta, J.; Phinney, D.G.; Sensebe, L. Mesenchymal Stem versus Stromal Cells: International Society for Cell & Gene Therapy (ISCT®) Mesenchymal Stromal Cell Committee Position Statement on Nomenclature. Cytotherapy 2019, 21, 1019–1024. [Google Scholar] [CrossRef]
- Dazzi, F.; Ramasamy, R.; Glennie, S.; Jones, S.P.; Roberts, I. The Role of Mesenchymal Stem Cells in Haemopoiesis. Blood Rev. 2006, 20, 161–171. [Google Scholar] [CrossRef]
- Colombo, M.; Mirandola, L.; Platonova, N.; Apicella, L.; Basile, A.; Figueroa, A.J.; Cobos, E.; Chiriva-Internati, M.; Chiaramonte, R. Notch-Directed Microenvironment Reprogramming in Myeloma: A Single Path to Multiple Outcomes. Leukemia 2013, 27, 1009–1018. [Google Scholar] [CrossRef]
- D’Souza, S.; Kurihara, N.; Shiozawa, Y.; Joseph, J.; Taichman, R.; Galson, D.L.; Roodman, G.D. Annexin II Interactions with the Annexin II Receptor Enhance Multiple Myeloma Cell Adhesion and Growth in the Bone Marrow Microenvironment. Blood 2012, 119, 1888–1896. [Google Scholar] [CrossRef]
- Hideshima, T.; Bergsagel, P.L.; Kuehl, W.M.; Anderson, K.C. Advances in Biology of Multiple Myeloma: Clinical Applications. Blood 2004, 104, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Simons, M.; Raposo, G. Exosomes–Vesicular Carriers for Intercellular Communication. Curr. Opin. Cell Biol. 2009, 21, 575–581. [Google Scholar] [CrossRef]
- Gargiulo, E.; Paggetti, J.; Moussay, E. Hematological Malignancy-Derived Small Extracellular Vesicles and Tumor Microenvironment: The Art of Turning Foes into Friends. Cells 2019, 8, 511. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Moscvin, M.; Bianchi, G. Exosomes in the Pathogenesis and Treatment of Multiple Myeloma in the Context of the Bone Marrow Microenvironment. Front. Oncol. 2020, 10, 608815. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Faict, S.; Maes, K.; Bruyne, E.D.; Valckenborgh, E.V.; Schots, R.; Vanderkerken, K.; Menu, E. Extracellular Vesicle Cross-Talk in the Bone Marrow Microenvironment: Implications in Multiple Myeloma. Oncotarget 2016, 7, 38927–38945. [Google Scholar] [CrossRef] [PubMed]
- André, T.; Meuleman, N.; Stamatopoulos, B.; De Bruyn, C.; Pieters, K.; Bron, D.; Lagneaux, L. Evidences of Early Senescence in Multiple Myeloma Bone Marrow Mesenchymal Stromal Cells. PLoS ONE 2013, 8, e59756. [Google Scholar] [CrossRef]
- Corre, J.; Mahtouk, K.; Attal, M.; Gadelorge, M.; Huynh, A.; Fleury-Cappellesso, S.; Danho, C.; Laharrague, P.; Klein, B.; Rème, T.; et al. Bone Marrow Mesenchymal Stem Cells Are Abnormal in Multiple Myeloma. Leukemia 2007, 21, 1079–1088. [Google Scholar] [CrossRef]
- Xu, S.; Evans, H.; Buckle, C.; De Veirman, K.; Hu, J.; Xu, D.; Menu, E.; De Becker, A.; Vande Broek, I.; Leleu, X.; et al. Impaired Osteogenic Differentiation of Mesenchymal Stem Cells Derived from Multiple Myeloma Patients Is Associated with a Blockade in the Deactivation of the Notch Signaling Pathway. Leukemia 2012, 26, 2546–2549. [Google Scholar] [CrossRef]
- Li, B.; Shi, M.; Li, J.; Zhang, H.; Chen, B.; Chen, L.; Gao, W.; Giuliani, N.; Zhao, R.C. Elevated Tumor Necrosis Factor-Alpha Suppresses TAZ Expression and Impairs Osteogenic Potential of Flk-1+ Mesenchymal Stem Cells in Patients with Multiple Myeloma. Stem Cells Dev. 2007, 16, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Garderet, L.; Mazurier, C.; Chapel, A.; Ernou, I.; Boutin, L.; Holy, X.; Gorin, N.C.; Lopez, M.; Doucet, C.; Lataillade, J.-J. Mesenchymal Stem Cell Abnormalities in Patients with Multiple Myeloma. Leuk. Lymphoma 2007, 48, 2032–2041. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Z.; Yao, C. Angiogenic Activity of Mesenchymal Stem Cells in Multiple Myeloma. Cancer Invest. 2011, 29, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Arnulf, B.; Lecourt, S.; Soulier, J.; Ternaux, B.; Lacassagne, M.-N.; Crinquette, A.; Dessoly, J.; Sciaini, A.-K.; Benbunan, M.; Chomienne, C.; et al. Phenotypic and Functional Characterization of Bone Marrow Mesenchymal Stem Cells Derived from Patients with Multiple Myeloma. Leukemia 2007, 21, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Fu, J.; Chen, P.; Zhuang, W. Impairment in Immunomodulatory Function of Mesenchymal Stem Cells from Multiple Myeloma Patients. Arch. Med. Res. 2010, 41, 623–633. [Google Scholar] [CrossRef]
- Zdzisińska, B.; Bojarska-Junak, A.; Dmoszyńska, A.; Kandefer-Szerszeń, M. Abnormal Cytokine Production by Bone Marrow Stromal Cells of Multiple Myeloma Patients in Response to RPMI8226 Myeloma Cells. Arch. Immunol. Ther. Exp. 2008, 56, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Todoerti, K.; Lisignoli, G.; Storti, P.; Agnelli, L.; Novara, F.; Manferdini, C.; Codeluppi, K.; Colla, S.; Crugnola, M.; Abeltino, M.; et al. Distinct Transcriptional Profiles Characterize Bone Microenvironment Mesenchymal Cells Rather than Osteoblasts in Relationship with Multiple Myeloma Bone Disease. Exp. Hematol. 2010, 38, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Garayoa, M.; Garcia, J.L.; Santamaria, C.; Garcia-Gomez, A.; Blanco, J.F.; Pandiella, A.; Hernández, J.M.; Sanchez-Guijo, F.M.; del Cañizo, M.-C.; Gutiérrez, N.C.; et al. Mesenchymal Stem Cells from Multiple Myeloma Patients Display Distinct Genomic Profile as Compared with Those from Normal Donors. Leukemia 2009, 23, 1515–1527. [Google Scholar] [CrossRef]
- Berenstein, R.; Blau, O.; Nogai, A.; Waechter, M.; Slonova, E.; Schmidt-Hieber, M.; Kunitz, A.; Pezzutto, A.; Doerken, B.; Blau, I.W. Multiple Myeloma Cells Alter the Senescence Phenotype of Bone Marrow Mesenchymal Stromal Cells under Participation of the DLK1-DIO3 Genomic Region. BMC Cancer 2015, 15, 68. [Google Scholar] [CrossRef]
- Campioni, D.; Bardi, M.A.; Cavazzini, F.; Tammiso, E.; Pezzolo, E.; Pregnolato, E.; Volta, E.; Cuneo, A.; Lanza, F. Cytogenetic and Molecular Cytogenetic Profile of Bone Marrow-Derived Mesenchymal Stromal Cells in Chronic and Acute Lymphoproliferative Disorders. Ann. Hematol. 2012, 91, 1563–1577. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Jiang, Y.; Li, A.; Liu, X.; Xing, X.; Guo, Y.; Xu, Y.; Hao, Y.; Zheng, C. Telomere Length Is Positively Associated with the Expression of IL-6 and MIP-1α in Bone Marrow Mesenchymal Stem Cells of Multiple Myeloma. Mol. Med. Rep. 2017, 16, 2497–2504. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mazziotta, C.; Lanzillotti, C.; Iaquinta, M.R.; Taraballi, F.; Torreggiani, E.; Rotondo, J.C.; Otòn-Gonzalez, L.; Mazzoni, E.; Frontini, F.; Bononi, I.; et al. MicroRNAs Modulate Signaling Pathways in Osteogenic Differentiation of Mesenchymal Stem Cells. Int. J. Mol. Sci. 2021, 22, 2362. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Cecilia Santini, G.; De Veirman, K.; Vande Broek, I.; Leleu, X.; De Becker, A.; Van Camp, B.; Vanderkerken, K.; Van Riet, I. Upregulation of MiR-135b Is Involved in the Impaired Osteogenic Differentiation of Mesenchymal Stem Cells Derived from Multiple Myeloma Patients. PLoS ONE 2013, 8, e79752. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, S.; Løvendorf, M.B.; Park, J.; Salem, K.Z.; Reagan, M.R.; Manier, S.; Zavidij, O.; Rahmat, M.; Huynh, D.; Takagi, S.; et al. Inhibition of MicroRNA-138 Enhances Bone Formation in Multiple Myeloma Bone Marrow Niche. Leukemia 2018, 32, 1739–1750. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, L.; De Luca, A.; Giavaresi, G.; Raimondo, S.; Gallo, A.; Taiana, E.; Alessandro, R.; Rossi, M.; Neri, A.; Viglietto, G.; et al. Non-Coding RNAs in Multiple Myeloma Bone Disease Pathophysiology. Non-Coding RNA 2020, 6, 37. [Google Scholar] [CrossRef] [PubMed]
- Papanota, A.-M.; Karousi, P.; Kontos, C.K.; Ntanasis-Stathopoulos, I.; Scorilas, A.; Terpos, E. Multiple Myeloma Bone Disease: Implication of MicroRNAs in Its Molecular Background. Int. J. Mol. Sci. 2021, 22, 2375. [Google Scholar] [CrossRef]
- Alameda, D.; Saez, B.; Lara-Astiaso, D.; Sarvide, S.; Lasa, M.; Alignani, D.; Rodriguez, I.; Garate, S.; Vilas, A.; Paiva, B.; et al. Characterization of Freshly Isolated Bone Marrow Mesenchymal Stromal Cells from Healthy Donors and Patients with Multiple Myeloma: Transcriptional Modulation of the Microenvironment. Haematologica 2020, 105, e470–e473. [Google Scholar] [CrossRef]
- Schinke, C.; Qu, P.; Mehdi, S.J.; Hoering, A.; Epstein, J.; Johnson, S.K.; van Rhee, F.; Zangari, M.; Thanendrarajan, S.; Barlogie, B.; et al. The Pattern of Mesenchymal Stem Cell Expression Is an Independent Marker of Outcome in Multiple Myeloma. Clin. Cancer Res. 2018, 24, 2913–2919. [Google Scholar] [CrossRef]
- Kanehira, M.; Fujiwara, T.; Nakajima, S.; Okitsu, Y.; Onishi, Y.; Fukuhara, N.; Ichinohasama, R.; Okada, Y.; Harigae, H. A Lysophosphatidic Acid Receptors 1 and 3 Axis Governs Cellular Senescence of Mesenchymal Stromal Cells and Promotes Growth and Vascularization of Multiple Myeloma: Mesenchymal Stromal Cells and Multiple Myeloma. Stem Cells 2017, 35, 739–753. [Google Scholar] [CrossRef]
- Guo, J.; Zhao, Y.; Fei, C.; Zhao, S.; Zheng, Q.; Su, J.; Wu, D.; Li, X.; Chang, C. Dicer1 Downregulation by Multiple Myeloma Cells Promotes the Senescence and Tumor-Supporting Capacity and Decreases the Differentiation Potential of Mesenchymal Stem Cells. Cell Death Dis. 2018, 9, 512. [Google Scholar] [CrossRef]
- Garcia-Gomez, A.; De Las Rivas, J.; Ocio, E.M.; Díaz-Rodríguez, E.; Montero, J.C.; Martín, M.; Blanco, J.F.; Sanchez-Guijo, F.M.; Pandiella, A.; San Miguel, J.F.; et al. Transcriptomic Profile Induced in Bone Marrow Mesenchymal Stromal Cells after Interaction with Multiple Myeloma Cells: Implications in Myeloma Progression and Myeloma Bone Disease. Oncotarget 2014, 5, 8284–8305. [Google Scholar] [CrossRef]
- Dotterweich, J.; Schlegelmilch, K.; Keller, A.; Geyer, B.; Schneider, D.; Zeck, S.; Tower, R.J.J.; Ebert, R.; Jakob, F.; Schütze, N. Contact of Myeloma Cells Induces a Characteristic Transcriptome Signature in Skeletal Precursor Cells -Implications for Myeloma Bone Disease. Bone 2016, 93, 155–166. [Google Scholar] [CrossRef]
- Knowles, H.J. Multiple Roles of Angiopoietin-Like 4 in Osteolytic Disease. Front. Endocrinol. 2017, 8, 80. [Google Scholar] [CrossRef] [PubMed]
- Abdi, J.; Qiu, L.; Chang, H. Micro-RNAs, New Performers in Multiple Myeloma Bone Marrow Microenvironment. Biomark Res. 2014, 2, 10. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, L.; De Luca, A.; Morelli, E.; Giavaresi, G.; Tagliaferri, P.; Tassone, P.; Amodio, N. MicroRNAs: Novel Crossroads between Myeloma Cells and the Bone Marrow Microenvironment. Biomed. Res. Int. 2016, 2016, 6504593. [Google Scholar] [CrossRef] [PubMed]
- Pitari, M.R.; Rossi, M.; Amodio, N.; Botta, C.; Morelli, E.; Federico, C.; Gullà, A.; Caracciolo, D.; Di Martino, M.T.; Arbitrio, M.; et al. Inhibition of MiR-21 Restores RANKL/OPG Ratio in Multiple Myeloma-Derived Bone Marrow Stromal Cells and Impairs the Resorbing Activity of Mature Osteoclasts. Oncotarget 2015, 6, 27343–27358. [Google Scholar] [CrossRef]
- De Veirman, K.; Wang, J.; Xu, S.; Leleu, X.; Himpe, E.; Maes, K.; De Bruyne, E.; Van Valckenborgh, E.; Vanderkerken, K.; Menu, E.; et al. Induction of MiR-146a by Multiple Myeloma Cells in Mesenchymal Stromal Cells Stimulates Their pro-Tumoral Activity. Cancer Lett. 2016, 377, 17–24. [Google Scholar] [CrossRef]
- Cheng, Q.; Li, X.; Liu, J.; Ye, Q.; Chen, Y.; Tan, S.; Liu, J. Multiple Myeloma-Derived Exosomes Regulate the Functions of Mesenchymal Stem Cells Partially via Modulating MiR-21 and MiR-146a. Stem Cells Int. 2017, 2017, 9012152. [Google Scholar] [CrossRef] [PubMed]
- Raimondo, S.; Urzì, O.; Conigliaro, A.; Lo Bosco, G.; Parisi, S.; Carlisi, M.; Siragusa, S.; Raimondi, L.; De Luca, A.; Giavaresi, G.; et al. Extracellular Vesicle MicroRNAs Contribute to the Osteogenic Inhibition of Mesenchymal Stem Cells in Multiple Myeloma. Cancers 2020, 12, 449. [Google Scholar] [CrossRef]
- Raimondo, S.; Urzì, O.; Conigliaro, A.; Raimondi, L.; Amodio, N.; Alessandro, R. Emerging Insights on the Biological Impact of Extracellular Vesicle-Associated NcRNAs in Multiple Myeloma. Noncoding RNA 2020, 6, 30. [Google Scholar] [CrossRef]
- Adamik, J.; Jin, S.; Sun, Q.; Zhang, P.; Weiss, K.R.; Anderson, J.L.; Silbermann, R.; Roodman, G.D.; Galson, D.L. EZH2 or HDAC1 Inhibition Reverses Multiple Myeloma-Induced Epigenetic Suppression of Osteoblast Differentiation. Mol. Cancer Res. 2017, 15, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Adamik, J.; Galson, D.L.; Roodman, G.D. Osteoblast Suppression in Multiple Myeloma Bone Disease. J. Bone Oncol. 2018, 13, 62–70. [Google Scholar] [CrossRef]
- Garcia-Gomez, A.; Li, T.; de la Calle-Fabregat, C.; Rodríguez-Ubreva, J.; Ciudad, L.; Català-Moll, F.; Godoy-Tena, G.; Martín-Sánchez, M.; San-Segundo, L.; Muntión, S.; et al. Targeting Aberrant DNA Methylation in Mesenchymal Stromal Cells as a Treatment for Myeloma Bone Disease. Nat. Commun. 2021, 12, 421. [Google Scholar] [CrossRef] [PubMed]
- Silbermann, R.; Roodman, G.D. Current Controversies in the Management of Myeloma Bone Disease. J. Cell. Physiol. 2016, 231, 2374–2379. [Google Scholar] [CrossRef]
- Adamik, J.; Roodman, G.D.; Galson, D.L. Epigenetic-Based Mechanisms of Osteoblast Suppression in Multiple Myeloma Bone Disease. JBMR Plus 2019, 3, e10183. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.M.; Chen, C.S. Deconstructing the Third Dimension: How 3D Culture Microenvironments Alter Cellular Cues. J. Cell Sci. 2012, 125, 3015–3024. [Google Scholar] [CrossRef] [PubMed]
- Zdzisińska, B.; Roliński, J.; Piersiak, T.; Kandefer-Szerszeń, M. A Comparison of Cytokine Production in 2-Dimensional and 3-Dimensional Cultures of Bone Marrow Stromal Cells of Multiple Myeloma Patients in Response to RPMI8226 Myeloma Cells. Folia Histochem. Cytobiol. 2009, 47, 69–74. [Google Scholar] [CrossRef]
- Reagan, M.R.; Mishima, Y.; Glavey, S.V.; Zhang, Y.; Manier, S.; Lu, Z.N.; Memarzadeh, M.; Zhang, Y.; Sacco, A.; Aljawai, Y.; et al. Investigating Osteogenic Differentiation in Multiple Myeloma Using a Novel 3D Bone Marrow Niche Model. Blood 2014, 124, 3250–3259. [Google Scholar] [CrossRef]
- Roccaro, A.M.; Sacco, A.; Maiso, P.; Azab, A.K.; Tai, Y.-T.; Reagan, M.; Azab, F.; Flores, L.M.; Campigotto, F.; Weller, E.; et al. BM Mesenchymal Stromal Cell-Derived Exosomes Facilitate Multiple Myeloma Progression. J. Clin. Invest. 2013, 123, 1542–1555. [Google Scholar] [CrossRef]
- Belloni, D.; Heltai, S.; Ponzoni, M.; Villa, A.; Vergani, B.; Pecciarini, L.; Marcatti, M.; Girlanda, S.; Tonon, G.; Ciceri, F.; et al. Modeling Multiple Myeloma-Bone Marrow Interactions and Response to Drugs in a 3D Surrogate Microenvironment. Haematologica 2018, 103, 707–716. [Google Scholar] [CrossRef]
- Braham, M.V.J.; Alblas, J.; Dhert, W.J.A.; Öner, F.C.; Minnema, M.C. Possibilities and Limitations of an in Vitro Three-Dimensional Bone Marrow Model for the Prediction of Clinical Responses in Patients with Relapsed Multiple Myeloma. Haematologica 2019, 104, e523–e526. [Google Scholar] [CrossRef]
- de la Puente, P.; Muz, B.; Gilson, R.C.; Azab, F.; Luderer, M.; King, J.; Achilefu, S.; Vij, R.; Azab, A.K. 3D Tissue-Engineered Bone Marrow as a Novel Model to Study Pathophysiology and Drug Resistance in Multiple Myeloma. Biomaterials 2015, 73, 70–84. [Google Scholar] [CrossRef]
- Marcus, H.; Attar-Schneider, O.; Dabbah, M.; Zismanov, V.; Tartakover-Matalon, S.; Lishner, M.; Drucker, L. Mesenchymal Stem Cells Secretomes’ Affect Multiple Myeloma Translation Initiation. Cell. Signal. 2016, 28, 620–630. [Google Scholar] [CrossRef] [PubMed]
- Hideshima, T.; Anderson, K.C. Signaling Pathway Mediating Myeloma Cell Growth and Survival. Cancers 2021, 13, 216. [Google Scholar] [CrossRef] [PubMed]
- Hiroshi, Y.; Teru, H.; Paul, G.R.; Kenneth, C.A. Recent Advances in the Treatment of Multiple Myeloma. Curr. Pharm. Biotechnol. 2006, 7, 381–393. [Google Scholar]
- Neri, P.; Bahlis, N.J. Targeting of Adhesion Molecules as a Therapeutic Strategy in Multiple Myeloma. Curr. Cancer Drug Targets 2012, 12, 776–796. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, Z.; Yao, C. Survivin Is Upregulated in Myeloma Cell Lines Cocultured with Mesenchymal Stem Cells. Leuk. Res. 2010, 34, 1325–1329. [Google Scholar] [CrossRef]
- Radtke, F.; Raj, K. The Role of Notch in Tumorigenesis: Oncogene or Tumour Suppressor? Nat. Rev. Cancer 2003, 3, 756–767. [Google Scholar] [CrossRef]
- Nefedova, Y.; Cheng, P.; Alsina, M.; Dalton, W.S.; Gabrilovich, D.I. Involvement of Notch-1 Signaling in Bone Marrow Stroma-Mediated de Novo Drug Resistance of Myeloma and Other Malignant Lymphoid Cell Lines. Blood 2004, 103, 3503–3510. [Google Scholar] [CrossRef]
- Gunn, W.G.; Conley, A.; Deininger, L.; Olson, S.D.; Prockop, D.J.; Gregory, C.A. A Crosstalk Between Myeloma Cells and Marrow Stromal Cells Stimulates Production of DKK1 and Interleukin-6: A Potential Role in the Development of Lytic Bone Disease and Tumor Progression in Multiple Myeloma. Stem Cells 2006, 24, 986–991. [Google Scholar] [CrossRef]
- Dabbah, M.; Attar-Schneider, O.; Tartakover Matalon, S.; Shefler, I.; Jarchwsky Dolberg, O.; Lishner, M.; Drucker, L. Microvesicles Derived from Normal and Multiple Myeloma Bone Marrow Mesenchymal Stem Cells Differentially Modulate Myeloma Cells’ Phenotype and Translation Initiation. Carcinogenesis 2017, 38, 708–716. [Google Scholar] [CrossRef]
- Dabbah, M.; Jarchowsky-Dolberg, O.; Attar-Schneider, O.; Tartakover Matalon, S.; Pasmanik-Chor, M.; Drucker, L.; Lishner, M. Multiple Myeloma BM-MSCs Increase the Tumorigenicity of MM Cells via Transfer of VLA4-Enriched Microvesicles. Carcinogenesis 2020, 41, 100–110. [Google Scholar] [CrossRef]
- Deng, M.; Yuan, H.; Liu, S.; Hu, Z.; Xiao, H. Exosome-Transmitted LINC00461 Promotes Multiple Myeloma Cell Proliferation and Suppresses Apoptosis by Modulating MicroRNA/BCL-2 Expression. Cytotherapy 2019, 21, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Umezu, T.; Imanishi, S.; Yoshizawa, S.; Kawana, C.; Ohyashiki, J.H.; Ohyashiki, K. Induction of Multiple Myeloma Bone Marrow Stromal Cell Apoptosis by Inhibiting Extracellular Vesicle MiR-10a Secretion. Blood Adv. 2019, 3, 3228–3240. [Google Scholar] [CrossRef]
- Bellamy, W.T.; Dalton, W.S.; Gleason, M.C.; Grogan, T.M.; Trent, J.M. Development and Characterization of a Melphalan-Resistant Human Multiple Myeloma Cell Line. Cancer Res. 1991, 51, 995–1002. [Google Scholar] [PubMed]
- Goldie, J.H.; Coldman, A.J. A Mathematic Model for Relating the Drug Sensitivity of Tumors to Their Spontaneous Mutation Rate. Cancer Treat. Rep. 1979, 63, 1727–1733. [Google Scholar] [PubMed]
- Teicher, B.A.; Herman, T.S.; Holden, S.A.; Wang, Y.Y.; Pfeffer, M.R.; Crawford, J.W.; Frei, E. Tumor Resistance to Alkylating Agents Conferred by Mechanisms Operative Only in Vivo. Science 1990, 247, 1457–1461. [Google Scholar] [CrossRef]
- Damiano, J.S.; Cress, A.E.; Hazlehurst, L.A.; Shtil, A.A.; Dalton, W.S. Cell Adhesion Mediated Drug Resistance (CAM-DR): Role of Integrins and Resistance to Apoptosis in Human Myeloma Cell Lines. Blood 1999, 93, 1658–1667. [Google Scholar] [CrossRef] [PubMed]
- Meads, M.B.; Hazlehurst, L.A.; Dalton, W.S. The Bone Marrow Microenvironment as a Tumor Sanctuary and Contributor to Drug Resistance. Clin. Cancer Res. 2008, 14, 2519–2526. [Google Scholar] [CrossRef] [PubMed]
- Cordes, N.; Seidler, J.; Durzok, R.; Geinitz, H.; Brakebusch, C. β 1-Integrin-Mediated Signaling Essentially Contributes to Cell Survival after Radiation-Induced Genotoxic Injury. Oncogene 2006, 25, 1378–1390. [Google Scholar] [CrossRef]
- Hazlehurst, L.A.; Argilagos, R.F.; Dalton, W.S. Β1 Integrin Mediated Adhesion Increases Bim Protein Degradation and Contributes to Drug Resistance in Leukaemia Cells. Br. J. Haematol. 2007, 136, 269–275. [Google Scholar] [CrossRef]
- Shain, K.H.; Landowski, T.H.; Dalton, W.S. Adhesion-Mediated Intracellular Redistribution of c-Fas-Associated Death Domain-Like IL-1-Converting Enzyme-Like Inhibitory Protein-Long Confers Resistance to CD95-Induced Apoptosis in Hematopoietic Cancer Cell Lines. J. Immunol. 2002, 168, 2544–2553. [Google Scholar] [CrossRef]
- Lwin, T.; Hazlehurst, L.A.; Dessureault, S.; Lai, R.; Bai, W.; Sotomayor, E.; Moscinski, L.C.; Dalton, W.S.; Tao, J. Cell Adhesion Induces P27Kip1-Associated Cell-Cycle Arrest through down-Regulation of the SCFSkp2 Ubiquitin Ligase Pathway in Mantle-Cell and Other Non-Hodgkin B-Cell Lymphomas. Blood 2007, 110, 1631–1638. [Google Scholar] [CrossRef] [PubMed]
- Hardin, J.; MacLeod, S.; Grigorieva, I.; Chang, R.; Barlogie, B.; Xiao, H.; Epstein, J. Interleukin-6 Prevents Dexamethasone-Induced Myeloma Cell Death. Blood 1994, 84, 3063–3070. [Google Scholar] [CrossRef]
- Chauhan, D.; Pandey, P.; Hideshima, T.; Treon, S.; Raje, N.; Davies, F.E.; Shima, Y.; Tai, Y.-T.; Rosen, S.; Avraham, S.; et al. SHP2 Mediates the Protective Effect of Interleukin-6 against Dexamethasone-Induced Apoptosis in Multiple Myeloma Cells. J. Biol. Chem. 2000, 275, 27845–27850. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Denu, R.A.; Dollar, B.A.; Escalante, L.E.; Kuether, J.P.; Callander, N.S.; Asimakopoulos, F.; Hematti, P. Macrophages and Mesenchymal Stromal Cells Support Survival and Proliferation of Multiple Myeloma Cells. Br. J. Haematol. 2012, 158, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Michigami, T.; Shimizu, N.; Williams, P.J.; Niewolna, M.; Dallas, S.L.; Mundy, G.R.; Yoneda, T. Cell–Cell Contact between Marrow Stromal Cells and Myeloma Cells via VCAM-1 and A4β1-Integrin Enhances Production of Osteoclast-Stimulating Activity. Blood 2000, 96, 1953–1960. [Google Scholar] [CrossRef] [PubMed]
- Delmore, J.E.; Issa, G.C.; Lemieux, M.E.; Rahl, P.B.; Shi, J.; Jacobs, H.M.; Kastritis, E.; Gilpatrick, T.; Paranal, R.M.; Qi, J.; et al. BET Bromodomain Inhibition as a Therapeutic Strategy to Target C-Myc. Cell 2011, 146, 904–917. [Google Scholar] [CrossRef]
- Noll, J.E.; Williams, S.A.; Tong, C.M.; Wang, H.; Quach, J.M.; Purton, L.E.; Pilkington, K.; To, L.B.; Evdokiou, A.; Gronthos, S.; et al. Myeloma Plasma Cells Alter the Bone Marrow Microenvironment by Stimulating the Proliferation of Mesenchymal Stromal Cells. Haematologica 2014, 99, 163–171. [Google Scholar] [CrossRef]
- Qiang, Y.-W.; Kopantzev, E.; Rudikoff, S. Insulinlike Growth Factor–I Signaling in Multiple Myeloma: Downstream Elements, Functional Correlates, and Pathway Cross-Talk. Blood 2002, 99, 4138–4146. [Google Scholar] [CrossRef]
- Mitsiades, C.S.; Mitsiades, N.; Poulaki, V.; Schlossman, R.; Akiyama, M.; Chauhan, D.; Hideshima, T.; Treon, S.P.; Munshi, N.C.; Richardson, P.G.; et al. Activation of NF-ΚB and Upregulation of Intracellular Anti-Apoptotic Proteins via the IGF-1/Akt Signaling in Human Multiple Myeloma Cells: Therapeutic Implications. Oncogene 2002, 21, 5673–5683. [Google Scholar] [CrossRef]
- Akiyama, M.; Hideshima, T.; Hayashi, T.; Tai, Y.-T.; Mitsiades, C.S.; Mitsiades, N.; Chauhan, D.; Richardson, P.; Munshi, N.C.; Anderson, K.C. Cytokines Modulate Telomerase Activity in a Human Multiple Myeloma Cell Line. Cancer Res. 2002, 62, 3876–3882. [Google Scholar]
- Hatano, K.; Kikuchi, J.; Takatoku, M.; Shimizu, R.; Wada, T.; Ueda, M.; Nobuyoshi, M.; Oh, I.; Sato, K.; Suzuki, T.; et al. Bortezomib Overcomes Cell Adhesion-Mediated Drug Resistance through Downregulation of VLA-4 Expression in Multiple Myeloma. Oncogene 2009, 28, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Landowski, T.H.; Olashaw, N.E.; Agrawal, D.; Dalton, W.S. Cell Adhesion-Mediated Drug Resistance (CAM-DR) Is Associated with Activation of NF- κ B (RelB/P50) in Myeloma Cells. Oncogene 2003, 22, 2417–2421. [Google Scholar] [CrossRef] [PubMed]
- Calabro, A.; Oken, M.M.; Hascall, V.C.; Masellis, A.M. Characterization of Hyaluronan Synthase Expression and Hyaluronan Synthesis in Bone Marrow Mesenchymal Progenitor Cells: Predominant Expression of HAS1 MRNA and up-Regulated Hyaluronan Synthesis in Bone Marrow Cells Derived from Multiple Myeloma Patients. Blood 2002, 100, 2578–2585. [Google Scholar] [CrossRef] [PubMed]
- Vincent, T.; Molina, L.; Espert, L.; Mechti, N. Hyaluronan, a Major Non-Protein Glycosaminoglycan Component of the Extracellular Matrix in Human Bone Marrow, Mediates Dexamethasone Resistance in Multiple Myeloma. Br. J. Haematol. 2003, 121, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Tajima, K.; Ohashi, R.; Sekido, Y.; Hida, T.; Nara, T.; Hashimoto, M.; Iwakami, S.; Minakata, K.; Yae, T.; Takahashi, F.; et al. Osteopontin-Mediated Enhanced Hyaluronan Binding Induces Multidrug Resistance in Mesothelioma Cells. Oncogene 2010, 29, 1941–1951. [Google Scholar] [CrossRef]
- Wang, J.; Hendrix, A.; Hernot, S.; Lemaire, M.; De Bruyne, E.; Van Valckenborgh, E.; Lahoutte, T.; De Wever, O.; Vanderkerken, K.; Menu, E. Bone Marrow Stromal Cell-Derived Exosomes as Communicators in Drug Resistance in Multiple Myeloma Cells. Blood 2014, 124, 555–566. [Google Scholar] [CrossRef]
- Zhang, L.; Pan, L.; Xiang, B.; Zhu, H.; Wu, Y.; Chen, M.; Guan, P.; Zou, X.; Valencia, C.A.; Dong, B.; et al. Potential Role of Exosome-Associated MicroRNA Panels and in Vivo Environment to Predict Drug Resistance for Patients with Multiple Myeloma. Oncotarget 2016, 7, 30876–30891. [Google Scholar] [CrossRef]
- Xu, H.; Han, H.; Song, S.; Yi, N.; Qian, C.; Qiu, Y.; Zhou, W.; Hong, Y.; Zhuang, W.; Li, Z.; et al. Exosome-Transmitted PSMA3 and PSMA3-AS1 Promotes Proteasome Inhibitors Resistance in Multiple Myeloma. Clin. Cancer Res. 2019, 25, 1923–1935. [Google Scholar] [CrossRef]
- Paiva, B.; Paino, T.; Sayagues, J.-M.; Garayoa, M.; San-Segundo, L.; Martín, M.; Mota, I.; Sanchez, M.-L.; Bárcena, P.; Aires-Mejia, I.; et al. Detailed Characterization of Multiple Myeloma Circulating Tumor Cells Shows Unique Phenotypic, Cytogenetic, Functional, and Circadian Distribution Profile. Blood 2013, 122, 3591–3598. [Google Scholar] [CrossRef]
- Chaidos, A.; Barnes, C.P.; Cowan, G.; May, P.C.; Melo, V.; Hatjiharissi, E.; Papaioannou, M.; Harrington, H.; Doolittle, H.; Terpos, E.; et al. Clinical Drug Resistance Linked to Interconvertible Phenotypic and Functional States of Tumor-Propagating Cells in Multiple Myeloma. Blood 2013, 121, 318–328. [Google Scholar] [CrossRef]
- Nowakowski, G.S.; Witzig, T.E.; Dingli, D.; Tracz, M.J.; Gertz, M.A.; Lacy, M.Q.; Lust, J.A.; Dispenzieri, A.; Greipp, P.R.; Kyle, R.A.; et al. Circulating Plasma Cells Detected by Flow Cytometry as a Predictor of Survival in 302 Patients with Newly Diagnosed Multiple Myeloma. Blood 2005, 106, 2276–2279. [Google Scholar] [CrossRef] [PubMed]
- Ghobrial, I.M. Myeloma as a Model for the Process of Metastasis: Implications for Therapy. Blood 2012, 120, 20–30. [Google Scholar] [CrossRef]
- Hideshima, T.; Chauhan, D.; Hayashi, T.; Podar, K.; Akiyama, M.; Gupta, D.; Richardson, P.; Munshi, N.; Anderson, K.C. The Biological Sequelae of Stromal Cell-Derived Factor-1α in Multiple Myeloma 1 This Work Was Supported by Multiple Myeloma Research Foundation Senior Awards (to T. H. and D. C.), Leukemia and Lymphoma Society Scholar Award (to N. M.), NIH Grant PO-1 78378, and the Doris Duke Distinguished Clinical Research Scientist Award (to K. A.). Mol. Cancer Ther. 2002, 1, 539–544. [Google Scholar] [PubMed]
- Alsayed, Y.; Ngo, H.; Runnels, J.; Leleu, X.; Singha, U.K.; Pitsillides, C.M.; Spencer, J.A.; Kimlinger, T.; Ghobrial, J.M.; Jia, X.; et al. Mechanisms of Regulation of CXCR4/SDF-1 (CXCL12)–Dependent Migration and Homing in Multiple Myeloma. Blood 2006, 109, 2708–2717. [Google Scholar] [CrossRef] [PubMed]
- Roccaro, A.M.; Sacco, A.; Purschke, W.G.; Moschetta, M.; Buchner, K.; Maasch, C.; Zboralski, D.; Zöllner, S.; Vonhoff, S.; Mishima, Y.; et al. SDF-1 Inhibition Targets the Bone Marrow Niche for Cancer Therapy. Cell Rep. 2014, 9, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Lapidot, T. Mechanism of Human Stem Cell Migration and Repopulation of NOD/SCID and B2mnull NOD/SCID Mice. Ann. N. Y. Acad. Sci. 2001, 938, 83–95. [Google Scholar] [CrossRef]
- Cipolleschi, M.G.; Sbarba, P.D.; Olivotto, M. The Role of Hypoxia in the Maintenance of Hematopoietic Stem Cells. Blood 1993, 82, 2031–2037. [Google Scholar] [CrossRef]
- Azab, A.K.; Hu, J.; Quang, P.; Azab, F.; Pitsillides, C.; Awwad, R.; Thompson, B.; Maiso, P.; Sun, J.D.; Hart, C.P.; et al. Hypoxia Promotes Dissemination of Multiple Myeloma through Acquisition of Epithelial to Mesenchymal Transition-like Features. Blood 2012, 119, 5782–5794. [Google Scholar] [CrossRef]
- Sanz-Rodríguez, F.; Hidalgo, A.; Teixidó, J. Chemokine Stromal Cell-Derived Factor-1α Modulates VLA-4 Integrin-Mediated Multiple Myeloma Cell Adhesion to CS-1/Fibronectin and VCAM-1. Blood 2001, 97, 346–351. [Google Scholar] [CrossRef]
- Azab, A.K.; Azab, F.; Blotta, S.; Pitsillides, C.M.; Thompson, B.; Runnels, J.M.; Roccaro, A.M.; Ngo, H.T.; Melhem, M.R.; Sacco, A.; et al. RhoA and Rac1 GTPases Play Major and Differential Roles in Stromal Cell–Derived Factor-1–Induced Cell Adhesion and Chemotaxis in Multiple Myeloma. Blood 2009, 114, 619–629. [Google Scholar] [CrossRef]
- Roodman, G.D. Osteoblast Function in Myeloma. Bone 2011, 48, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Longo, V.; Brunetti, O.; D’Oronzo, S.; Dammacco, F.; Silvestris, F. Therapeutic Approaches to Myeloma Bone Disease: An Evolving Story. Cancer Treat. Rev. 2012, 38, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Sonmez, M.; Akagun, T.; Topbas, M.; Cobanoglu, U.; Sonmez, B.; Yilmaz, M.; Ovali, E.; Omay, S.B. Effect of Pathologic Fractures on Survival in Multiple Myeloma Patients: A Case Control Study. J. Exp. Clin. Cancer Res. 2008, 27, 11. [Google Scholar] [CrossRef]
- Garcia-Gomez, A.; Quwaider, D.; Canavese, M.; Ocio, E.M.; Tian, Z.; Blanco, J.F.; Berger, A.J.; Ortiz-de-Solorzano, C.; Hernández-Iglesias, T.; Martens, A.C.M.; et al. Preclinical Activity of the Oral Proteasome Inhibitor MLN9708 in Myeloma Bone Disease. Clin. Cancer Res. 2014, 20, 1542–1554. [Google Scholar] [CrossRef] [PubMed]
- Hurchla, M.; Garcia-Gomez, A.; Hornick, M.; Ocio, E.; Li, A.; Blanco, J.; Collins, L.; Kirk, C.; Piwnica-Worms, D.; Vij, R.; et al. The Epoxyketone-Based Proteasome Inhibitors Carfilzomib and Orally Bioavailable Oprozomib Have Anti-Resorptive and Bone-Anabolic Activity in Addition to Anti-Myeloma Effects. Leukemia 2013, 27, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Terpos, E.; Ntanasis-Stathopoulos, I.; Gavriatopoulou, M.; Dimopoulos, M.A. Pathogenesis of Bone Disease in Multiple Myeloma: From Bench to Bedside. Blood Cancer J. 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.L.; Hankenson, K.D. Integration of BMP, Wnt, and Notch Signaling Pathways in Osteoblast Differentiation. J. Cell. Biochem. 2011, 112, 3491–3501. [Google Scholar] [CrossRef]
- Galli, C.; Piemontese, M.; Lumetti, S.; Manfredi, E.; Macaluso, G.M.; Passeri, G. The Importance of WNT Pathways for Bone Metabolism and Their Regulation by Implant Topography. Eur. Cell Mater. 2012, 24, 46–59. [Google Scholar] [CrossRef] [PubMed]
- Tian, E.; Zhan, F.; Walker, R.; Rasmussen, E.; Ma, Y.; Barlogie, B.; Shaughnessy, J.D.J. The Role of the Wnt-Signaling Antagonist DKK1 in the Development of Osteolytic Lesions in Multiple Myeloma. Available online: https://www.nejm.org/doi/10.1056/NEJMoa030847 (accessed on 14 March 2021).
- Terpos, E.; Christoulas, D.; Katodritou, E.; Bratengeier, C.; Gkotzamanidou, M.; Michalis, E.; Delimpasi, S.; Pouli, A.; Meletis, J.; Kastritis, E.; et al. Elevated Circulating Sclerostin Correlates with Advanced Disease Features and Abnormal Bone Remodeling in Symptomatic Myeloma: Reduction Post-Bortezomib Monotherapy. Int. J. Cancer 2012, 131, 1466–1471. [Google Scholar] [CrossRef]
- Qiang, Y.-W.; Barlogie, B.; Rudikoff, S.; Shaughnessy, J.D. Dkk1-Induced Inhibition of Wnt Signaling in Osteoblast Differentiation Is an Underlying Mechanism of Bone Loss in Multiple Myeloma. Bone 2008, 42, 669–680. [Google Scholar] [CrossRef]
- Balemans, W.; Ebeling, M.; Patel, N.; Van Hul, E.; Olson, P.; Dioszegi, M.; Lacza, C.; Wuyts, W.; Van Den Ende, J.; Willems, P.; et al. Increased Bone Density in Sclerosteosis Is Due to the Deficiency of a Novel Secreted Protein (SOST). Hum. Mol. Genet. 2001, 10, 537–544. [Google Scholar] [CrossRef]
- Winkler, D.G.; Sutherland, M.K.; Geoghegan, J.C.; Yu, C.; Hayes, T.; Skonier, J.E.; Shpektor, D.; Jonas, M.; Kovacevich, B.R.; Staehling-Hampton, K.; et al. Osteocyte Control of Bone Formation via Sclerostin, a Novel BMP Antagonist. EMBO J. 2003, 22, 6267–6276. [Google Scholar] [CrossRef] [PubMed]
- Colucci, S.; Brunetti, G.; Oranger, A.; Mori, G.; Sardone, F.; Specchia, G.; Rinaldi, E.; Curci, P.; Liso, V.; Passeri, G.; et al. Myeloma Cells Suppress Osteoblasts through Sclerostin Secretion. Blood Cancer J. 2011, 1, e27. [Google Scholar] [CrossRef] [PubMed]
- Faraahi, Z.; Baud’huin, M.; Croucher, P.I.; Eaton, C.; Lawson, M.A. Sostdc1: A Soluble BMP and Wnt Antagonist That Is Induced by the Interaction between Myeloma Cells and Osteoblast Lineage Cells. Bone 2019, 122, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, V.; Bryant, H.U.; MacDougald, O.A. Regulation of Bone Mass by Wnt Signaling. J. Clin. Investig.. 2006, 116, 1202–1209. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, N.; Morandi, F.; Tagliaferri, S.; Lazzaretti, M.; Donofrio, G.; Bonomini, S.; Sala, R.; Mangoni, M.; Rizzoli, V. Production of Wnt Inhibitors by Myeloma Cells: Potential Effects on Canonical Wnt Pathway in the Bone Microenvironment. Cancer Res. 2007, 67, 7665–7674. [Google Scholar] [CrossRef]
- Kristensen, I.B.; Haaber, J.; Lyng, M.B.; Knudsen, L.M.; Rasmussen, T.; Ditzel, H.J.; Abildgaard, N. Myeloma Plasma Cell Expression of Osteoblast Regulatory Genes: Overexpression of SFRP3 Correlates with Clinical Bone Involvement at Diagnosis. Leuk. Lymphoma 2013, 54, 425–427. [Google Scholar] [CrossRef]
- Chen, D.; Zhao, M.; Mundy, G.R. Bone Morphogenetic Proteins. Growth Factors 2004, 22, 233–241. [Google Scholar] [CrossRef]
- Vallet, S.; Mukherjee, S.; Vaghela, N.; Hideshima, T.; Fulciniti, M.; Pozzi, S.; Santo, L.; Cirstea, D.; Patel, K.; Sohani, A.R.; et al. Activin A Promotes Multiple Myeloma-Induced Osteolysis and Is a Promising Target for Myeloma Bone Disease. Proc. Natl. Acad. Sci. USA 2010, 107, 5124–5129. [Google Scholar] [CrossRef]
- Maeda, S.; Hayashi, M.; Komiya, S.; Imamura, T.; Miyazono, K. Endogenous TGF-β Signaling Suppresses Maturation of Osteoblastic Mesenchymal Cells. EMBO J. 2004, 23, 552–563. [Google Scholar] [CrossRef]
- Takeuchi, K.; Abe, M.; Hiasa, M.; Oda, A.; Amou, H.; Kido, S.; Harada, T.; Tanaka, O.; Miki, H.; Nakamura, S.; et al. TGF-β Inhibition Restores Terminal Osteoblast Differentiation to Suppress Myeloma Growth. PLoS ONE 2010, 5, e9870. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-H.; Kim, Y.-J.; Kim, H.-J.; Park, H.-D.; Kang, A.-R.; Kyung, H.-M.; Sung, J.-H.; Wozney, J.M.; Kim, H.-J.; Ryoo, H.-M. BMP-2-Induced Runx2 Expression Is Mediated by Dlx5, and TGF-Β1 Opposes the BMP-2-Induced Osteoblast Differentiation by Suppression of Dlx5 Expression. J. Biol. Chem. 2003, 278, 34387–34394. [Google Scholar] [CrossRef] [PubMed]
- Alliston, T.; Choy, L.; Ducy, P.; Karsenty, G.; Derynck, R. TGF-β-Induced Repression of CBFA1 by Smad3 Decreases Cbfa1 and Osteocalcin Expression and Inhibits Osteoblast Differentiation. EMBO J. 2001, 20, 2254–2272. [Google Scholar] [CrossRef] [PubMed]
- Standal, T.; Abildgaard, N.; Fagerli, U.-M.; Stordal, B.; Hjertner, Ø.; Borset, M.; Sundan, A. HGF Inhibits BMP-Induced Osteoblastogenesis: Possible Implications for the Bone Disease of Multiple Myeloma. Blood 2007, 109, 3024–3030. [Google Scholar] [CrossRef] [PubMed]
- Gooding, S.; Olechnowicz, S.W.Z.; Morris, E.V.; Armitage, A.E.; Arezes, J.; Frost, J.; Repapi, E.; Edwards, J.R.; Ashley, N.; Waugh, C.; et al. Transcriptomic Profiling of the Myeloma Bone-Lining Niche Reveals BMP Signalling Inhibition to Improve Bone Disease. Nat. Commun. 2019, 10, 4533. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Chung, H.Y.; Ehrlich, L.A.; Jelinek, D.F.; Callander, N.S.; Roodman, G.D.; Choi, S.J. IL-3 Expression by Myeloma Cells Increases Both Osteoclast Formation and Growth of Myeloma Cells. Blood 2004, 103, 2308–2315. [Google Scholar] [CrossRef]
- Giuliani, N.; Colla, S.; Sala, R.; Moroni, M.; Lazzaretti, M.; La Monica, S.; Bonomini, S.; Hojden, M.; Sammarelli, G.; Barillè, S.; et al. Human Myeloma Cells Stimulate the Receptor Activator of Nuclear Factor-ΚB Ligand (RANKL) in T Lymphocytes: A Potential Role in Multiple Myeloma Bone Disease. Blood 2002, 100, 4615–4621. [Google Scholar] [CrossRef]
- Giuliani, N.; Colla, S.; Morandi, F.; Lazzaretti, M.; Sala, R.; Bonomini, S.; Grano, M.; Colucci, S.; Svaldi, M.; Rizzoli, V. Myeloma Cells Block RUNX2/CBFA1 Activity in Human Bone Marrow Osteoblast Progenitors and Inhibit Osteoblast Formation and Differentiation. Blood 2005, 106, 2472–2483. [Google Scholar] [CrossRef]
- D’Souza, S.; del Prete, D.; Jin, S.; Sun, Q.; Huston, A.J.; Kostov, F.E.; Sammut, B.; Hong, C.-S.; Anderson, J.L.; Patrene, K.D.; et al. Gfi1 Expressed in Bone Marrow Stromal Cells Is a Novel Osteoblast Suppressor in Patients with Multiple Myeloma Bone Disease. Blood 2011, 118, 6871–6880. [Google Scholar] [CrossRef]
- Giuliani, N.; Morandi, F.; Tagliaferri, S.; Colla, S.; Bonomini, S.; Sammarelli, G.; Rizzoli, V. Interleukin-3 (IL-3) Is Overexpressed by T Lymphocytes in Multiple Myeloma Patients. Blood 2006, 107, 841–842. [Google Scholar] [CrossRef]
- Ehrlich, L.A.; Chung, H.Y.; Ghobrial, I.; Choi, S.J.; Morandi, F.; Colla, S.; Rizzoli, V.; Roodman, G.D.; Giuliani, N. IL-3 Is a Potential Inhibitor of Osteoblast Differentiation in Multiple Myeloma. Blood 2005, 106, 1407–1414. [Google Scholar] [CrossRef] [PubMed]
- Vallet, S.; Pozzi, S.; Patel, K.; Vaghela, N.; Fulciniti, M.T.; Veiby, P.; Hideshima, T.; Santo, L.; Cirstea, D.; Scadden, D.T.; et al. A Novel Role for CCL3 (MIP-1α) in Myeloma-Induced Bone Disease via Osteocalcin Downregulation and Inhibition of Osteoblast Function. Leukemia 2011, 25, 1174–1181. [Google Scholar] [CrossRef] [PubMed]
- Vallet, S.; Anderson, K.C. CCR1 as a Target for Multiple Myeloma. Expert Opin. Ther. Targets 2011, 15, 1037–1047. [Google Scholar] [CrossRef]
- Lu, X.; Gilbert, L.; He, X.; Rubin, J.; Nanes, M.S. Transcriptional Regulation of the Osterix (Osx, Sp7) Promoter by Tumor Necrosis Factor Identifies Disparate Effects of Mitogen-Activated Protein Kinase and NF Kappa B Pathways. J. Biol. Chem. 2006, 281, 6297–6306. [Google Scholar] [CrossRef] [PubMed]
- Olfa, G.; Christophe, C.; Philippe, L.; Romain, S.; Khaled, H.; Pierre, H.; Odile, B.; Jean-Christophe, D. RUNX2 Regulates the Effects of TNFα on Proliferation and Apoptosis in SaOs-2 Cells. Bone 2010, 46, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Teramachi, J.; Silbermann, R.; Yang, P.; Zhao, W.; Mohammad, K.S.; Guo, J.; Anderson, J.L.; Zhou, D.; Feng, R.; Myint, K.-Z.; et al. Blocking the ZZ Domain of Sequestosome1/P62 Suppresses Myeloma Growth and Osteoclast Formation in Vitro and Induces Dramatic Bone Formation in Myeloma-Bearing Bones in Vivo. Leukemia 2016, 30, 390–398. [Google Scholar] [CrossRef]
- Vincent, C.; Findlay, D.M.; Welldon, K.J.; Wijenayaka, A.R.; Zheng, T.S.; Haynes, D.R.; Fazzalari, N.L.; Evdokiou, A.; Atkins, G.J. Pro-Inflammatory Cytokines TNF-Related Weak Inducer of Apoptosis (TWEAK) and TNFα Induce the Mitogen-Activated Protein Kinase (MAPK)-Dependent Expression of Sclerostin in Human Osteoblasts. J. Bone Miner. Res. 2009, 24, 1434–1449. [Google Scholar] [CrossRef]
- Brunetti, G.; Rizzi, R.; Oranger, A.; Gigante, I.; Mori, G.; Taurino, G.; Mongelli, T.; Colaianni, G.; Benedetto, A.D.; Tamma, R.; et al. LIGHT/TNFSF14 Increases Osteoclastogenesis and Decreases Osteoblastogenesis in Multiple Myeloma-Bone Disease. Oncotarget 2014, 5, 12950–12967. [Google Scholar] [CrossRef]
- Silvestris, F.; Cafforio, P.; Matteo, M.D.; Calvani, N.; Frassanito, M.A.; Dammacco, F. Negative Regulation of the Osteoblast Function in Multiple Myeloma through the Repressor Gene E4BP4 Activated by Malignant Plasma Cells. Clin. Cancer Res. 2008, 14, 6081–6091. [Google Scholar] [CrossRef]
- Engin, F.; Lee, B. NOTCHing the Bone: Insights into Multi-Functionality. Bone 2010, 46, 274–280. [Google Scholar] [CrossRef]
- Engin, F.; Yao, Z.; Yang, T.; Zhou, G.; Bertin, T.; Jiang, M.M.; Chen, Y.; Wang, L.; Zheng, H.; Sutton, R.E.; et al. Dimorphic Effects of Notch Signaling in Bone Homeostasis. Nat. Med. 2008, 14, 299–305. [Google Scholar] [CrossRef]
- Zhao, C.; Irie, N.; Takada, Y.; Shimoda, K.; Miyamoto, T.; Nishiwaki, T.; Suda, T.; Matsuo, K. Bidirectional EphrinB2-EphB4 Signaling Controls Bone Homeostasis. Cell Metab. 2006, 4, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Pennisi, A.; Ling, W.; Li, X.; Khan, S.; Shaughnessy, J.D., Jr.; Barlogie, B.; Yaccoby, S. The EphrinB2/EphB4 Axis Is Dysregulated in Osteoprogenitors from Myeloma Patients and Its Activation Affects Myeloma Bone Disease and Tumor Growth. Blood 2009, 114, 1803–1812. [Google Scholar] [CrossRef]
- Baksh, D.; Tuan, R.S. Canonical and Non-Canonical Wnts Differentially Affect the Development Potential of Primary Isolate of Human Bone Marrow Mesenchymal Stem Cells. J. Cell. Physiol. 2007, 212, 817–826. [Google Scholar] [CrossRef]
- Billiard, J.; Way, D.S.; Seestaller-Wehr, L.M.; Moran, R.A.; Mangine, A.; Bodine, P.V.N. The Orphan Receptor Tyrosine Kinase Ror2 Modulates Canonical Wnt Signaling in Osteoblastic Cells. Mol. Endocrinol. 2005, 19, 90–101. [Google Scholar] [CrossRef]
- Bolzoni, M.; Donofrio, G.; Storti, P.; Guasco, D.; Toscani, D.; Lazzaretti, M.; Bonomini, S.; Agnelli, L.; Capocefalo, A.; Dalla Palma, B.; et al. Myeloma Cells Inhibit Non-Canonical Wnt Co-Receptor Ror2 Expression in Human Bone Marrow Osteoprogenitor Cells: Effect of Wnt5a/Ror2 Pathway Activation on the Osteogenic Differentiation Impairment Induced by Myeloma Cells. Leukemia 2013, 27, 451–463. [Google Scholar] [CrossRef]
- Liu, Y.; Rubin, B.; Bodine, P.V.N.; Billiard, J. Wnt5a Induces Homodimerization and Activation of Ror2 Receptor Tyrosine Kinase. J. Cell. Biochem. 2008, 105, 497–502. [Google Scholar] [CrossRef]
- Faict, S.; Muller, J.; De Veirman, K.; De Bruyne, E.; Maes, K.; Vrancken, L.; Heusschen, R.; De Raeve, H.; Schots, R.; Vanderkerken, K.; et al. Exosomes Play a Role in Multiple Myeloma Bone Disease and Tumor Development by Targeting Osteoclasts and Osteoblasts. Blood Cancer J. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Raimondo, S.; Saieva, L.; Vicario, E.; Pucci, M.; Toscani, D.; Manno, M.; Raccosta, S.; Giuliani, N.; Alessandro, R. Multiple Myeloma-Derived Exosomes Are Enriched of Amphiregulin (AREG) and Activate the Epidermal Growth Factor Pathway in the Bone Microenvironment Leading to Osteoclastogenesis. J. Hematol. Oncol. 2019, 12, 2. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Xu, H.; Han, H.; Song, S.; Zhang, X.; Ouyang, L.; Qian, C.; Hong, Y.; Qiu, Y.; Zhou, W.; et al. Exosome-Mediated Transfer of LncRUNX2-AS1 from Multiple Myeloma Cells to MSCs Contributes to Osteogenesis. Oncogene 2018, 37, 5508–5519. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, Y.; Geng, C.; Zhou, H.; Gao, W.; Chen, W. Serum Exosomal MicroRNAs as Novel Biomarkers for Multiple Myeloma. Hematol. Oncol. 2019, 37, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Tagliaferri, P.; Tassone, P. MicroRNAs in Multiple Myeloma and Related Bone Disease. Ann. Transl. Med. 2015, 3, 13. [Google Scholar] [CrossRef]
- Giuliani, N.; Bataille, R.; Mancini, C.; Lazzaretti, M.; Barillé, S. Myeloma Cells Induce Imbalance in the Osteoprotegerin/Osteoprotegerin Ligand System in the Human Bone Marrow Environment. Blood 2001, 98, 3527–3533. [Google Scholar] [CrossRef]
- Sezer, O.; Heider, U.; Zavrski, I.; Kühne, C.A.; Hofbauer, L.C. RANK Ligand and Osteoprotegerin in Myeloma Bone Disease. Blood 2003, 101, 2094–2098. [Google Scholar] [CrossRef]
- Pearse, R.N.; Sordillo, E.M.; Yaccoby, S.; Wong, B.R.; Liau, D.F.; Colman, N.; Michaeli, J.; Epstein, J.; Choi, Y. Multiple Myeloma Disrupts the TRANCE/ Osteoprotegerin Cytokine Axis to Trigger Bone Destruction and Promote Tumor Progression. Proc. Natl. Acad. Sci. USA 2001, 98, 11581–11586. [Google Scholar] [CrossRef]
- Chantry, A.D.; Heath, D.; Mulivor, A.W.; Pearsall, S.; Baud’huin, M.; Coulton, L.; Evans, H.; Abdul, N.; Werner, E.D.; Bouxsein, M.L.; et al. Inhibiting Activin-A Signaling Stimulates Bone Formation and Prevents Cancer-Induced Bone Destruction in Vivo. J. Bone Miner. Res. 2010, 25, 2633–2646. [Google Scholar] [CrossRef] [PubMed]
- Maeda, K.; Kobayashi, Y.; Udagawa, N.; Uehara, S.; Ishihara, A.; Mizoguchi, T.; Kikuchi, Y.; Takada, I.; Kato, S.; Kani, S.; et al. Wnt5a-Ror2 Signaling between Osteoblast-Lineage Cells and Osteoclast Precursors Enhances Osteoclastogenesis. Nat. Med. 2012, 18, 405–412. [Google Scholar] [CrossRef]
- Yaccoby, S. Advances in the Understanding of Myeloma Bone Disease and Tumour Growth. Br. J. Haematol. 2010, 149, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Fairfield, H.; Falank, C.; Avery, L.; Reagan, M.R. Multiple Myeloma in the Marrow: Pathogenesis and Treatments. Ann. N. Y. Acad. Sci. 2016, 1364, 32–51. [Google Scholar] [CrossRef] [PubMed]
- Reagan, M.R.; Liaw, L.; Rosen, C.J.; Ghobrial, I.M. Dynamic Interplay between Bone and Multiple Myeloma: Emerging Roles of the Osteoblast. Bone 2015, 75, 161–169. [Google Scholar] [CrossRef]
- Savopoulos, C.; Dokos, C.; Kaiafa, G.; Hatzitolios, A. Adipogenesis and Osteoblastogenesis: Trans-Differentiation in the Pathophysiology of Bone Disorders. Hippokratia 2011, 15, 18–21. [Google Scholar]
- Liu, Z.; Liu, H.; He, J.; Lin, P.; Tong, Q.; Yang, J. Myeloma Cells Shift Osteoblastogenesis to Adipogenesis by Inhibiting the Ubiquitin Ligase MURF1 in Mesenchymal Stem Cells. Sci. Signal. 2020, 13. [Google Scholar] [CrossRef]
- Fairfield, H.; Falank, C.; Harris, E.; Demambro, V.; McDonald, M.; Pettitt, J.A.; Mohanty, S.T.; Croucher, P.; Kramer, I.; Kneissel, M.; et al. The Skeletal Cell-Derived Molecule Sclerostin Drives Bone Marrow Adipogenesis. J. Cell. Physiol. 2018, 233, 1156–1167. [Google Scholar] [CrossRef]
- Ruan, J.; Trotter, T.N.; Nan, L.; Luo, R.; Javed, A.; Sanderson, R.D.; Suva, L.J.; Yang, Y. Heparanase Inhibits Osteoblastogenesis and Shifts Bone Marrow Progenitor Cell Fate in Myeloma Bone Disease. Bone 2013, 57, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Christodoulides, C.; Laudes, M.; Cawthorn, W.P.; Schinner, S.; Soos, M.; O’Rahilly, S.; Sethi, J.K.; Vidal-Puig, A. The Wnt Antagonist Dickkopf-1 and Its Receptors Are Coordinately Regulated during Early Human Adipogenesis. J. Cell Sci. 2006, 119, 2613–2620. [Google Scholar] [CrossRef]
- Fairfield, H.; Dudakovic, A.; Khatib, C.M.; Farrell, M.; Costa, S.; Falank, C.; Hinge, M.; Murphy, C.S.; DeMambro, V.; Pettitt, J.A.; et al. Myeloma-Modified Adipocytes Exhibit Metabolic Dysfunction and a Senescence-Associated Secretory Phenotype. Cancer Res. 2021, 81, 634–647. [Google Scholar] [CrossRef]
- Falank, C.; Fairfield, H.; Reagan, M.R. Reflections on Cancer in the Bone Marrow: Adverse Roles of Adipocytes. Curr Mol Biol Rep 2017, 3, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Trotter, T.N.; Gibson, J.T.; Sherpa, T.L.; Gowda, P.S.; Peker, D.; Yang, Y. Adipocyte-Lineage Cells Support Growth and Dissemination of Multiple Myeloma in Bone. Am. J. Pathol. 2016, 186, 3054–3063. [Google Scholar] [CrossRef]
- Fowler, J.A.; Lwin, S.T.; Drake, M.T.; Edwards, J.R.; Kyle, R.A.; Mundy, G.R.; Edwards, C.M. Host-Derived Adiponectin Is Tumor-Suppressive and a Novel Therapeutic Target for Multiple Myeloma and the Associated Bone Disease. Blood 2011, 118, 5872–5882. [Google Scholar] [CrossRef] [PubMed]
- Müller, L.; Tunger, A.; Wobus, M.; von Bonin, M.; Towers, R.; Bornhäuser, M.; Dazzi, F.; Wehner, R.; Schmitz, M. Immunomodulatory Properties of Mesenchymal Stromal Cells: An Update. Front. Cell Dev. Biol. 2021, 9. [Google Scholar] [CrossRef]
- Gao, M.; Yao, H.; Dong, Q.; Zhang, H.; Yang, Z.; Yang, Y.; Zhu, J.; Xu, M.; Xu, R. Tumourigenicity and Immunogenicity of Induced Neural Stem Cell Grafts Versus Induced Pluripotent Stem Cell Grafts in Syngeneic Mouse Brain. Sci. Rep. 2016, 6, 29955. [Google Scholar] [CrossRef] [PubMed]
- Roemeling-van Rhijn, M.; Khairoun, M.; Korevaar, S.S.; Lievers, E.; Leuning, D.G.; IJzermans, J.N.; Betjes, M.G.; Genever, P.G.; van Kooten, C.; de Fijter, H.J.; et al. Human Bone Marrow- and Adipose Tissue-Derived Mesenchymal Stromal Cells Are Immunosuppressive In Vitro and in a Humanized Allograft Rejection Model. J. Stem Cell Res. Ther. 2013, Suppl 6, 20780. [Google Scholar] [CrossRef]
- André, T.; Najar, M.; Stamatopoulos, B.; Pieters, K.; Pradier, O.; Bron, D.; Meuleman, N.; Lagneaux, L. Immune Impairments in Multiple Myeloma Bone Marrow Mesenchymal Stromal Cells. Cancer Immunol. Immunother. 2015, 64, 213–224. [Google Scholar] [CrossRef]
- Prabhala, R.H.; Pelluru, D.; Fulciniti, M.; Prabhala, H.K.; Nanjappa, P.; Song, W.; Pai, C.; Amin, S.; Tai, Y.-T.; Richardson, P.G.; et al. Elevated IL-17 Produced by Th17 Cells Promotes Myeloma Cell Growth and Inhibits Immune Function in Multiple Myeloma. Blood 2010, 115, 5385–5392. [Google Scholar] [CrossRef]
- de Jong, M.M.E.; Kellermayer, Z.; Papazian, N.; Duin, M.; Broyl, A.; Sonneveld, P.; Cupedo, T. Single Cell Transcriptomic Analysis of the Multiple Myeloma Bone Marrow Identifies a Unique Inflammatory Stromal Cell Population Associated with TNF Signaling. Blood 2019, 134, 690. [Google Scholar] [CrossRef]
- Díaz-Tejedor, A.; Lorenzo-Mohamed, M.; Puig, N.; García-Sanz, R.; Mateos, M.-V.; Garayoa, M.; Paíno, T. Immune System Alterations in Multiple Myeloma: Molecular Mechanisms and Therapeutic Strategies to Reverse Immunosuppression. Cancers 2021, 13, 1353. [Google Scholar] [CrossRef]
- Romano, A.; Conticello, C.; Cavalli, M.; Vetro, C.; La Fauci, A.; Parrinello, N.L.; Di Raimondo, F. Immunological Dysregulation in Multiple Myeloma Microenvironment. Biomed. Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- De Veirman, K.; Menu, E.; Maes, K.; De Beule, N.; De Smedt, E.; Maes, A.; Vlummens, P.; Fostier, K.; Kassambara, A.; Moreaux, J.; et al. Myeloid-Derived Suppressor Cells Induce Multiple Myeloma Cell Survival by Activating the AMPK Pathway. Cancer Lett. 2019, 442, 233–241. [Google Scholar] [CrossRef]
- Leone, P.; Solimando, A.G.; Malerba, E.; Fasano, R.; Buonavoglia, A.; Pappagallo, F.; De Re, V.; Argentiero, A.; Silvestris, N.; Vacca, A.; et al. Actors on the Scene: Immune Cells in the Myeloma Niche. Front. Oncol. 2020, 10, 599098. [Google Scholar] [CrossRef] [PubMed]
- Yen, B.L.; Yen, M.-L.; Hsu, P.-J.; Liu, K.-J.; Wang, C.-J.; Bai, C.-H.; Sytwu, H.-K. Multipotent Human Mesenchymal Stromal Cells Mediate Expansion of Myeloid-Derived Suppressor Cells via Hepatocyte Growth Factor/c-Met and STAT3. Stem Cell Rep. 2013, 1, 139–151. [Google Scholar] [CrossRef]
- Giallongo, C.; Tibullo, D.; Parrinello, N.L.; La Cava, P.; Di Rosa, M.; Bramanti, V.; Di Raimondo, C.; Conticello, C.; Chiarenza, A.; Palumbo, G.A.; et al. Granulocyte-like Myeloid Derived Suppressor Cells (G-MDSC) Are Increased in Multiple Myeloma and Are Driven by Dysfunctional Mesenchymal Stem Cells (MSC). Oncotarget 2016, 7, 85764–85775. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maiso, P.; Mogollón, P.; Ocio, E.M.; Garayoa, M. Bone Marrow Mesenchymal Stromal Cells in Multiple Myeloma: Their Role as Active Contributors to Myeloma Progression. Cancers 2021, 13, 2542. https://doi.org/10.3390/cancers13112542
Maiso P, Mogollón P, Ocio EM, Garayoa M. Bone Marrow Mesenchymal Stromal Cells in Multiple Myeloma: Their Role as Active Contributors to Myeloma Progression. Cancers. 2021; 13(11):2542. https://doi.org/10.3390/cancers13112542
Chicago/Turabian StyleMaiso, Patricia, Pedro Mogollón, Enrique M. Ocio, and Mercedes Garayoa. 2021. "Bone Marrow Mesenchymal Stromal Cells in Multiple Myeloma: Their Role as Active Contributors to Myeloma Progression" Cancers 13, no. 11: 2542. https://doi.org/10.3390/cancers13112542
APA StyleMaiso, P., Mogollón, P., Ocio, E. M., & Garayoa, M. (2021). Bone Marrow Mesenchymal Stromal Cells in Multiple Myeloma: Their Role as Active Contributors to Myeloma Progression. Cancers, 13(11), 2542. https://doi.org/10.3390/cancers13112542






