LIMK2-NKX3.1 Engagement Promotes Castration-Resistant Prostate Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
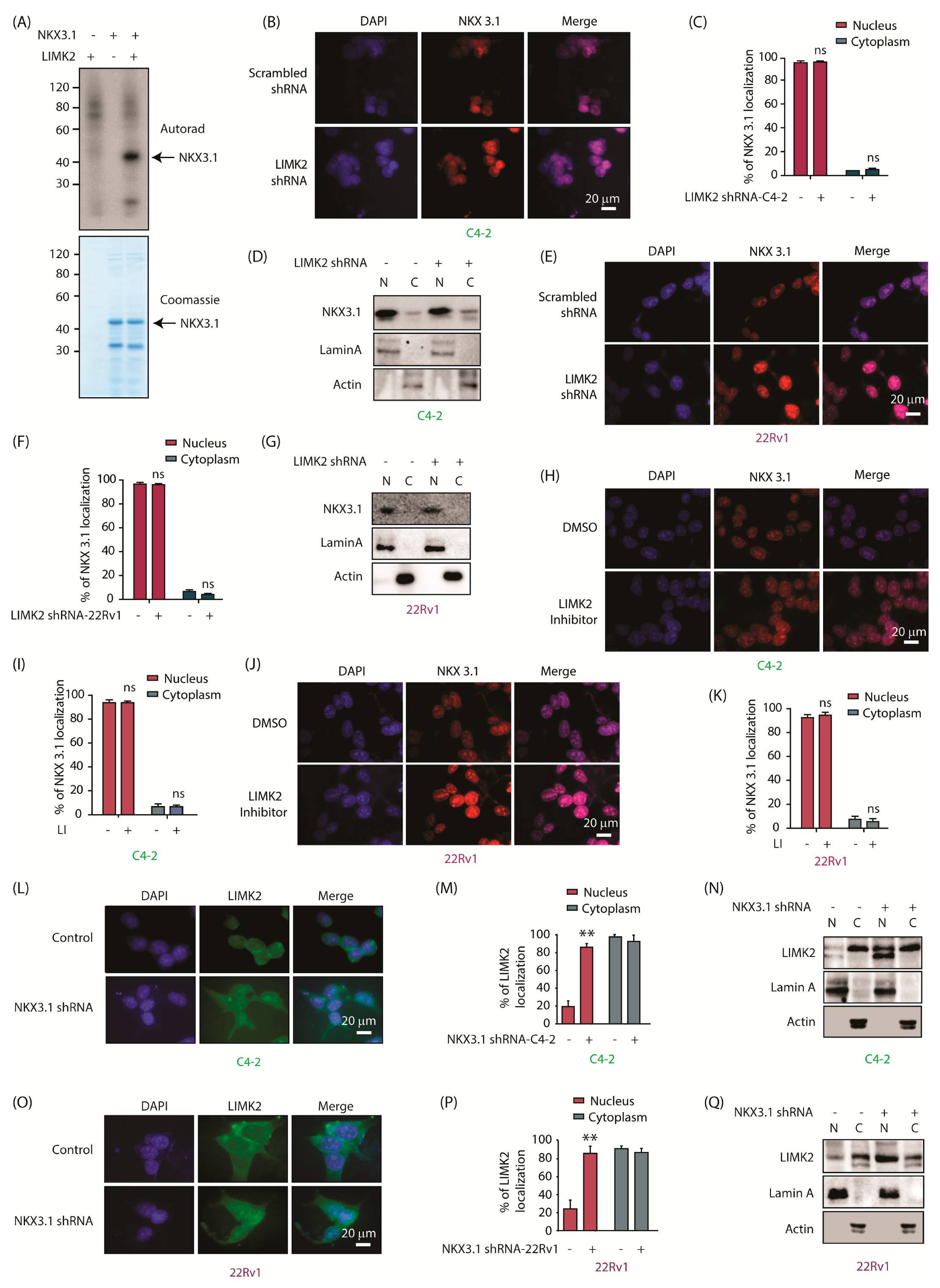
2.1. Analog-Sensitive LIMK2 (LIMK2-as7) Identifies NKX3.1 as a Direct Target
2.2. LIMK2 Does Not Alter NKX3.1 Subcellular Localization, but NKX3.1 Promotes Cytoplasmic Localization of LIMK2
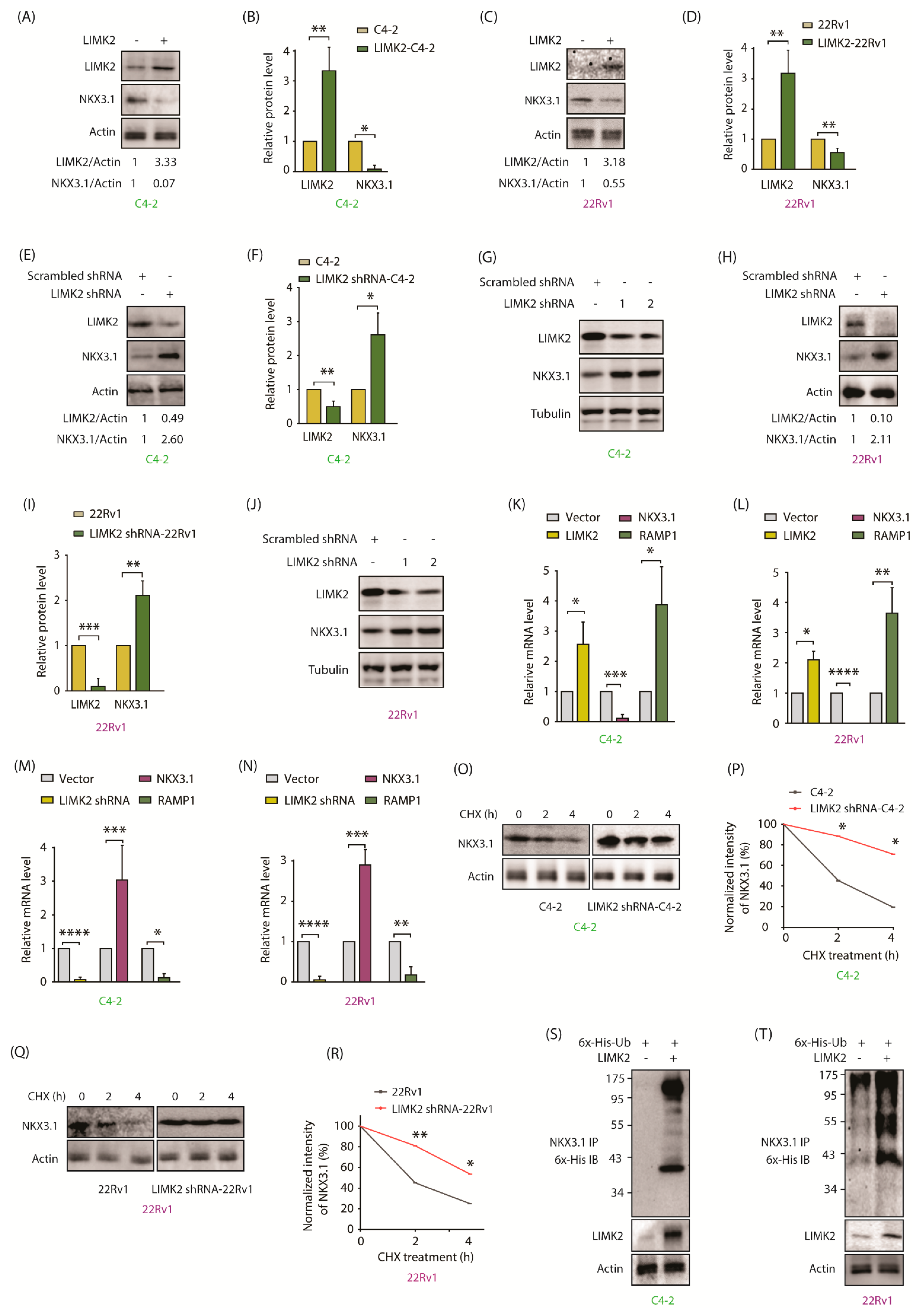
2.3. LIMK2 Negatively Regulates NKX3.1
2.4. LIMK2 Negatively Regulates NKX3.1 mRNA Levels and Decreases It’s Stability by Enhanced Ubiquitylation
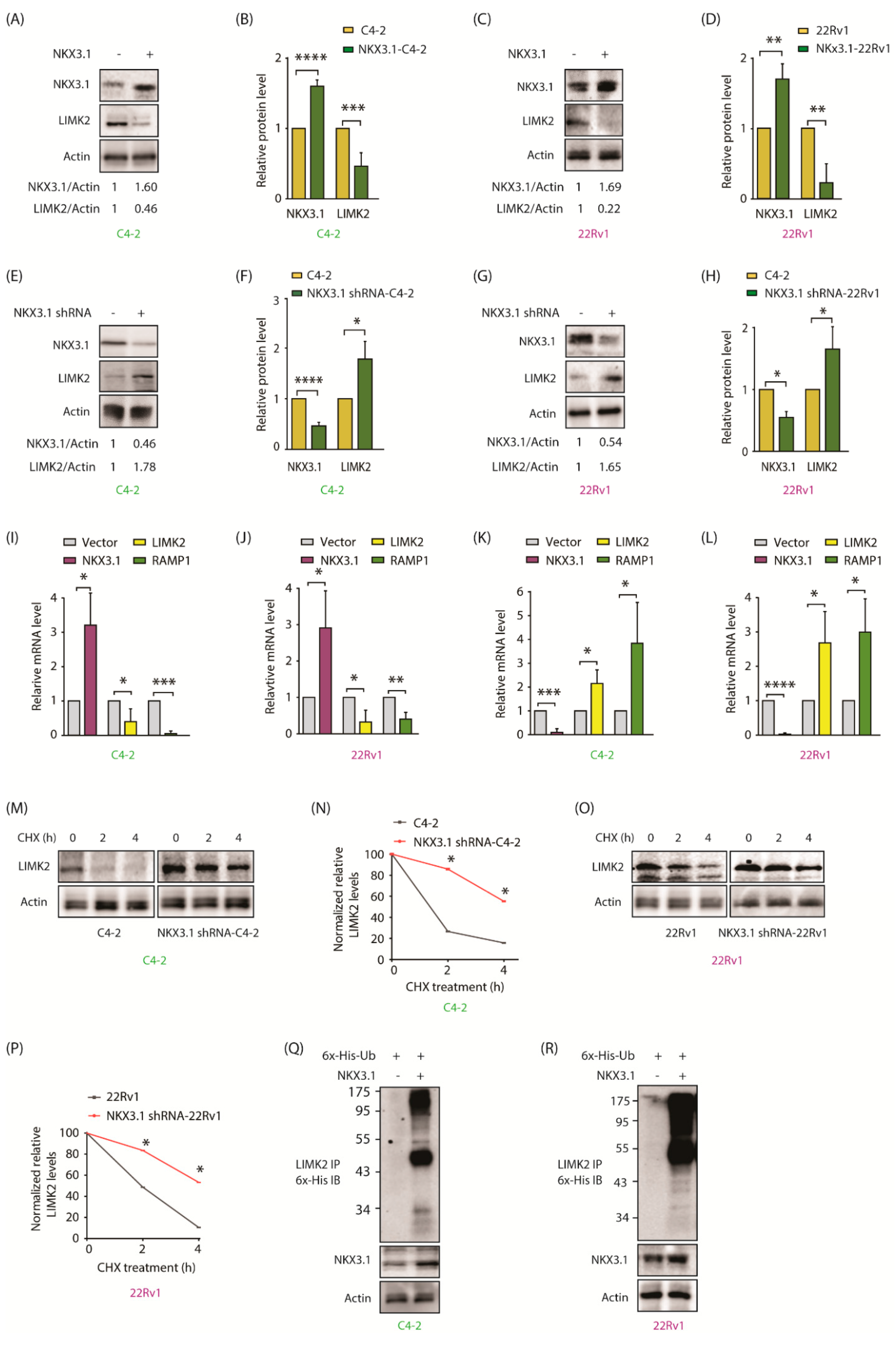
2.5. NKX3.1 Negatively Regulates LIMK2 at mRNA and Protein Levels
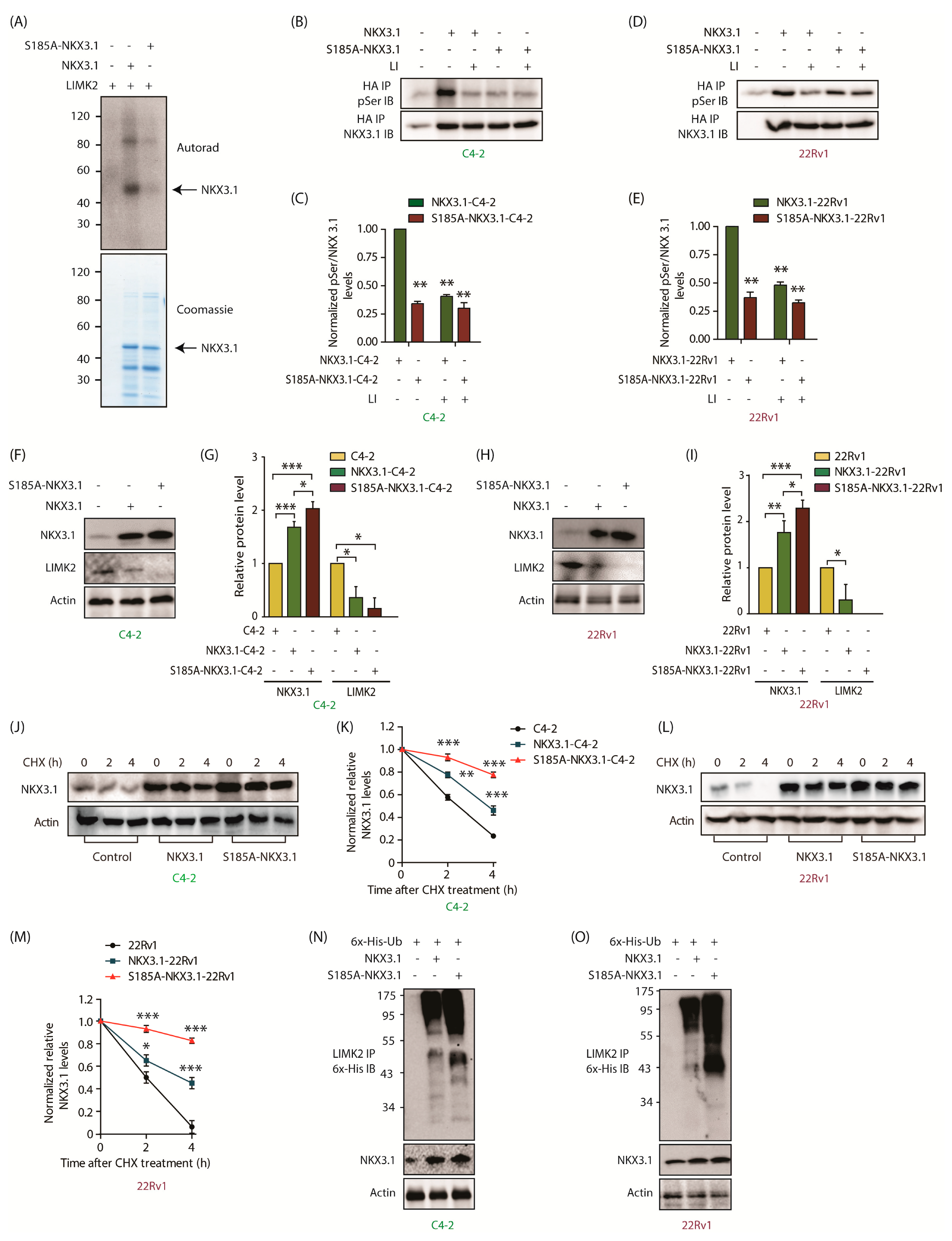
2.6. LIMK2 Phosphorylates NKX3.1 at S185 Triggering Its Degradation
2.7. LIMK2-Mediated NKX3.1 Phosphorylation Is Critical for Promoting Aggressive Phenotypes
2.8. The Phosphomimetic S185D-NKX3.1 Decreases NKX3.1 Stability and Enhances LIMK2 Protein Level by Inhibiting Its Ubiquitylation
2.9. The Phosphomimetic S185D-NKX3.1 Promotes Cell Proliferation, Colony Formation, and Chemotaxis
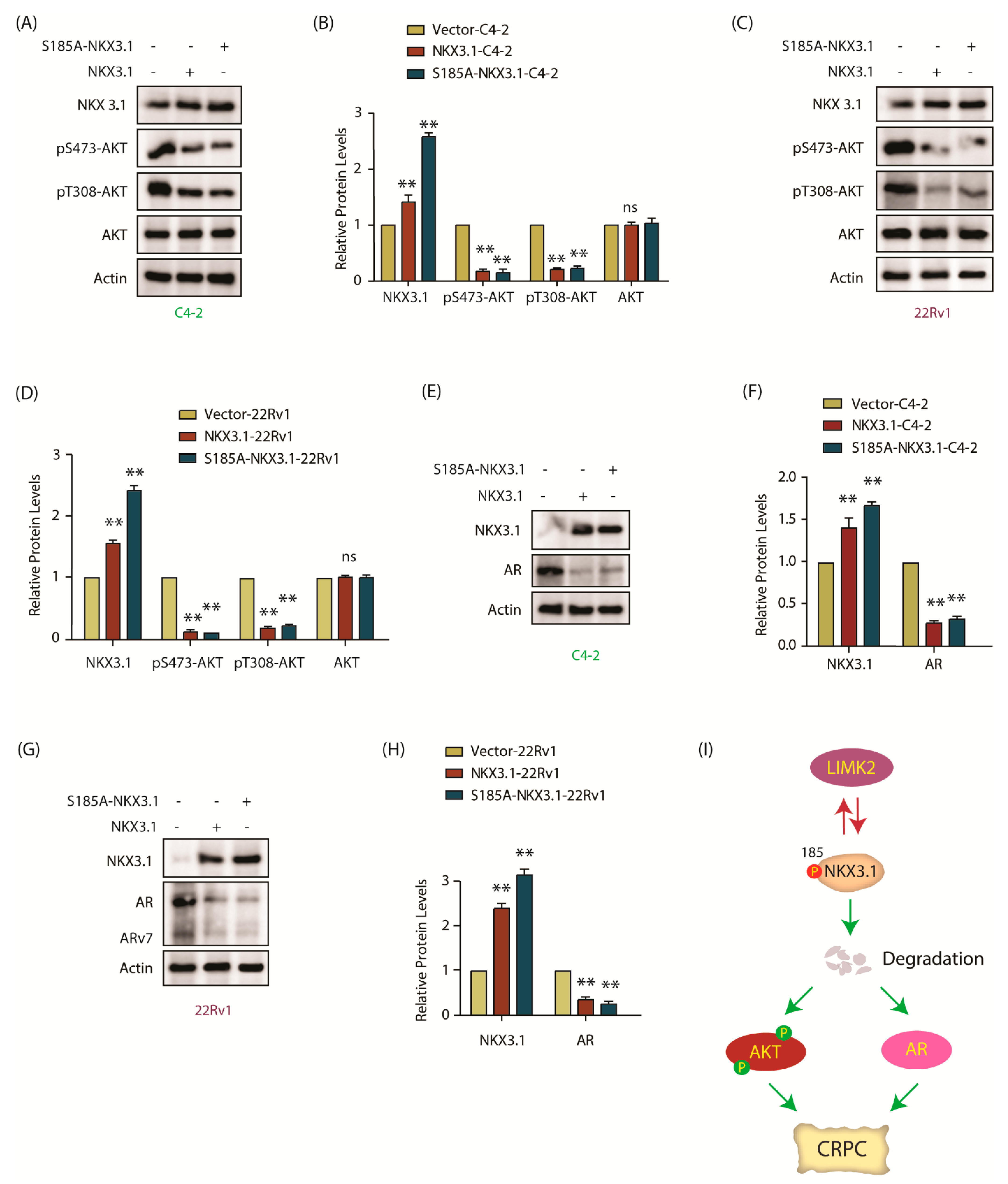
2.10. NKX3.1 Downregulates AKT Activation and Decreases AR and ARv7 Levels in CRPC Cells
2.11. NKX3.1 Suppresses Tumor Growth In Vivo
3. Discussion
4. Materials and Methods
4.1. Cell Lines, Antibodies, and Chemicals
4.2. LIMK2 and NKX3.1 shRNAs
4.3. Site-Directed Mutagenesis, Expression, and Purification of LIMK2, Wild-Type (WT), and S185A-NKX3.1 Mutant
4.4. In Vitro Kinase Assays
4.5. Transfection and Retroviral Infection
4.6. Western Blotting
4.7. Isolation of Cytosolic and Nuclear Fractions
4.8. Cell Viability Assay
4.9. Dose-Response Assay with LIMK2 Inhibitor
4.10. Clonogenic Assay
4.11. Migration Assay
4.12. Ubiquitylation Assay
4.13. LIMK2 Inhibitor
4.14. qPCR Assay
4.15. C4-2 Xenografts in Nude Mice
4.16. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Teo, M.Y.; Rathkopf, D.E.; Kantoff, P. Treatment of Advanced Prostate Cancer. Annu. Rev. Med. 2019, 70, 479–499. [Google Scholar] [CrossRef]
- Stavridi, F.; Karapanagiotou, E.M.; Syrigos, K.N. Targeted therapeutic approaches for hormone-refractory prostate cancer. Cancer Treat. Rev. 2010, 36, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Capoun, O.; Mikulova, V.; Jancikova, M.; Honova, H.; Kolostova, K.; Sobotka, R.; Michael, P.; Zima, T.; Hanus, T.; Soukup, V. Prognosis of Castration-resistant Prostate Cancer Patients—Use of the AdnaTest(R) System for Detection of Circulating Tumor Cells. Anticancer Res. 2016, 36, 2019–2026. [Google Scholar] [PubMed]
- Nikhil, K.; Chang, L.; Viccaro, K.; Jacobsen, M.; McGuire, C.; Satapathy, S.R.; Tandiary, M.; Broman, M.M.; Cresswell, G.; He, Y.J.; et al. Identification of LIMK2 as a therapeutic target in castration resistant prostate cancer. Cancer Lett. 2019, 448, 182–196. [Google Scholar] [CrossRef]
- Johnson, E.O.; Chang, K.-H.; Ghosh, S.; Venkatesh, C.; Giger, K.; Low, P.S.; Shah, K. LIMK2 is a crucial regulator and effector of Aurora-A-kinase-mediated malignancy. J. Cell Sci. 2012, 125, 1204–1216. [Google Scholar] [CrossRef]
- Wang, W.; Yang, C.; Nie, H.; Qiu, X.; Zhang, L.; Xiao, Y.; Zhou, W.; Zeng, Q.; Zhang, X.; Wu, Y.; et al. LIMK2 acts as an oncogene in bladder cancer and its functional SNP in the microRNA-135a binding site affects bladder cancer risk. Int. J. Cancer 2019, 144, 1345–1355. [Google Scholar] [CrossRef]
- Chen, J.; Ananthanarayanan, B.; Springer, K.S.; Wolf, K.J.; Sheyman, S.M.; Tran, V.D.; Kumar, S. Suppression of LIM Kinase 1 and LIM Kinase 2 Limits Glioblastoma Invasion. Cancer Res. 2020, 80, 69–78. [Google Scholar] [CrossRef]
- Wang, S.; Ren, T.; Jiao, G.; Huang, Y.; Bao, X.; Zhang, F.; Liu, K.; Zheng, B.; Sun, K.; Guo, W. BMPR2 promotes invasion and metastasis via the RhoA-ROCK-LIMK2 pathway in human osteosarcoma cells. Oncotarget 2017, 8, 58625–58641. [Google Scholar] [CrossRef]
- Nikhil, K.; Haymour, H.S.; Kamra, M.; Shah, K. Phosphorylation-dependent regulation of SPOP by LIMK2 promotes castration-resistant prostate cancer. Br. J. Cancer. 2021, 124, 995–1008. [Google Scholar] [CrossRef] [PubMed]
- Nikhil, K.; Kamra, M.; Raza, A.; Shah, K. Negative cross talk between LIMK2 and PTEN promotes castration resistant prostate cancer pathogenesis in cells and in vivo. Cancer Lett. 2021, 498, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Shah, K.; Vincent, F. Divergent roles of c-Src in controlling platelet-derived growth factor- dependent signaling in fibroblasts. Mol. Biol. Cell 2005, 16, 5418–5432. [Google Scholar] [CrossRef]
- Johnson, E.O.; Chang, K.-H.; de Pablo, Y.; Ghosh, S.; Mehta, R.; Badve, S.; Shah, K. PHLDA1 is a crucial negative regulator and effector of aurora A kinase in breast cancer. J. Cell Sci. 2011, 124, 2711–2722. [Google Scholar] [CrossRef]
- Sun, K.H.; de Pablo, Y.; Vincent, F.; Shah, K. Deregulated Cdk5 promotes oxidative stress and mitochondrial dysfunction. J. Neurochem. 2008, 107, 265–278. [Google Scholar] [CrossRef]
- Bhatia-Gaur, R.; Donjacour, A.A.; Sciavolino, P.J.; Kim, M.; Desai, N.; Young, P.; Norton, C.R.; Gridley, T.; Cardiff, R.D.; Cunha, G.R.; et al. Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 1999, 13, 966–977. [Google Scholar] [CrossRef]
- Asatiani, E.; Huang, W.X.; Wang, A.; Rodriguez Ortner, E.; Cavalli, L.R.; Haddad, B.R.; Gelmann, E.P. Deletion, methylation, and expression of the NKX3.1 suppressor gene in primary human prostate cancer. Cancer Res. 2005, 65, 1164–1173. [Google Scholar] [CrossRef] [PubMed]
- Abdulkadir, S.A.; Magee, J.A.; Peters, T.J.; Kaleem, Z.; Naughton, C.K.; Humphrey, P.A.; Milbrandt, J. Conditional loss of Nkx3.1 in adult mice induces prostatic intraepithelial neoplasia. Mol. Cell Biol. 2002, 22, 1495–1503. [Google Scholar] [CrossRef]
- Zheng, S.L.; Ju, J.H.; Chang, B.L.; Ortner, E.; Sun, J.; Isaacs, S.D.; Sun, J.; Wiley, K.E.; Liu, W.; Zemedkun, M.; et al. Germ-line mutation of NKX3.1 cosegregates with hereditary prostate cancer and alters the homeodomain structure and function. Cancer Res. 2006, 66, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Bowen, C.; Bubendorf, L.; Voeller, H.J.; Slack, R.; Willi, N.; Sauter, G.; Gasser, T.C.; Koivisto, P.; Lack, E.E.; Kononen, J.; et al. Loss of NKX3.1 expression in human prostate cancers correlates with tumor progression. Cancer Res. 2000, 60, 6111–6115. [Google Scholar] [PubMed]
- Magee, J.A.; Abdulkadir, S.A.; Milbrandt, J. Haploinsufficiency at the Nkx3.1 locus. A paradigm for stochastic, dosage-sensitive gene regulation during tumor initiation. Cancer Cell 2003, 3, 273–283. [Google Scholar] [CrossRef]
- Shah, K.; Kim, H. The significant others: Global search for direct kinase substrates using chemical approaches. IUBMB Life 2019, 71, 721–737. [Google Scholar] [CrossRef]
- Wang, J.; Nikhil, K.; Viccaro, K.; Chang, L.; Jacobsen, M.; Sandusky, G.; Shah, K. The Aurora-A-Twist1 axis promotes highly aggressive phenotypes in pancreatic carcinoma. J. Cell Sci. 2017, 130, 1078–1093. [Google Scholar] [CrossRef]
- Wang, J.; Nikhil, K.; Viccaro, K.; Chang, L.; White, J.; Shah, K. Phosphorylation-dependent regulation of ALDH1A1 by Aurora kinase A: Insights on their synergistic relationship in pancreatic cancer. BMC Biol. 2017, 15, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Nikhil, K.; Raza, A.; Haymour, H.S.; Flueckiger, B.V.; Chu, J.; Shah, K. Aurora Kinase A-YBX1 Synergy Fuels Aggressive Oncogenic Phenotypes and Chemoresistance in Castration-Resistant Prostate Cancer. Cancers 2020, 12, 660. [Google Scholar] [CrossRef] [PubMed]
- Nikhil, K.; Kamra, M.; Raza, A.; Haymour, H.S.; Shah, K. Molecular Interplay between AURKA and SPOP Dictates CRPC Pathogenesis via Androgen Receptor. Cancers 2020, 12, 3247. [Google Scholar] [CrossRef]
- Goodwin, N.C.; Cianchetta, G.; Burgoon, H.A.; Healy, J.; Mabon, R.; Strobel, E.D.; Allen, J.; Wang, S.; Hamman, B.D.; Rawlins, D.B. Discovery of a type III inhibitor of LIM kinase 2 that binds in a DFG-out conformation. ACS Med. Chem. Lett. 2014, 6, 53–57. [Google Scholar] [CrossRef]
- Bieberich, C.J.; Fujita, K.; He, W.W.; Jay, G. Prostate-specific and androgen-dependent expression of a novel homeobox gene. J. Biol. Chem. 1996, 271, 31779–31782. [Google Scholar] [CrossRef]
- Logan, M.; Anderson, P.D.; Saab, S.T.; Hameed, O.; Abdulkadir, S.A. RAMP1 is a direct NKX3.1 target gene up-regulated in prostate cancer that promotes tumorigenesis. Am. J. Pathol. 2013, 183, 951–963. [Google Scholar] [CrossRef] [PubMed]
- Dutta, A.; Le Magnen, C.; Mitrofanova, A.; Ouyang, X.; Califano, A.; Abate-Shen, C. Identification of an NKX3.1-G9a-UTY transcriptional regulatory network that controls prostate differentiation. Science 2016, 352, 1576–1580. [Google Scholar] [CrossRef]
- Gurel, B.; Ali, T.Z.; Montgomery, E.A.; Begum, S.; Hicks, J.; Goggins, M.; Eberhart, C.G.; Clark, D.P.; Bieberich, C.J.; Epstein, J.I.; et al. NKX3.1 as a marker of prostatic origin in metastatic tumors. Am. J. Surg. Pathol. 2010, 34, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- Bethel, C.R.; Faith, D.; Li, X.; Guan, B.; Hicks, J.L.; Lan, F.; Jenkins, R.B.; Bieberich, C.J.; de Marzo, A.M. Decreased NKX3.1 protein expression in focal prostatic atrophy, prostatic intraepithelial neoplasia, and adenocarcinoma: Association with gleason score and chromosome, 8p deletion. Cancer Res. 2006, 66, 10683–10690. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell 2015, 163, 1011–1025. [Google Scholar] [CrossRef] [PubMed]
- Baca, S.C.; Prandi, D.; Lawrence, M.S.; Mosquera, J.M.; Romanel, A.; Drier, Y.; Park, K.; Kitabayashi, N.; MacDonald, T.Y.; Ghandi, M.; et al. Punctuated evolution of prostate cancer genomes. Cell 2013, 153, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.S.; Schultz, N.; Hieronymus, H.; Gopalan, A.; Xiao, Y.; Carver, B.S.; Arora, V.K.; Kaushik, P.; Cerami, E.; Reva, B.; et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010, 18, 11–22. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Li, X.; Guan, B.; Maghami, S.; Bieberich, C.J. NKX3.1 is regulated by protein kinase CK2 in prostate tumor cells. Mol. Cell Biol. 2006, 26, 3008–3017. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, A.; Gosc, E.B.; Bieberich, C.J. Stabilization of the prostate-specific tumor suppressor NKX3.1 by the oncogenic protein kinase Pim-1 in prostate cancer cells. J. Cell Biochem. 2013, 114, 1050–1057. [Google Scholar] [CrossRef]
- Song, L.N.; Silva, J.; Koller, A.; Rosenthal, A.; Chen, E.I.; Gelmann, E.P. The tumor suppressor NKX3.1 is targeted for degradation by DYRK1B kinase. Mol. Cancer Res. 2015, 13, 913–922. [Google Scholar] [CrossRef] [PubMed]
- Markowski, M.C.; Bowen, C.; Gelmann, E.P. Inflammatory cyto-kines induce phosphorylation and ubiquitination of prostatesuppressor protein NKX3.1. Cancer Res. 2008, 68, 6896–6901. [Google Scholar] [CrossRef]
- Bowen, C.; Ju, J.H.; Lee, J.H.; Paull, T.T.; Gelmann, E.P. Functionalactivation of ATM by the prostate cancer suppressor NKX3.1. Cell Rep. 2013, 4, 516–529. [Google Scholar] [CrossRef]
- Erbaykent-Tepedelen, B.; Karamil, S.; Gonen-Korkmaz, C.; Korkmaz, K.S. DNA damage response (DDR) via NKX3.1 expression inprostate cells. J. Steroid Biochem. Mol. Biol. 2014, 141, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Gelmann, E.P.; Steadman, D.J.; Ma, J.; Ahronovitz, N.; Voeller, H.J.; Swope, S.; Abbaszadegan, M.; Brown, K.M.; Strand, K.; Hayes, R.B.; et al. Occurrence of NKX3.1 C154T polymorphism inmen with and without prostate cancer and studies of its effecton protein function. Cancer Res. 2002, 62, 2654–2659. [Google Scholar]
- Emmert-Buck, M.; Vocke, C.D.; Pozzatti, R.O.; Duray, P.H.; Jennings, S.B.; Florence, C.D.; Zhuang, Z.; Bostwick, D.G.; Liotta, L.A.; Linehan, W.M. Allelic loss on chromosome 8p12-21 in microdissected prostatic intraepithelial neoplasia. Cancer Res. 1995, 55, 2959–2962. [Google Scholar]
- Kim, M.J.; Cardiff, R.D.; Desai, N.; Banach-Petrosky, W.A.; Parsons, R.; Shen, M.M.; Abate-Shen, C. Cooperativity of Nkx3.1 and Pten loss of function in a mouse model of prostate carcinogenesis. Proc. Natl. Acad. Sci. USA 2002, 99, 2884–2889. [Google Scholar] [CrossRef]
- Bowen, C.; Ostrowski, M.C.; Leone, G.; Gelmann, E.P. Loss of PTEN Accelerates NKX3.1 Degradation to Promote Prostate Cancer Progression. Cancer Res. 2019, 79, 4124–4134. [Google Scholar] [CrossRef]
- Moffat, J.; Grueneberg, D.A.; Yang, X.; Kim, S.Y.; Kloepfer, A.M.; Hinkle, G.; Piqani, B.; Eisenhaure, T.M.; Luo, B.; Grenier, J.K.; et al. A Lentiviral RNAi Library for Human and Mouse Genes Applied to an Arrayed Viral High-Content Screen. Cell 2006, 124, 1283–1298. [Google Scholar] [CrossRef] [PubMed]
- Nagahara, H.; Vocero-Akbani, A.M.; Snyder, E.L.; Ho, A.; Latham, D.G.; Lissy, N.A.; Becker-Hapak, M.; Ezhevsky, S.A.; Dowdy, S.F. Transduction of full-length TAT fusion proteins into mammalian cells: TAT-p27Kip1 induces cell migration. Nat. Med. 1998, 4, 1449–1452. [Google Scholar] [CrossRef]
- Sun, K.H.; Lee, H.G.; Smith, M.A.; Shah, K. Direct and indirect roles of cyclin-dependent kinase 5 as an upstream regulator in the c-Jun NH2-terminal kinase cascade: Relevance to neurotoxic insults in Alzheimer’s disease. Mol. Biol. Cell 2009, 20, 4611–4619. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Viccaro, K.; Lee, H.G.; Shah, K. Cdk5-FOXO3a axis: Initially neuroprotective, eventually neurodegenerative in Alzheimer’s disease models. J. Cell Sci. 2016, 129, 1815–1830. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.H.; Multani, P.S.; Sun, K.H.; Vincent, F.; de Pablo, Y.; Ghosh, S.; Gupta, R.; Lee, H.P.; Lee, H.G.; Smith, M.A.; et al. Nuclear envelope dispersion triggered by deregulated Cdk5 precedes neuronal death. Mol. Biol. Cell. 2011, 22, 1452–1462. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sooreshjani, M.A.; Nikhil, K.; Kamra, M.; Nguyen, D.N.; Kumar, D.; Shah, K. LIMK2-NKX3.1 Engagement Promotes Castration-Resistant Prostate Cancer. Cancers 2021, 13, 2324. https://doi.org/10.3390/cancers13102324
Sooreshjani MA, Nikhil K, Kamra M, Nguyen DN, Kumar D, Shah K. LIMK2-NKX3.1 Engagement Promotes Castration-Resistant Prostate Cancer. Cancers. 2021; 13(10):2324. https://doi.org/10.3390/cancers13102324
Chicago/Turabian StyleSooreshjani, Moloud A., Kumar Nikhil, Mohini Kamra, Dung N. Nguyen, Dinesh Kumar, and Kavita Shah. 2021. "LIMK2-NKX3.1 Engagement Promotes Castration-Resistant Prostate Cancer" Cancers 13, no. 10: 2324. https://doi.org/10.3390/cancers13102324
APA StyleSooreshjani, M. A., Nikhil, K., Kamra, M., Nguyen, D. N., Kumar, D., & Shah, K. (2021). LIMK2-NKX3.1 Engagement Promotes Castration-Resistant Prostate Cancer. Cancers, 13(10), 2324. https://doi.org/10.3390/cancers13102324






