Current Views on the Interplay between Tyrosine Kinases and Phosphatases in Chronic Myeloid Leukemia
Abstract
Simple Summary
Abstract
1. Introduction
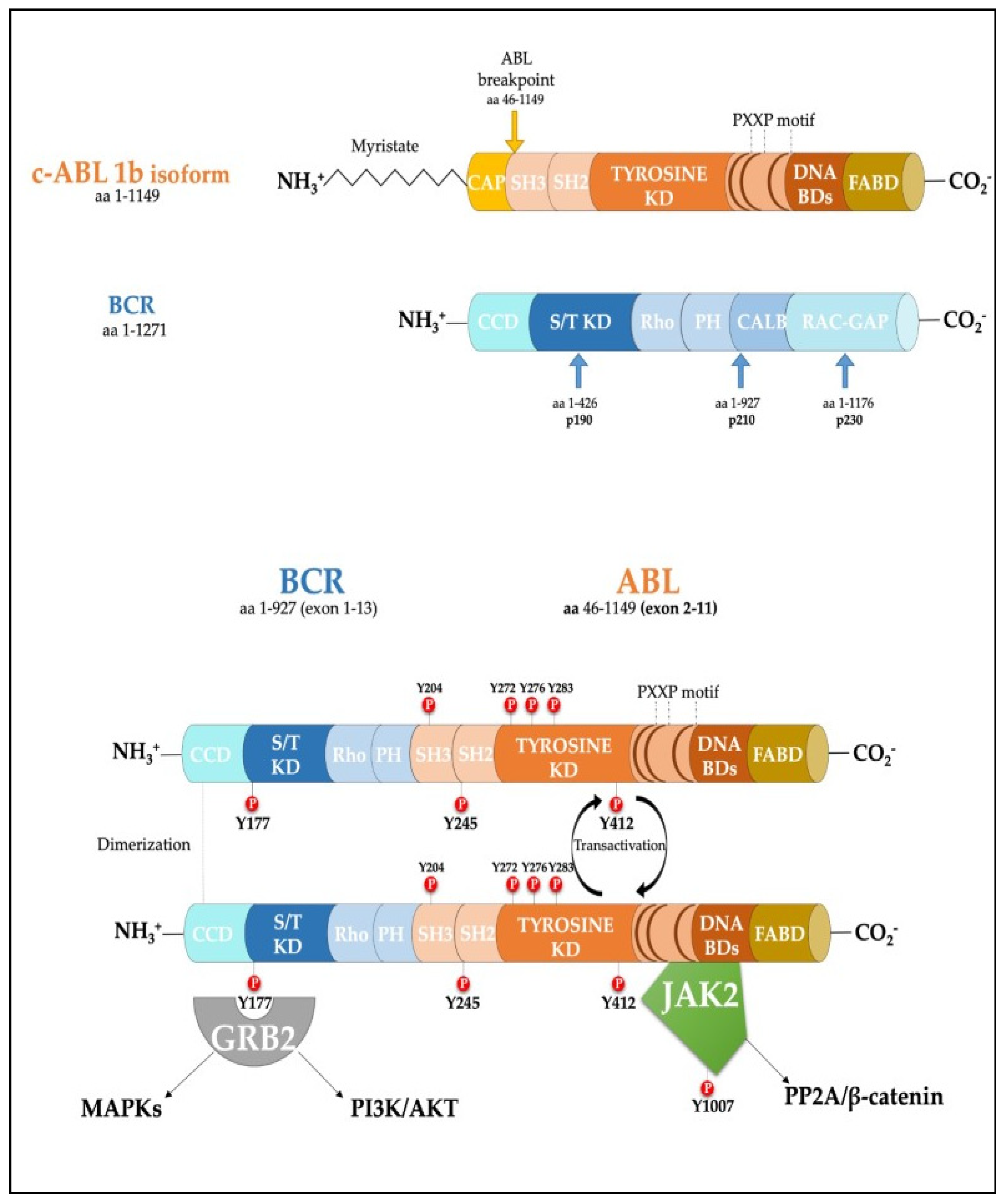
2. Role of Phosphatases in Chronic Myeloid Leukemia
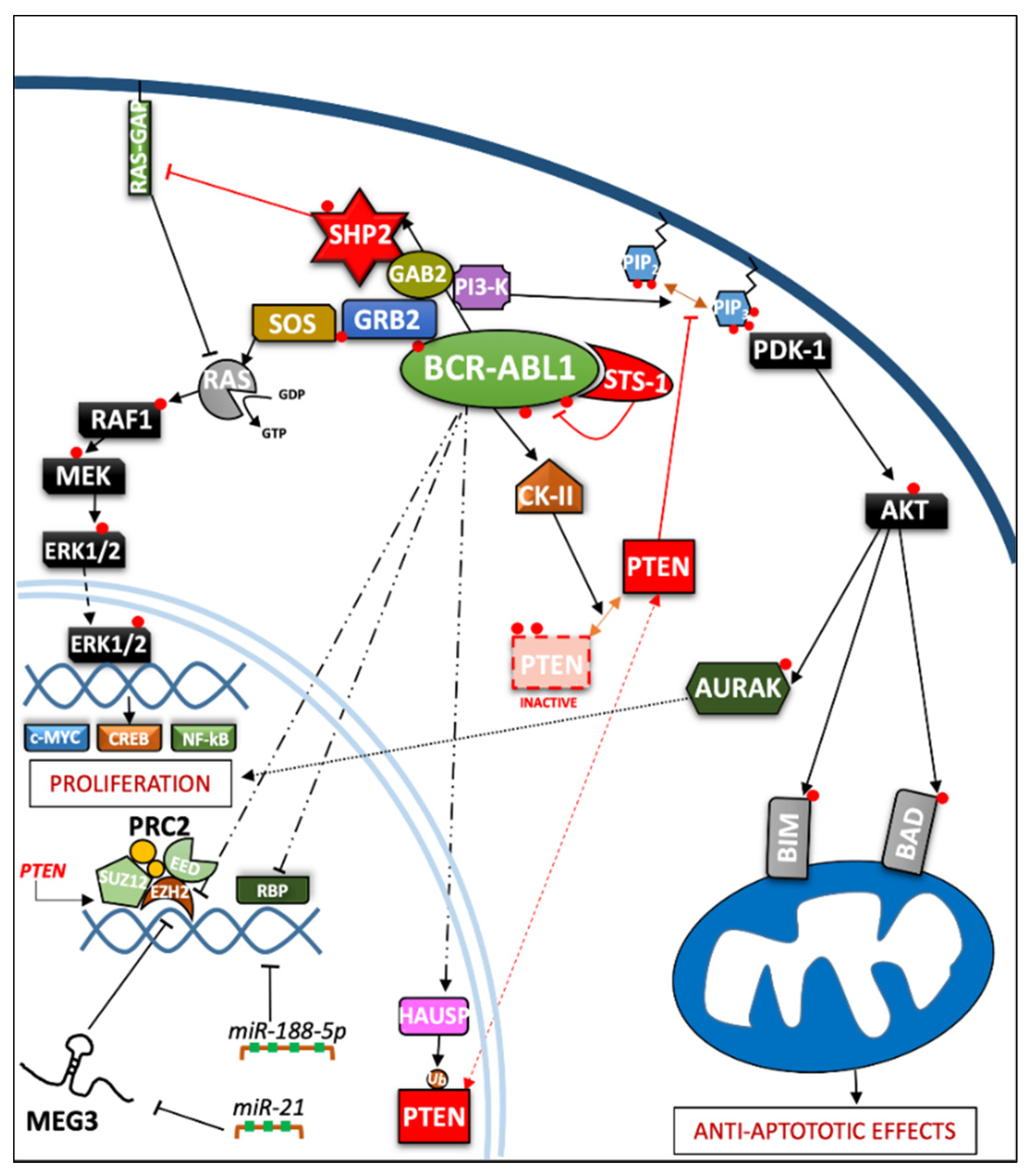
2.1. Role of Phosphatases in the Regulation of Cell Proliferation
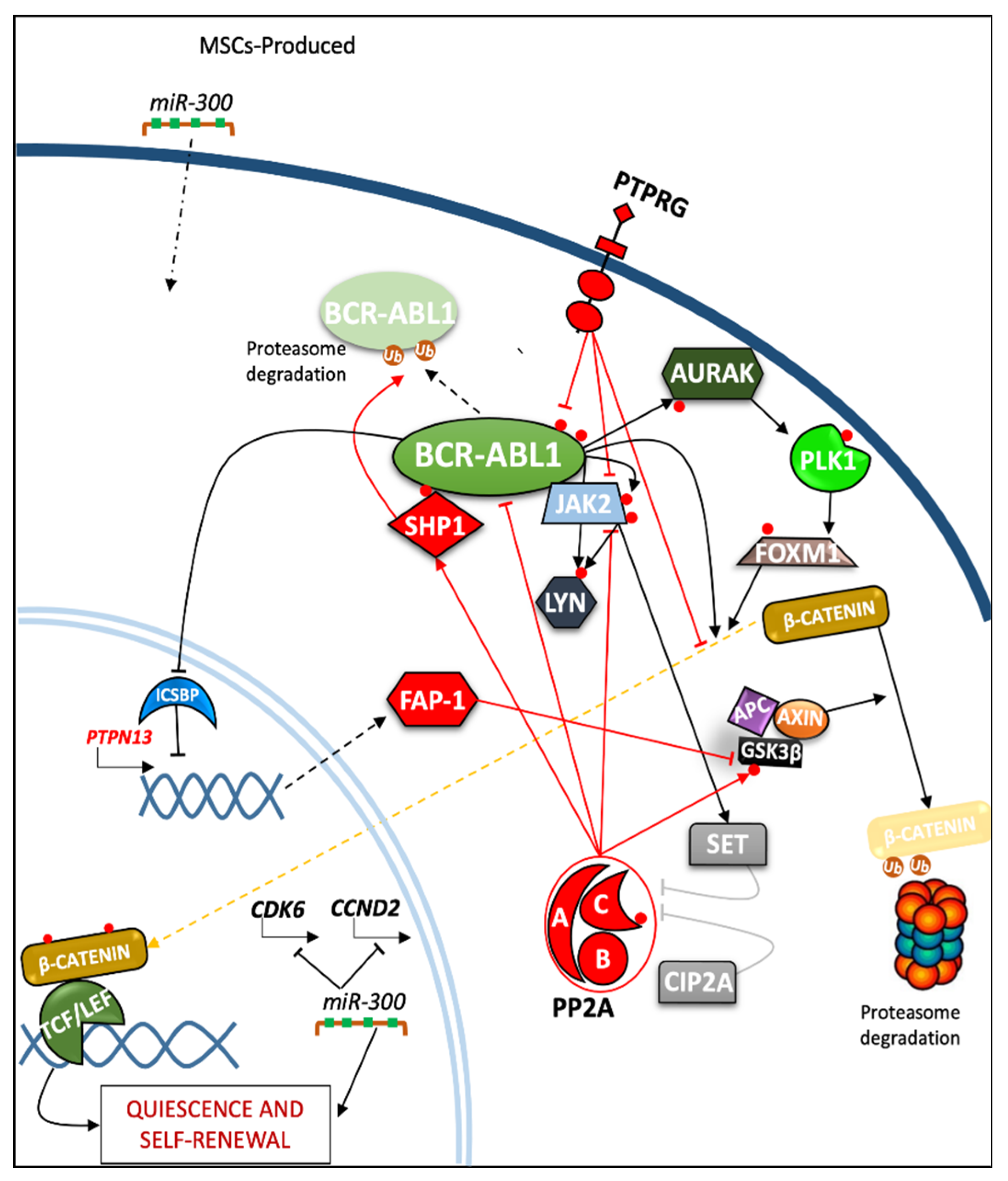
2.2. Role of Phosphatases in Regulation of Quiescence and Self-Renewal in Progenitor and Leukemia Stem Cells (LSC)
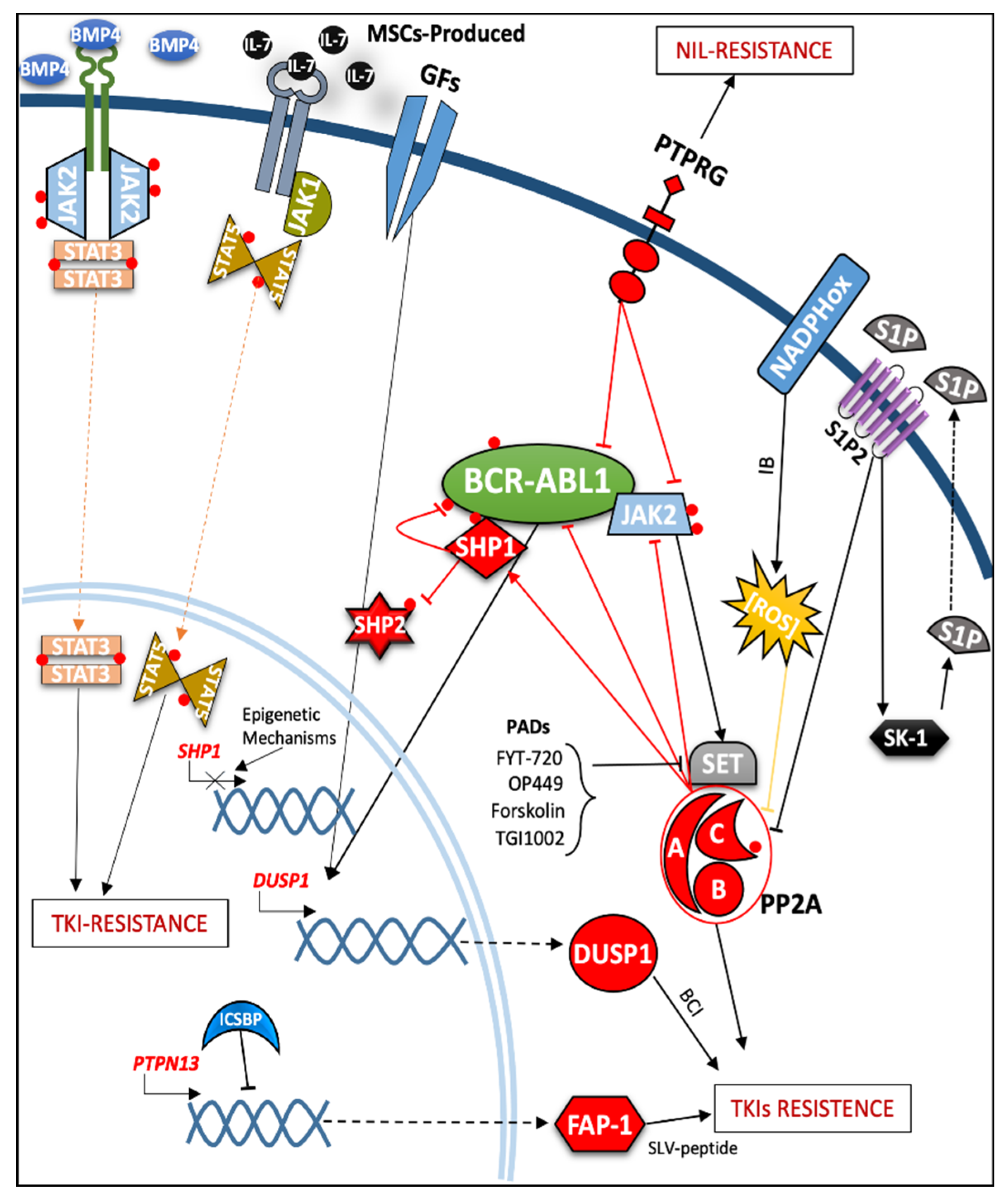
2.3. Role of Protein Phosphatases in the Modulation of TKIs Response and Resistance Mechanisms
3. Pathogenetic Role of Protein Phosphatases Requiring Validation in Primary CML Cells or Mouse Leukemia Models
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| PTK | Protein tyrosine kinase |
| TKIs | Tyrosine Kinase Inhibitors |
| PPs | Protein Phosphatases |
| PTPs | Protein Tyrosine Phosphatases |
| PTKRs | Protein Tyrosine Kinase Receptors |
| PTKNRs | Protein Tyrosine Kinase Non Receptors |
| CML | Chronic Myeloid Leukemia |
| CP | Chronic Phase |
| AP | Accelerated phase |
| BP | Blast phase |
| ALL | Acute Lymphoblastic Leukemia |
| HSCs | Hematopoietic Stem Cells |
| LICs | Leukemia Initiating Cells |
| LSCs | Leukemia Stem Cells |
| MSCs | Mesenchymal Stem cells |
| LSK | Lin−/Sca-1+/c-Kit+ cells |
| IM | Imatinib |
| MAPKs | Mitogen-activated protein kinases |
| GRB2 | Growth Factor Receptor Bound Protein-2 |
| GAB2 | GRB2-associated binding protein 2 |
| RAS | Rat sarcoma; SOS Son of Sevenless |
| PI3K | Phosphoinositide-3 kinase |
| PtdIns3P | Phosphatidylinositol (3,4,5)-trisphosphate |
| PTEN | Phosphatase and tensin homologue |
| SHP | Src homology region 2 domain-containing phosphatase |
| PP2A | Protein phosphatase 2 |
| PADs | PP2A-activating drugs |
| PIDs | PP2A-inhibiting drugs |
| CIP2A | Cancerous inhibitor of Protein Phosphatase 2A |
| PTPRG | Protein Tyrosine Phosphatase Receptor Type γ |
| FAP-1 | Fas-Associated Phosphatase-1 |
| DUSP | dual-specificity phosphatase |
| LMW-PTP | Low molecular weight protein tyrosine phosphatase |
| NSG | Non-obese diabetic (NOD)/severe combined immunodeficient (SCID) interleukin-2 receptor γ-chain–deficient |
| PPFIA1 | Receptor type, f polypeptide, leukocyte common antigen (LAR) interacting protein (liprin), alpha 1 |
| DNMT | DNA (cytosine-5)-methyltransferase |
| ICSBP | Interferon Consensus Sequence-Binding Protein |
| LTC-ICs | Long-Term Culture-Initiating Cells |
References
- Hunter, T. Protein kinases and phosphatases: The Yin and Yang of protein phosphorylation and signaling. Cell 1995, 80, 225–236. [Google Scholar] [CrossRef]
- Ubersax, J.A.; Ferrell, J.E. Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 2007, 8, 530–541. [Google Scholar] [CrossRef]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The Protein Kinase Complement of the Human Genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef]
- Robinson, D.R.; Wu, Y.-M.; Lin, S.-F. The protein tyrosine kinase family of the human genome. Oncogene 2000, 19, 5548–5557. [Google Scholar] [CrossRef]
- Lemmon, M.A.; Schlessinger, J. Cell Signaling by Receptor Tyrosine Kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef]
- Shah, N.H.; Amacher, J.F.; Nocka, L.M.; Kuriyan, J. The Src module: An ancient scaffold in the evolution of cytoplasmic tyrosine kinases. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 535–563. [Google Scholar] [CrossRef]
- Thomas, S.M.; Brugge, J.S. Cellular Functions Regulated by Src Family Kinases. Annu. Rev. Cell Dev. Biol. 1997, 13, 513–609. [Google Scholar] [CrossRef]
- Roskoski, R. A historical overview of protein kinases and their targeted small molecule inhibitors. Pharmacol. Res. 2015, 100, 1–23. [Google Scholar] [CrossRef]
- Blume-Jensen, P.; Hunter, T. Oncogenic kinase signalling. Nature 2001, 411, 355–365. [Google Scholar] [CrossRef]
- Nagar, B.; Hantschel, O.; Seeliger, M.; Davies, J.M.; Weis, W.I.; Superti-Furga, G.; Kuriyan, J. Organization of the SH3-SH2 Unit in Active and Inactive Forms of the c-Abl Tyrosine Kinase. Mol. Cell 2006, 21, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Dorey, K.; Engen, J.R.; Kretzschmar, J.; Wilm, M.; Neubauer, G.; Schindler, T.; Superti-Furga, G. Phosphorylation and structure-based functional studies reveal a positive and a negative role for the activation loop of the c-Abl tyrosine kinase. Oncogene 2001, 20, 8075–8084. [Google Scholar] [CrossRef]
- McWhirter, J.R.; Galasso, D.L.; Wang, J.Y. A coiled-coil oligomerization domain of Bcr is essential for the transforming function of Bcr-Abl oncoproteins. Mol. Cell. Biol. 1993, 13, 7587–7595. [Google Scholar] [CrossRef]
- Colicelli, J. ABL Tyrosine Kinases: Evolution of Function, Regulation, and Specificity. Sci. Signal. 2010, 3, re6. [Google Scholar] [CrossRef]
- Deininger, M.W.; Goldman, J.M.; Melo, J.V. The molecular biology of chronic myeloid leukemia. Blood 2000, 96, 3343–3356. [Google Scholar] [CrossRef]
- Daley, G.Q.; Van Etten, R.A.; Baltimore, D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science 1990, 247, 824–830. [Google Scholar] [CrossRef]
- Druker, B.J. Translation of the Philadelphia chromosome into therapy for CML. Blood 2008, 112, 4808–4817. [Google Scholar] [CrossRef]
- Apperley, J.F. Part I: Mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 2007, 8, 1018–1029. [Google Scholar] [CrossRef]
- Branford, S.; Rudzki, Z.; Walsh, S.; Parkinson, I.; Grigg, A.; Szer, J.; Taylor, K.; Herrmann, R.; Seymour, J.F.; Arthur, C.; et al. Detection of BCR-ABL mutations in patients with CML treated with imatinib is virtually always accompanied by clinical resistance, and mutations in the ATP phosphate-binding loop (P-loop) are associated with a poor prognosis. Blood 2003, 102, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.; Jabbour, E.; Kantarjian, H.; Yin, C.C.; Shan, J.; O’Brien, S.; Garcia-Manero, G.; Giles, F.; Breeden, M.; Reeves, N.; et al. Dynamics of BCR-ABL kinase domain mutations in chronic myeloid leukemia after sequential treatment with multiple tyrosine kinase inhibitors. Blood 2007, 110, 4005–4011. [Google Scholar] [CrossRef]
- Hughes, T.P.; Saglio, G.; Quintás-Cardama, A.; Mauro, M.J.; Kim, D.-W.; Lipton, J.H.; Bradley-Garelik, M.B.; Ukropec, J.; Hochhaus, A. BCR-ABL1 mutation development during first-line treatment with dasatinib or imatinib for chronic myeloid leukemia in chronic phase. Leukemia 2015, 29, 1832–1838. [Google Scholar] [CrossRef]
- Labbe, D.P.; Hardy, S.; Tremblay, M.L. Protein tyrosine phosphatases in cancer: Friends and foes! Prog. Mol. Biol. Transl. Sci. 2012, 106, 253–306. [Google Scholar] [PubMed]
- Bononi, A.; Agnoletto, C.; De Marchi, E.; Marchi, S.; Patergnani, S.; Bonora, M.; Giorgi, C.; Missiroli, S.; Poletti, F.; Rimessi, A.; et al. Protein Kinases and Phosphatases in the Control of Cell Fate. Enzym. Res. 2011, 2011, 1–26. [Google Scholar] [CrossRef]
- Ruvolo, P.P. Role of protein phosphatases in the cancer microenvironment. Biochim. Biophys. Acta Bioenerg. 2019, 1866, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Notari, M.; Neviani, P.; Santhanam, R.; Blaser, B.W.; Chang, J.-S.; Galietta, A.; Willis, A.E.; Roy, D.C.; Caligiuri, M.A.; Marcucci, G.; et al. A MAPK/HNRPK pathway controls BCR/ABL oncogenic potential by regulating MYC mRNA translation. Blood 2006, 107, 2507–2516. [Google Scholar] [CrossRef]
- Platanias, L.C. Map kinase signaling pathways and hematologic malignancies. Blood 2003, 101, 4667–4679. [Google Scholar] [CrossRef]
- Kang, C.-D.; Yoo, S.-D.; Hwang, B.-W.; Kim, K.-W.; Kim, D.-W.; Kim, C.-M.; Kim, S.-H.; Chung, B.-S. The inhibition of ERK/MAPK not the activation of JNK/SAPK is primarily required to induce apoptosis in chronic myelogenous leukemic K562 cells. Leuk. Res. 2000, 24, 527–534. [Google Scholar] [CrossRef]
- Juric, D.; Lacayo, N.J.; Ramsey, M.C.; Racevskis, J.; Wiernik, P.H.; Rowe, J.M.; Goldstone, A.H.; O’Dwyer, P.J.; Paietta, E.; Sikic, B.I. Differential Gene Expression Patterns and Interaction Networks in BCR-ABL–Positive and –Negative Adult Acute Lymphoblastic Leukemias. J. Clin. Oncol. 2007, 25, 1341–1349. [Google Scholar] [CrossRef]
- Cutler, J.A.; Tahir, R.; Sreenivasamurthy, S.K.; Mitchell, C.; Renuse, S.; Nirujogi, R.S.; Patil, A.H.; Heydarian, M.; Wong, X.; Wu, X.; et al. Differential signaling through p190 and p210 BCR-ABL fusion proteins revealed by interactome and phosphoproteome analysis. Leukemia 2017, 31, 1513–1524. [Google Scholar] [CrossRef] [PubMed]
- Brehme, M.; Hantschel, O.; Colinge, J.; Kaupe, I.; Planyavsky, M.; Köcher, T.; Mechtler, K.; Bennett, K.L.; Superti-Furga, G. Charting the molecular network of the drug target Bcr-Abl. Proc. Natl. Acad. Sci. USA 2009, 106, 7414–7419. [Google Scholar] [CrossRef]
- Mian, A.A.; Baumann, I.; Liebermann, M.; Grebien, F.; Superti-Furga, G.; Ruthardt, M.; Ottmann, O.G.; Hantschel, O. The phosphatase UBASH3B/Sts-1 is a negative regulator of Bcr-Abl kinase activity and leukemogenesis. Leukemia 2019, 33, 2319–2323. [Google Scholar] [CrossRef]
- Pendergast, A.M.; Quilliam, L.A.; Cripe, L.D.; Bassing, C.H.; Dai, Z.; Li, N.; Batzer, A.; Rabun, K.M.; Der, C.J.; Schlessinger, J. BCR-ABL-induced oncogenesis is mediated by direct interaction with the SH2 domain of the GRB-2 adaptor protein. Cell 1993, 75, 175–185. [Google Scholar] [CrossRef]
- Sattler, M.; Mohi, M.; Pride, Y.B.; Quinnan, L.R.; Malouf, N.A.; Podar, K.; Gesbert, F.; Iwasaki, H.; Li, S.; Van Etten, R.A.; et al. Critical role for Gab2 in transformation by BCR/ABL. Cancer Cell 2002, 1, 479–492. [Google Scholar] [CrossRef]
- Modi, H.; Li, L.; Chu, S.; Rossi, J.; Yee, J.-K.; Bhatia, R. Inhibition of Grb2 expression demonstrates an important role in BCR–ABL-mediated MAPK activation and transformation of primary human hematopoietic cells. Leukemia 2010, 25, 305–312. [Google Scholar] [CrossRef]
- Chen, M.; Turhan, A.G.; Ding, H.; Lin, Q.; Meng, K.; Jiang, X. Targeting BCR-ABL+ stem/progenitor cells and BCR-ABL-T315I mutant cells by effective inhibition of the BCR-ABL-Tyr177-GRB2 complex. Oncotarget 2017, 8, 43662–43677. [Google Scholar] [CrossRef][Green Version]
- Gu, H.; Pratt, J.C.; Burakoff, S.J.; Neel, B.G. Cloning of p97/Gab2, the Major SHP2-Binding Protein in Hematopoietic Cells, Reveals a Novel Pathway for Cytokine-Induced Gene Activation. Mol. Cell 1998, 2, 729–740. [Google Scholar] [CrossRef]
- Zhang, Y.; Diaz-Flores, E.; Li, G.; Wang, Z.; Kang, Z.; Haviernikova, E.; Rowe, S.; Qu, C.-K.; Tse, W.; Shannon, K.M.; et al. Abnormal hematopoiesis in Gab2 mutant mice. Blood 2007, 110, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Qu, C.-K.; Nguyen, S.; Chen, J.; Feng, G.-S. Requirement of Shp-2 tyrosine phosphatase in lymphoid and hematopoietic cell development. Blood 2001, 97, 911–914. [Google Scholar] [CrossRef]
- Dance, M.; Montagner, A.; Salles, J.P.; Yart, A.; Raynal, P. The molecular functions of Shp2 in the Ras/Mitogen-activated protein kinase (ERK1/2) pathway. Cell. Signal. 2008, 20, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.; Saxena, M.; Kapur, R. Role of SHP2 in hematopoiesis and leukemogenesis. Curr. Opin. Hematol. 2017, 24, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Scherr, M.; Chaturvedi, A.; Battmer, K.; Dallmann, I.; Schultheis, B.; Ganser, A.; Eder, M. Enhanced sensitivity to inhibition of SHP2, STAT5, and Gab2 expression in chronic myeloid leukemia (CML). Blood 2006, 107, 3279–3287. [Google Scholar] [CrossRef][Green Version]
- Xu, R.; Yu, Y.; Zheng, S.; Zhao, X.; Dong, Q.; He, Z.; Liang, Y.; Lu, Q.; Fang, Y.; Gan, X.; et al. Overexpression of Shp2 tyrosine phosphatase is implicated in leukemogenesis in adult human leukemia. Blood 2005, 106, 3142–3149. [Google Scholar] [CrossRef]
- Gu, S.; Chan, W.W.; Mohi, G.; Rosenbaum, J.; Sayad, A.; Lu, Z.; Virtanen, C.; Li, S.; Neel, B.G.; Van Etten, R.A. Distinct GAB2 signaling pathways are essential for myeloid and lymphoid transformation and leukemogenesis by BCR-ABL1. Blood 2016, 127, 1803–1813. [Google Scholar] [CrossRef]
- Gu, S.; Sayad, A.; Chan, G.; Yang, W.; Lu, Z.; Virtanen, C.; Van Etten, R.A.; Neel, B.G. SHP2 is required for BCR-ABL1-induced hematologic neoplasia. Leukemia 2018, 32, 203–213. [Google Scholar] [CrossRef]
- Esposito, N.; Colavita, I.; Quintarelli, C.; Sica, A.R.; Peluso, A.L.; Luciano, L.; Picardi, M.; Del Vecchio, L.; Buonomo, T.; Hughes, T.P.; et al. SHP-1 expression accounts for resistance to imatinib treatment in Philadelphia chromosome–positive cells derived from patients with chronic myeloid leukemia. Blood 2011, 118, 3634–3644. [Google Scholar] [CrossRef][Green Version]
- Montagner, A.; Yart, A.; Dance, M.; Perret, B.; Salles, J.-P.; Raynal, P. A Novel Role for Gab1 and SHP2 in Epidermal Growth Factor-induced Ras Activation. J. Biol. Chem. 2005, 280, 5350–5360. [Google Scholar] [CrossRef]
- Kharas, M.G.; Fruman, D.A. ABL Oncogenes and Phosphoinositide 3-Kinase: Mechanism of Activation and Downstream Effectors. Cancer Res. 2005, 65, 2047–2053. [Google Scholar] [CrossRef]
- Tasian, S.K.; Teachey, D.T.; Rheingold, S.R. Targeting the PI3K/mTOR Pathway in Pediatric Hematologic Malignancies. Front. Oncol. 2014, 4, 108. [Google Scholar] [CrossRef]
- Juntilla, M.M.; Patil, V.D.; Calamito, M.; Joshi, R.P.; Birnbaum, M.J.; Koretzky, G.A. AKT1 and AKT2 maintain hematopoietic stem cell function by regulating reactive oxygen species. Blood 2010, 115, 4030–4038. [Google Scholar] [CrossRef]
- Nie, Z.Y.; Yang, L.; Liu, X.; Yang, Z.; Yang, G.; Zhou, J.; Qin, Y.; Yu, J.; Jiang, L.; Wen, J.; et al. Morin Inhibits Proliferation and Induces Apoptosis by Modulating the miR-188-5p/PTEN/AKT Regulatory Pathway in CML Cells. Mol. Cancer Ther. 2019, 18, 2296–2307. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, T.; Du, J.; Li, Y.; Wang, X.; Zhou, Y.; Yu, X.; Fan, W.; Zhu, Q.; Tong, X.; et al. The Critical Role of PTEN/PI3K/AKT Signaling Pathway in Shikonin-Induced Apoptosis and Proliferation Inhibition of Chronic Myeloid Leukemia. Cell. Physiol. Biochem. 2018, 47, 981–993. [Google Scholar] [CrossRef]
- Skorski, T.; Kanakaraj, P.; Nieborowska-Skorska, M.; Ratajczak, M.Z.; Wen, S.C.; Zon, G.; Gewirtz, A.M.; Perussia, B.; Calabretta, B. Phosphatidylinositol-3 kinase activity is regulated by BCR/ABL and is required for the growth of Philadelphia chromosome-positive cells. Blood 1995, 86, 726–736. [Google Scholar] [CrossRef]
- Skorski, T.; Bellacosa, A.; Nieborowska-Skorska, M.; Majewski, M.; Martinez, R.; Choi, J.K.; Trotta, R.; Wlodarski, P.; Perrotti, D.; Chan, T.O.; et al. Transformation of hematopoietic cells by BCR/ABL requires activation of a PI-3k/Akt-dependent pathway. EMBO J. 1997, 16, 6151–6161. [Google Scholar] [CrossRef]
- Yang, J.; Ikezoe, T.; Nishioka, C.; Udaka, K.; Yokoyama, A. Bcr-Abl activates AURKA and AURKB in chronic myeloid leukemia cells via AKT signaling. Int. J. Cancer 2013, 134, 1183–1194. [Google Scholar] [CrossRef]
- Hamzah, H.G.; Pierce, A.; Stewart, W.A.; Downes, C.P.; Gray, A.; Irvine, A.; Spooncer, E.; Whetton, A.D. Chronic myeloid leukemia CD34+ cells have elevated levels of phosphatidylinositol 3,4,5 trisphosphate (PtdIns(3,4,5)P3) and lack a PtdIns(3,4,5)P3 response to cytokines and chemotactic factors; effects reversed by imatinib. Leukemia 2005, 19, 1851–1853. [Google Scholar] [CrossRef][Green Version]
- Xin, P.; Li, C.; Zheng, Y.; Peng, Q.; Xiao, H.; Huang, Y.; Zhu, X. Efficacy of the dual PI3K and mTOR inhibitor NVP-BEZ235 in combination with imatinib mesylate against chronic myelogenous leukemia cell lines. Drug Des. Dev. Ther. 2017, 11, 1115–1126. [Google Scholar] [CrossRef]
- Schuster, K.; Zheng, J.; Arbini, A.A.; Zhang, C.C.; Scaglioni, P.P. Selective targeting of the mTORC1/2 protein kinase complexes leads to antileukemic effects in vitro and in vivo. Blood Cancer J. 2011, 1, e34. [Google Scholar] [CrossRef]
- Trimboli, A.J.; Cantemir-Stone, C.Z.; Li, F.; Wallace, J.A.; Merchant, A.; Creasap, N.; Thompson, J.C.; Caserta, E.; Wang, H.; Chong, J.-L.; et al. Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nat. Cell Biol. 2009, 461, 1084–1091. [Google Scholar] [CrossRef]
- Nishioka, C.; Ikezoe, T.; Yang, J.; Yokoyama, A. Long-term exposure of leukemia cells to multi-targeted tyrosine kinase inhibitor induces activations of AKT, ERK and STAT5 signaling via epigenetic silencing of the PTEN gene. Leukemia 2010, 24, 1631–1640. [Google Scholar] [CrossRef]
- Peng, C.; Chen, Y.; Yang, Z.; Zhang, H.; Osterby, L.; Rosmarin, A.G.; Li, S. PTEN is a tumor suppressor in CML stem cells and BCR-ABL–induced leukemias in mice. Blood 2010, 115, 626–635. [Google Scholar] [CrossRef] [PubMed]
- Panuzzo, C.; Crivellaro, S.; Carra, G.; Guerrasio, A.; Saglio, G.; Morotti, A. BCR-ABL Promotes PTEN Downregulation in Chronic Myeloid Leukemia. PLoS ONE 2014, 9, e110682. [Google Scholar] [CrossRef]
- Bassi, C.; Ho, J.; Srikumar, T.; Dowling, R.J.O.; Gorrini, C.; Miller, S.J.; Mak, T.W.; Neel, B.G.; Raught, B.; Stambolic, V. Nuclear PTEN Controls DNA Repair and Sensitivity to Genotoxic Stress. Science 2013, 341, 395–399. [Google Scholar] [CrossRef]
- Song, M.S.; Salmena, L.; Carracedo, A.; Egia, A.; Lo-Coco, F.; Teruya-Feldstein, J.; Pandolfi, P.P. The deubiquitinylation and localization of PTEN are regulated by a HAUSP–PML network. Nat. Cell Biol. 2008, 455, 813–817. [Google Scholar] [CrossRef]
- Morotti, A.; Panuzzo, C.; Crivellaro, S.; Pergolizzi, B.; Familiari, U.; Berger, A.H.; Saglio, G.; Pandolfi, P.P. BCR-ABL disrupts PTEN nuclear-cytoplasmic shuttling through phosphorylation-dependent activation of HAUSP. Leukemia 2014, 28, 1326–1333. [Google Scholar] [CrossRef] [PubMed]
- Morotti, A.; Panuzzo, C.; Crivellaro, S.; Carrà, G.; Fava, C.; Guerrasio, A.; Pandolfi, P.P.; Saglio, G. BCR-ABL inactivates cytosolic PTEN through Casein Kinase II mediated tail phosphorylation. Cell Cycle 2015, 14, 973–979. [Google Scholar] [CrossRef] [PubMed]
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef]
- Xie, H.; Peng, C.; Huang, J.; Li, B.E.; Kim, W.; Smith, E.C.; Fujiwara, Y.; Qi, J.; Cheloni, G.; Das, P.P.; et al. Chronic Myelogenous Leukemia–Initiating Cells Require Polycomb Group Protein EZH2. Cancer Discov. 2016, 6, 1237–1247. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.T.; Korfi, K.; Saffrey, P.; Hopcroft, L.E.; Kinstrie, R.; Pellicano, F.; Guenther, C.; Gallipoli, P.; Cruz, M.; Dunn, K.; et al. Epigenetic Reprogramming Sensitizes CML Stem Cells to Combined EZH2 and Tyrosine Kinase Inhibition. Cancer Discov. 2016, 6, 1248–1257. [Google Scholar] [CrossRef]
- Zhou, J.; Nie, D.; Li, J.; Du, X.; Lu, Y.; Li, Y.; Liu, C.; Dai, W.; Wang, Y.; Jin, Y.; et al. PTEN Is Fundamental for Elimination of Leukemia Stem Cells Mediated by GSK126 Targeting EZH2 in Chronic Myelogenous Leukemia. Clin. Cancer Res. 2017, 24, 145–157. [Google Scholar] [CrossRef]
- Zhou, X.; Yuan, P.; Liu, Q.; Liu, Z. LncRNA MEG3 Regulates Imatinib Resistance in Chronic Myeloid Leukemia via Suppressing MicroRNA-21. Biomol. Ther. 2017, 25, 490–496. [Google Scholar] [CrossRef]
- Li, Z.; Yang, L.; Liu, X.; Nie, Z.; Luo, J. Long noncoding RNA MEG3 inhibits proliferation of chronic myeloid leukemia cells by sponging microRNA21. Biomed. Pharmacother. 2018, 104, 181–192. [Google Scholar] [CrossRef]
- Yin, X.; Zhou, M.; Fu, Y.; Yang, L.; Xu, M.; Sun, T.; Wang, X.; Huang, T.; Chen, C. Histone demethylase RBP2 mediates the blast crisis of chronic myeloid leukemia through an RBP2/PTEN/BCR-ABL cascade. Cell. Signal. 2019, 63, 109360. [Google Scholar] [CrossRef]
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef]
- Hope, K.J.; Jin, L.; Dick, J.E. Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self-renewal capacity. Nat. Immunol. 2004, 5, 738–743. [Google Scholar] [CrossRef]
- Lapidot, T.; Sirard, C.; Vormoor, J.; Murdoch, B.; Hoang, T.; Caceres-Cortes, J.; Minden, M.; Paterson, B.; Caligiuri, M.A.; Dick, J.E. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nat. Cell Biol. 1994, 367, 645–648. [Google Scholar] [CrossRef]
- Holyoake, T.L.; Vetrie, D. The chronic myeloid leukemia stem cell: Stemming the tide of persistence. Blood 2017, 129, 1595–1606. [Google Scholar] [CrossRef]
- Fang, B.; Zheng, C.; Liao, L.; Han, Q.; Sun, Z.; Jiang, X.; Zhao, R.C.H. Identification of human chronic myelogenous leukemia progenitor cells with hemangioblastic characteristics. Blood 2005, 105, 2733–2740. [Google Scholar] [CrossRef] [PubMed]
- O’Hare, T.; Zabriskie, M.S.; Eiring, A.M.; Deininger, M.W. Pushing the limits of targeted therapy in chronic myeloid leukaemia. Nat. Rev. Cancer 2012, 12, 513–526. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, H.; Sadovnik, I.; Cerny-Reiterer, S.; Rülicke, T.; Stefanzl, G.; Willmann, M.; Hoermann, G.; Bilban, M.; Blatt, K.; Herndlhofer, S.; et al. Dipeptidylpeptidase IV (CD26) defines leukemic stem cells (LSC) in chronic myeloid leukemia. Blood 2014, 123, 3951–3962. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, H.; Sadovnik, I.; Eisenwort, G.; Rulicke, T.; Blatt, K.; Herndlhofer, S.; Willmann, M.; Stefanzl, G.; Baumgartner, S.; Greiner, G.; et al. Delineation of target expression profiles in CD34+/CD38- and CD34+/CD38+ stem and progenitor cells in AML and CML. Blood Adv. 2020, 4, 5118–5132. [Google Scholar] [CrossRef]
- Sadovnik, I.; Hoelbl-Kovacic, A.; Herrmann, H.; Eisenwort, G.; Cerny-Reiterer, S.; Warsch, W.; Hoermann, G.; Greiner, G.; Blatt, K.; Peter, B.; et al. Identification of CD25 as STAT5-Dependent Growth Regulator of Leukemic Stem Cells in Ph+ CML. Clin. Cancer Res. 2016, 22, 2051–2061. [Google Scholar] [CrossRef]
- Lemoli, R.M.; Salvestrini, V.; Bianchi, E.; Bertolini, F.; Fogli, M.; Amabile, M.; Tafuri, A.; Salati, S.; Zini, R.; Testoni, N.; et al. Molecular and functional analysis of the stem cell compartment of chronic myelogenous leukemia reveals the presence of a CD34− cell population with intrinsic resistance to imatinib. Blood 2009, 114, 5191–5200. [Google Scholar] [CrossRef] [PubMed]
- Taussig, D.C.; Miraki-Moud, F.; Anjos-Afonso, F.; Pearce, D.J.; Allen, K.; Ridler, C.; Lillington, D.; Oakervee, H.; Cavenagh, J.; Agrawal, S.G.; et al. Anti-CD38 antibody–mediated clearance of human repopulating cells masks the heterogeneity of leukemia-initiating cells. Blood 2008, 112, 568–575. [Google Scholar] [CrossRef]
- Perrotti, D.; Silvestri, G.; Stramucci, L.; Yu, J.; Trotta, R. Cellular and Molecular Networks in Chronic Myeloid Leukemia: The Leukemic Stem, Progenitor and Stromal Cell Interplay. Curr. Drug Targets 2017, 18, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Blum, J.; Chen, A.; Kwon, H.Y.; Jung, S.H.; Cook, J.M.; Lagoo, A.; Reya, T. Loss of beta-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer Cell 2007, 12, 528–541. [Google Scholar] [CrossRef]
- Minami, Y.; Stuart, S.A.; Ikawa, T.; Jiang, Y.; Banno, A.; Hunton, I.C.; Young, D.J.; Naoe, T.; Murre, C.; Jamieson, C.H.M.; et al. BCR-ABL-transformed GMP as myeloid leukemic stem cells. Proc. Natl. Acad. Sci. USA 2008, 105, 17967–17972. [Google Scholar] [CrossRef]
- Coluccia, A.M.; Vacca, A.; Dunach, M.; Mologni, L.; Redaelli, S.; Bustos, V.H.; Benati, D.; Pinna, L.A.; Gambacorti-Passerini, C. Bcr-Abl stabilizes beta-catenin in chronic myeloid leukemia through its tyrosine phosphorylation. EMBO J. 2007, 26, 1456–1466. [Google Scholar] [CrossRef] [PubMed]
- Mancini, M.; De Santis, S.; Monaldi, C.; Bavaro, L.; Martelli, M.; Castagnetti, F.; Gugliotta, G.; Rosti, G.; Santucci, M.A.; Martinelli, G.; et al. Hyper-activation of Aurora kinase a-polo-like kinase 1-FOXM1 axis promotes chronic myeloid leukemia resistance to tyrosine kinase inhibitors. J. Exp. Clin. Cancer Res. 2019, 38, 216. [Google Scholar] [CrossRef]
- Walz, C.; Ahmed, W.; Lazarides, K.; Betancur, M.; Patel, N.; Hennighausen, L.; Zaleskas, V.M.; Van Etten, R.A. Essential role for Stat5a/b in myeloproliferative neoplasms induced by BCR-ABL1 and JAK2V617F in mice. Blood 2012, 119, 3550–3560. [Google Scholar] [CrossRef] [PubMed]
- Samanta, A.K.; Lin, H.; Sun, T.; Kantarjian, H.; Arlinghaus, R.B. Janus Kinase 2: A Critical Target in Chronic Myelogenous Leukemia. Cancer Res. 2006, 66, 6468–6472. [Google Scholar] [CrossRef]
- Chen, M.; Gallipoli, P.; DeGeer, D.; Sloma, I.; Forrest, D.L.; Chan, M.; Lai, D.; Jorgensen, H.; Ringrose, A.; Wang, H.M.; et al. Targeting Primitive Chronic Myeloid Leukemia Cells by Effective Inhibition of a New AHI-1–BCR-ABL–JAK2 Complex. J. Natl. Cancer Inst. 2013, 105, 405–423. [Google Scholar] [CrossRef]
- Xie, S.; Wang, Y.; Liu, J.; Sun, T.; Wilson, M.B.; Smithgall, T.E.; Arlinghaus, R.B. Involvement of Jak2 tyrosine phosphorylation in Bcr–Abl transformation. Oncogene 2001, 20, 6188–6195. [Google Scholar] [CrossRef] [PubMed]
- Samanta, A.; Perazzona, B.; Chakraborty, S.; Sun, X.; Modi, H.R.; Bhatia, R.; Priebe, W.; Arlinghaus, R.B. Janus kinase 2 regulates Bcr–Abl signaling in chronic myeloid leukemia. Leukemia 2010, 25, 463–472. [Google Scholar] [CrossRef]
- Janssens, V.; Goris, J.; Van Hoof, C. PP2A: The expected tumor suppressor. Curr. Opin. Genet. Dev. 2005, 15, 34–41. [Google Scholar] [CrossRef]
- McCubrey, J.A.; Steelman, L.S.; Bertrand, F.E.; Davis, N.M.; Abrams, S.L.; Montalto, G.; D’Assoro, A.B.; Libra, M.; Nicoletti, F.; Maestro, R.; et al. Multifaceted roles of GSK-3 and Wnt/beta-catenin in hematopoiesis and leukemogenesis: Opportunities for therapeutic intervention. Leukemia 2014, 28, 15–33. [Google Scholar] [CrossRef]
- Thompson, J.J.; Williams, C.S. Protein Phosphatase 2A in the Regulation of Wnt Signaling, Stem Cells, and Cancer. Genes 2018, 9, 121. [Google Scholar] [CrossRef] [PubMed]
- Neviani, P.; Santhanam, R.; Trotta, R.; Notari, M.; Blaser, B.W.; Liu, S.; Mao, H.; Chang, J.S.; Galietta, A.; Uttam, A.; et al. The tumor suppressor PP2A is functionally inactivated in blast crisis CML through the inhibitory activity of the BCR/ABL-regulated SET protein. Cancer Cell 2005, 8, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Samanta, A.K.; Chakraborty, S.N.; Wang, Y.; Kantarjian, H.; Sun, X.; Hood, J.; Perrotti, D.; Arlinghaus, R.B. Jak2 inhibition deactivates Lyn kinase through the SET–PP2A–SHP1 pathway, causing apoptosis in drug-resistant cells from chronic myelogenous leukemia patients. Oncogene 2009, 28, 1669–1681. [Google Scholar] [CrossRef] [PubMed]
- Neviani, P.; Santhanam, R.; Oaks, J.J.; Eiring, A.M.; Notari, M.; Blaser, B.W.; Liu, S.; Trotta, R.; Muthusamy, N.; Gambacorti-Passerini, C.; et al. FTY720, a new alternative for treating blast crisis chronic myelogenous leukemia and Philadelphia chromosome–positive acute lymphocytic leukemia. J. Clin. Investig. 2007, 117, 2408–2421. [Google Scholar] [CrossRef]
- Lucas, C.M.; Harris, R.J.; Giannoudis, A.; Copland, M.; Slupsky, J.R.; Clark, R.E. Cancerous inhibitor of PP2A (CIP2A) at diagnosis of chronic myeloid leukemia is a critical determinant of disease progression. Blood 2011, 117, 6660–6668. [Google Scholar] [CrossRef] [PubMed]
- Lucas, C.M.; Milani, M.; Butterworth, M.; Carmell, N.; Scott, L.J.; Clark, R.E.; Cohen, G.M.; Varadarajan, S. High CIP2A levels correlate with an antiapoptotic phenotype that can be overcome by targeting BCL-XL in chronic myeloid leukemia. Leukemia 2016, 30, 1273–1281. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Edwards, C.A.; Mungall, A.J.; Matthews, L.; Ryder, E.; Gray, D.J.; Pask, A.J.; Shaw, G.; Graves, J.A.; Rogers, J.; Dunham, I.; et al. The Evolution of the DLK1-DIO3 Imprinted Domain in Mammals. PLoS Biol. 2008, 6, e135. [Google Scholar] [CrossRef] [PubMed]
- Benetatos, L.; Hatzimichael, E.; Londin, E.; Vartholomatos, G.; Loher, P.; Rigoutsos, I.; Briasoulis, E. The microRNAs within the DLK1-DIO3 genomic region: Involvement in disease pathogenesis. Cell. Mol. Life Sci. 2012, 70, 795–814. [Google Scholar] [CrossRef]
- Silvestri, G.; Trotta, R.; Stramucci, L.; Ellis, J.J.; Harb, J.G.; Neviani, P.; Wang, S.; Eisfeld, A.K.; Walker, C.J.; Zhang, B.; et al. Persistence of Drug-Resistant Leukemic Stem Cells and Impaired NK Cell Immunity in CML Patients Depend on MIR300 Antiproliferative and PP2A-Activating Functions. Blood Cancer Discov. 2020, 1, 48–67. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, J.; Takahashi, M.; Kanki, H.; Yano-Yanagisawa, H.; Tazunoki, T.; Sawa, E.; Nishitoba, T.; Kamishohara, M.; Kobayashi, E.; Kataoka, S.; et al. The molecular interaction of Fas and FAP-1. A tripeptide blocker of human Fas interaction with FAP-1 promotes Fas-induced apoptosis. J. Biol. Chem. 1997, 272, 8539–8545. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Zhu, C.; Wang, H.; Horvath, E.; Eklund, E.A. The Interferon Consensus Sequence-binding Protein (ICSBP/IRF8) Represses PTPN13 Gene Transcription in Differentiating Myeloid Cells. J. Biol. Chem. 2008, 283, 7921–7935. [Google Scholar] [CrossRef]
- Hao, S.X.; Ren, R.; Jahn, T.; Seipel, P.; Urschel, S.; Peschel, C.; Duyster, J. Expression of Interferon Consensus Sequence Binding Protein (ICSBP) Is Downregulated in Bcr-Abl-Induced Murine Chronic Myelogenous Leukemia-Like Disease, and Forced Coexpression of ICSBP Inhibits Bcr-Abl-Induced Myeloproliferative Disorder. Mol. Cell. Biol. 2000, 20, 979–991. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schmidt, M.; Nagel, S.; Proba, J.; Thiede, C.; Ritter, M.; Waring, J.F.; Rosenbauer, F.; Huhn, D.; Wittig, B.; Horak, I.; et al. Lack of interferon consensus sequence binding protein (ICSBP) transcripts in human myeloid leukemias. Blood 1998, 91, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sapienza, P.J.; Ke, H.; Chang, A.; Hengel, S.R.; Wang, H.; Phillips, G.N.; Lee, A.L. Crystallographic and Nuclear Magnetic Resonance Evaluation of the Impact of Peptide Binding to the Second PDZ Domain of Protein Tyrosine Phosphatase 1E. Biochemistry 2010, 49, 9280–9291. [Google Scholar] [CrossRef]
- Huang, W.; Bei, L.; Eklund, E.A. Fas-associated phosphatase 1 (Fap1) influences betacatenin activity in myeloid progenitor cells expressing the Bcr-abl oncogene. J. Biol. Chem. 2013, 288, 12766–12776. [Google Scholar] [CrossRef]
- Huang, W.; Bei, L.; Eklund, E.A. Fas-associated phosphatase 1 mediates Fas resistance in myeloid progenitor cells expressing the Bcr–abl oncogene. Leuk. Lymphoma 2012, 54, 619–630. [Google Scholar] [CrossRef]
- van Niekerk, C.C.; Poels, L.G. Reduced expression of protein tyrosine phosphatase gamma in lung and ovarian tumors. Cancer Lett. 1999, 137, 61–73. [Google Scholar] [CrossRef]
- Lissandrini, D.; Vermi, W.; Vezzalini, M.; Sozzani, S.; Facchetti, F.; Bellone, G.; Mafficini, A.; Gentili, F.; Ennas, M.G.; Tecchio, C.; et al. Receptor-type protein tyrosine phosphatase gamma (PTPγ), a new identifier for myeloid dendritic cells and specialized macrophages. Blood 2006, 108, 4223–4231. [Google Scholar] [CrossRef]
- Sorio, C.; Melotti, P.; D’Arcangelo, D.; Mendrola, J.; Calabretta, B.; Croce, C.M.; Huebner, K. Receptor protein tyrosine phosphatase gamma, Ptp gamma, regulates hematopoietic differentiation. Blood 1997, 90, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Mafficini, A.; Vezzalini, M.; Zamai, L.; Galeotti, L.; Bergamini, G.; Della Peruta, M.; Melotti, P.; Sorio, C. Protein Tyrosine Phosphatase Gamma (PTPgamma) is a Novel Leukocyte Marker Highly Expressed by CD34 Precursors. Biomark. Insights 2007, 2, 218–225. [Google Scholar] [CrossRef]
- Mirenda, M.; Toffali, L.; Montresor, A.; Scardoni, G.; Sorio, C.; Laudanna, C. Protein tyrosine phosphatase receptor type gamma is a JAK phosphatase and negatively regulates leukocyte integrin activation. J. Immunol. 2015, 194, 2168–2179. [Google Scholar] [CrossRef] [PubMed]
- Della Peruta, M.; Martinelli, G.; Moratti, E.; Pintani, D.; Vezzalini, M.; Mafficini, A.; Grafone, T.; Iacobucci, I.; Soverini, S.; Murineddu, M.; et al. Protein tyrosine phosphatase receptor type {gamma} is a functional tumor suppressor gene specifically downregulated in chronic myeloid leukemia. Cancer Res. 2010, 70, 8896–8906. [Google Scholar] [CrossRef]
- Vezzalini, M.; Mafficini, A.; Tomasello, L.; Lorenzetto, E.; Moratti, E.; Fiorini, Z.; Holyoake, T.L.; Pellicano, F.; Krampera, M.; Tecchio, C.; et al. A new monoclonal antibody detects downregulation of protein tyrosine phosphatase receptor type gamma in chronic myeloid leukemia patients. J. Hematol. Oncol. 2017, 10, 129. [Google Scholar] [CrossRef] [PubMed]
- Tomasello, L.; Vezzalini, M.; Boni, C.; Bonifacio, M.; Scaffidi, L.; Yassin, M.; Al-Dewik, N.; Takam Kamga, P.; Krampera, M.; Sorio, C. Regulative Loop between beta-catenin and Protein Tyrosine Receptor Type gamma in Chronic Myeloid Leukemia. Int. J. Mol. Sci. 2020, 21, 2298. [Google Scholar] [CrossRef]
- van Doorn, R.; Zoutman, W.H.; Dijkman, R.; de Menezes, R.X.; Commandeur, S.; Mulder, A.A.; van der Velden, P.A.; Vermeer, M.H.; Willemze, R.; Yan, P.S.; et al. Epigenetic profiling of cutaneous T-cell lymphoma: Promoter hypermethylation of multiple tumor suppressor genes including BCL7a, PTPRG, and p73. J. Clin. Oncol. 2005, 23, 3886–3896. [Google Scholar] [CrossRef]
- Ismail, M.A.; Samara, M.; Al Sayab, A.; Alsharshani, M.; Yassin, M.A.; Varadharaj, G.; Vezzalini, M.; Tomasello, L.; Monne, M.; Morsi, H.; et al. Aberrant DNA methylation of PTPRG as one possible mechanism of its under-expression in CML patients in the State of Qatar. Mol. Genet. Genom. Med. 2020, 8, 1319. [Google Scholar] [CrossRef] [PubMed]
- Van Etten, R.A. Mechanisms of transformation by the BCR-ABL oncogene: New perspectives in the post-imatinib era. Leuk. Res. 2004, 28, 21–28. [Google Scholar] [CrossRef]
- Jabbour, E.; Kantarjian, H.M.; Jones, D.; Talpaz, M.; Bekele, N.; Obrien, S.J.; Zhou, X.; Luthra, R.; Garciamanero, G.; Giles, F.J.; et al. Frequency and clinical significance of BCR-ABL mutations in patients with chronic myeloid leukemia treated with imatinib mesylate. Leukemia 2006, 20, 1767–1773. [Google Scholar] [CrossRef] [PubMed]
- Sattler, M.; Verma, S.; Shrikhande, G.; Byrne, C.H.; Pride, Y.B.; Winkler, T.; Greenfield, E.A.; Salgia, R.; Griffin, J.D. The BCR/ABL Tyrosine Kinase Induces Production of Reactive Oxygen Species in Hematopoietic Cells. J. Biol. Chem. 2000, 275, 24273–24278. [Google Scholar] [CrossRef] [PubMed]
- White, D.L.; Saunders, V.A.; Dang, P.; Engler, J.; Zannettino, A.C.W.; Cambareri, A.C.; Quinn, S.R.; Manley, P.W.; Hughes, T.P. OCT-1-mediated influx is a key determinant of the intracellular uptake of imatinib but not nilotinib (AMN107): Reduced OCT-1 activity is the cause of low in vitro sensitivity to imatinib. Blood 2006, 108, 697–704. [Google Scholar] [CrossRef]
- Barouch-Bentov, R.; Sauer, K. Mechanisms of drug resistance in kinases. Expert Opin. Investig. Drugs 2011, 20, 153–208. [Google Scholar] [CrossRef] [PubMed]
- Vianello, F.; Villanova, F.; Tisato, V.; Lymperi, S.; Ho, K.-K.; Gomes, A.R.; Marin, D.; Bonnet, D.; Apperley, J.; Lam, E.W.-F.; et al. Bone marrow mesenchymal stromal cells non-selectively protect chronic myeloid leukemia cells from imatinib-induced apoptosis via the CXCR4/CXCL12 axis. Haematologica 2010, 95, 1081–1089. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, M.; McDonald, T.; Holyoake, T.L.; Moon, R.T.; Campana, D.; Shultz, L.; Bhatia, R. Microenvironmental protection of CML stem and progenitor cells from tyrosine kinase inhibitors through N-cadherin and Wnt-beta-catenin signaling. Blood 2013, 121, 1824–1838. [Google Scholar] [CrossRef]
- Gallipoli, P.; Cook, A.; Rhodes, S.; Hopcroft, L.; Wheadon, H.; Whetton, A.D.; Jørgensen, H.G.; Bhatia, R.; Holyoake, T.L. JAK2/STAT5 inhibition by nilotinib with ruxolitinib contributes to the elimination of CML CD34+ cells in vitro and in vivo. Blood 2014, 124, 1492–1501. [Google Scholar] [CrossRef]
- Jeanpierre, S.; Arizkane, K.; Thongjuea, S.; Grockowiak, E.; Geistlich, K.; Barral, L.; Voeltzel, T.; Guillemin, A.; Gonin-Giraud, S.; Gandrillon, O.; et al. The quiescent fraction of chronic myeloid leukemic stem cells depends on BMPR1B, Stat3 and BMP4-niche signals to persist in patients in remission. Haematologica 2020, 106, 111–122. [Google Scholar] [CrossRef]
- Zhang, X.; Tu, H.; Yang, Y.; Jiang, X.; Hu, X.; Luo, Q.; Li, J. Bone marrow-derived mesenchymal stromal cells promote resistance to tyrosine kinase inhibitors in chronic myeloid leukemia via the IL-7/JAK1/STAT5 pathway. J. Biol. Chem. 2019, 294, 12167–12179. [Google Scholar] [CrossRef]
- Mancini, M.; De Santis, S.; Monaldi, C.; Bavaro, L.; Martelli, M.; Gugliotta, G.; Castagnetti, F.; Rosti, G.; Santucci, M.A.; Martinelli, G.; et al. Ponatinib treatment in chronic myeloid leukemia cell lines targets aurora kinase A/FOXM1 axis. Hematol. Oncol. 2020, 38, 201–203. [Google Scholar] [CrossRef] [PubMed]
- Slack, D.N.; Seternes, O.-M.; Gabrielsen, M.; Keyse, S.M. Distinct Binding Determinants for ERK2/p38α and JNK MAP Kinases Mediate Catalytic Activation and Substrate Selectivity of MAP Kinase Phosphatase-1. J. Biol. Chem. 2001, 276, 16491–16500. [Google Scholar] [CrossRef]
- Kesarwani, M.; Kincaid, Z.; Gomaa, A.; Huber, E.; Rohrabaugh, S.; Siddiqui, Z.; Bouso, M.F.; Latif, T.; Xu, M.; Komurov, K.; et al. Targeting c-FOS and DUSP1 abrogates intrinsic resistance to tyrosine-kinase inhibitor therapy in BCR-ABL-induced leukemia. Nat. Med. 2017, 23, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Ismail, M.A.; Vezzalini, M.; Morsi, H.; Abujaber, A.; Al Sayab, A.; Siveen, K.; Yassin, M.A.; Monne, M.; Samara, M.; Cook, R.; et al. Predictive value of tyrosine phosphatase receptor gamma for the response to treatment tyrosine kinase inhibitors in chronic myeloid leukemia patients. Sci. Rep. 2021, 11, 8833. [Google Scholar] [CrossRef]
- Drube, J.; Ernst, T.; Pfirrmann, M.; Albert, B.V.; Drube, S.; Reich, D.; Kresinsky, A.; Halfter, K.; Sorio, C.; Fabisch, C.; et al. PTPRG and PTPRC modulate nilotinib response in chronic myeloid leukemia cells. Oncotarget 2018, 9, 9442–9455. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Guo, X.; Liu, X.; Luo, J. DNA methyltransferase 1 mediated aberrant methylation and silencing of SHP-1 gene in chronic myelogenous leukemia cells. Leuk. Res. 2017, 58, 9–13. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, L.; Liu, X.; Nie, Z.; Wang, X.; Pan, Y.; Luo, J. Research on the epigenetic regulation mechanism of thePTPN6gene in advanced chronic myeloid leukaemia. Br. J. Haematol. 2017, 178, 728–738. [Google Scholar] [CrossRef]
- Wöhrle, F.U.; Halbach, S.; Aumann, K.; Schwemmers, S.; Braun, S.; Auberger, P.; Schramek, D.; Penninger, J.M.; Laßmann, S.; Werner, M.; et al. Gab2 signaling in chronic myeloid leukemia cells confers resistance to multiple Bcr-Abl inhibitors. Leukemia 2012, 27, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Pang, J.; Xue, W.; Wang, Y.; Tian, T.; Elgehama, A.; Wu, X.; Wu, X.; Sun, Y.; Qiu, H.; et al. Inducible SHP-2 activation confers resistance to imatinib in drug-tolerant chronic myeloid leukemia cells. Toxicol. Appl. Pharmacol. 2018, 360, 249–256. [Google Scholar] [CrossRef]
- Neviani, P.; Harb, J.G.; Oaks, J.J.; Santhanam, R.; Walker, C.J.; Ellis, J.J.; Ferenchak, G.; Dorrance, A.M.; Paisie, C.A.; Eiring, A.M.; et al. PP2A-activating drugs selectively eradicate TKI-resistant chronic myeloid leukemic stem cells. J. Clin. Investig. 2013, 123, 4144–4157. [Google Scholar] [CrossRef]
- Wang, S.; Xie, W.; Wang, D.; Peng, Z.; Zheng, Y.; Liu, N.; Dai, W.; Wang, Y.; Wang, Z.; Yang, Y.; et al. Discovery of a small molecule targeting SET-PP2A interaction to overcome BCR-ABLT315I mutation of chronic myeloid leukemia. Oncotarget 2015, 6, 12128–12140. [Google Scholar] [CrossRef]
- Agarwal, A.; MacKenzie, R.J.; Pippa, R.; Eide, C.A.; Oddo, J.; Tyner, J.W.; Sears, R.; Vitek, M.P.; Odero, M.D.; Christensen, D.J.; et al. Antagonism of SET Using OP449 Enhances the Efficacy of Tyrosine Kinase Inhibitors and Overcomes Drug Resistance in Myeloid Leukemia. Clin. Cancer Res. 2014, 20, 2092–2103. [Google Scholar] [CrossRef] [PubMed]
- Baran, Y.; Salas, A.; Senkal, C.E.; Gunduz, U.; Bielawski, J.; Obeid, L.M.; Ogretmen, B. Alterations of Ceramide/Sphingosine 1-Phosphate Rheostat Involved in the Regulation of Resistance to Imatinib-induced Apoptosis in K562 Human Chronic Myeloid Leukemia Cells*. J. Biol. Chem. 2007, 282, 10922–10934. [Google Scholar] [CrossRef] [PubMed]
- Salas, A.; Ponnusamy, S.; Senkal, C.E.; Meyers-Needham, M.; Selvam, S.P.; Saddoughi, S.A.; Apohan, E.; Sentelle, R.D.; Smith, C.; Gault, C.R.; et al. Sphingosine kinase-1 and sphingosine 1-phosphate receptor 2 mediate Bcr-Abl1 stability and drug resistance by modulation of protein phosphatase 2A. Blood 2011, 117, 5941–5952. [Google Scholar] [CrossRef]
- Laidlaw, K.M.E.; Berhan, S.; Liu, S.; Silvestri, G.; Holyoake, T.L.; Frank, D.A.; Aggarwal, B.; Bonner, M.Y.; Perrotti, D.; Jørgensen, H.G.; et al. Cooperation of imipramine blue and tyrosine kinase blockade demonstrates activity against chronic myeloid leukemia. Oncotarget 2016, 7, 51651–51664. [Google Scholar] [CrossRef]
- Lai, D.; Chen, M.; Su, J.; Liu, X.; Rothe, K.; Hu, K.; Forrest, D.L.; Eaves, C.J.; Morin, G.B.; Jiang, X. PP2A inhibition sensitizes cancer stem cells to ABL tyrosine kinase inhibitors in BCR-ABL+human leukemia. Sci. Transl. Med. 2018, 10, eaan8735. [Google Scholar] [CrossRef]
- Perrotti, D.; Agarwal, A.; Lucas, C.M.; Narla, G.; Neviani, P.; Odero, M.D.; Ruvolo, P.P.; Verrills, N.M. Comment on “PP2A inhibition sensitizes cancer stem cells to ABL tyrosine kinase inhibitors in BCR-ABL human leukemia”. Sci. Transl. Med. 2019, 11, eaau0416. [Google Scholar] [CrossRef]
- Lai, D.; Chen, M.; Su, J.; Liu, X.; Rothe, K.; Hu, K.; Forrest, D.L.; Eaves, C.J.; Morin, G.B.; Jiang, X. Response to Comment on “PP2A inhibition sensitizes cancer stem cells to ABL tyrosine kinase inhibitors in BCR-ABL+ human leukemia”. Sci. Transl. Med. 2019, 11, eaav0819. [Google Scholar] [CrossRef]
- Huang, W.; Luan, C.-H.; Hjort, E.E.; Bei, L.; Mishra, R.; Sakamoto, K.M.; Platanias, L.C.; Eklund, E.A. The role of Fas-associated phosphatase 1 in leukemia stem cell persistence during tyrosine kinase inhibitor treatment of chronic myeloid leukemia. Leukemia 2016, 30, 1502–1509. [Google Scholar] [CrossRef]
- Través, P.G.; Pardo, V.; Pimentel-Santillana, M.; González-Rodríguez, Á.; Mojena, M.; Rico, D.; Montenegro, Y.; Calés, C.; Martín-Sanz, P.; Valverde, A.M.; et al. Pivotal role of protein tyrosine phosphatase 1B (PTP1B) in the macrophage response to pro-inflammatory and anti-inflammatory challenge. Cell Death Dis. 2014, 5, e1125. [Google Scholar] [CrossRef]
- Le Sommer, S.; Morrice, N.; Pesaresi, M.; Thompson, D.; Vickers, M.A.; Murray, G.I.; Mody, N.; Neel, B.G.; Bence, K.K.; Wilson, H.M.; et al. Deficiency in Protein Tyrosine Phosphatase PTP1B Shortens Lifespan and Leads to Development of Acute Leukemia. Cancer Res. 2018, 78, 75–87. [Google Scholar] [CrossRef]
- Alvira, D.; Naughton, R.; Bhatt, L.; Tedesco, S.; Landry, W.D.; Cotter, T.G. Inhibition of Protein-tyrosine Phosphatase 1B (PTP1B) Mediates Ubiquitination and Degradation of Bcr-Abl Protein. J. Biol. Chem. 2011, 286, 32313–32323. [Google Scholar] [CrossRef] [PubMed]
- LaMontagne, K.R.; Hannon, G.; Tonks, N.K. Protein tyrosine phosphatase PTP1B suppresses p210 bcr-abl-induced transformation of Rat-1 fibroblasts and promotes differentiation of K562 cells. Proc. Natl. Acad. Sci. USA 1998, 95, 14094–14099. [Google Scholar] [CrossRef]
- Elgehama, A.; Chen, W.; Pang, J.; Mi, S.; Li, J.; Guo, W.; Wang, X.; Gao, J.; Yu, B.; Shen, Y.; et al. Blockade of the interaction between Bcr-Abl and PTB1B by small molecule SBF-1 to overcome imatinib-resistance of chronic myeloid leukemia cells. Cancer Lett. 2016, 372, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Koyama, N.; Koschmieder, S.; Tyagi, S.; Portero-Robles, I.; Chromic, J.; Myloch, S.; Nürnberger, H.; Rossmanith, T.; Hofmann, W.-K.; Hoelzer, D.; et al. Inhibition of Phosphotyrosine Phosphatase 1B Causes Resistance in BCR-ABL-Positive Leukemia Cells to the ABL Kinase Inhibitor STI571. Clin. Cancer Res. 2006, 12, 2025–2031. [Google Scholar] [CrossRef] [PubMed]
- Chien, W.; Tidow, N.; Williamson, E.A.; Shih, L.-Y.; Krug, U.; Kettenbach, A.; Fermin, A.C.; Roifman, C.M.; Koeffler, H. Characterization of a Myeloid Tyrosine Phosphatase, Lyp, and Its Role in the Bcr-Abl Signal Transduction Pathway. J. Biol. Chem. 2003, 278, 27413–27420. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Liu, Y.; Yin, Z.; Yang, J.; Huang, G.; Zhu, X.; Li, Y.; Fei, J. Discovery of the Oncogenic Parp1, a Target of bcr-abl and a Potential Therapeutic, in mir-181a/PPFIA1 Signaling Pathway. Mol. Ther. Nucleic Acids 2019, 16, 1–14. [Google Scholar] [CrossRef]
- Dedinszki, D.; Kiss, A.; Márkász, L.; Márton, A.; Tóth, E.; Székely, L.; Erdődi, F. Inhibition of protein phosphatase-1 and -2A decreases the chemosensitivity of leukemic cells to chemotherapeutic drugs. Cell. Signal. 2015, 27, 363–372. [Google Scholar] [CrossRef]
- Naughton, R.; Quiney, C.; Turner, S.D.; Cotter, T.G. Bcr-Abl-mediated redox regulation of the PI3K/AKT pathway. Leukemia 2009, 23, 1432–1440. [Google Scholar] [CrossRef] [PubMed]
- Hoekstra, E.; Kodach, L.L.; Das, A.M.; Ruela-De-Sousa, R.R.; Ferreira, C.V.; Hardwick, J.C.; Van Der Woude, C.J.; Peppelenbosch, M.P.; Hagen, T.L.T.; Fuhler, G.M. Low molecular weight protein tyrosine phosphatase (LMWPTP) upregulation mediates malignant potential in colorectal cancer. Oncotarget 2015, 6, 8300–8312. [Google Scholar] [CrossRef]
- Faria, A.V.S.; Clerici, S.P.; de Souza Oliveira, P.F.; Queiroz, K.C.S.; Peppelenbosch, M.P.; Ferreira-Halder, C.V. LMWPTP modulates the antioxidant response and autophagy process in human chronic myeloid leukemia cells. Mol. Cell. Biochem. 2020, 466, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, P.A.; Ruela-De-Sousa, R.R.; Queiroz, K.C.S.; Souza, A.C.S.; Milani, R.; Pilli, R.A.; Peppelenbosch, M.P.; Hertog, J.D.; Ferreira, C.V. Knocking Down Low Molecular Weight Protein Tyrosine Phosphatase (LMW-PTP) Reverts Chemoresistance through Inactivation of Src and Bcr-Abl Proteins. PLoS ONE 2012, 7, e44312. [Google Scholar] [CrossRef] [PubMed]
- You-Ten, K.E.; Muise, E.S.; Itié, A.; Michaliszyn, E.; Wagner, J.; Jothy, S.; Lapp, W.S.; Tremblay, M.L. Impaired Bone Marrow Microenvironment and Immune Function in T Cell Protein Tyrosine Phosphatase–deficient Mice. J. Exp. Med. 1997, 186, 683–693. [Google Scholar] [CrossRef]
- Shimizu, T.; Miyakawa, Y.; Oda, A.; Kizaki, M.; Ikeda, Y. STI571-resistant KT-1 cells are sensitive to interferon-α accompanied by the loss of T-cell protein tyrosine phosphatase and prolonged phosphorylation of Stat1. Exp. Hematol. 2003, 31, 601–608. [Google Scholar] [CrossRef]
- Shimizu, T.; Miyakawa, Y.; Iwata, S.; Kuribara, A.; Tiganis, T.; Morimoto, C.; Ikeda, Y.; Kizaki, M. A novel mechanism for imatinib mesylate (STI571) resistance in CML cell line KT-1: Role of TC-PTP in modulating signals downstream from the BCR-ABL fusion protein. Exp. Hematol. 2004, 32, 1057–1063. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.; Sasikumar, K.; Parthasaradhi, B.; Radha, V. The tyrosine phosphatase TC48 interacts with and inactivates the oncogenic fusion protein BCR-Abl but not cellular Abl. Biochim. Biophys. Acta Mol. Basis Dis. 2013, 1832, 275–284. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Reckel, S.; Hamelin, R.; Georgeon, S.; Armand, F.; Jolliet, Q.; Chiappe, D.; Moniatte, M.; Hantschel, O. Differential signaling networks of Bcr–Abl p210 and p190 kinases in leukemia cells defined by functional proteomics. Leukemia 2017, 31, 1502–1512. [Google Scholar] [CrossRef]
- Matsuzawa, S.-I.; Cuddy, M.; Fukushima, T.; Reed, J.C. Method for targeting protein destruction by using a ubiquitin-independent, proteasome-mediated degradation pathway. Proc. Natl. Acad. Sci. USA 2005, 102, 14982–14987. [Google Scholar] [CrossRef]
- Gao, M.; Huang, Z.-L.; Tao, K.; Xiao, Q.; Wang, X.; Cao, W.-X.; Xu, M.; Hu, J.; Feng, W.-L. Depression of oncogenecity by dephosphorylating and degrading BCR-ABL. Oncotarget 2017, 8, 3304–3314. [Google Scholar] [CrossRef]
- Xu, M.; Ye, Y.; Ye, Z.; Xu, S.; Liu, W.; Xu, J.; Zhang, Y.; Liu, Q.; Huang, Z.; Zhang, W. Human BCR/ABL1 induces chronic myeloid leukemia-like disease in zebrafish. Haematologica 2019, 105, 674–686. [Google Scholar] [CrossRef]
- Zizioli, D.; Bernardi, S.; Varinelli, M.; Farina, M.; Mignani, L.; Bosio, K.; Finazzi, D.; Monti, E.; Polverelli, N.; Malagola, M.; et al. Development of BCR-ABL1 Transgenic Zebrafish Model Reproducing Chronic Myeloid Leukemia (CML) Like-Disease and Providing a New Insight into CML Mechanisms. Cells 2021, 10, 445. [Google Scholar] [CrossRef] [PubMed]
- La Rosée, P.; Holm-Eriksen, S.; Konig, H.; Härtel, N.; Ernst, T.; Debatin, J.; Mueller, M.C.; Erben, P.; Binckebanck, A.; Wunderle, L.; et al. Phospho-CRKL monitoring for the assessment of BCR-ABL activity in imatinib-resistant chronic myeloid leukemia or Ph+ acute lymphoblastic leukemia patients treated with nilotinib. Haematologica 2008, 93, 765–769. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lucas, C.M.; Harris, R.J.; Giannoudis, A.; Knight, K.; Watmough, S.J.; Clark, R.E. BCR-ABL1 tyrosine kinase activity at diagnosis, as determined via the pCrkL/CrkL ratio, is predictive of clinical outcome in chronic myeloid leukaemia. Br. J. Haematol. 2010, 149, 458–460. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, A.; Elrick, L.; Myssina, S.; Copland, M.; Jørgensen, H.; Melo, J.V.; Holyoake, T. BCR-ABL activity and its response to drugs can be determined in CD34+ CML stem cells by CrkL phosphorylation status using flow cytometry. Leukemia 2006, 20, 1035–1039. [Google Scholar] [CrossRef]
- Simara, P.; Peterkova, M.; Stejskal, S.; Potesilova, M.; Koutna, I.; Racil, Z.; Razga, F.; Jurcek, T.; Dvorakova, D.; Mayer, J. BCR-ABL activity measured by 50% inhibitory concentration for imatinib, p-CrkL/CrkL ratio or p-CrkL ratio in CD34+ cells of patients with chronic myeloid leukemia does not predict treatment response. Leuk. Lymphoma 2012, 53, 1627–1629. [Google Scholar] [CrossRef]
- White, D.; Saunders, V.; Grigg, A.; Arthur, C.; Filshie, R.; Leahy, M.F.; Lynch, K.; To, L.B.; Hughes, T. Measurement of In Vivo BCR-ABL Kinase Inhibition to Monitor Imatinib-Induced Target Blockade and Predict Response in Chronic Myeloid Leukemia. J. Clin. Oncol. 2007, 25, 4445–4451. [Google Scholar] [CrossRef]
- Calin, G.A.; Ferracin, M.; Cimmino, A.; Di Leva, G.; Shimizu, M.; Wojcik, S.E.; Iorio, M.V.; Visone, R.; Sever, N.I.; Fabbri, M.; et al. A MicroRNA Signature Associated with Prognosis and Progression in Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2005, 353, 1793–1801. [Google Scholar] [CrossRef]
- Lovat, F.; Nigita, G.; Distefano, R.; Nakamura, T.; Gasparini, P.; Tomasello, L.; Fadda, P.; Ibrahimova, N.; Catricala, S.; Palamarchuk, A.; et al. Combined loss of function of two different loci of miR-15/16 drives the pathogenesis of acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2020, 117, 12332–12340. [Google Scholar] [CrossRef]
- Lovat, F.; Gasparini, P.; Nigita, G.; Larkin, K.; Byrd, J.C.; Minden, M.D.; Andreeff, M.; Carter, B.Z.; Croce, C.M. Loss of expression of both miR-15/16 loci in CML transition to blast crisis. Proc. Natl. Acad. Sci. USA 2021, 118, e2101566118. [Google Scholar] [CrossRef]
| Protein | Coding Gene | Role in CML | References |
|---|---|---|---|
| Data verified in primary CML cells or in leukemia mouse models | |||
| DUSP1 | DUSP1 | Implicated in TKI-response | [133] |
| STS-1 | UBASH3B | Decreases cell proliferation Direct BCR-ABL1 regulation | [27,30] |
| FAP-1 | PTPN13 | Regulation of β-catenin functions Decreases TKI sensitivity | [105,106,107,109,110,149] |
| PTPRG | PTPRG | Regulation of β-catenin functions Implicated in TKI response | [112,113,114,116,117,118,120,134,135] |
| SHP1 | PTPN6 | Acts through the PP2A on BCR-ABL1 Regulates BCR-ABL1-independent IM resistance | [44,96,97,136,137,144] |
| SHP2 | PTPN11 | Increases cell proliferation Implicated in TKI resistance | [35,37,38,39,40,41,42,43,44] |
| PP2A | PPP2CA | Quiescence and Self-renewal regulation Governs TKI-response | [96,97,98,99,100,103,140,141,142,144,145,146,147,148] |
| PTEN | PTEN | Control of cell proliferation | [49,59,60,61,63,64,66,67,68,70,71] |
| Data obtained only in CML cell lines | |||
| PTP1B | PTPN1 | Reduces cell viability Correlated with IM response | [150,151,152,153,154,155] |
| LAR –LIPRIN Α1 | PPFIA1 | Mitigates BCR-ABL1 leukemogenesis | [157] |
| LYP | PTPN22 | Decreases IM sensitivity | [156] |
| LMW-PTP | ACP1 | Regulates autophagy process Correlated with IM resistance | [161,162] |
| PP1Α | PPP1CA | Improves cell survival and apoptosis resistance | [159] |
| TC45/TC48 | TC-PTP | Implicated in IM- and INFα-resistance Regulation of proliferation and apoptosis | [164,165,166] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boni, C.; Sorio, C. Current Views on the Interplay between Tyrosine Kinases and Phosphatases in Chronic Myeloid Leukemia. Cancers 2021, 13, 2311. https://doi.org/10.3390/cancers13102311
Boni C, Sorio C. Current Views on the Interplay between Tyrosine Kinases and Phosphatases in Chronic Myeloid Leukemia. Cancers. 2021; 13(10):2311. https://doi.org/10.3390/cancers13102311
Chicago/Turabian StyleBoni, Christian, and Claudio Sorio. 2021. "Current Views on the Interplay between Tyrosine Kinases and Phosphatases in Chronic Myeloid Leukemia" Cancers 13, no. 10: 2311. https://doi.org/10.3390/cancers13102311
APA StyleBoni, C., & Sorio, C. (2021). Current Views on the Interplay between Tyrosine Kinases and Phosphatases in Chronic Myeloid Leukemia. Cancers, 13(10), 2311. https://doi.org/10.3390/cancers13102311







