Genetic Variants in Immune Related Genes as Predictors of Responsiveness to BCG Immunotherapy in Metastatic Melanoma Patients
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
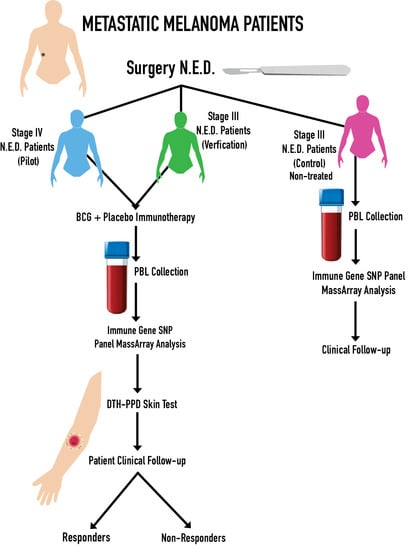
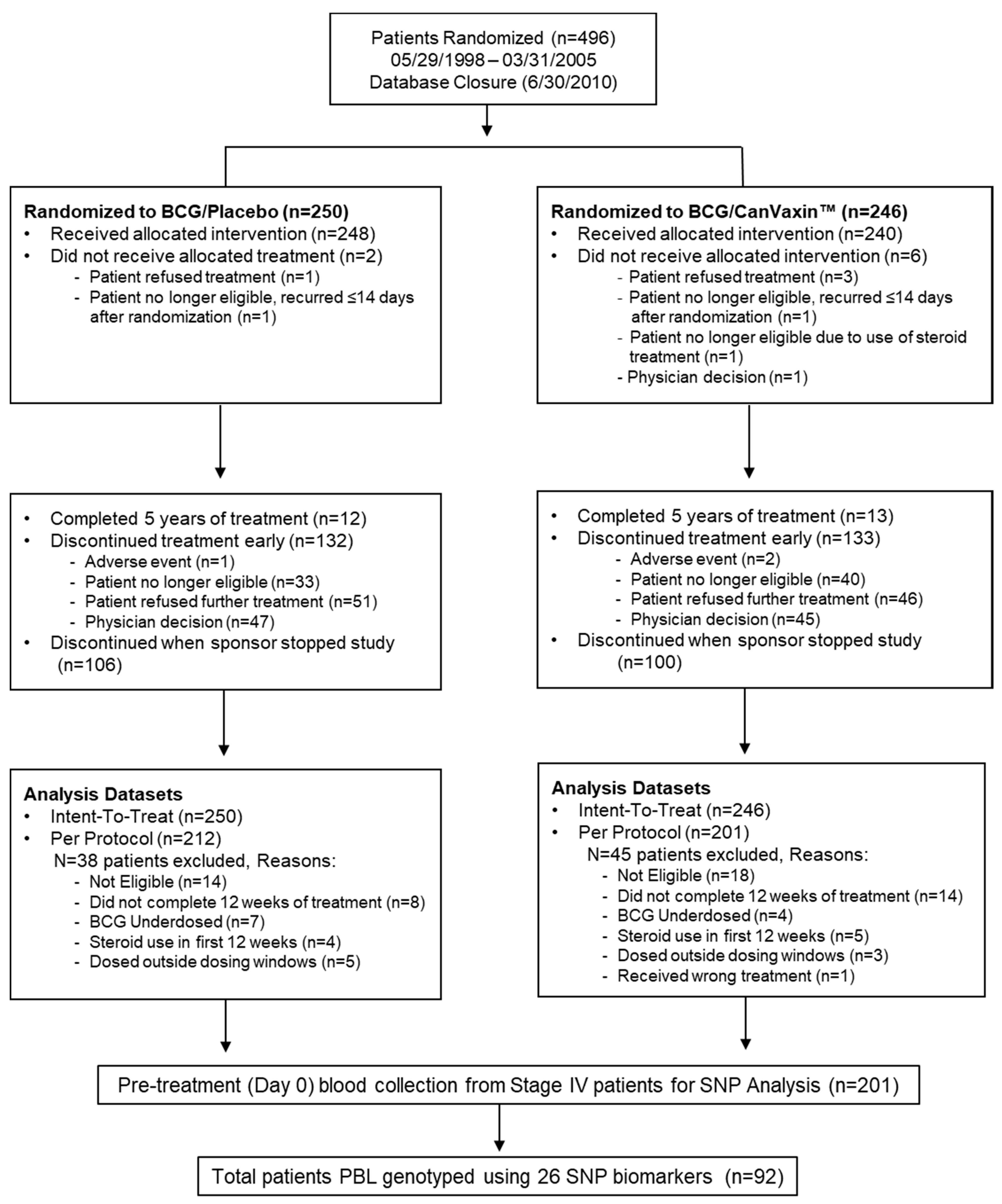
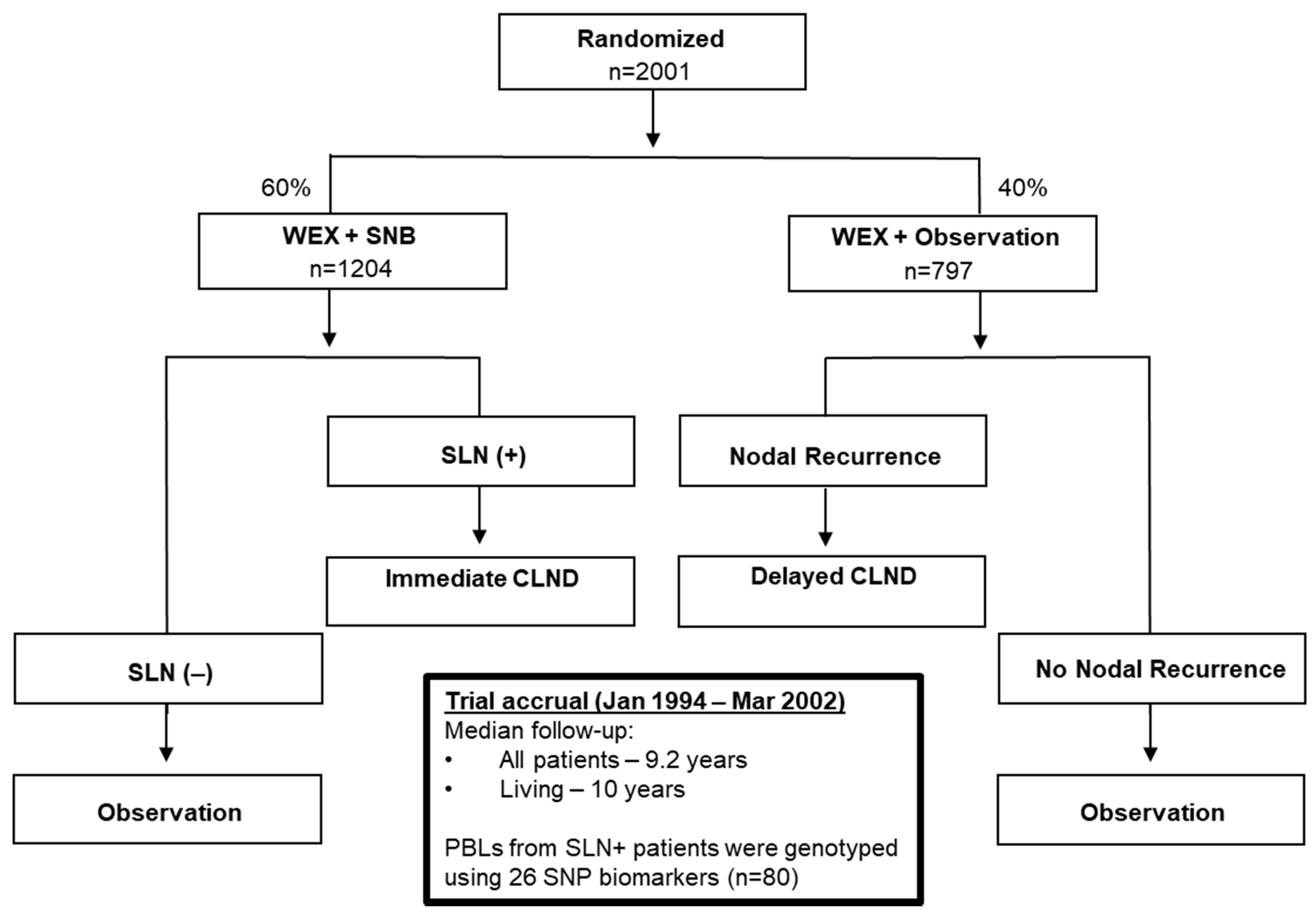
2.1. Patient Characteristics
2.2. Immune Gene SNP Selection
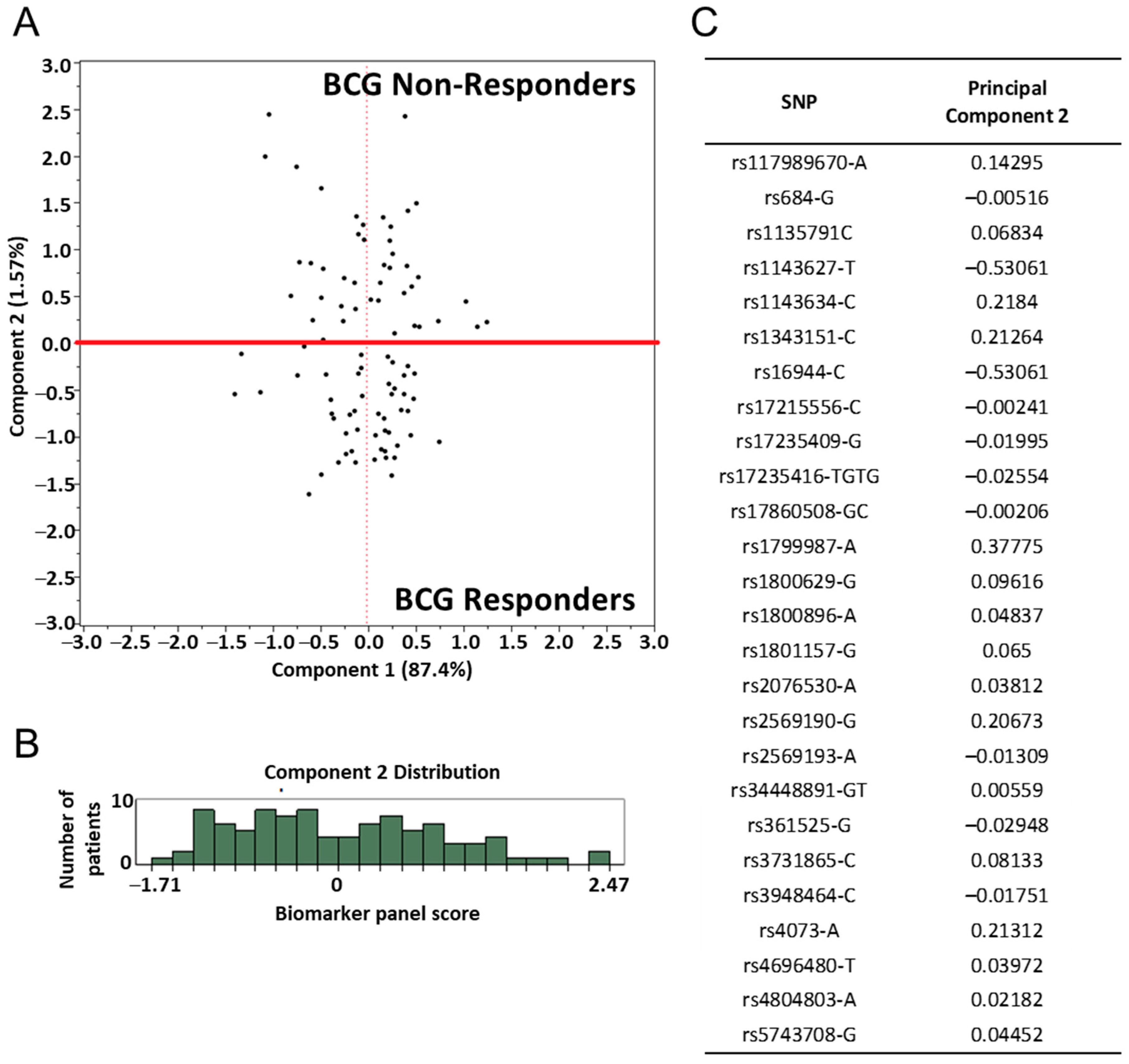
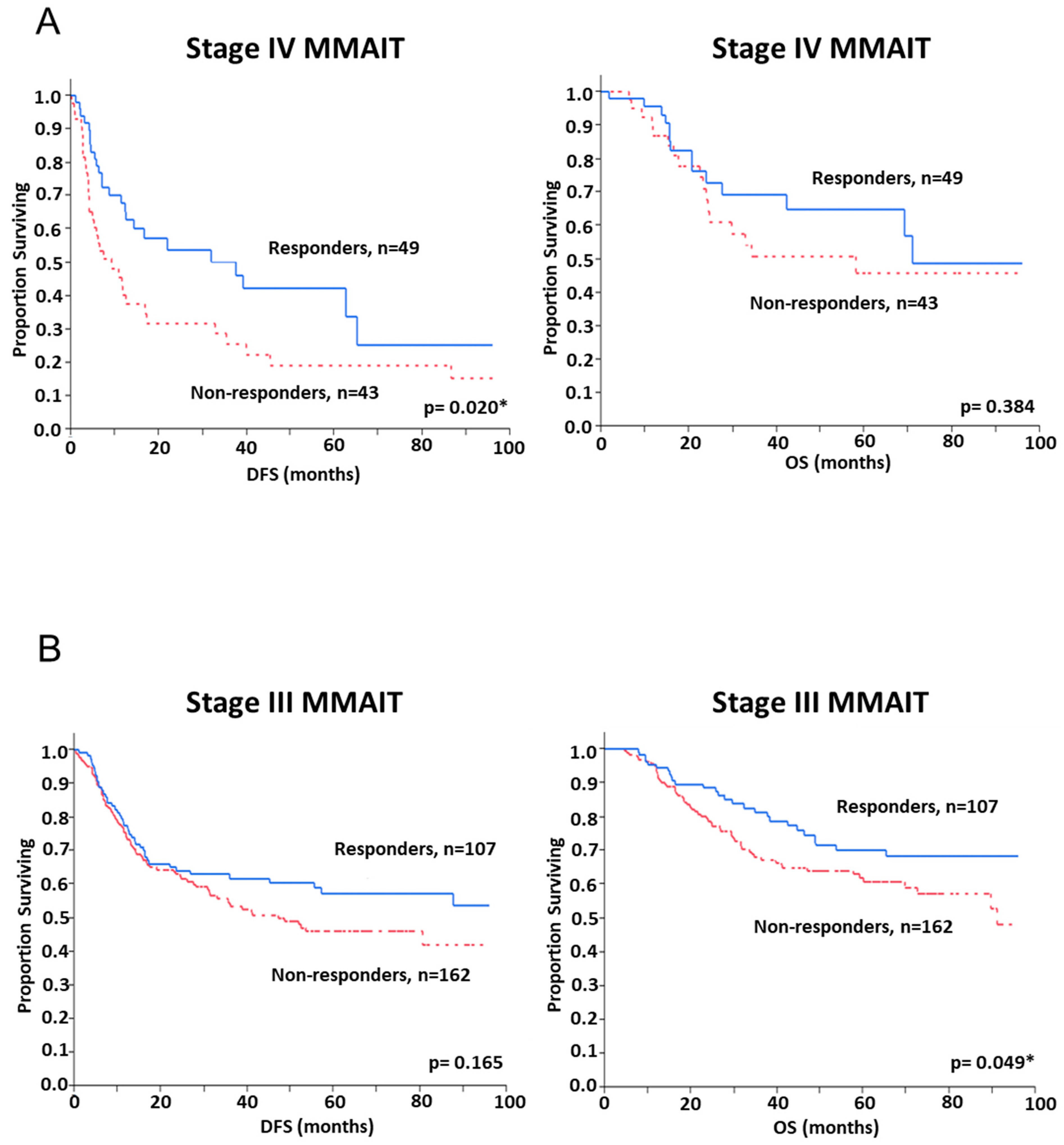
2.3. Assessment of SNP and Disease Outcome Analysis
2.4. BCG Treated Immune SNP Panel Correlation to Patients’ DTH Response to PPD
3. Discussion
4. Materials and Methods
4.1. Melanoma Patients
4.2. DTH-PPD Test
4.3. Peripheral Blood Leukocytes (PBLs) Collection
4.4. Experimental Approach
4.5. Biostatistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AJCC | American Joint Committee on Cancer |
| SNP | Single Nucleotide Polymorphism |
| BCG | Bacillus Calmette–Guérin |
| SLN | Sentinel lymph node |
| DFS | Disease free survival |
| HR | Hazard ratio |
| OS | Overall survival |
| IL | Interleukin |
| ICI | Immune Checkpoint Inhibitors |
| GM-CSF | Granulocyte-macrophage colony-stimulating factor |
| TLR | Toll-Like Receptor |
| MSLT-I | Multicenter Selective Lymphadectomy |
| MMAIT | Malignant Melanoma Active Immunotherapy Trial |
| TNF | Tumor necrosis factor |
| CCR5 | C-C chemokine receptor type 5 |
| CXCL12 | C-X-C motif chemokine 12 |
| NRAMP1 | natural resistance-associated macrophage protein 1 |
| SLC11A1 | solute carrier protein 11A1 |
| CD209 | Cluster of Differentiation 209 |
| DC-SIGN | Dendritic Cell-Specific Intercellular adhesion molecule-3-Grabbing Non-integrin |
| BTNL2 | Butyrophilin Like 2 |
| PCA | Principal Component Analysis |
| DTH | Delayed Type Hypersensitivity |
| PPD | Purified Protein Derivative |
| VEGF | Vascular Endothelial Factor |
| pDC | Plasmacytoid Dendritic Cells |
| CT | Computed Tomography |
| MRI | Magnetic Resonance Imaging |
| ECOG | Eastern Cooperative Oncology Group |
| PBL | Peripheral Blood Leukocytes |
| IRB | Institutional Review Board |
| DTH | Delayed Type Hypersensitivity |
| MIP | Macrophage Inflammation Protein |
| TB | Tuberculosis |
| ECOG | Eastern Cooperative Oncology Group |
| CLND | Complete Lymph Node Dissection |
References
- Testori, A.A.E.; Chiellino, S.; van Akkooi, A.C.J. Adjuvant Therapy for Melanoma: Past, Current, and Future Developments. Cancers 2020, 12, 1994. [Google Scholar] [CrossRef] [PubMed]
- Kirkwood, J.M.; Tarhini, A.A.; Moschos, S.J.; Panelli, M.C. Adjuvant therapy with high-dose interferon alpha 2b in patients with high-risk stage IIB/III melanoma. Nat. Clin. Pract. Oncol. 2008, 5, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Kidner, T.B.; Morton, D.L.; Lee, D.J.; Hoban, M.; Foshag, L.J.; Turner, R.R.; Faries, M.B. Combined intralesional Bacille Calmette-Guérin (BCG) and topical imiquimod for in-transit melanoma. J. Immunother. 2012, 35, 716–720. [Google Scholar] [CrossRef] [PubMed]
- Herrscher, H.; Robert, C. Immune checkpoint inhibitors in melanoma in the metastatic, neoadjuvant, and adjuvant setting. Curr. Opin. Oncol. 2020, 32, 106–113. [Google Scholar] [CrossRef]
- Blank, C.U.; Rozeman, E.A.; Fanchi, L.F.; Sikorska, K.; van de Wiel, B.; Kvistborg, P.; Krijgsman, O.; van den Braber, M.; Philips, D.; Broeks, A.; et al. Neoadjuvant versus adjuvant ipilimumab plus nivolumab in macroscopic stage III melanoma. Nat. Med. 2018, 24, 1655–1661. [Google Scholar] [CrossRef]
- Eggermont, A.M.; Robert, C. Melanoma: Smart therapeutic strategies in immuno-oncology. Nat. Rev. Clin. Oncol. 2014, 11, 181–182. [Google Scholar] [CrossRef]
- Blankenstein, S.A.; Aarts, M.J.B.; van den Berkmortel, F.W.P.J.; Boers-Sonderen, M.J.; van den Eertwegh, A.J.M.; Franken, M.G.; de Groot, J.W.B.; Haanen, J.B.A.G.; Hospers, G.A.P.; Kapiteijn, E.; et al. Surgery for Unresectable Stage IIIC and IV Melanoma in the Era of New Systemic Therapy. Cancers 2020, 12, 1176. [Google Scholar] [CrossRef]
- Callahan, M.K.; Postow, M.A.; Wolchok, J.D. Immunomodulatory therapy for melanoma: Ipilimumab and beyond. Clin. Dermatol. 2013, 31, 191–199. [Google Scholar] [CrossRef]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef]
- Brahmer, J.R.; Tykodi, S.S.; Chow, L.Q.; Hwu, W.J.; Topalian, S.L.; Hwu, P.; Drake, C.G.; Camacho, L.H.; Kauh, J.; Odunsi, K.; et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 2012, 366, 2455–2465. [Google Scholar] [CrossRef] [PubMed]
- Wada-Ohno, M.; Ito, T.; Furue, M. Adjuvant Therapy for Melanoma. Curr. Treat. Options Oncol. 2019, 20, 63. [Google Scholar] [CrossRef] [PubMed]
- Askeland, E.; Newton, M.; O’Donnell, M.; Luo, Y. Bladder Cancer Immunotherapy: BCG and Beyond. Adv. Urol. 2012, 2012, 181987. [Google Scholar] [CrossRef] [PubMed]
- Redelman-Sidi, G.; Glickman, M.S.; Bochner, B.H. The mechanism of action of BCG therapy for bladder cancer—A current perspective. Nat. Rev. Urol. 2014, 11, 153–162. [Google Scholar] [CrossRef]
- Benitez, M.L.R.; Bender, C.B.; Oliveira, T.L.; Schachtschneider, K.M.; Collares, T.; Seixas, F.K. Mycobacterium bovis BCG in metastatic melanoma therapy. Appl. Microbiol. Biotechnol. 2019, 103, 7903–7916. [Google Scholar] [CrossRef]
- Hoshimoto, S.; Faries, M.B.; Morton, D.L.; Shingai, T.; Kuo, C.; Wang, H.J.; Elashoff, R.; Mozzillo, N.; Kelley, M.C.; Thompson, J.F.; et al. Assessment of prognostic circulating tumor cells in a phase III trial of adjuvant immunotherapy after complete resection of stage IV melanoma. Ann. Surg. 2012, 255, 357–362. [Google Scholar] [CrossRef]
- Hoshimoto, S.; Shingai, T.; Morton, D.L.; Kuo, C.; Faries, M.B.; Chong, K.; Elashoff, D.; Wang, H.J.; Elashoff, R.M.; Hoon, D.S. Association between circulating tumor cells and prognosis in patients with stage III melanoma with sentinel lymph node metastasis in a phase III international multicenter trial. J. Clin. Oncol. 2012, 30, 3819–3826. [Google Scholar] [CrossRef]
- Hsueh, E.C.; Famatiga, E.; Shu, S.; Ye, X.; Morton, D.L. Peripheral blood CD4+ T-cell response before postoperative active immunotherapy correlates with clinical outcome in metastatic melanoma. Ann. Surg. Oncol. 2004, 11, 892–899. [Google Scholar] [CrossRef]
- Eilber, F.R.; Morton, D.L.; Holmes, E.C.; Sparks, F.C.; Ramming, K.P. Adjuvant immunotherapy with BCG in treatment of regional-lymph-node metastases from malignant melanoma. N. Engl. J. Med. 1976, 294, 237–240. [Google Scholar] [CrossRef]
- Podaza, E.; Carri, I.; Aris, M.; von Euw, E.; Bravo, A.I.; Blanco, P.; Ortiz Wilczyñski, J.M.; Koile, D.; Yankilevich, P.; Nielsen, M.; et al. Evaluation of T-Cell Responses Against Shared Melanoma Associated Antigens and Predicted Neoantigens in Cutaneous Melanoma Patients Treated With the CSF-470 Allogeneic Cell Vaccine Plus BCG and GM-CSF. Front. Immunol. 2020, 11, 1147. [Google Scholar] [CrossRef]
- Mordoh, J.; Pampena, M.B.; Aris, M.; Blanco, P.A.; Lombardo, M.; von Euw, E.M.; Mac Keon, S.; Yépez Crow, M.; Bravo, A.I.; O’Connor, J.M.; et al. Phase II Study of Adjuvant Immunotherapy with the CSF-470 Vaccine Plus Bacillus Calmette-Guerin Plus Recombinant Human Granulocyte Macrophage-Colony Stimulating Factor vs Medium-Dose Interferon Alpha 2B in Stages IIB, IIC, and III Cutaneous Melanoma Patients: A Single Institution, Randomized Study. Front. Immunol. 2017, 8, 625. [Google Scholar] [CrossRef] [PubMed]
- Gutterman, J.U.; Mavligit, G.; McBride, C.; Frei, E., 3rd; Freireich, E.J.; Hersh, E.M. Active immunotherapy with B.C.G. for recurrent malignant melanoma. Lancet 1973, 1, 1208–1212. [Google Scholar] [CrossRef]
- Veronesi, U.; Adamus, J.; Aubert, C.; Bajetta, E.; Beretta, G.; Bonadonna, G.; Bufalino, R.; Cascinelli, N.; Cocconi, G.; Durand, J.; et al. A randomized trial of adjuvant chemotherapy and immunotherapy in cutaneous melanoma. N. Engl. J. Med. 1982, 307, 913–916. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.W.; Chen-Harris, H.; Mayba, O.; Lianoglou, S.; Wuster, A.; Bhangale, T.; Khan, Z.; Mariathasan, S.; Daemen, A.; Reeder, J. Germline genetic polymorphisms influence tumor gene expression and immune cell infiltration. Proc. Natl. Acad. Sci. USA 2018, 115, E11701–E11710. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Consortium, U.-D.; De Wolf, H.; Knezevic, B.; Burnham, K.L.; Osgood, J.; Sanniti, A.; Lledó Lara, A.; Kasela, S.; De Cesco, S.; et al. A genetics-led approach defines the drug target landscape of 30 immune-related traits. Nat. Genet. 2019, 51, 1082–1091. [Google Scholar] [CrossRef] [PubMed]
- Faries, M.B.; Mozzillo, N.; Kashani-Sabet, M.; Thompson, J.F.; Kelley, M.C.; DeConti, R.C.; Lee, J.E.; Huth, J.F.; Wagner, J.; Dalgleish, A.; et al. Long-Term Survival after Complete Surgical Resection and Adjuvant Immunotherapy for Distant Melanoma Metastases. Ann. Surg. Oncol. 2017, 24, 3991–4000. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Netea, M.G. Trained Innate Immunity, Epigenetics, and Covid-19. N. Engl. J. Med. 2020, 383, 1078–1080. [Google Scholar] [CrossRef]
- Escobar, L.E.; Molina-Cruz, A.; Barillas-Mury, C. BCG vaccine protection from severe coronavirus disease 2019 (COVID-19). Proc. Natl. Acad. Sci. USA 2020, 117, 17720. [Google Scholar] [CrossRef]
- Roden, D.M.; McLeod, H.L.; Relling, M.V.; Williams, M.S.; Mensah, G.A.; Peterson, J.F.; Van Driest, S.L. Pharmacogenomics. Lancet 2019, 394, 521–532. [Google Scholar] [CrossRef]
- Cho, P.; Gelinas, L.; Corbett, N.P.; Tebbutt, S.J.; Turvey, S.E.; Fortuno, E.S.; Kollmann, T.R. Association of common single-nucleotide polymorphisms in innate immune genes with differences in TLR-induced cytokine production in neonates. Genes Immun. 2013, 14, 199–211. [Google Scholar] [CrossRef]
- Powell, D.A.; Frelinger, J.A. Efficacy of Resistance to Francisella Imparted by ITY/NRAMP/SLC11A1 Depends on Route of Infection. Front. Immunol. 2017, 8, 206. [Google Scholar] [CrossRef] [PubMed]
- Zamani, F.; Zare Shahneh, F.; Aghebati-Maleki, L.; Baradaran, B. Induction of CD14 Expression and Differentiation to Monocytes or Mature Macrophages in Promyelocytic Cell Lines: New Approach. Adv. Pharm. Bull. 2013, 3, 329–332. [Google Scholar] [CrossRef]
- Souza, C.O.S.; Espíndola, M.S.; Fontanari, C.; Prado, M.K.B.; Frantz, F.G.; Rodrigues, V.; Gardinassi, L.G.; Faccioli, L.H. CD18 Regulates Monocyte Hematopoiesis and Promotes Resistance to Experimental Schistosomiasis. Front. Immunol. 2018, 9, 1970. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Guo, Z.; Yao, K.; Miao, Y.; Liang, S.; Liu, F.; Wang, Y.; Zhang, Y. The Transcriptional Foundations of Sp110-mediated Macrophage (RAW264.7) Resistance to Mycobacterium tuberculosis H37Ra. Sci. Rep. 2016, 6, 22041. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Pabst, S.; Lokhande, S.; Grohé, C.; Wollnik, B. Extended genetic analysis of BTNL2 in sarcoidosis. Tissue Antigens 2009, 73, 59–61. [Google Scholar] [CrossRef] [PubMed]
- Morton, D.L.; Thompson, J.F.; Cochran, A.J.; Mozzillo, N.; Elashoff, R.; Essner, R.; Nieweg, O.E.; Roses, D.F.; Hoekstra, H.J.; Karakousis, C.P.; et al. Sentinel-Node Biopsy or Nodal Observation in Melanoma. N. Engl. J. Med. 2006, 355, 1307–1317. [Google Scholar] [CrossRef]
- Morton, D.L.; Thompson, J.F.; Cochran, A.J.; Mozzillo, N.; Nieweg, O.E.; Roses, D.F.; Hoekstra, H.J.; Karakousis, C.P.; Puleo, C.A.; Coventry, B.J.; et al. Final Trial Report of Sentinel-Node Biopsy versus Nodal Observation in Melanoma. N. Engl. J. Med. 2014, 370, 599–609. [Google Scholar] [CrossRef]
- Morton, D.L. Overview and update of the phase III Multicenter Selective Lymphadenectomy Trials (MSLT-I and MSLT-II) in melanoma. Clin. Exp. Met. 2012, 29, 699–706. [Google Scholar] [CrossRef]
- Hsueh, E.C.; Gupta, R.K.; Qi, K.; Morton, D.L. Correlation of specific immune responses with survival in melanoma patients with distant metastases receiving polyvalent melanoma cell vaccine. J. Clin. Oncol. 1998, 16, 2913–2920. [Google Scholar] [CrossRef]
- Bingle, L.; Brown, N.; Lewis, C.E. The role of tumour-associated macrophages in tumour progression: Implications for new anticancer therapies. J. Pathol. J. Path 2002, 196, 254–265. [Google Scholar] [CrossRef]
- Alter-Koltunoff, M.; Goren, S.; Nousbeck, J.; Feng, C.G.; Sher, A.; Ozato, K.; Azriel, A.; Levi, B.-Z. Innate immunity to intraphagosomal pathogens is mediated by interferon regulatory factor 8 (IRF-8) that stimulates the expression of macrophage-specific Nramp1 through antagonizing repression by c-Myc. J. Biol. Chem. 2008, 283, 2724–2733. [Google Scholar] [CrossRef] [PubMed]
- Buu, N.; Sánchez, F.; Schurr, E. The Bcg Host-Resistance Gene. Clin. Infect. Dis. 2000, 31, S81–S85. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Badawy, A.A.; Yahya, R.S.; Awad, S.I.; Al-Sawah, G.A.; Kizilbash, N.A. Relationship between NRAMP1 gene polymorphism and efficacy of BCG vaccine in a helminth-infected population. Genet. Mol. Res. 2013, 12, 3048–3056. [Google Scholar] [CrossRef]
- Bellamy, R.; Ruwende, C.; Corrah, T.; McAdam, K.P.; Whittle, H.C.; Hill, A.V. Variations in the NRAMP1 gene and susceptibility to tuberculosis in West Africans. N. Engl. J. Med. 1998, 338, 640–644. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, B.A.; Archer, N.S.; Simpson, A.M.; Torpy, F.R.; Nassif, N.T. Association of SLC11A1 promoter polymorphisms with the incidence of autoimmune and inflammatory diseases: A meta-analysis. J. Autoimmun. 2008, 31, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Decobert, M.; LaRue, H.; Bergeron, A.; Harel, F.; Pfister, C.; Rousseau, F.; Lacombe, L.; Fradet, Y. Polymorphisms of the human NRAMP1 gene are associated with response to bacillus Calmette-Guerin immunotherapy for superficial bladder cancer. J. Urol. 2006, 175, 1506–1511. [Google Scholar] [CrossRef]
- Guallar-Garrido, S.; Julián, E. Bacillus Calmette-Guérin (BCG) Therapy for Bladder Cancer: An Update. Immunotargets 2020, 9, 1–11. [Google Scholar] [CrossRef]
- Arts, R.J.W.; Moorlag, S.J.C.F.M.; Novakovic, B.; Li, Y.; Wang, S.-Y.; Oosting, M.; Kumar, V.; Xavier, R.J.; Wijmenga, C.; Joosten, L.A.B.; et al. BCG Vaccination Protects against Experimental Viral Infection in Humans through the Induction of Cytokines Associated with Trained Immunity. Cell Host Microbe 2018, 23, 89–100. [Google Scholar] [CrossRef]
- Kaneko, N.; Kurata, M.; Yamamoto, T.; Morikawa, S.; Masumoto, J. The role of interleukin-1 in general pathology. Inflamm. Regen. 2019, 39, 12. [Google Scholar] [CrossRef]
- Eskan, M.A.; Benakanakere, M.R.; Rose, B.G.; Zhang, P.; Zhao, J.; Stathopoulou, P.; Fujioka, D.; Kinane, D.F. Interleukin-1β Modulates Proinflammatory Cytokine Production in Human Epithelial Cells. Infect. Immun. 2008, 76, 2080–2089. [Google Scholar] [CrossRef]
- Ma, X.; Reich, R.A.; Wright, J.A.; Tooker, H.R.; Teeter, L.D.; Musser, J.M.; Graviss, E.A. Association between Interleukin-8 Gene Alleles and Human Susceptibility to Tuberculosis Disease. J. Infect. Dis. 2003, 188, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Morahan, G.; Huang, D.; Ymer, S.I.; Cancilla, M.R.; Stephen, K.; Dabadghao, P.; Werther, G.; Tait, B.D.; Harrison, L.C.; Colman, P.G. Linkage disequilibrium of a type 1 diabetes susceptibility locus with a regulatory IL12B allele. Nat. Genet. 2001, 27, 218–221. [Google Scholar] [CrossRef] [PubMed]
- Awomoyi, A.A.; Charurat, M.; Marchant, A.; Miller, E.N.; Blackwell, J.M.; McAdam, K.P.; Newport, M.J. Polymorphism in IL1B: IL1B—511 association with tuberculosis and decreased lipopolysaccharide-induced IL-1β in IFN-γ primed ex-vivo whole blood assay. J. Endotoxin Res. 2005, 11, 281–286. [Google Scholar] [PubMed]
- Correa, P.A.; Gomez, L.M.; Cadena, J.; Anaya, J.-M. Autoimmunity and tuberculosis. Opposite association with TNF polymorphism. J. Rheumatol. 2005, 32, 219–224. [Google Scholar]
- Redford, P.S.; Murray, P.J.; O’Garra, A. The role of IL-10 in immune regulation during M. tuberculosis infection. Mucosal Immunol. 2011, 4, 261–270. [Google Scholar] [CrossRef]
- Luo, Y. Blocking IL-10 enhances bacillus Calmette-Guérin induced T helper Type 1 immune responses and anti-bladder cancer immunity. Oncoimmunology 2012, 1, 1183–1185. [Google Scholar] [CrossRef][Green Version]
- Geijtenbeek, T.B.H.; Krooshoop, D.J.E.B.; Bleijs, D.A.; van Vliet, S.J.; van Duijnhoven, G.C.F.; Grabovsky, V.; Alon, R.; Figdor, C.G.; van Kooyk, Y. DC-SIGN–ICAM-2 interaction mediates dendritic cell trafficking. Nat. Immunol. 2000, 1, 353–357. [Google Scholar] [CrossRef]
- Geijtenbeek, T.B.H.; Torensma, R.; van Vliet, S.J.; van Duijnhoven, G.C.F.; Adema, G.J.; van Kooyk, Y.; Figdor, C.G. Identification of DC-SIGN, a Novel Dendritic Cell–Specific ICAM-3 Receptor that Supports Primary Immune Responses. Cell 2000, 100, 575–585. [Google Scholar] [CrossRef]
- Tailleux, L.; Schwartz, O.; Herrmann, J.-L.; Pivert, E.; Jackson, M.; Amara, A.; Legres, L.; Dreher, D.; Nicod, L.P.; Gluckman, J.C.; et al. DC-SIGN Is the Major Mycobacterium tuberculosis Receptor on Human Dendritic Cells. J. Exp. Med. 2002, 197, 121–127. [Google Scholar] [CrossRef]
- Schröder, N.W.J.; Schumann, R.R. Single nucleotide polymorphisms of Toll-like receptors and susceptibility to infectious disease. Lancet Infect. Dis. 2005, 5, 156–164. [Google Scholar] [CrossRef]
- Ogus, A.C.; Yoldas, B.; Ozdemir, T.; Uguz, A.; Olcen, S.; Keser, I.; Coskun, M.; Cilli, A.; Yegin, O. The Arg753Gln polymorphism of the human Toll-like receptor 2 gene in tuberculosis disease. Eur. Respir. 2004, 23, 219–223. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Qu, S.; Chen, X.; Wu, Q.; Shi, M. Promising Targets for Cancer Immunotherapy: TLRs, RLRs, and STING-Mediated Innate Immune Pathways. Int. J. Mol. Sci. 2017, 18, 404. [Google Scholar] [CrossRef] [PubMed]
- Underhill, D.M.; Ozinsky, A.; Smith, K.D.; Aderem, A. Toll-like receptor-2 mediates mycobacteria-induced proinflammatory signaling in macrophages. Proc. Natl. Acad. Sci. USA 1999, 96, 14459–14463. [Google Scholar] [CrossRef] [PubMed]
- Quesniaux, V.J.; Nicolle, D.M.; Torres, D.; Kremer, L.; Guérardel, Y.; Nigou, J.; Puzo, G.; Erard, F.; Ryffel, B. Toll-Like Receptor 2 (TLR2)-Dependent-Positive and TLR2-Independent-Negative Regulation of Proinflammatory Cytokines by Mycobacterial Lipomannans. J. Immunol. 2004, 172, 4425–4434. [Google Scholar] [CrossRef] [PubMed]
- Nagarsheth, N.; Wicha, M.S.; Zou, W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 2017, 17, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Aldinucci, D.; Borghese, C.; Casagrande, N. The CCL5/CCR5 Axis in Cancer Progression. Cancers 2020, 12, 1765. [Google Scholar] [CrossRef]
- Zhou, W.; Guo, S.; Liu, M.; Burow, M.E.; Wang, G. Targeting CXCL12/CXCR4 Axis in Tumor Immunotherapy. Curr. Med. Chem. 2019, 26, 3026–3041. [Google Scholar] [CrossRef]
- Méndez-Samperio, P. Expression and regulation of chemokines in mycobacterial infection. J. Infect. 2008, 57, 374–384. [Google Scholar] [CrossRef]
- Ugurel, S.; Schrama, D.; Keller, G.; Schadendorf, D.; Bröcker, E.-B.; Houben, R.; Zapatka, M.; Fink, W.; Kaufman, H.L.; Becker, J.C. Impact of the CCR5 gene polymorphism on the survival of metastatic melanoma patients receiving immunotherapy. Cancer Immunol. Immunother. 2008, 57, 685–691. [Google Scholar] [CrossRef]
- Fox, G.J.; Sy, D.N.; Nhung, N.V.; Yu, B.; Ellis, M.K.; Van Hung, N.; Cuong, N.K.; Thi Lien, L.; Marks, G.B.; Saunders, B.M.; et al. Polymorphisms of SP110 are associated with both pulmonary and extra-pulmonary tuberculosis among the Vietnamese. PLoS ONE 2014, 9, e99496. [Google Scholar] [CrossRef]
- Wu, Y.; Guo, Z.; Liu, F.; Yao, K.; Gao, M.; Luo, Y.; Zhang, Y. Sp110 enhances macrophage resistance to Mycobacterium tuberculosis via inducing endoplasmic reticulum stress and inhibiting anti-apoptotic factors. Oncotarget 2017, 8, 64050–64065. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zimmermann, A.; Knecht, H.; Häsler, R.; Zissel, G.; Gaede, K.I.; Hofmann, S.; Nebel, A.; Müller-Quernheim, J.; Schreiber, S.; Fischer, A. Atopobium and Fusobacterium as novel candidates for sarcoidosis-associated microbiota. Eur. Respir. J. 2017, 50, 1600746. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Liu, X.K.; Zhang, Y.L.; Dong, C. BTNL2, a butyrophilin-like molecule that functions to inhibit T cell activation. J. Immunol. 2006, 176, 7354–7360. [Google Scholar] [CrossRef] [PubMed]
- Arnett, H.A.; Escobar, S.S.; Gonzalez-Suarez, E.; Budelsky, A.L.; Steffen, L.A.; Boiani, N.; Zhang, M.; Siu, G.; Brewer, A.W.; Viney, J.L. BTNL2, a Butyrophilin/B7-Like Molecule, Is a Negative Costimulatory Molecule Modulated in Intestinal Inflammation. J. Immunol. 2007, 178, 1523–1533. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Kaneda, K.; Kasama, T. Immunopathogenesis of delayed-type hypersensitivity. Microsc. Res. Tech. 2001, 53, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.Q.; Field, M.; Andrew, E.; Haskard, D.; Feldmann, M.; Maini, R.N. Detection of cytokines at the site of tuberculin-induced delayed-type hypersensitivity in man. Clin. Exp. Immunol. 1992, 90, 522–529. [Google Scholar] [CrossRef]
- Redelman-Sidi, G. Could BCG be used to protect against COVID-19? Nat. Rev. Urol. 2020, 17, 316–317. [Google Scholar] [CrossRef]
- Giamarellos-Bourboulis, E.J.; Tsilika, M.; Moorlag, S.; Antonakos, N.; Kotsaki, A.; Domínguez-Andrés, J.; Kyriazopoulou, E.; Gkavogianni, T.; Adami, M.-E.; Damoraki, G. Activate: Randomized Clinical Trial of BCG Vaccination Against Infection in the Elderly. Cell 2020, 183, 315–323. [Google Scholar] [CrossRef]
- Netea, M.G.; Joosten, L.A.; Latz, E.; Mills, K.H.; Natoli, G.; Stunnenberg, H.G.; O’Neill, L.A.; Xavier, R.J. Trained immunity: A program of innate immune memory in health and disease. Science 2016, 352, aaf1098. [Google Scholar] [CrossRef]
- O’Neill, L.A.J.; Netea, M.G. BCG-induced trained immunity: Can it offer protection against COVID-19? Nat. Rev. Immunol. 2020, 20, 335–337. [Google Scholar] [CrossRef]
- Kleinnijenhuis, J.; Quintin, J.; Preijers, F.; Benn, C.S.; Joosten, L.A.; Jacobs, C.; van Loenhout, J.; Xavier, R.J.; Aaby, P.; van der Meer, J.W.; et al. Long-lasting effects of BCG vaccination on both heterologous Th1/Th17 responses and innate trained immunity. J. Innate Immun. 2014, 6, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Moorlag, S.; van Deuren, R.C.; van Werkhoven, C.H.; Jaeger, M.; Debisarun, P.; Taks, E.; Mourits, V.P.; Koeken, V.; de Bree, L.C.J.; Ten Doesschate, T.; et al. Safety and COVID-19 Symptoms in Individuals Recently Vaccinated with BCG: A Retrospective Cohort Study. Cell Rep. Med. 2020, 1, 100073. [Google Scholar] [CrossRef] [PubMed]
- Jirjees, F.J.; Dallal Bashi, Y.H.; Al-Obaidi, H.J. COVID-19 Death and BCG Vaccination Programs Worldwide. Tuberc. Respir. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Noval Rivas, M.; Ebinger, J.E.; Wu, M.; Sun, N.; Braun, J.; Sobhani, K.; Van Eyk, J.E.; Cheng, S.; Arditi, M. BCG vaccination history associates with decreased SARS-CoV-2 seroprevalence across a diverse cohort of healthcare workers. J. Clin. Investig. 2020. [Google Scholar] [CrossRef] [PubMed]
- Joy, M.; Malavika, B.; Asirvatham, E.S.; Sudarsanam, T.D.; Jeyaseelan, L. Is BCG associated with reduced incidence of COVID-19? A meta-regression of global data from 160 countries. Clin. Epidemiol. Glob. Health 2020. [Google Scholar] [CrossRef]
- Faries, M.B.; Thompson, J.F.; Cochran, A.J.; Andtbacka, R.H.; Mozzillo, N.; Zager, J.S.; Jahkola, T.; Bowles, T.L.; Testori, A.; Beitsch, P.D.; et al. Completion Dissection or Observation for Sentinel-Node Metastasis in Melanoma. N. Engl. J. Med. 2017, 376, 2211–2222. [Google Scholar] [CrossRef]
- Morton, D.L.; Hsueh, E.C.; Essner, R.; Foshag, L.J.; O’Day, S.J.; Bilchik, A.; Gupta, R.K.; Hoon, D.S.B.; Ravindranath, M.; Nizze, J.A.; et al. Prolonged survival of patients receiving active immunotherapy with Canvaxin therapeutic polyvalent vaccine after complete resection of melanoma metastatic to regional lymph nodes. Ann. Surg. 2002, 236, 438–449. [Google Scholar] [CrossRef]
- Wang, J.; Huang, S.K.; Marzese, D.M.; Hsu, S.C.; Kawas, N.P.; Chong, K.K.; Long, G.V.; Menzies, A.M.; Scolyer, R.A.; Izraely, S.; et al. Epigenetic Changes of EGFR Have an Important Role in BRAF Inhibitor–Resistant Cutaneous Melanomas. J. Investig. Dermatol. 2015, 135, 532–541. [Google Scholar] [CrossRef]
- Hoshimoto, S.; Kuo, C.T.; Chong, K.K.; Takeshima, T.-L.; Takei, Y.; Li, M.W.; Huang, S.K.; Sim, M.-S.; Morton, D.L.; Hoon, D.S.B. AIM1 and LINE-1 epigenetic aberrations in tumor and serum relate to melanoma progression and disease outcome. J. Investig. Dermatol. 2012, 132, 1689–1697. [Google Scholar] [CrossRef]
- Gabriel, S.; Ziaugra, L.; Tabbaa, D. SNP Genotyping Using the Sequenom MassARRAY iPLEX Platform. Curr. Protoc. Hum. Genet. 2009, 60, 2–12. [Google Scholar] [CrossRef]
- Kathiresan, S.; Voight, B.F.; Purcell, S.; Musunuru, K.; Ardissino, D.; Mannucci, P.M.; Anand, S.; Engert, J.C.; Samani, N.J.; Schunkert, H.; et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 2009, 41, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.M.; Knight, J. Introduction to genetic association studies. Cold Spring Harb. Protoc. 2012, 2012, 297–306. [Google Scholar] [CrossRef] [PubMed]

| SNP | Gene | Chromosome | Position GRCh37 | Minor Allele Frequency (dbSNP) |
|---|---|---|---|---|
| Macrophage | ||||
| rs17215556 | SLC11A1/NRAMP | 2 | 219,258,856 | 0.0060 (T > C) |
| rs17235409 | SLC11A1/NRAMP | 2 | 219,259,732 | 0.0702 (G > A) |
| rs17235416 | SLC11A1/NRAMP | 2 | 219,259,814 | 0.1024 (del TGTG) |
| rs34448891 | SLC11A1/NRAMP | 2 | 219,246,649 | MAF not reported (ins GT) |
| rs3731865 | SLC11A1/NRAMP | 2 | 219,250,003 | 0.1680 (G > C) |
| rs2569190 | CD14 | 5 | 140,012,916 | 0.4734 (A > G) |
| rs2569193 | CD14 | 5 | 140,015,495 | 0.2172 (G > A) |
| rs684 | CD18 | 21 | 46,306,161 | 0.1639 (G > A) |
| rs117989670 | CD18 | 21 | 46,305,913 | 0.0069 (A > C) |
| Dendritic cells | ||||
| rs4804803 | CD209 | 19 | 7,812,733 | 0.2117 (A > G) |
| Toll-like receptor | ||||
| rs4696480 | TLR2 | 4 | 154,607,126 | 0.4601 (T > A) |
| rs5743708 | TLR2 | 4 | 154,626,317 | 0.0119 (G > A) |
| Inflammatory cytokines | ||||
| rs1800629 | TNF | 6 | 31,543,031 | 0.0955 (G > A) |
| rs361525 | TNF | 6 | 31,543,101 | 0.0505 (G > A) |
| rs1143627 | IL1B | 2 | 113,594,387 | 0.4803 (C > T) |
| rs1143634 | IL1B | 2 | 113,590,390 | 0.1455 (C > T) |
| rs16944 | IL1B | 2 | 113,594,867 | 0.4651 (A > G) |
| rs4073 | IL8 | 4 | 74,606,024 | 0.4972 (A > T) |
| rs1800896 | IL10 | 1 | 206,946,897 | 0.3026 (A > G) |
| rs17860508 | IL12B-near 5′ | 5 | 158,760,200 | not reported |
| rs1343151 | IL23R | 1 | 67,719,129 | 0.3237 (G > A) |
| Chemokines/Chemokine receptor | ||||
| rs1799987 | CCR5 | 3 | 46,411,935 | 0.4871 (A > G) |
| rs1801157 | CXCL12 | 10 | 44,868,257 | 0.2080 (G > A) |
| Others | ||||
| rs3948464 | SP110 | 2 | 231,050,715 | 0.1028 (C > T) |
| rs1135791 | SP110 | 2 | 231,042,276 | 0.3375 (T > C) |
| rs2076530 | BTNL2 | 6 | 32,363,816 | 0.3779 (A > G) |
| Variable | Disease-Free Survival | Overall Survival | ||
|---|---|---|---|---|
| Multivariable Analysis | Multivariable Analysis | |||
| * p-Value | HR (95% CI) | * p-Value | HR (95% CI) | |
| Immune-gene Panel SNP Score (Responder vs. Non-Responder) | 0.021 | 1.84 (1.09–3.13) | ||
| Age | ||||
| Gender | ||||
| Male/Female | ||||
| Lymph Node Positive | ||||
| 1 | ||||
| 0 | ||||
| 2–3 | ||||
| ≥4 | ||||
| Unknown | ||||
| Primary Site | ||||
| Extremity | ||||
| Head/Neck | ||||
| Mucosal | ||||
| Trunk | ||||
| Unknown | ||||
| M-Stage | ||||
| M1a | ||||
| M1b | ||||
| M1c | ||||
| Baseline LDH | ||||
| Normal /Elevated | ||||
| Number of Metastasis | ||||
| 1 | ||||
| 2–3 | ||||
| 4–5 | ||||
| ECOG Performance Status | ||||
| 0 | ||||
| 1 | ||||
| Unknown | ||||
| Prior Diagnosis of Stage III Melanoma | ||||
| Yes/No | ||||
| Previous Treatment for Melanoma | ||||
| Yes/No | ||||
| Variable | Disease-Free Survival | Overall Survival | ||
|---|---|---|---|---|
| Multivariate Analysis | Multivariate Analysis | |||
| * p-Value | HR (95% CI) | * p-Value | HR (95% CI) | |
| Immune-gene Panel SNP Score (Responder vs. Non-Responder) | 0.158 | 1.31 (0.90–1.93) | 0.025 | 1.67 (1.07–2.67) |
| Age | 0.007 | 1.02 (1.01–1.03) | ||
| Gender | 0.040 | 1.64 (1.02–2.71) | ||
| Male/Female | ||||
| Breslow | ||||
| ≤1.00 mm | 1.00 (Reference) | |||
| 1.01–2.00 mm | 0.71 (0.40–1.30) | |||
| 2.01–4.00 mm | 0.59 (0.32–1.11) | |||
| >4.00 mm | 0.98 (0.52–1.85) | |||
| Unknown | 0.55 (0.29–1.04) | |||
| Lymph Node Positive | 0.048 | |||
| 1 | 1.00 (Reference) | |||
| 2–3 | 0.014 | 1.81 (1.13–2.89) | ||
| ≥4 | 1.31 (0.67–2.44) | |||
| Primary Site | ||||
| Extremity | ||||
| Head/Neck | ||||
| Mucosal | ||||
| Trunk | ||||
| Unknown | ||||
| Palpable Lymph Node | ||||
| Yes/No | 1.33 (0.90–1.97) | 0.029 | 1.68 (1.05–2.69) | |
| Ulceration | 0.016 | |||
| No | 1.00 (Reference) | 1.00 (Reference) | ||
| Yes | 0.061 | 1.60 (0.98–2.63) | 0.008 | 2.08 (1.21–3.65) |
| Unknown | 1.02 (0.64–1.65) | 1.13 (0.65–2.00) | ||
| Gene | SNP | p Value |
|---|---|---|
| IL1β | rs1143627 | 0.003 |
| NRAMP1/SLC11A1 | rs17215556 | 0.029 |
| IL-1β | rs16944 | 0.001 |
| CXCL12 | rs1801157 | 0.050 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramos, R.I.; Shaw, M.A.; Foshag, L.; Stern, S.L.; Rahimzadeh, N.; Elashoff, D.; Hoon, D.S.B. Genetic Variants in Immune Related Genes as Predictors of Responsiveness to BCG Immunotherapy in Metastatic Melanoma Patients. Cancers 2021, 13, 91. https://doi.org/10.3390/cancers13010091
Ramos RI, Shaw MA, Foshag L, Stern SL, Rahimzadeh N, Elashoff D, Hoon DSB. Genetic Variants in Immune Related Genes as Predictors of Responsiveness to BCG Immunotherapy in Metastatic Melanoma Patients. Cancers. 2021; 13(1):91. https://doi.org/10.3390/cancers13010091
Chicago/Turabian StyleRamos, Romela Irene, Misa A. Shaw, Leland Foshag, Stacey L. Stern, Negin Rahimzadeh, David Elashoff, and Dave S. B. Hoon. 2021. "Genetic Variants in Immune Related Genes as Predictors of Responsiveness to BCG Immunotherapy in Metastatic Melanoma Patients" Cancers 13, no. 1: 91. https://doi.org/10.3390/cancers13010091
APA StyleRamos, R. I., Shaw, M. A., Foshag, L., Stern, S. L., Rahimzadeh, N., Elashoff, D., & Hoon, D. S. B. (2021). Genetic Variants in Immune Related Genes as Predictors of Responsiveness to BCG Immunotherapy in Metastatic Melanoma Patients. Cancers, 13(1), 91. https://doi.org/10.3390/cancers13010091