STAT3 Stabilizes IKKα Protein through Direct Interaction in Transformed and Cancerous Human Breast Epithelial Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
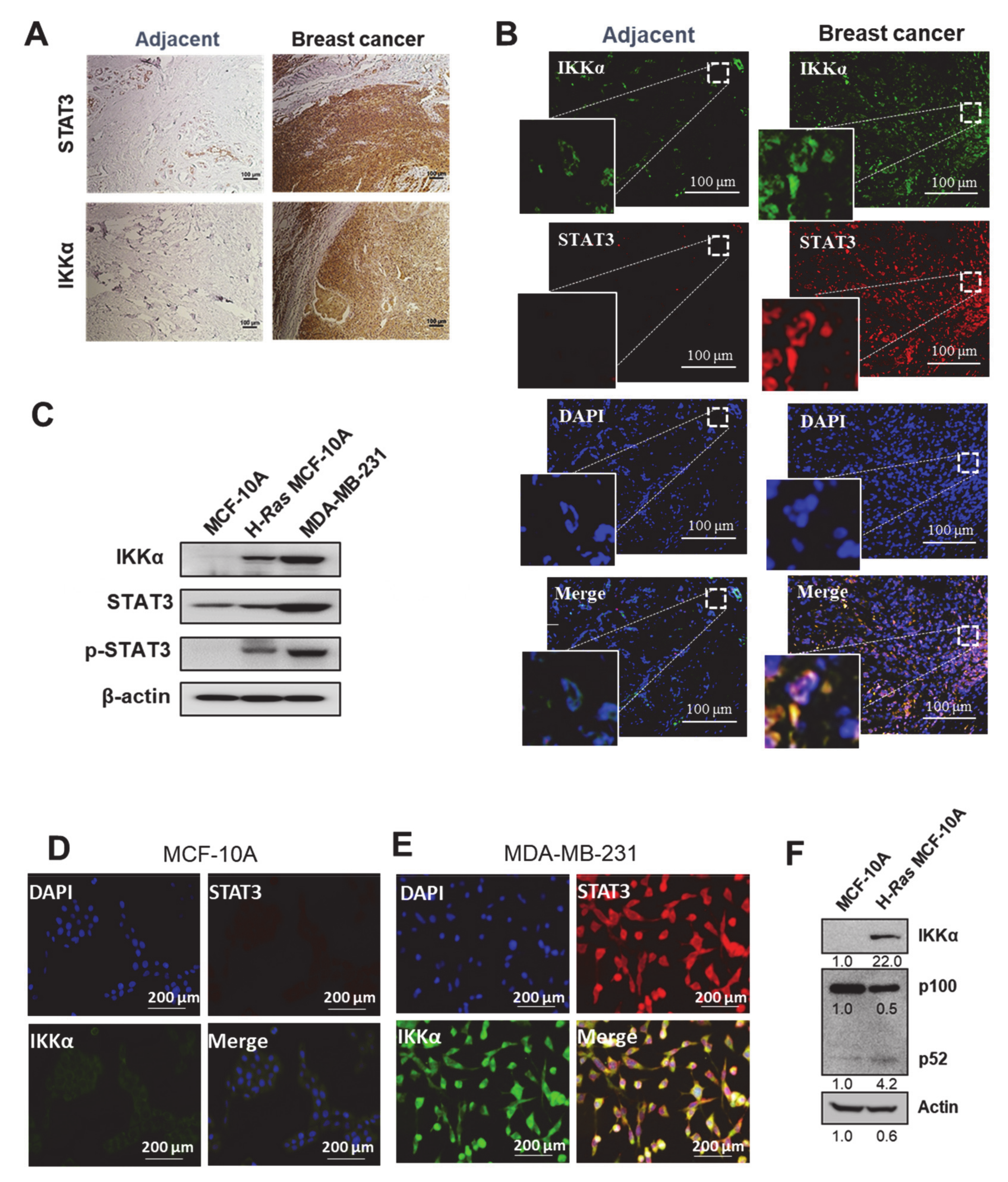
2.1. IKKα Is Co-Expressed and Co-Localizes with STAT3 in Human Breast Carcinoma
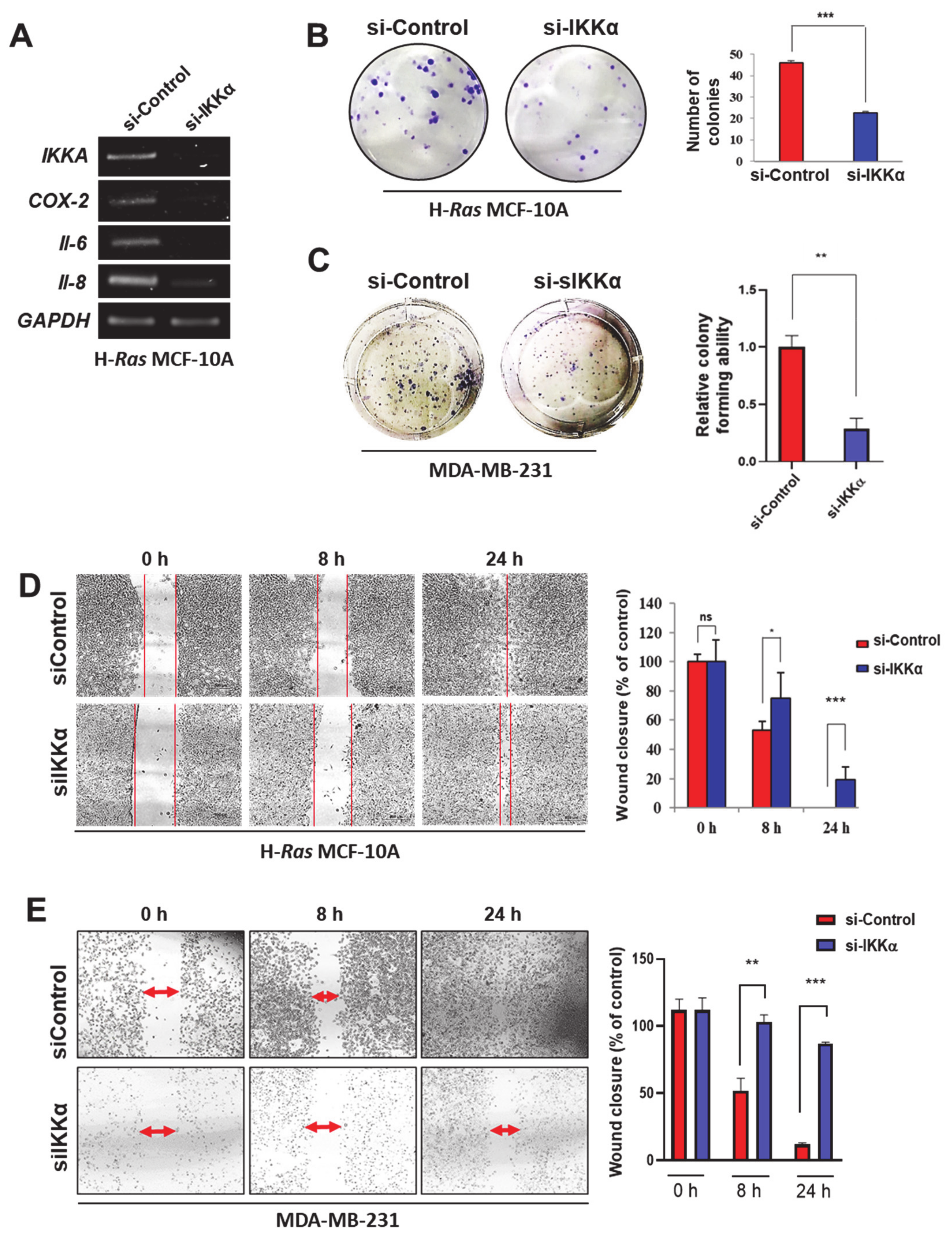
2.2. IKKα May Promote Human Breast Cancer Progression through Non-Canonical NF-κB Activation
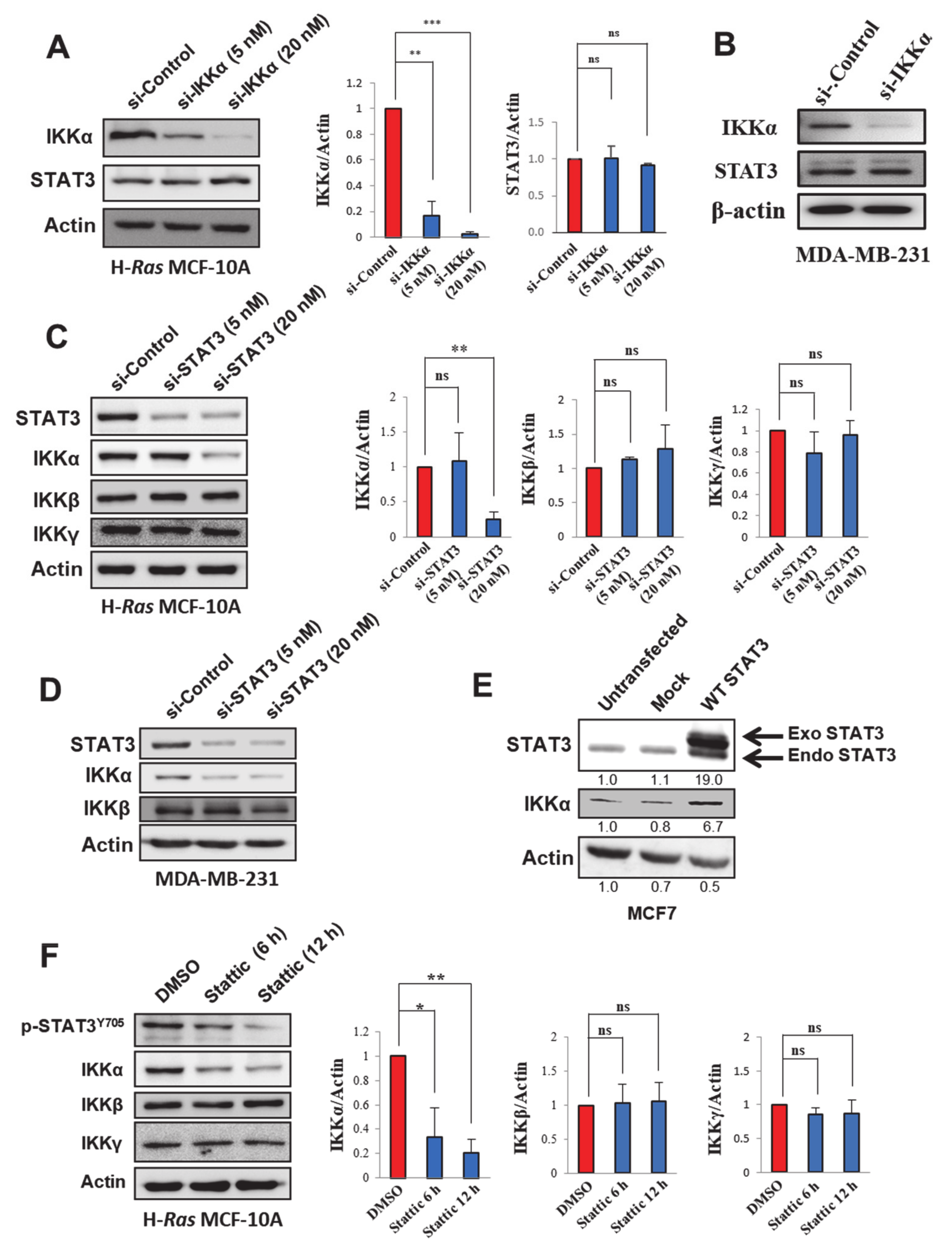
2.3. STAT3 Regulates IKKα Expression in H-Ras Transformed and Cancerous Human Breast Epithelial Cell Lines
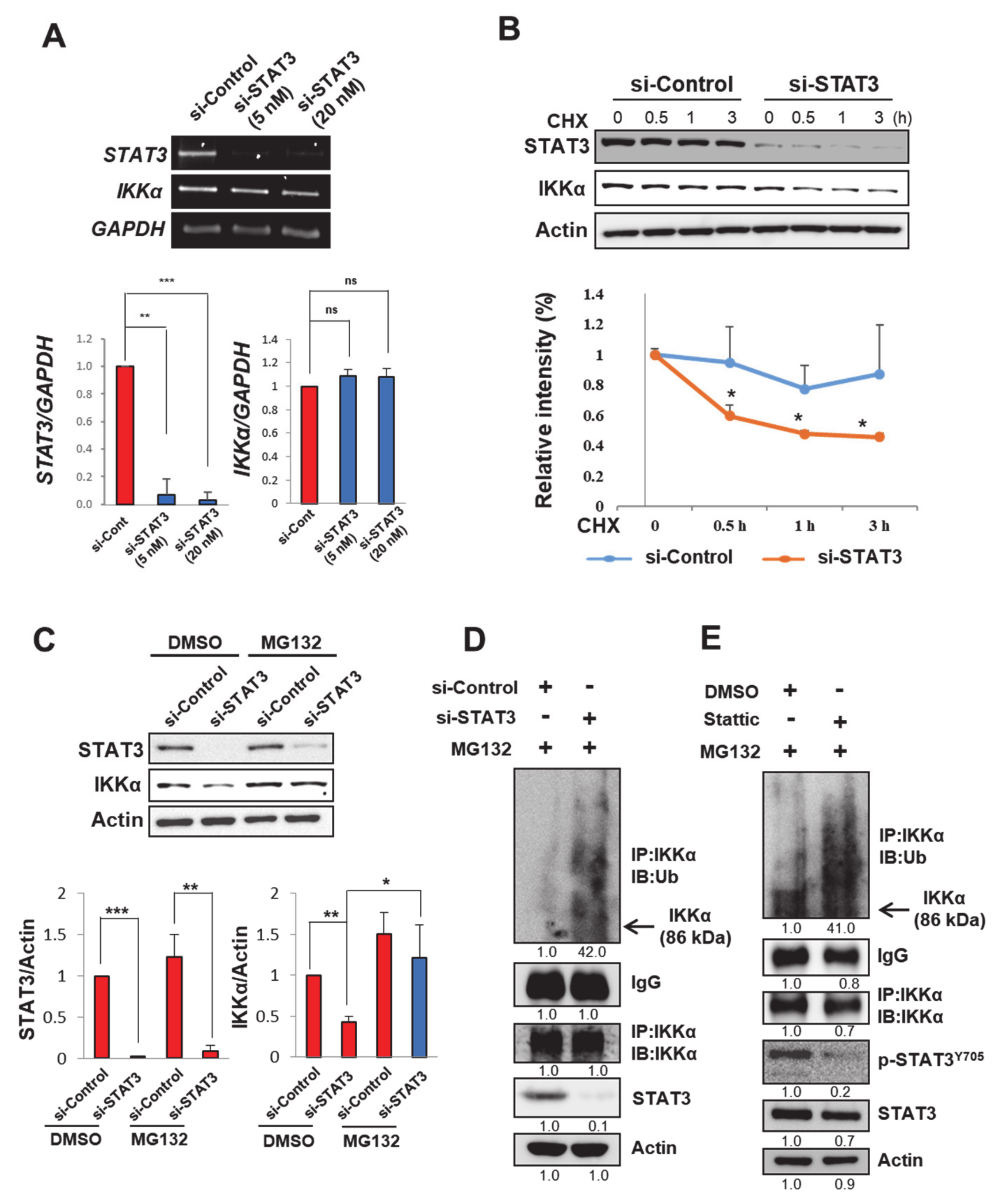
2.4. STAT3 Stabilizes IKKα Protein in H-Ras MCF-10A Cells
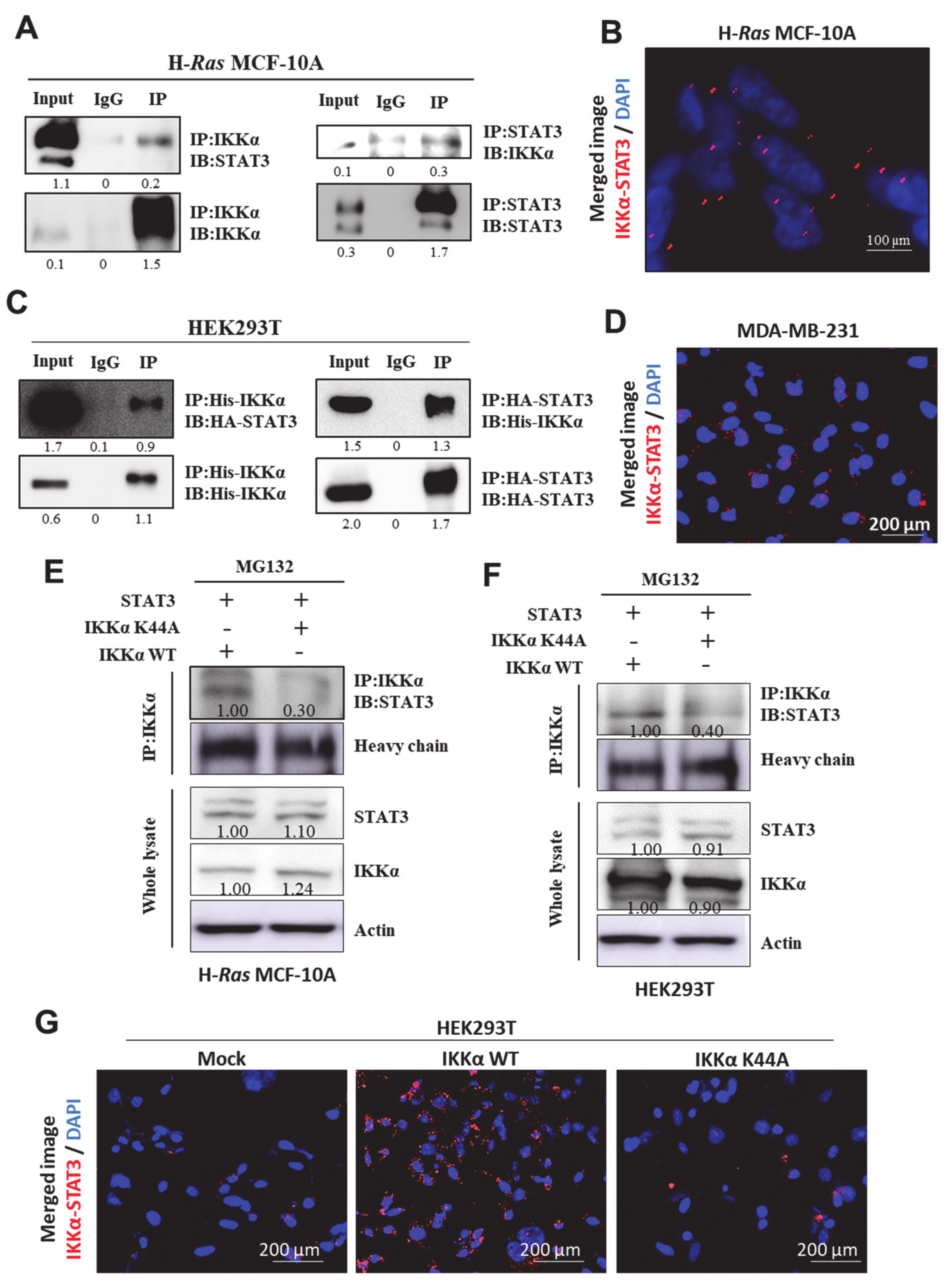
2.5. STAT3 Physically Interacts with IKKα, Presumably at Lys44
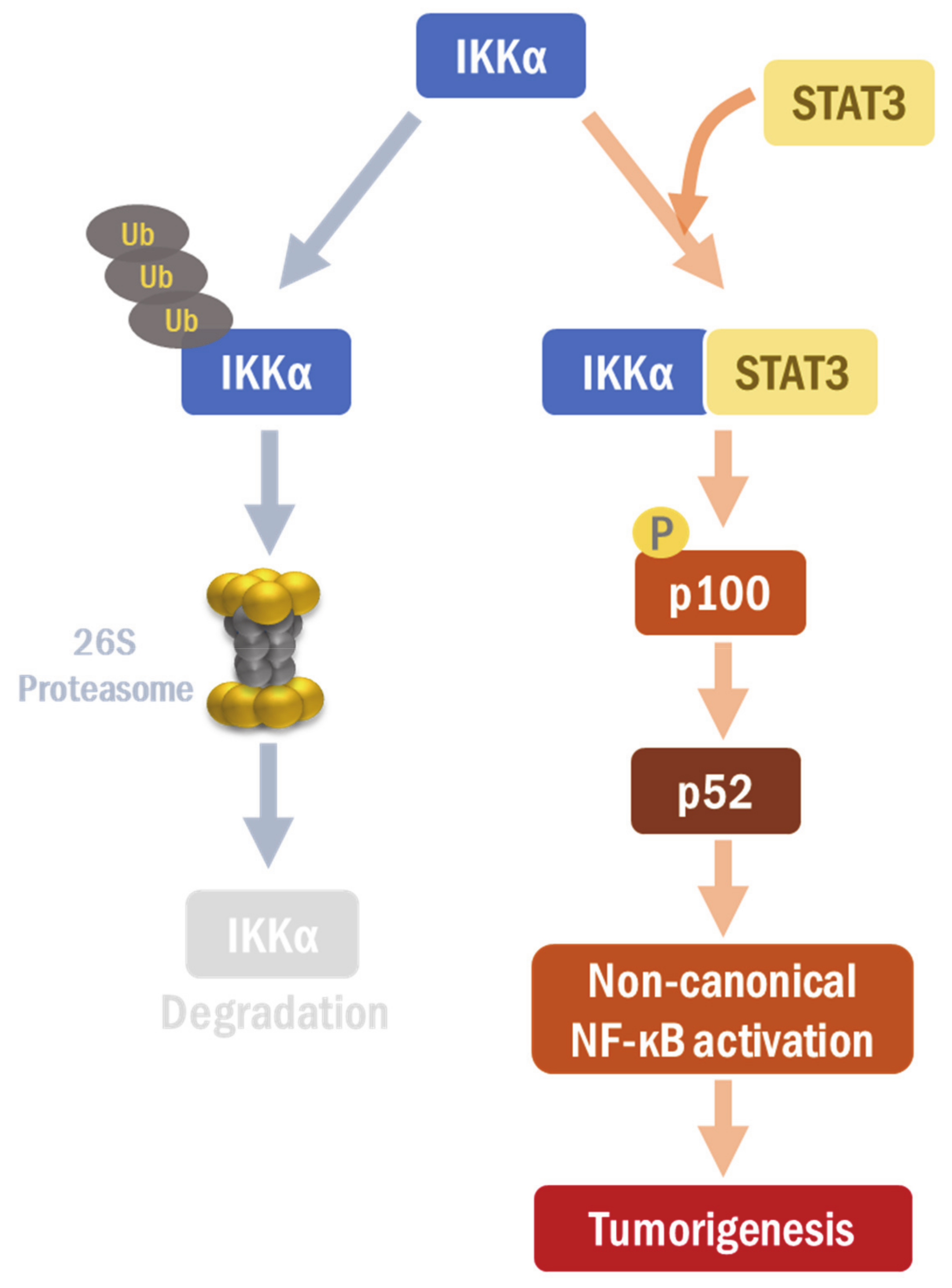
3. Discussion
4. Materials and Methods
4.1. Tissues, Cell Culture, and siRNA Knockdown of Gene Expression
4.2. Reagents and Antibodies
4.3. Public Data Resources and Survival Analysis
4.4. Immunoblot and Immunoprecipitaion Assays
4.5. Immunohistochemistry and Immunofluorescence Staining
4.6. Reverse Transcription Polymerase Chain Reaction (RT-PCR)
4.7. Soft Agar Colony Forming and Migration Assays
4.8. CHX Chase Assay
4.9. PLA
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bowman, T.; Garcia, R.; Turkson, J.; Jove, R. STATs in oncogenesis. Oncogene 2000, 19, 2474–2488. [Google Scholar] [CrossRef] [PubMed]
- Bromberg, J.F.; Wrzeszczynska, M.H.; Devgan, G.; Zhao, Y.; Pestell, R.G.; Albanese, C.; Darnell, J.E., Jr. Stat3 as an oncogene. Cell 1999, 98, 295–303. [Google Scholar] [CrossRef]
- Karin, M. NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harb. Perspect. Biol. 2009, 1, a000141. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Wu, P.; Zhao, L.; Huang, L.; Zhang, Z.; Zhao, S.; Huang, J. NF-kappaB Expression and Outcomes in Solid Tumors: A Systematic Review and Meta-Analysis. Medicine 2015, 94, e1687. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Wu, D.; Zhao, L.; Huang, L.; Shen, G.; Huang, J.; Chai, Y. Prognostic role of STAT3 in solid tumors: A systematic review and meta-analysis. Oncotarget 2016, 7, 19863–19883. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef]
- Sun, S.C. The non-canonical NF-kappaB pathway in immunity and inflammation. Nat. Rev. Immunol. 2017, 17, 545–558. [Google Scholar] [CrossRef]
- Senftleben, U.; Cao, Y.; Xiao, G.; Greten, F.R.; Krahn, G.; Bonizzi, G.; Chen, Y.; Hu, Y.; Fong, A.; Sun, S.C.; et al. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science 2001, 293, 1495–1499. [Google Scholar] [CrossRef]
- Lee, H.; Herrmann, A.; Deng, J.H.; Kujawski, M.; Niu, G.; Li, Z.; Forman, S.; Jove, R.; Pardoll, D.M.; Yu, H. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell 2009, 15, 283–293. [Google Scholar] [CrossRef]
- Kesanakurti, D.; Chetty, C.; Rajasekhar Maddirela, D.; Gujrati, M.; Rao, J.S. Essential role of cooperative NF-kappaB and Stat3 recruitment to ICAM-1 intronic consensus elements in the regulation of radiation-induced invasion and migration in glioma. Oncogene 2013, 32, 5144–5155. [Google Scholar] [CrossRef]
- Taniguchi, K.; Karin, M. NF-kappaB, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol. 2018, 18, 309–324. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Mao, R.; Yang, J. NF-kappaB and STAT3 signaling pathways collaboratively link inflammation to cancer. Protein Cell 2013, 4, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Grivennikov, S.I.; Karin, M. Dangerous liaisons: STAT3 and NF-kappaB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev. 2010, 21, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Espinoza-Sanchez, N.A.; Gyorffy, B.; Fuentes-Panana, E.M.; Gotte, M. Differential impact of classical and non-canonical NF-kappaB pathway-related gene expression on the survival of breast cancer patients. J. Cancer 2019, 10, 5191–5211. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Shi, Z.; Liu, J.; Zheng, S.; Yang, J.; Liu, Y.; Yang, Y.; Chang, F.; Yu, W. Clinicopathological and Prognostic Role of STAT3/p-STAT3 in Breast Cancer Patients in China: A Meta-Analysis. Sci. Rep. 2019, 9, 11243. [Google Scholar] [CrossRef]
- Rojo, F.; Gonzalez-Perez, A.; Furriol, J.; Nicolau, M.J.; Ferrer, J.; Burgues, O.; Sabbaghi, M.; Gonzalez-Navarrete, I.; Cristobal, I.; Serrano, L.; et al. Non-canonical NF-kappaB pathway activation predicts outcome in borderline oestrogen receptor positive breast carcinoma. Br. J. Cancer 2016, 115, 322–331. [Google Scholar] [CrossRef]
- Hahn, Y.I.; Kim, S.J.; Choi, B.Y.; Cho, K.C.; Bandu, R.; Kim, K.P.; Kim, D.H.; Kim, W.; Park, J.S.; Han, B.W.; et al. Curcumin interacts directly with the Cysteine 259 residue of STAT3 and induces apoptosis in H-Ras transformed human mammary epithelial cells. Sci. Rep. 2018, 8, 6409. [Google Scholar] [CrossRef]
- Gong, J.; Xie, J.; Bedolla, R.; Rivas, P.; Chakravarthy, D.; Freeman, J.W.; Reddick, R.; Kopetz, S.; Peterson, A.; Wang, H.; et al. Combined targeting of STAT3/NF-kappaB/COX-2/EP4 for effective management of pancreatic cancer. Clin. Cancer Res. 2014, 20, 1259–1273. [Google Scholar] [CrossRef]
- Lee, C.; Cheung, S.T. STAT3: An Emerging Therapeutic Target for Hepatocellular Carcinoma. Cancers 2019, 11, 1646. [Google Scholar] [CrossRef]
- Schust, J.; Sperl, B.; Hollis, A.; Mayer, T.U.; Berg, T. Stattic: A small-molecule inhibitor of STAT3 activation and dimerization. Chem. Biol. 2006, 13, 1235–1242. [Google Scholar] [CrossRef]
- Chen, G.; Cao, P.; Goeddel, D.V. TNF-induced recruitment and activation of the IKK complex require Cdc37 and Hsp90. Mol. Cell 2002, 9, 401–410. [Google Scholar] [CrossRef]
- Israel, A. The IKK complex, a central regulator of NF-kappaB activation. Cold Spring Harb. Perspect. Biol. 2010, 2, a000158. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Jiang, Q.; Willette-Brown, J.; Xi, S.; Zhu, F.; Burkett, S.; Back, T.; Song, N.Y.; Datla, M.; Sun, Z.; et al. The pivotal role of IKKalpha in the development of spontaneous lung squamous cell carcinomas. Cancer Cell 2013, 23, 527–540. [Google Scholar] [CrossRef] [PubMed]
- He, G.; Karin, M. NF-kappaB and STAT3-key players in liver inflammation and cancer. Cell Res. 2011, 21, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, K.; Resat, H. Constitutive activation of STAT3 in breast cancer cells: A review. Int. J. Cancer 2016, 138, 2570–2578. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, M.M.; Sung, B.; Yadav, V.R.; Kannappan, R.; Aggarwal, B.B. NF-kappaB addiction and its role in cancer: ‘one size does not fit all’. Oncogene 2011, 30, 1615–1630. [Google Scholar] [CrossRef]
- Kim, D.J.; Kataoka, K.; Rao, D.; Kiguchi, K.; Cotsarelis, G.; Digiovanni, J. Targeted disruption of stat3 reveals a major role for follicular stem cells in skin tumor initiation. Cancer Res. 2009, 69, 7587–7594. [Google Scholar] [CrossRef]
- Nguyen, A.V.; Wu, Y.Y.; Liu, Q.; Wang, D.; Nguyen, S.; Loh, R.; Pang, J.; Friedman, K.; Orlofsky, A.; Augenlicht, L.; et al. STAT3 in epithelial cells regulates inflammation and tumor progression to malignant state in colon. Neoplasia 2013, 15, 998–1008. [Google Scholar] [CrossRef]
- Basseres, D.S.; Ebbs, A.; Levantini, E.; Baldwin, A.S. Requirement of the NF-kappaB subunit p65/RelA for K-Ras-induced lung tumorigenesis. Cancer Res. 2010, 70, 3537–3546. [Google Scholar] [CrossRef]
- Greten, F.R.; Eckmann, L.; Greten, T.F.; Park, J.M.; Li, Z.W.; Egan, L.J.; Kagnoff, M.F.; Karin, M. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell 2004, 118, 285–296. [Google Scholar] [CrossRef]
- Kaltschmidt, B.; Greiner, J.F.W.; Kadhim, H.M.; Kaltschmidt, C. Subunit-Specific Role of NF-kappaB in Cancer. Biomedicines 2018, 6, 44. [Google Scholar] [CrossRef] [PubMed]
- Connelly, L.; Robinson-Benion, C.; Chont, M.; Saint-Jean, L.; Li, H.; Polosukhin, V.V.; Blackwell, T.S.; Yull, F.E. A transgenic model reveals important roles for the NF-kappa B alternative pathway (p100/p52) in mammary development and links to tumorigenesis. J. Biol. Chem. 2007, 282, 10028–10035. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liao, X.; Agarwal, M.K.; Barnes, L.; Auron, P.E.; Stark, G.R. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev. 2007, 21, 1396–1408. [Google Scholar] [CrossRef] [PubMed]
- Tegowski, M.; Baldwin, A. Noncanonical NF-kappaB in Cancer. Biomedicines 2018, 6, 66. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hahn, Y.-I.; Saeidi, S.; Kim, S.-J.; Park, S.-Y.; Song, N.-Y.; Zheng, J.; Kim, D.-H.; Lee, H.-B.; Han, W.; Noh, D.-Y.; et al. STAT3 Stabilizes IKKα Protein through Direct Interaction in Transformed and Cancerous Human Breast Epithelial Cells. Cancers 2021, 13, 82. https://doi.org/10.3390/cancers13010082
Hahn Y-I, Saeidi S, Kim S-J, Park S-Y, Song N-Y, Zheng J, Kim D-H, Lee H-B, Han W, Noh D-Y, et al. STAT3 Stabilizes IKKα Protein through Direct Interaction in Transformed and Cancerous Human Breast Epithelial Cells. Cancers. 2021; 13(1):82. https://doi.org/10.3390/cancers13010082
Chicago/Turabian StyleHahn, Young-Il, Soma Saeidi, Su-Jung Kim, Se-Young Park, Na-Young Song, Jie Zheng, Do-Hee Kim, Han-Byoel Lee, Wonshik Han, Dong-Young Noh, and et al. 2021. "STAT3 Stabilizes IKKα Protein through Direct Interaction in Transformed and Cancerous Human Breast Epithelial Cells" Cancers 13, no. 1: 82. https://doi.org/10.3390/cancers13010082
APA StyleHahn, Y.-I., Saeidi, S., Kim, S.-J., Park, S.-Y., Song, N.-Y., Zheng, J., Kim, D.-H., Lee, H.-B., Han, W., Noh, D.-Y., Na, H.-K., & Surh, Y.-J. (2021). STAT3 Stabilizes IKKα Protein through Direct Interaction in Transformed and Cancerous Human Breast Epithelial Cells. Cancers, 13(1), 82. https://doi.org/10.3390/cancers13010082





