The Interactome of Cancer-Related Lysyl Oxidase and Lysyl Oxidase-Like Proteins
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
2.1. The Interaction Dataset of the Lysyl Oxidase Family
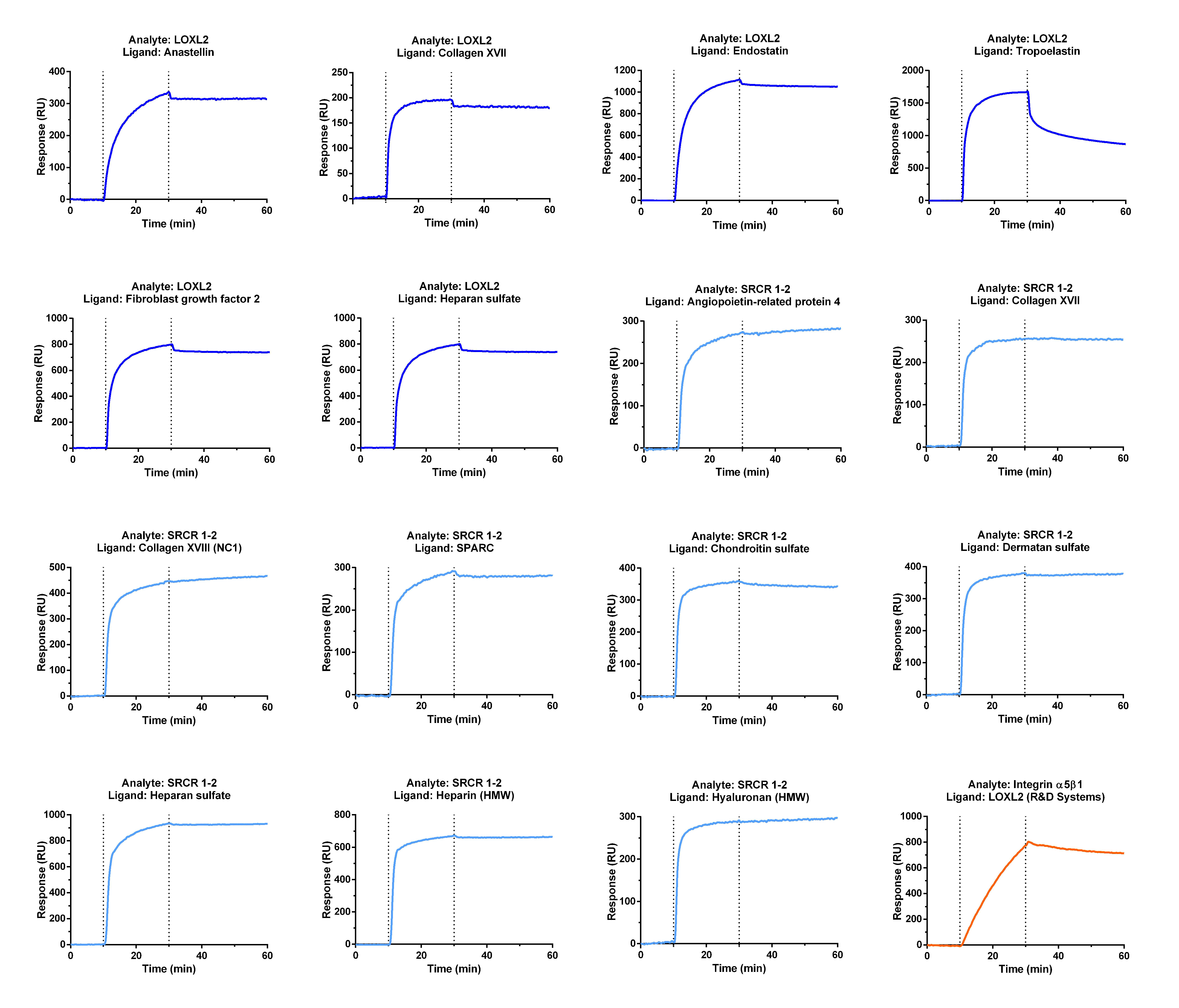
2.1.1. Identification of New Partners of LOXL2 by In Vitro Binding Assays
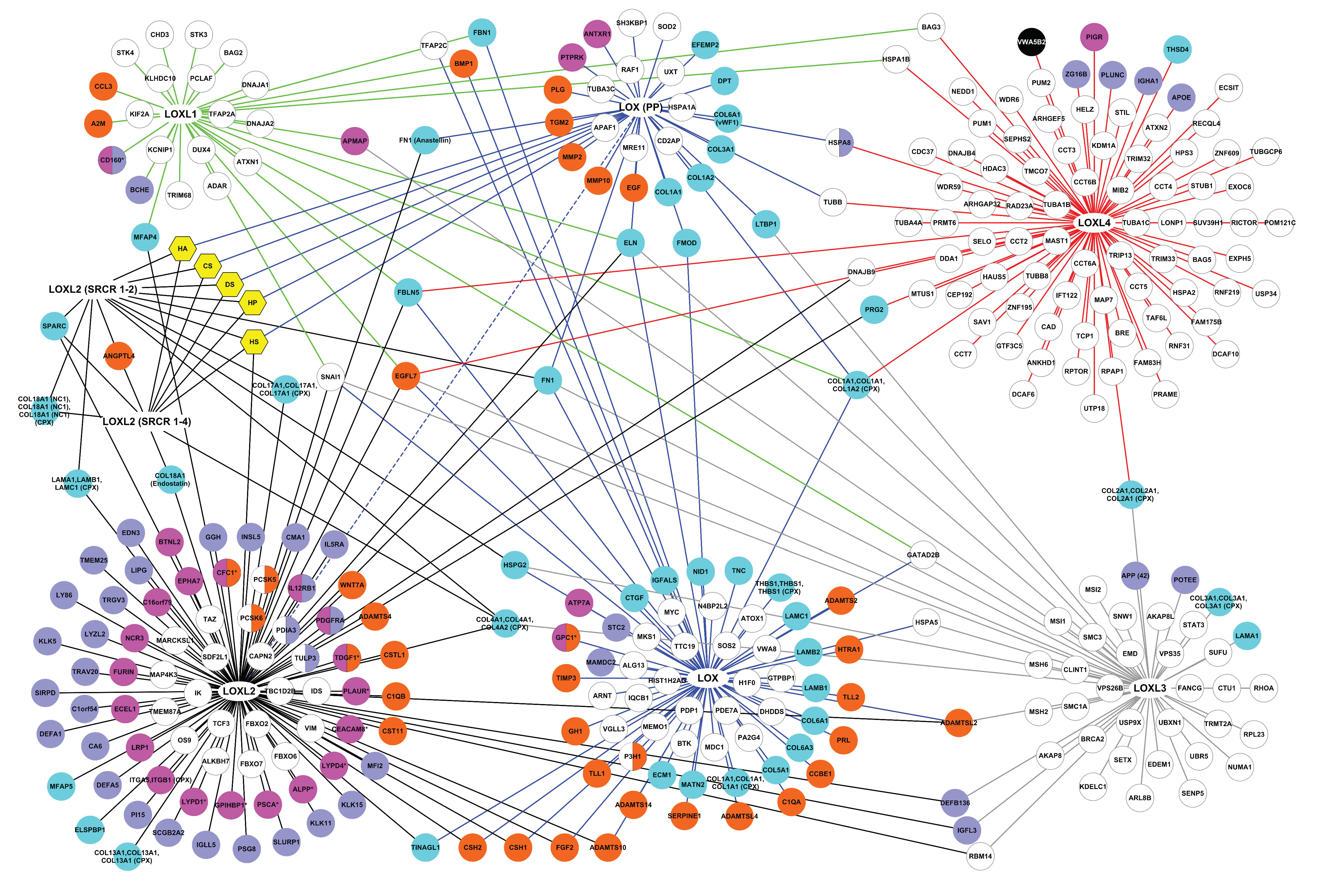
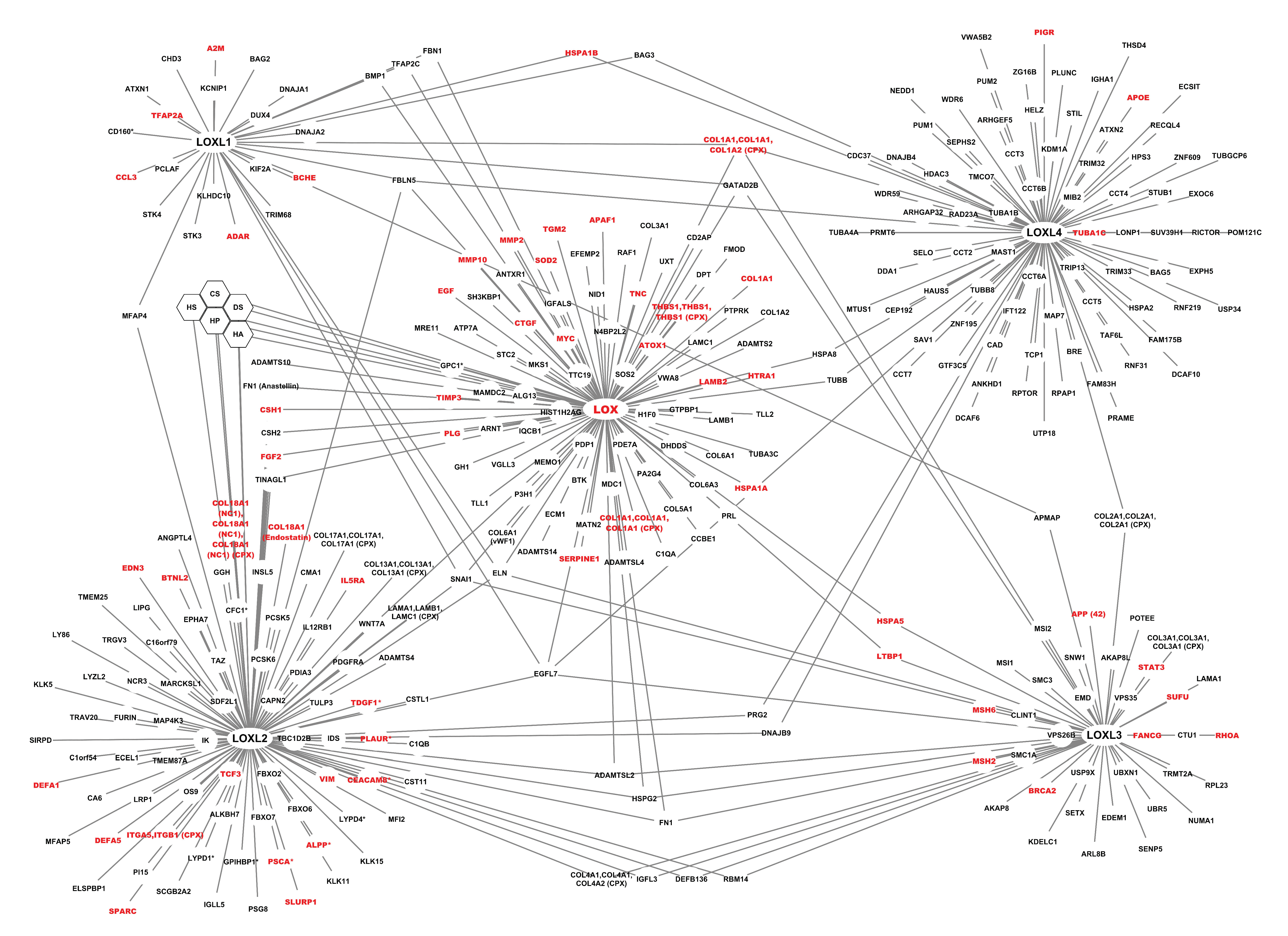
2.1.2. The Interactome of the Lysyl Oxidase Family
2.2. Analyses of the Interactome of the LOX Family
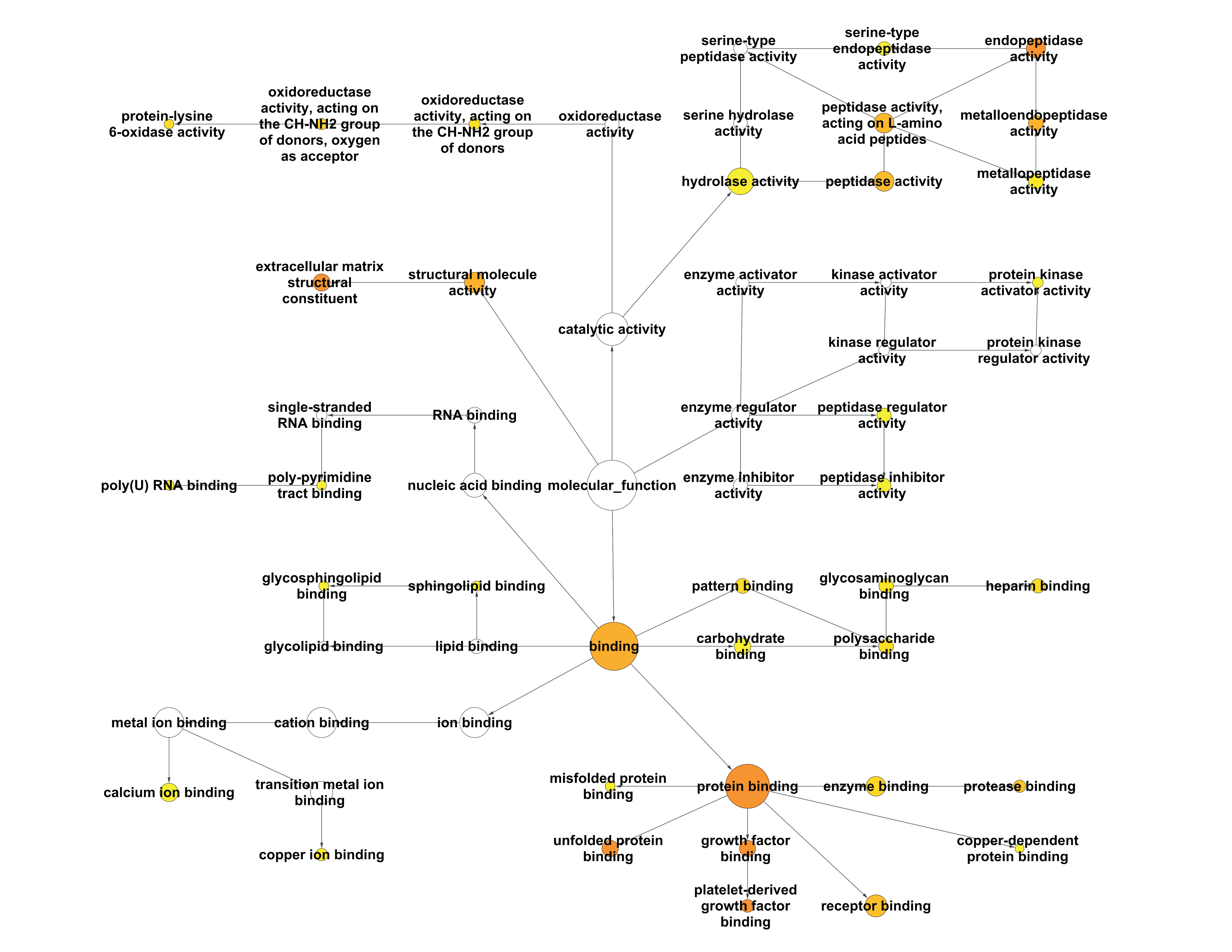
2.2.1. Enrichment Analysis of the Interactome of LOX Family Using BiNGO
2.2.2. Enrichment Analysis of the Interactome of LOX Family Using FunRich
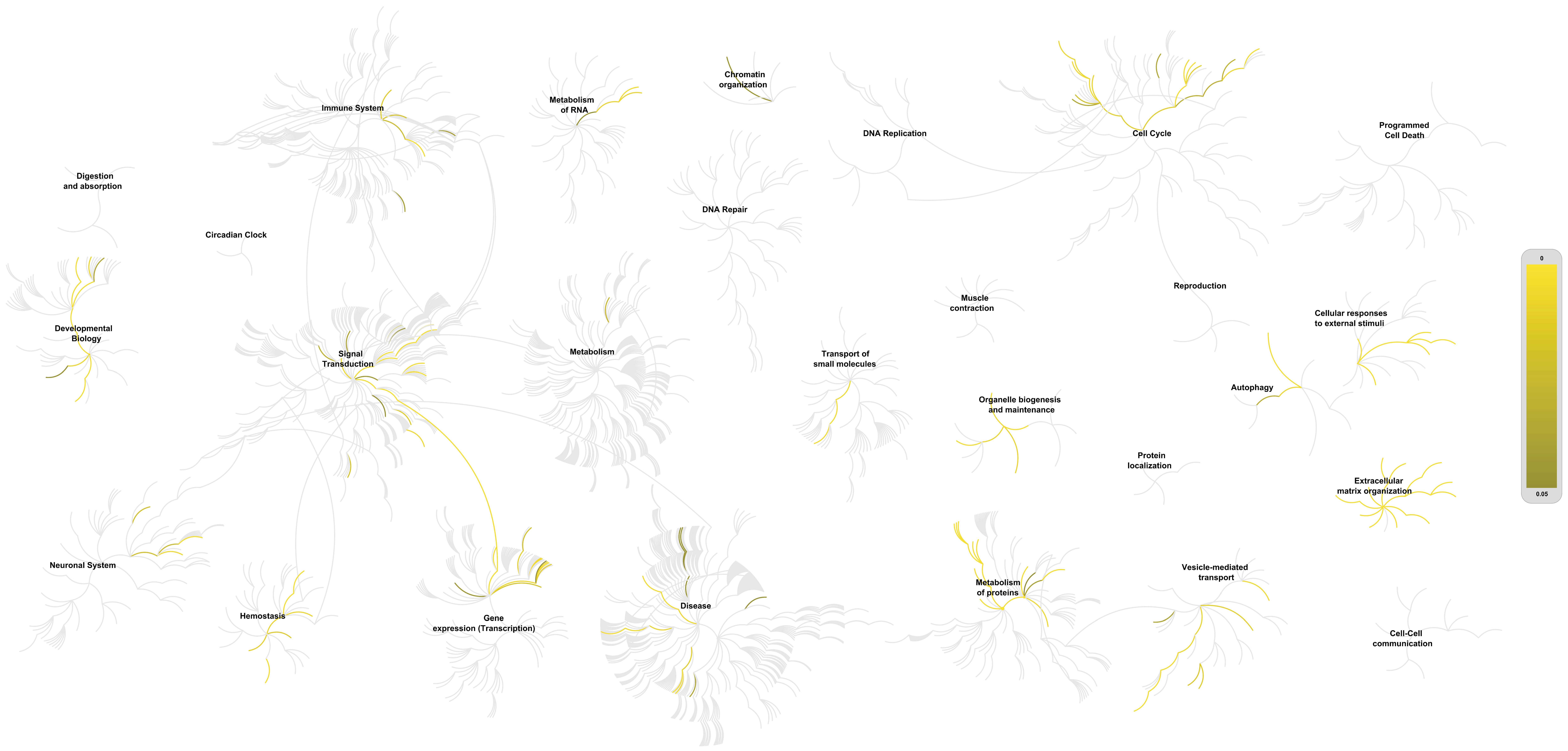
2.2.3. Pathway Analysis of the Interactome of LOX Family Using Reactome
2.3. The Interactome of the LOX Family in Cancer
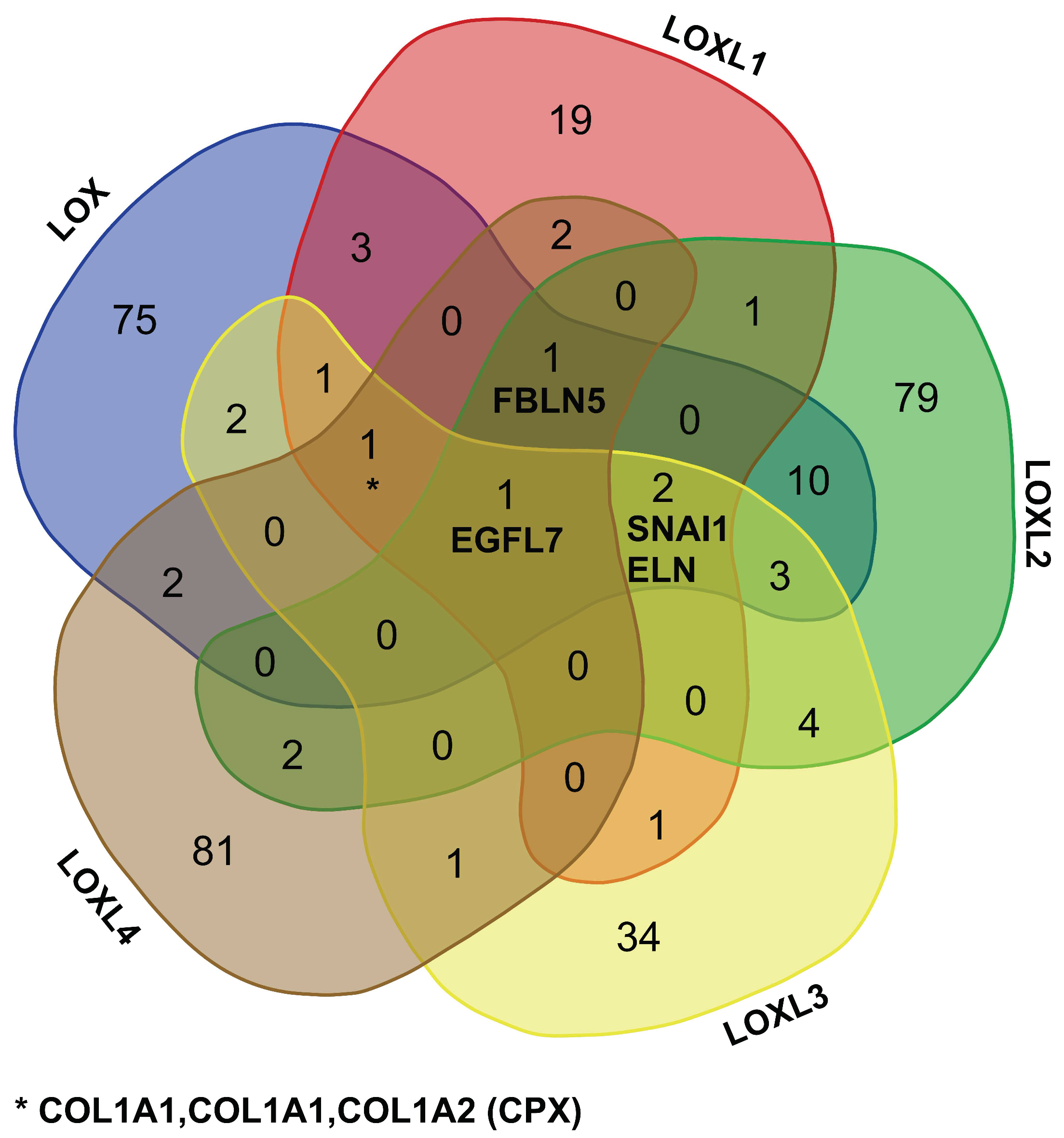
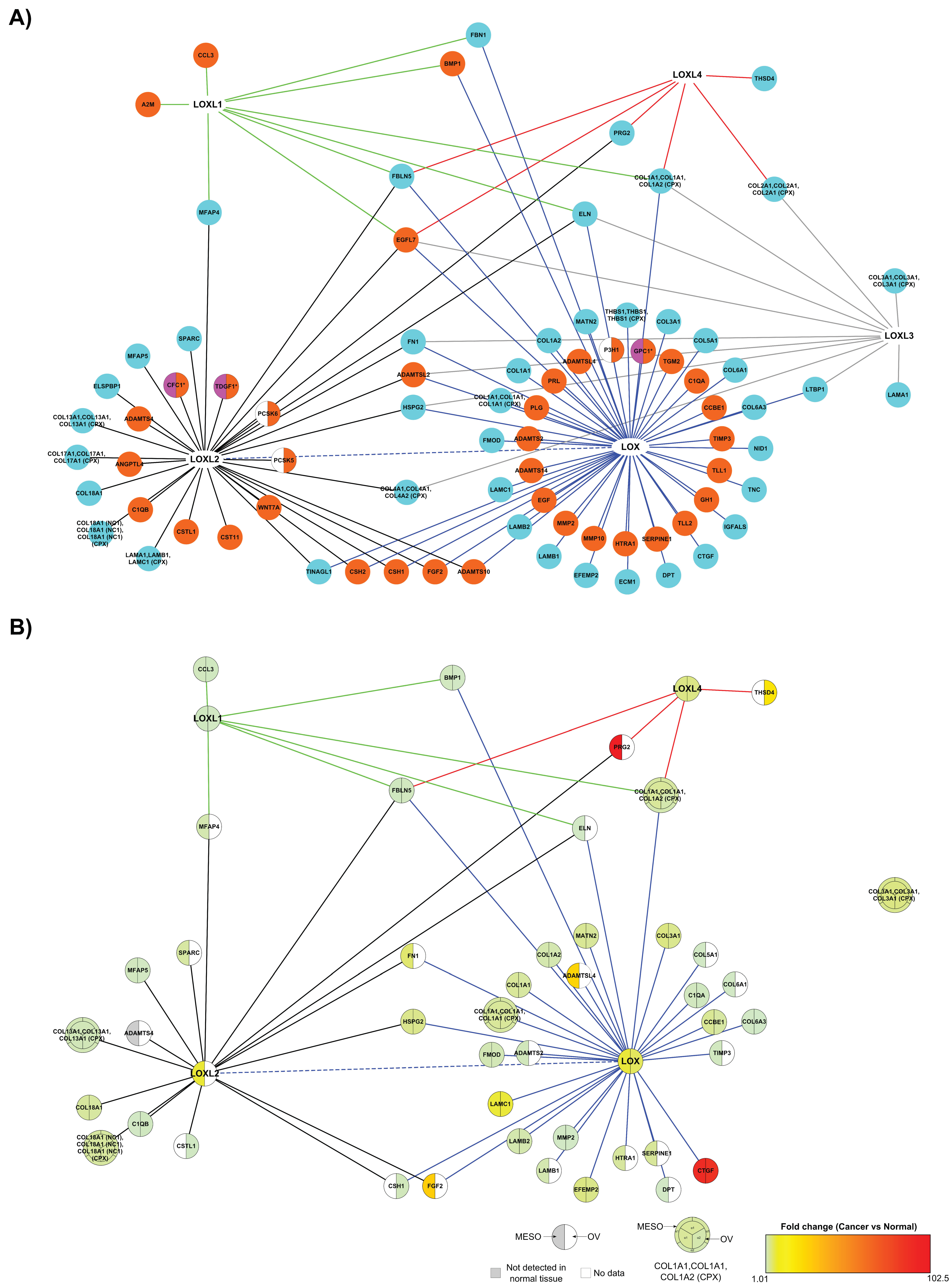
2.3.1. Comparison of the LOX Family Interactome in HEK 293 and HCT 116 Cells
2.3.2. The Matrisome Part of LOX Family Interactome in Cancer
3. Discussion
4. Materials and Methods
4.1. Surface Plasmon Resonance (SPR) Binding Assays
4.2. Bio-Layer Interferometry (BLI) Binding Assays
4.3. Querying Interaction Databases and Large-Scale Datasets
4.4. Analyses of the Interaction Network of the LOX Family
4.5. Data Visualization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pinnell, S.R.; Martin, G.R. The cross-linking of collagen and elastin: Enzymatic conversion of lysine in peptide linkage to alpha-aminoadipic-delta-semialdehyde (allysine) by an extract from bone. Proc. Natl. Acad. Sci. USA 1968, 61, 708–716. [Google Scholar] [CrossRef] [PubMed]
- Vallet, S.D.; Ricard-Blum, S. Lysyl oxidases: From enzyme activity to extracellular matrix cross-links. Essays Biochem. 2019, 63, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Greene, A.G.; Eivers, S.B.; Dervan, E.W.J.; O’Brien, C.J.; Wallace, D.M. Lysyl Oxidase Like 1: Biological roles and regulation. Exp. Eye Res. 2020, 193, 107975. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.-J.; Finney, J.; Ronnebaum, T.; Mure, M. Human lysyl oxidase-like 2. Bioorg. Chem. 2014, 57, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Laurentino, T.D.; Soares, R.D.; Marie, S.K.; Oba-Shinjo, S.M. LOXL3 Function beyond Amino Oxidase and Role in Pathologies, Including Cancer. Int. J. Mol. Sci. 2019, 20, 3587. [Google Scholar] [CrossRef] [PubMed]
- Mäki, J.M.; Tikkanen, H.; Kivirikko, K.I. Cloning and characterization of a fifth human lysyl oxidase isoenzyme: The third member of the lysyl oxidase-related subfamily with four scavenger receptor cysteine-rich domains. Matrix Biol. 2001, 20, 493–496. [Google Scholar] [CrossRef]
- Grau-Bové, X.; Ruiz-Trillo, I.; Rodriguez-Pascual, F. Origin and evolution of lysyl oxidases. Sci. Rep. 2015, 5, 10568. [Google Scholar] [CrossRef]
- Trackman, P.C. Enzymatic and non-enzymatic functions of the lysyl oxidase family in bone. Matrix Biol. 2016, 52–54, 7–18. [Google Scholar] [CrossRef]
- Bignon, M.; Pichol-Thievend, C.; Hardouin, J.; Malbouyres, M.; Bréchot, N.; Nasciutti, L.; Barret, A.; Teillon, J.; Guillon, E.; Etienne, E.; et al. Lysyl oxidase-like protein-2 regulates sprouting angiogenesis and type IV collagen assembly in the endothelial basement membrane. Blood 2011, 118, 3979–3989. [Google Scholar] [CrossRef]
- Almacellas-Rabaiget, O.; Monaco, P.; Huertas-Martinez, J.; García-Monclús, S.; Chicón-Bosch, M.; Maqueda-Marcos, S.; Fabra-Heredia, I.; Herrero-Martín, D.; Rello-Varona, S.; de Alava, E.; et al. LOXL2 promotes oncogenic progression in alveolar rhabdomyosarcoma independently of its catalytic activity. Cancer Lett. 2020, 474, 1–14. [Google Scholar] [CrossRef]
- Wei, S.; Gao, L.; Wu, C.; Qin, F.; Yuan, J. Role of the lysyl oxidase family in organ development (Review). Exp. Ther. Med. 2020, 20, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Song, Y.; Pan, S.; Chu, M.; Wang, Z.-W.; Zhu, X. Evolving roles of lysyl oxidase family in tumorigenesis and cancer therapy. Pharmacol. Ther. 2020, 107633. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wu, C.; Gao, L.; Qin, F.; Wei, Q.; Yuan, J. Lysyl oxidase family members in urological tumorigenesis and fibrosis. Oncotarget 2018, 9, 20156–20164. [Google Scholar] [CrossRef] [PubMed]
- Johnston, K.A.; Lopez, K.M. Lysyl oxidase in cancer inhibition and metastasis. Cancer Lett. 2018, 417, 174–181. [Google Scholar] [CrossRef]
- Li, R.; Wang, Y.; Zhang, X.; Feng, M.; Ma, J.; Li, J.; Yang, X.; Fang, F.; Xia, Q.; Zhang, Z.; et al. Exosome-mediated secretion of LOXL4 promotes hepatocellular carcinoma cell invasion and metastasis. Mol. Cancer 2019, 18, 18. [Google Scholar] [CrossRef]
- Palmieri, V.; Lazaris, A.; Mayer, T.Z.; Petrillo, S.K.; Alamri, H.; Rada, M.; Jarrouj, G.; Park, W.-Y.; Gao, Z.-H.; McDonald, P.P.; et al. Neutrophils expressing lysyl oxidase-like 4 protein are present in colorectal cancer liver metastases resistant to anti-angiogenic therapy. J. Pathol. 2020, 251, 213–223. [Google Scholar] [CrossRef]
- Xiao, Q.; Ge, G. Lysyl oxidase, extracellular matrix remodeling and cancer metastasis. Cancer Microenviron. 2012, 5, 261–273. [Google Scholar] [CrossRef]
- Nishioka, T.; Eustace, A.; West, C. Lysyl oxidase: From basic science to future cancer treatment. Cell Struct. Funct. 2012, 37, 75–80. [Google Scholar] [CrossRef]
- Cox, T.R.; Erler, J.T. Lysyl oxidase in colorectal cancer. Am J. Physiol. Gastrointest Liver Physiol. 2013, 305, G659–G666. [Google Scholar] [CrossRef]
- Perryman, L.; Erler, J.T. Lysyl oxidase in cancer research. Future Oncol. 2014, 10, 1709–1717. [Google Scholar] [CrossRef]
- Wu, L.; Zhu, Y. The function and mechanisms of action of LOXL2 in cancer (Review). Int. J. Mol. Med. 2015, 36, 1200–1204. [Google Scholar] [CrossRef] [PubMed]
- Añazco, C.; Delgado-López, F.; Araya, P.; González, I.; Morales, E.; Pérez-Castro, R.; Romero, J.; Rojas, A. Lysyl oxidase isoforms in gastric cancer. Biomark Med. 2016, 10, 987–998. [Google Scholar] [CrossRef] [PubMed]
- Trackman, P.C. Lysyl Oxidase Isoforms and Potential Therapeutic Opportunities for Fibrosis and Cancer. Expert Opin. Ther. Targets 2016, 20, 935–945. [Google Scholar] [CrossRef]
- Cox, T.R.; Gartland, A.; Erler, J.T. Lysyl Oxidase, a Targetable Secreted Molecule Involved in Cancer Metastasis. Cancer Res. 2016, 76, 188–192. [Google Scholar] [CrossRef] [PubMed]
- Chopra, V.; Sangarappillai, R.M.; Romero-Canelón, I.; Jones, A.M. Lysyl Oxidase Like-2 (LOXL2): An Emerging Oncology Target. Adv. Ther. 2020, 3, 1900119. [Google Scholar] [CrossRef]
- Ma, L.; Huang, C.; Wang, X.-J.; Xin, D.E.; Wang, L.-S.; Zou, Q.C.; Zhang, Y.-N.S.; Tan, M.-D.; Wang, Y.-M.; Zhao, T.C.; et al. Lysyl Oxidase 3 Is a Dual-Specificity Enzyme Involved in STAT3 Deacetylation and Deacetylimination Modulation. Mol. Cell 2017, 65, 296–309. [Google Scholar] [CrossRef] [PubMed]
- Iturbide, A.; García de Herreros, A.; Peiró, S. A new role for LOX and LOXL2 proteins in transcription regulation. FEBS J. 2015, 282, 1768–1773. [Google Scholar] [CrossRef]
- Tenti, P.; Vannucci, L. Lysyl oxidases: Linking structures and immunity in the tumor microenvironment. Cancer Immunol. Immunother. 2020, 69, 223–235. [Google Scholar] [CrossRef]
- Chitty, J.L.; Setargew, Y.F.I.; Cox, T.R. Targeting the lysyl oxidases in tumour desmoplasia. Biochem. Soc. Trans. 2019, 47, 1661–1678. [Google Scholar] [CrossRef]
- Chen, W.; Yang, A.; Jia, J.; Popov, Y.V.; Schuppan, D.; You, H. Lysyl oxidase (LOX) family members: Rationale and their potential as therapeutic targets for liver fibrosis. Hepatology 2020. [Google Scholar] [CrossRef]
- Puente, A.; Fortea, J.I.; Cabezas, J.; Arias Loste, M.T.; Iruzubieta, P.; Llerena, S.; Huelin, P.; Fábrega, E.; Crespo, J. LOXL2-A New Target in Antifibrogenic Therapy? Int. J. Mol. Sci. 2019, 20, 1634. [Google Scholar] [CrossRef] [PubMed]
- Klepfish, M.; Gross, T.; Vugman, M.; Afratis, N.A.; Havusha-Laufer, S.; Brazowski, E.; Solomonov, I.; Varol, C.; Sagi, I. LOXL2 Inhibition Paves the Way for Macrophage-Mediated Collagen Degradation in Liver Fibrosis. Front. Immunol. 2020, 11, 480. [Google Scholar] [CrossRef] [PubMed]
- Vallet, S.D.; Miele, A.E.; Uciechowska-Kaczmarzyk, U.; Liwo, A.; Duclos, B.; Samsonov, S.A.; Ricard-Blum, S. Insights into the structure and dynamics of lysyl oxidase propeptide, a flexible protein with numerous partners. Sci. Rep. 2018, 8, 11768. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Huang, P.; Wu, B.; Chen, S.; Huang, Z.; Wang, J.; Sun, H.; Wu, J.; Xie, L.; Cheng, Y.; et al. Clinical significance of LOXL4 expression and features of LOXL4-associated protein-protein interaction network in esophageal squamous cell carcinoma. Amino Acids 2019, 51, 813–828. [Google Scholar] [CrossRef] [PubMed]
- Huttlin, E.L.; Bruckner, R.J.; Navarrete-Perea, J.; Cannon, J.R.; Baltier, K.; Gebreab, F.; Gygi, M.P.; Thornock, A.; Zarraga, G.; Tam, S.; et al. Dual Proteome-scale Networks Reveal Cell-specific Remodeling of the Human Interactome. bioRxiv 2020. [Google Scholar] [CrossRef]
- Debaugny, R.E.; Skok, J.A. CTCF and CTCFL in cancer. Curr. Opin. Genet. Dev. 2020, 61, 44–52. [Google Scholar] [CrossRef]
- Chautard, E.; Ballut, L.; Thierry-Mieg, N.; Ricard-Blum, S. MatrixDB, a database focused on extracellular protein-protein and protein-carbohydrate interactions. Bioinformatics 2009, 25, 690–691. [Google Scholar] [CrossRef]
- Chautard, E.; Fatoux-Ardore, M.; Ballut, L.; Thierry-Mieg, N.; Ricard-Blum, S. MatrixDB, the extracellular matrix interaction database. Nucleic Acids Res. 2011, 39, D235–D240. [Google Scholar] [CrossRef]
- Launay, G.; Salza, R.; Multedo, D.; Thierry-Mieg, N.; Ricard-Blum, S. MatrixDB, the extracellular matrix interaction database: Updated content, a new navigator and expanded functionalities. Nucleic Acids Res. 2015, 43, D321–D327. [Google Scholar] [CrossRef]
- Clerc, O.; Deniaud, M.; Vallet, S.D.; Naba, A.; Rivet, A.; Perez, S.; Thierry-Mieg, N.; Ricard-Blum, S. MatrixDB: Integration of new data with a focus on glycosaminoglycan interactions. Nucleic Acids Res. 2019, 47, D376–D381. [Google Scholar] [CrossRef]
- Orchard, S.; Kerrien, S.; Abbani, S.; Aranda, B.; Bhate, J.; Bidwell, S.; Bridge, A.; Briganti, L.; Brinkman, F.S.L.; Brinkman, F.; et al. Protein interaction data curation: The International Molecular Exchange (IMEx) consortium. Nat. Methods 2012, 9, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Orchard, S.; Ammari, M.; Aranda, B.; Breuza, L.; Briganti, L.; Broackes-Carter, F.; Campbell, N.H.; Chavali, G.; Chen, C.; del-Toro, N.; et al. The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014, 42, D358–D363. [Google Scholar] [CrossRef] [PubMed]
- Porras, P.; Barrera, E.; Bridge, A.; del-Toro, N.; Cesareni, G.; Duesbury, M.; Hermjakob, H.; Iannuccelli, M.; Jurisica, I.; Kotlyar, M.; et al. Towards a unified open access dataset of molecular interactions. Nat. Commun. 2020, 11, 6144. [Google Scholar] [CrossRef]
- Huttlin, E.L.; Bruckner, R.J.; Paulo, J.A.; Cannon, J.R.; Ting, L.; Baltier, K.; Colby, G.; Gebreab, F.; Gygi, M.P.; Parzen, H.; et al. Architecture of the human interactome defines protein communities and disease networks. Nature 2017, 545, 505–509. [Google Scholar] [CrossRef]
- Faye, C.; Chautard, E.; Olsen, B.R.; Ricard-Blum, S. The first draft of the endostatin interaction network. J. Biol. Chem. 2009, 284, 22041–22047. [Google Scholar] [CrossRef] [PubMed]
- Salza, R.; Peysselon, F.; Chautard, E.; Faye, C.; Moschcovich, L.; Weiss, T.; Perrin-Cocon, L.; Lotteau, V.; Kessler, E.; Ricard-Blum, S. Extended interaction network of procollagen C-proteinase enhancer-1 in the extracellular matrix. Biochem. J. 2014, 457, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Nyström, A.; Bruckner-Tuderman, L. Matrix molecules and skin biology. Semin. Cell Dev. Biol. 2019, 89, 136–146. [Google Scholar] [CrossRef]
- Latvanlehto, A.; Fox, M.A.; Sormunen, R.; Tu, H.; Oikarainen, T.; Koski, A.; Naumenko, N.; Shakirzyanova, A.; Kallio, M.; Ilves, M.; et al. Muscle-derived collagen XIII regulates maturation of the skeletal neuromuscular junction. J. Neurosci. 2010, 30, 12230–12241. [Google Scholar] [CrossRef]
- Powell, A.M.; Sakuma-Oyama, Y.; Oyama, N.; Black, M.M. Collagen XVII/BP180: A collagenous transmembrane protein and component of the dermoepidermal anchoring complex. Clin. Exp. Dermatol. 2005, 30, 682–687. [Google Scholar] [CrossRef]
- Schmelzer, C.E.H.; Heinz, A.; Troilo, H.; Lockhart-Cairns, M.P.; Jowitt, T.A.; Marchand, M.F.; Bidault, L.; Bignon, M.; Hedtke, T.; Barret, A.; et al. Lysyl oxidase-like 2 (LOXL2)-mediated cross-linking of tropoelastin. FASEB J. 2019, 33, 5468–5481. [Google Scholar] [CrossRef]
- Trackman, P.C. Functional importance of lysyl oxidase family propeptide regions. J. Cell Commun. Signal 2018, 12, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Nareshkumar, R.N.; Sulochana, K.N.; Coral, K. Inhibition of angiogenesis in endothelial cells by Human Lysyl oxidase propeptide. Sci. Rep. 2018, 8, 10426. [Google Scholar] [CrossRef] [PubMed]
- Izzi, V.; Lakkala, J.; Devarajan, R.; Kääriäinen, A.; Koivunen, J.; Heljasvaara, R.; Pihlajaniemi, T. Pan-Cancer analysis of the expression and regulation of matrisome genes across 32 tumor types. Matrix Biol. Plus 2019, 1, 100004. [Google Scholar] [CrossRef]
- Moran-Jones, K.; Gloss, B.S.; Murali, R.; Chang, D.K.; Colvin, E.K.; Jones, M.D.; Yuen, S.; Howell, V.M.; Brown, L.M.; Wong, C.W.; et al. Connective tissue growth factor as a novel therapeutic target in high grade serous ovarian cancer. Oncotarget 2015, 6, 44551–44562. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, Q.; Wu, J.; Wang, J.; Shi, Y.; Liu, M. Crystal structure of human lysyl oxidase-like 2 (hLOXL2) in a precursor state. Proc. Natl. Acad. Sci. USA 2018, 115, 3828–3833. [Google Scholar] [CrossRef]
- Vallet, S.D.; Guéroult, M.; Belloy, N.; Dauchez, M.; Ricard-Blum, S. A Three-Dimensional Model of Human Lysyl Oxidase, a Cross-Linking Enzyme. ACS Omega 2019, 4, 8495–8505. [Google Scholar] [CrossRef]
- de Jong, O.G.; van Balkom, B.W.M.; Gremmels, H.; Verhaar, M.C. Exosomes from hypoxic endothelial cells have increased collagen crosslinking activity through up-regulation of lysyl oxidase-like 2. J. Cell Mol. Med. 2016, 20, 342–350. [Google Scholar] [CrossRef]
- Herum, K.M.; Lunde, I.G.; Skrbic, B.; Louch, W.E.; Hasic, A.; Boye, S.; Unger, A.; Brorson, S.-H.; Sjaastad, I.; Tønnessen, T.; et al. Syndecan-4 is a key determinant of collagen cross-linking and passive myocardial stiffness in the pressure-overloaded heart. Cardiovasc. Res. 2015, 106, 217–226. [Google Scholar] [CrossRef]
- Beishline, K.; Azizkhan-Clifford, J. Sp1 and the “hallmarks of cancer.”. FEBS J. 2015, 282, 224–258. [Google Scholar] [CrossRef]
- Jonsson, P.F.; Bates, P.A. Global topological features of cancer proteins in the human interactome. Bioinformatics 2006, 22, 2291–2297. [Google Scholar] [CrossRef]
- Boudiaf-Benmammar, C.; Cresteil, T.; Melki, R. The cytosolic chaperonin CCT/TRiC and cancer cell proliferation. PLoS ONE 2013, 8, e60895. [Google Scholar] [CrossRef] [PubMed]
- Umana-Diaz, C.; Pichol-Thievend, C.; Marchand, M.F.; Atlas, Y.; Salza, R.; Malbouyres, M.; Barret, A.; Teillon, J.; Ardidie-Robouant, C.; Ruggiero, F.; et al. Scavenger Receptor Cysteine-Rich domains of Lysyl Oxidase-Like2 regulate endothelial ECM and angiogenesis through non-catalytic scaffolding mechanisms. Matrix Biol. 2020, 88, 33–52. [Google Scholar] [CrossRef] [PubMed]
- Thomassin, L.; Werneck, C.C.; Broekelmann, T.J.; Gleyzal, C.; Hornstra, I.K.; Mecham, R.P.; Sommer, P. The Pro-regions of lysyl oxidase and lysyl oxidase-like 1 are required for deposition onto elastic fibers. J. Biol. Chem. 2005, 280, 42848–42855. [Google Scholar] [CrossRef] [PubMed]
- Bruckner-Tuderman, L.; Has, C. Disorders of the cutaneous basement membrane zone--the paradigm of epidermolysis bullosa. Matrix Biol. 2014, 33, 29–34. [Google Scholar] [CrossRef]
- Kober, K.I.; Cano, A.; Géraud, C.; Sipilä, K.; Mobasseri, S.A.; Philippeos, C.; Pisco, A.O.; Stannard, A.; Martin, A.; Salvador, F.; et al. Loxl2 is dispensable for dermal development, homeostasis and tumour stroma formation. PLoS ONE 2018, 13. [Google Scholar] [CrossRef]
- Zhang, H.; Fredericks, T.; Xiong, G.; Qi, Y.; Rychahou, P.G.; Li, J.-D.; Pihlajaniemi, T.; Xu, W.; Xu, R. Membrane associated collagen XIII promotes cancer metastasis and enhances anoikis resistance. Breast Cancer Res. 2018, 20. [Google Scholar] [CrossRef]
- Jones, V.A.; Patel, P.M.; Gibson, F.T.; Cordova, A.; Amber, K.T. The Role of Collagen XVII in Cancer: Squamous Cell Carcinoma and Beyond. Front. Oncol. 2020, 10, 352. [Google Scholar] [CrossRef]
- Amendola, P.G.; Reuten, R.; Erler, J.T. Interplay between LOX Enzymes and Integrins in the Tumor Microenvironment. Cancers 2019, 11, 729. [Google Scholar] [CrossRef]
- Laczko, R.; Csiszar, K. Lysyl Oxidase (LOX): Functional Contributions to Signaling Pathways. Biomolecules 2020, 10, 1093. [Google Scholar] [CrossRef]
- Spitaleri, A.; Mari, S.; Curnis, F.; Traversari, C.; Longhi, R.; Bordignon, C.; Corti, A.; Rizzardi, G.-P.; Musco, G. Structural basis for the interaction of isoDGR with the RGD-binding site of alphavbeta3 integrin. J. Biol. Chem. 2008, 283, 19757–19768. [Google Scholar] [CrossRef]
- Schaffner, F.; Ray, A.M.; Dontenwill, M. Integrin α5β1, the Fibronectin Receptor, as a Pertinent Therapeutic Target in Solid Tumors. Cancers 2013, 5, 27–47. [Google Scholar] [CrossRef] [PubMed]
- Vallet, S.D.; Deddens, L.; Vonarburg, A.; Salza, R.; Faye, C.; Aranyos, A.; Thierry-Mieg, N.; Ricard-Blum, S. Strategies for Building Protein–Glycosaminoglycan Interaction Networks Combining SPRi, SPR, and BLI. In Handbook of Surface Plasmon Resonance, 2nd ed.; The Royal Society of Chemistry: London, UK, 2017; pp. 398–414. ISBN 978-1-78262-730-2. [Google Scholar]
- Pathan, M.; Keerthikumar, S.; Ang, C.-S.; Gangoda, L.; Quek, C.Y.J.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef] [PubMed]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium the Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [CrossRef]
- UniProt Consortium UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [CrossRef]
- Naba, A.; Clauser, K.R.; Ding, H.; Whittaker, C.A.; Carr, S.A.; Hynes, R.O. The extracellular matrix: Tools and insights for the “omics” era. Matrix Biol. 2016, 49, 10–24. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vallet, S.D.; Berthollier, C.; Salza, R.; Muller, L.; Ricard-Blum, S. The Interactome of Cancer-Related Lysyl Oxidase and Lysyl Oxidase-Like Proteins. Cancers 2021, 13, 71. https://doi.org/10.3390/cancers13010071
Vallet SD, Berthollier C, Salza R, Muller L, Ricard-Blum S. The Interactome of Cancer-Related Lysyl Oxidase and Lysyl Oxidase-Like Proteins. Cancers. 2021; 13(1):71. https://doi.org/10.3390/cancers13010071
Chicago/Turabian StyleVallet, Sylvain D., Coline Berthollier, Romain Salza, Laurent Muller, and Sylvie Ricard-Blum. 2021. "The Interactome of Cancer-Related Lysyl Oxidase and Lysyl Oxidase-Like Proteins" Cancers 13, no. 1: 71. https://doi.org/10.3390/cancers13010071
APA StyleVallet, S. D., Berthollier, C., Salza, R., Muller, L., & Ricard-Blum, S. (2021). The Interactome of Cancer-Related Lysyl Oxidase and Lysyl Oxidase-Like Proteins. Cancers, 13(1), 71. https://doi.org/10.3390/cancers13010071




