In Vitro Spectroscopy-Based Profiling of Urothelial Carcinoma: A Fourier Transform Infrared and Raman Imaging Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
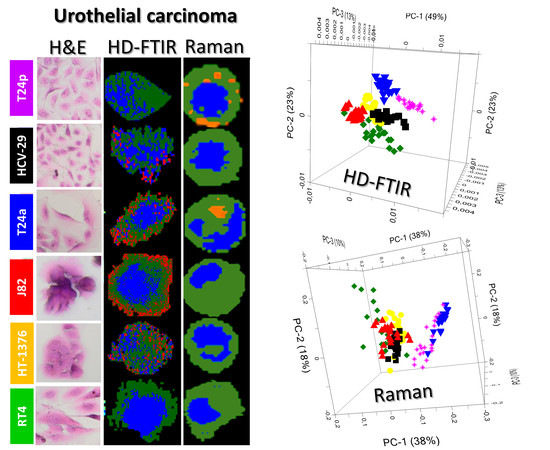
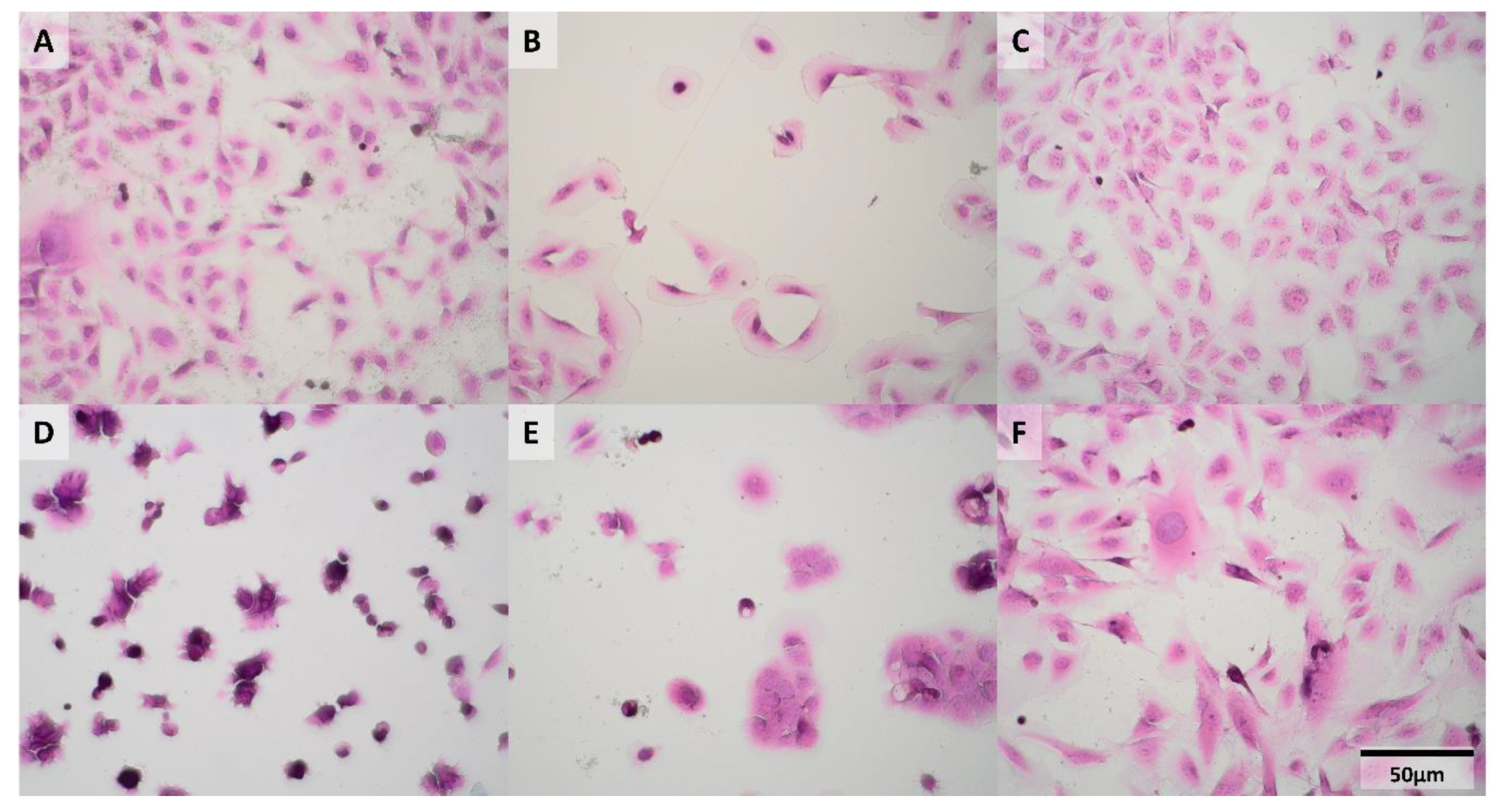
2.1. Morphology of Cells
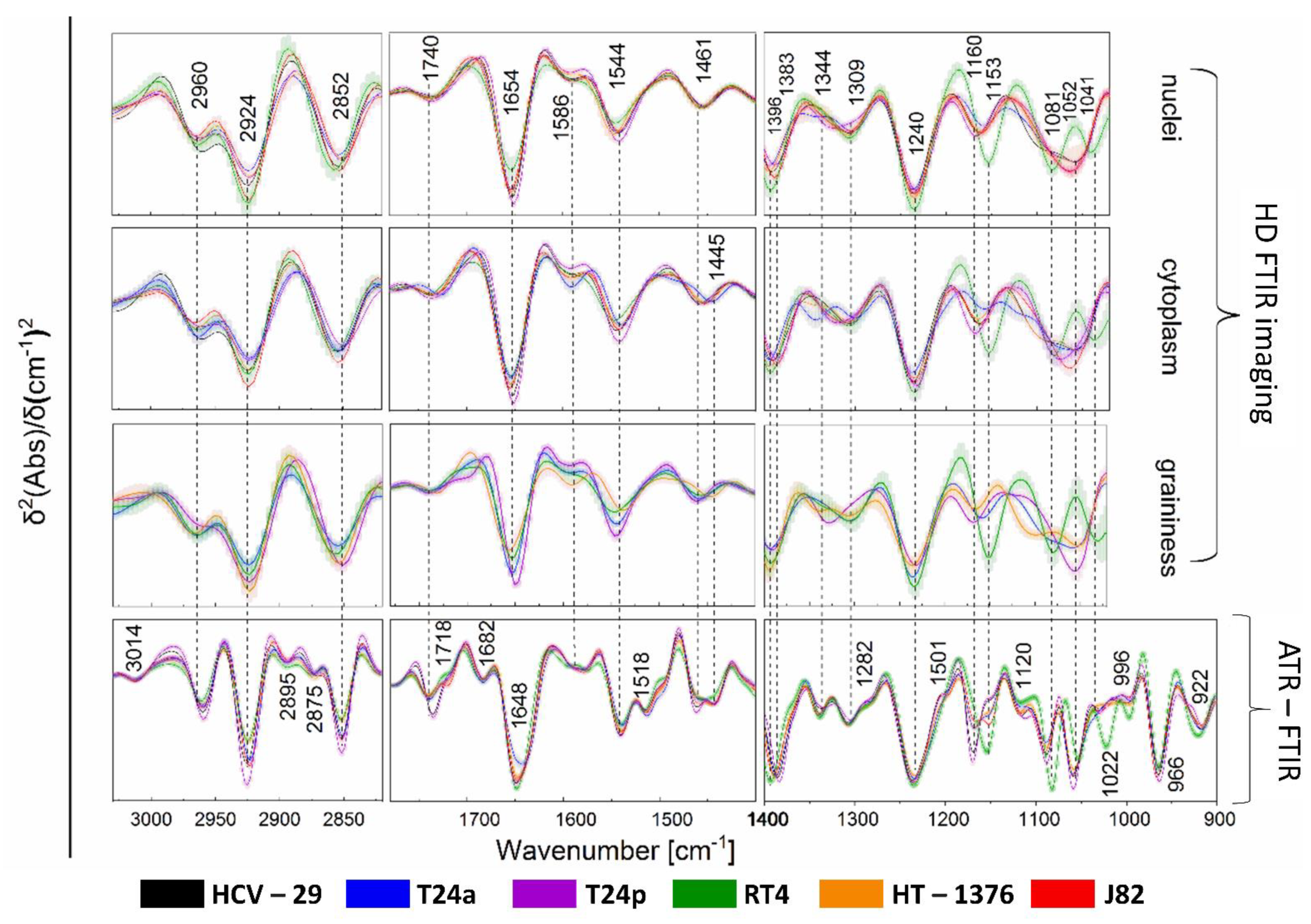
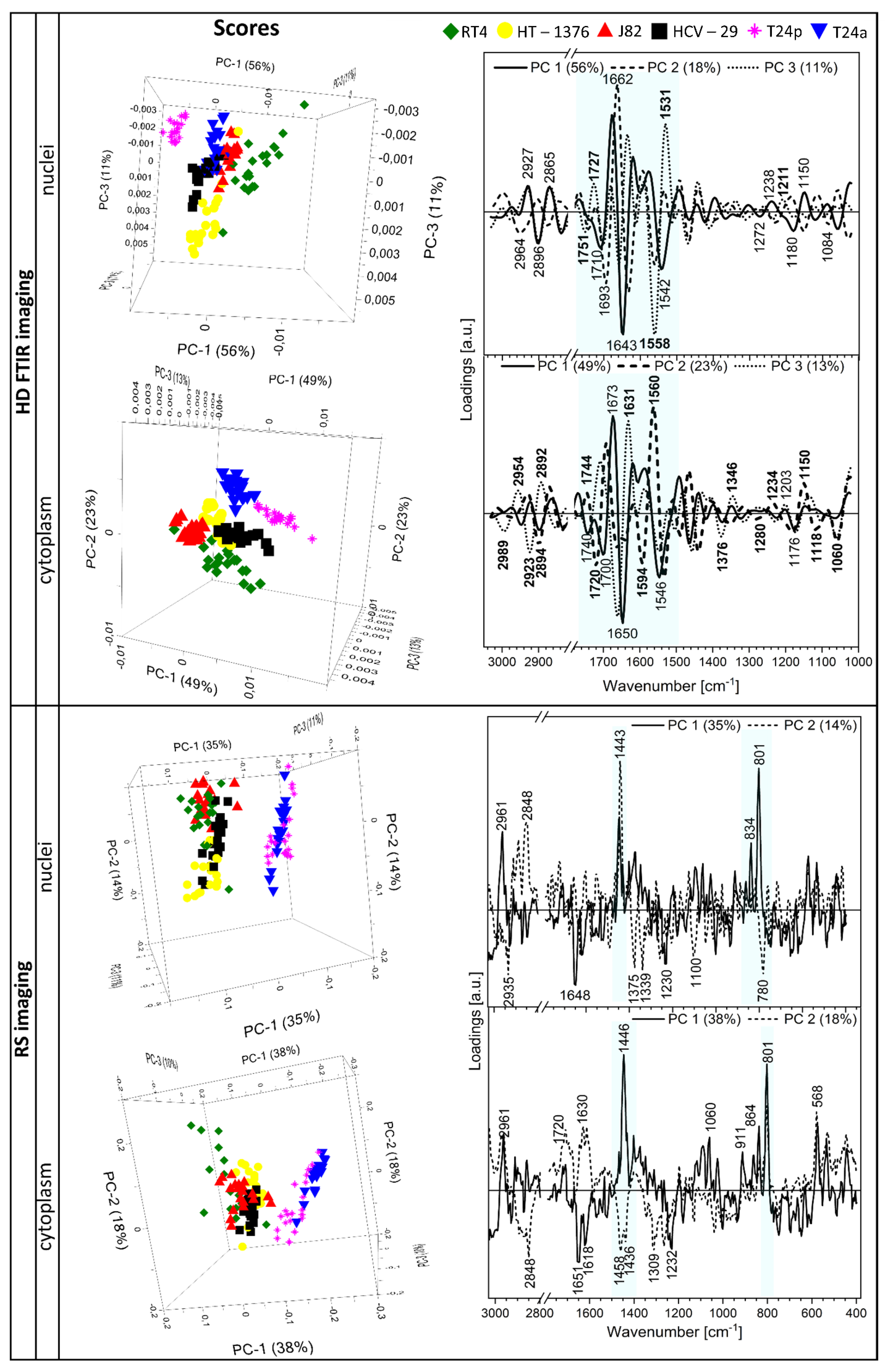
2.2. FTIR-Based Biocomposition of BC Cells
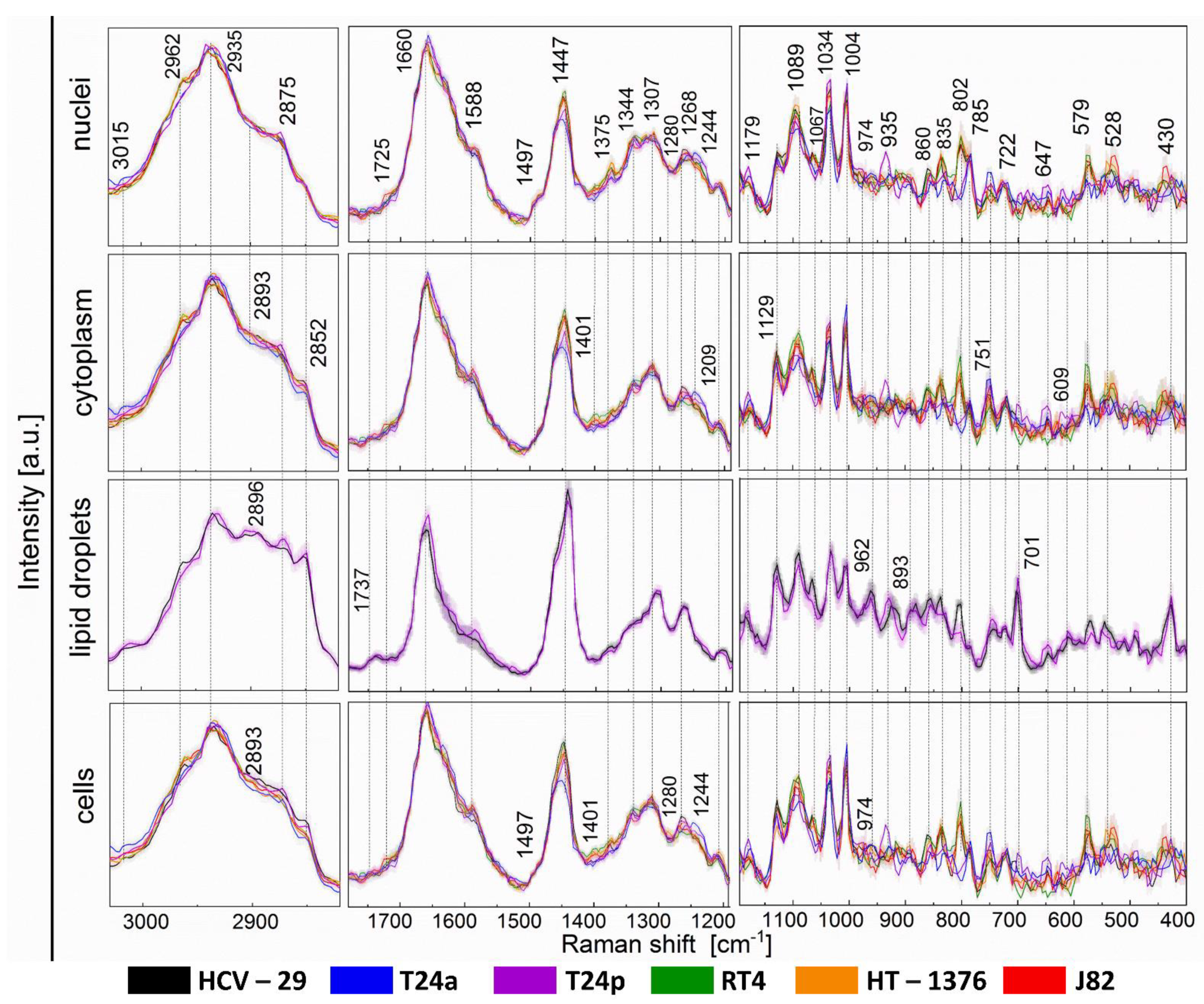
2.3. Raman-Based Biocomposition of BC Cells
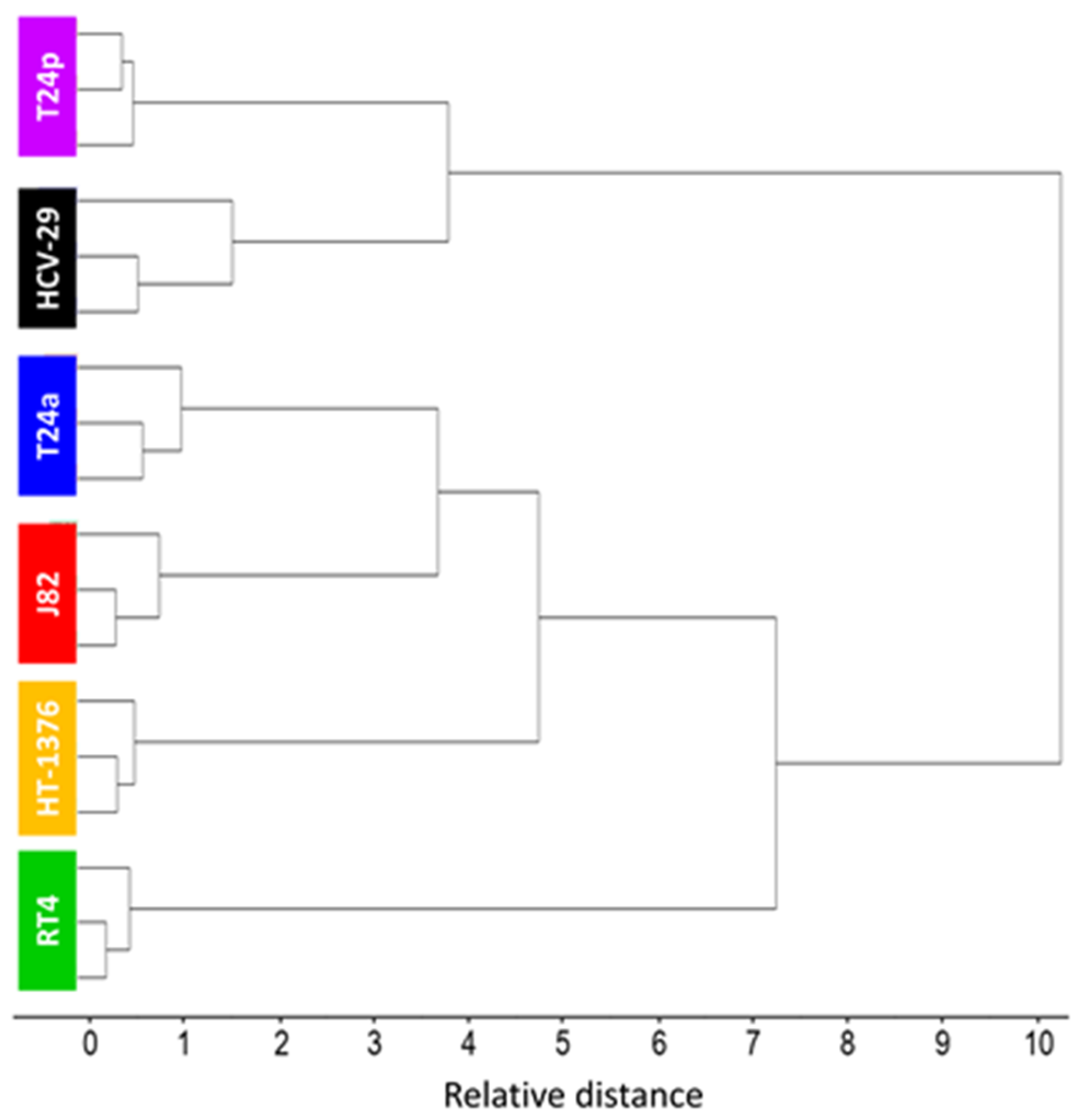
2.4. In Search of a Spectroscopic Model of Bladder Cancer Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Sample Preparation for Spectroscopic Measurements
4.2. FTIR Spectroscopy, Spectral Pre-Processing and Analysis
4.3. Raman Imaging, Spectral Pre-Processing and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, C.C.; Czerniak, B. Bladder cancer in the genomic era. Arch. Pathol. Lab. Med. 2019, 143, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Millis, S.Z.; Bryant, D.; Basu, G.; Bender, R.; Vranic, S.; Gatalica, Z.; Vogelzang, N.J. Molecular profiling of infiltrating urothelial carcinoma of bladder and nonbladder origin. Clin. Genitourin. Cancer 2015, 13, e37–e49. [Google Scholar] [CrossRef] [PubMed]
- Ward, D.G.; Gordon, N.S.; Boucher, R.H.; Pirrie, S.J.; Baxter, L.; Ott, S.; Silcock, L.; Whalley, C.M.; Stockton, J.D.; Beggs, A.D.; et al. Targeted deep sequencing of urothelial bladder cancers and associated urinary DNA: A 23-gene panel with utility for non-invasive diagnosis and risk stratification. BJU Int. 2019, 124, 532–544. [Google Scholar] [CrossRef] [PubMed]
- Garczyk, S.; Ortiz-Brüchle, N.; Schneider, U.; Lurje, I.; Guricova, K.; Gaisa, N.T.; Lorsy, E.; Lindemann-Docter, K.; Heidenreich, A.; Knüchel, R. Next-generation sequencing reveals potential predictive biomarkers and targets of therapy for urothelial carcinoma in situ of the urinary bladder. Am. J. Pathol. 2020, 190, 323–332. [Google Scholar] [CrossRef]
- Inamura, K. Bladder cancer: New insights into its molecular pathology. Cancers 2018, 10, 100. [Google Scholar] [CrossRef]
- Zuiverloon, T.C.M.; De Jong, F.C.; Costello, J.C.; Theodorescu, D. Systematic review: Characteristics and preclinical uses of bladder cancer cell lines. Bl. Cancer 2018, 4, 169–183. [Google Scholar] [CrossRef]
- Yoshida, T.; Sopko, N.A.; Kates, M.; Liu, X.; Joice, G.; McConkey, D.J.; Bivalacqua, T.J. Impact of spheroid culture on molecular and functional characteristics of bladder cancer cell lines. Oncol. Lett. 2019, 18, 4923–4929. [Google Scholar] [CrossRef]
- Lee, S.H.; Hu, W.; Matulay, J.T.; Silva, M.V.; Owczarek, T.B.; Kim, K.; Chua, C.W.; Barlow, L.M.J.; Kandoth, C.; Williams, A.B.; et al. Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell 2018, 173, 515–528.e17. [Google Scholar] [CrossRef]
- Rodrigues, D.; Jerónimo, C.; Henrique, R.; Belo, L.; De Lourdes Bastos, M.; De Pinho, P.G.; Carvalho, M. Biomarkers in bladder cancer: A metabolomic approach using in vitro and ex vivo model systems. Int. J. Cancer 2016, 139, 256–268. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, X.; Liu, X.; He, L.; Zhang, W.; Wang, Y.; Sun, W.; Ji, Z. Investigation of the urinary metabolic variations and the application in bladder cancer biomarker discovery. Int. J. Cancer 2018, 143, 408–418. [Google Scholar] [CrossRef]
- Diem, M.; Mazur, A.; Lenau, K.; Schubert, J.; Bird, B.; Miljković, M.; Krafft, C.; Popp, J. Molecular pathology via IR and Raman spectral imaging. J. Biophoton. 2013, 6, 855–886. [Google Scholar] [CrossRef] [PubMed]
- Wiercigroch, E.; Staniszewska-Slezak, E.; Szkaradek, K.; Wojcik, T.; Ozaki, Y.; Baranska, M.; Malek, K. FT-IR spectroscopic imaging of endothelial cells response to tumor necrosis factor-α: To follow markers of inflammation using standard and high-magnification resolution. Anal. Chem. 2018, 90, 3727–3736. [Google Scholar] [CrossRef] [PubMed]
- Harvey, T.J.; Hughes, C.; Ward, A.D.; Faria, E.C.; Henderson, A.; Clarke, N.W.; Brown, M.D.; Snook, R.D.; Gardner, P. Classification of fixed urological cells using Raman tweezers. J. Biophoton. 2009, 2, 47–69. [Google Scholar] [CrossRef] [PubMed]
- Kerr, L.T.; Adams, A.; O’Dea, S.; Domijan, K.; Cullen, I.; Hennelly, B.M. Classification of bladder cancer cell lines using Raman spectroscopy: A comparison of excitation wavelength, sample substrate and statistical algorithms. Biophoton. Photon. Solut. Better Health Care IV 2014, 9129, 91290E. [Google Scholar]
- Kerr, L.T.; Lynn, T.M.; Cullen, I.M.; Daly, P.J.; Shah, N.; O’Dea, S.; Malkin, A.; Hennelly, B.M. Methodologies for bladder cancer detection with Raman based urine cytology. Anal. Methods 2016, 8, 4991–5000. [Google Scholar] [CrossRef]
- Jen, C.P.; Huang, C.T.; Chen, Y.S.; Kuo, C.T.; Wang, H.C. Diagnosis of human bladder cancer cells at different stages using multispectral imaging microscopy. IEEE J. Sel. Top. Quantum Electron. 2014, 20, 81–88. [Google Scholar] [CrossRef]
- Canetta, E.; Mazilu, M.; De Luca, A.C.; Carruthers, A.E.; Dholakia, K.; Neilson, S.; Sargeant, H.; Briscoe, T.; Herrington, C.S.; Riches, A.C. Modulated Raman spectroscopy for enhanced identification of bladder tumor cells in urine samples. J. Biomed. Opt. 2011, 16, 037002. [Google Scholar] [CrossRef]
- Gok, S.; Aydin, O.Z.; Sural, Y.S.; Zorlu, F.; Bayol, U.; Severcan, F. Bladder cancer diagnosis from bladder wash by Fourier transform infrared spectroscopy as a novel test for tumor recurrence. J. Biophotonics 2016, 9, 967–975. [Google Scholar] [CrossRef]
- Yosef, H.K.; Krauß, S.D.; Lechtonen, T.; Jütte, H.; Tannapfel, A.; Käfferlein, H.U.; Brüning, T.; Roghmann, F.; Noldus, J.; Mosig, A.; et al. Noninvasive diagnosis of high-grade urothelial carcinoma in urine by Raman spectral imaging. Anal. Chem. 2017, 89, 6893–6899. [Google Scholar] [CrossRef]
- Hughes, C.; Iqbal-Wahid, J.; Brown, M.; Shanks, J.H.; Eustace, A.; Denley, H.; Hoskin, P.J.; West, C.; Clarke, N.W.; Gardner, P. FTIR microspectroscopy of selected rare diverse sub-variants of carcinoma of the urinary bladder. J. Biophotonics 2013, 6, 73–87. [Google Scholar] [CrossRef]
- Shapiro, A.; Gofrit, O.N.; Pizov, G.; Cohen, J.K.; Maier, J. Raman molecular imaging: A novel spectroscopic technique for diagnosis of bladder cancer in urine specimens. Eur. Urol. 2011, 59, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Crow, P.; Uff, J.S.; Farmer, J.A.; Wright, M.P.; Stone, N. The use of Raman spectroscopy to identify and characterize transitional cell carcinoma in vitro. BJU Int. 2004, 93, 1232–1236. [Google Scholar] [CrossRef] [PubMed]
- Stone, N.; Hart Prieto, M.C.; Crow, P.; Uff, J.; Ritchie, A.W. The use of Raman spectroscopy to provide an estimation of the gross biochemistry associated with urological pathologies. Anal. Bioanal. Chem. 2007, 387, 1657–1668. [Google Scholar] [CrossRef] [PubMed]
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix Metalloproteinases: Regulators of the tumor. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef] [PubMed]
- Krauß, S.D.; Yosef, H.K.; Lechtonen, T.; Jütte, H.; Tannapfel, A.; Käfferlein, H.U.; Brüning, T.; Roghmann, F.; Noldus, J.; El-Mashtoly, S.F.; et al. Integrating spatial, morphological, and textural information for improved cell type differentiation using Raman microscopy. J. Chemom. 2018, 32, 1–11. [Google Scholar] [CrossRef]
- Bongiovanni, G.A.; Eynard, A.R.; Calderón, R.O. Altered lipid profile and changes in uroplakin properties of rat urothelial plasma membrane with diets of different lipid composition. Mol. Cell. Biochem. 2005, 271, 69–75. [Google Scholar] [CrossRef]
- Bohnert, M. Tether Me, Tether me not—Dynamic organelle contact sites in metabolic rewiring. Dev. Cell 2020, 54, 212–225. [Google Scholar] [CrossRef]
- Sahu, R.K.; Argov, S.; Salman, A.; Huleihel, M.; Grossman, N.; Hammody, Z.; Kapelushnik, J.; Mordechai, S. Characteristic absorbance of nucleic acids in the Mid-IR region as possible common biomarkers for diagnosis of malignancy. Technol. Cancer Res. Treat. 2004, 3, 629–638. [Google Scholar]
- Staniszewska, E.; Malek, K.; Baranska, M. Rapid approach to analyze biochemical variation in rat organs by ATR FTIR spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 118, 981–986. [Google Scholar] [CrossRef]
- Banyay, M.; Sarkar, M.; Gräslund, A. A library of IR bands of nucleic acids in solution. Biophys. Chem. 2003, 104, 477–488. [Google Scholar]
- Whelan, D.R.; Bambery, K.R.; Heraud, P.; Tobin, M.J.; Diem, M.; McNaughton, D.; Wood, B.R. Monitoring the reversible B to A-like transition of DNA in eukaryotic cells using Fourier transform infrared spectroscopy. Nucleic Acids Res. 2011, 39, 5439–5448. [Google Scholar]
- Brandenburg, K.; Seydel, U. Infrared spectroscopy of glycolipids. Chem. Phys. Lipids 1998, 96, 23–40. [Google Scholar] [CrossRef]
- Bik, E.; Dorosz, A.; Mateuszuk, L.; Baranska, M.; Majzner, K. Fixed versus live endothelial cells: The effect of glutaraldehyde fixation manifested by characteristic bands on the Raman spectra of cells. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 240, 118460. [Google Scholar]
- Majzner, K.; Chlopicki, S.; Baranska, M. Lipid droplets formation in human endothelial cells in response to polyunsaturated fatty acids and 1-methyl-nicotinamide (MNA); confocal Raman imaging and fluorescence microscopy studies. J. Biophotonics 2016, 9, 396–405. [Google Scholar]
- Prescott, B.; Steinmetz, W.; Thomas, G.J. Characterization of DNA structures by laser Raman spectroscopy. Biopolymers 1984, 23, 235–256. [Google Scholar] [CrossRef]
- Brazhe, N.A.; Treiman, M.; Brazhe, A.R.; Find, N.L.; Maksimov, G.V.; Sosnovtseva, O.V. Mapping of Redox State of Mitochondrial Cytochromes in Live Cardiomyocytes Using Raman Microspectroscopy. PLoS ONE 2012, 7, e41990. [Google Scholar]
- Nawaz, H.; Garcia, A.; Meade, A.D.; Lyng, F.M.; Byrne, H.J. Raman micro spectroscopy study of the interaction of vincristine with A549 cells supported by expression analysis of bcl-2 protein. Analyst 2013, 138, 6177–6184. [Google Scholar] [CrossRef]
- Schulze, A.; Harris, A.L. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature 2012, 491, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Massari, F.; Ciccarese, C.; Santoni, M.; Iacovelli, R.; Mazzucchelli, R.; Piva, F.; Scarpelli, M.; Berardi, R.; Tortora, G.; Lopez-Beltran, A.; et al. Metabolic phenotype of bladder cancer. Cancer Treat. Rev. 2016, 45, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Gerresheim, G.K.; Roeb, E.; Michel, A.M.; Niepmann, M. Oxidative phosphorylation, reminiscent of the Warburg effect in cancer cells. Cells 2019, 8, 1410. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy Levy, J.M.; Thorburn, A. Autophagy in cancer: Moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ. 2020, 27, 843–857. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.; Li, J.; Lee, S.Y.; Lee, H.J.; Shao, T.; Song, B.; Cheng, L.; Masterson, T.A.; Liu, X.; Ratliff, T.L.; et al. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab. 2014, 19, 393–406. [Google Scholar] [CrossRef]
- Su, Y.; Zhao, B.; Zhou, L.; Zhang, Z.; Shen, Y.; Lv, H.; AlQudsy, L.H.H.; Shang, P. Ferroptosis, a novel pharmacological mechanism of anti-cancer drugs. Cancer Lett. 2020, 483, 127–136. [Google Scholar] [CrossRef]
- Lasch, P. CytospecTM. A Matlab Based Application for Infrared Imaging. Available online: http://www.cytospec.com (accessed on 15 February 2010).
- Bassan, P.; Kohler, A.; Martens, H.; Lee, J.; Byrne, H.J.; Dumas, P.; Gazi, E.; Brown, M.; Clarke, N.; Gardner, P. Resonant Mie Scattering (RMieS) correction of infrared spectra from highly scattering biological samples. Analyst 2010, 135, 268–277. [Google Scholar] [CrossRef]

| Cell Line | HCV-29 | T24a and T24p | RT4 | HT-1376 | J82 |
|---|---|---|---|---|---|
| Stage | na | Ta | T1 | T2 | T3 |
| Grade | na | high | low | high | high |
| Type | na | subtype dependent | luminal | mixed | basal |
| Mutations | na | HRAS, TERT, TP53 | TERT, CDKN2A, TP53 | FGFR3, PIK3CA, TERT, TP53 | PIK3CA, TERT, TP53 |
| Biocomponent | HCV-29 | Ta—T24a | Ta—T24p | T1-RT4 | T2—HT-1376 | T3—J82 |
|---|---|---|---|---|---|---|
| Nucleus | ||||||
| Lipid content (FTIR) | ↑ | ↑ | ↑ | ↑↑ | ↑ | ↑↑ |
| Elongated acyl chains (FTIR) | ↑ | ↑↑ | ↑↑ | ↓ | ↑↑ | ↑↑ |
| Free fatty acids (FTIR) | ↑ | |||||
| Phospholipids (RS) | ↑ | ↑ | ↑ | ↑ | ↑↑ | ↑↑ |
| Cholesterol and its esters (RS) | ↑↑ | ↑↑ | ||||
| Protein content (FTIR) | ↑ | |||||
| Tyr (RS) | ↑ | ↑ | ||||
| Uroplakin | ↑ | |||||
| Protein crosslinking (RS) | ↑ | ↓ | ↑↑ | ↓ | ↑ | ↑ |
| S-S bridge (RS) | ↑ | ↑ | ↑↑ | ↑↑ | ||
| DNA content (FTIR) | ↑ | |||||
| Glycogen (FTIR) | ↑ | |||||
| Carbohydrates (RS) | ↑ | ↑↑ | ↑↑ | ↑↑ | ||
| Cytoplasm | ||||||
| Lipid content (FTIR/RS) | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ |
| Esterified fatty acids and cholesterol (FTIR) | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ |
| Unsaturated fatty acids (RS) | ↑↑ | ↑ | ↑↑ | ↑ | ||
| Phospholipids (RS) | ↑ | ↑↑ | ↑ | ↑ | ||
| Protein content (FTIR) | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ |
| Cytochromes (RS) | ↑ | ↑↑ | ↑↑ | ↓ | ↑ | ↑ |
| S-S bridge (RS) | ↑ | ↑↑ | ↑↑ | ↑↑ | ||
| Carbohydrates (RS) | ↑ | ↑↑ | ↑ | ↑ | ||
| Graininess | ||||||
| Lipid content (FTIR) | ↑ | ↑ | ↑↑ | ↑↑ | ||
| Esterified fatty acids and cholesterol (FTIR) | ↑ | ↑ | ↑ | |||
| Protein content (FTIR) | ↑↑ | ↑↑ | ↑ | ↑ | ||
| Glycogen (FTIR) | ↑ | |||||
| Sugar moieties (FTIR; 1052 cm−1) | ↑ | ↑↑ | ↑ | |||
| Nucleic acids (FTIR) | ↑ | ↑ | ↑↑ | ↑ | ||
| RNA (FTIR) | ↑ | |||||
| Lipid droplets | ||||||
| Unsaturated fatty acids (RS) | ↑ | ↑↑ | ||||
| Gauche conformation of acyl chains (RS) | ↑ | |||||
| Phospholipids (RS) | ↑ | |||||
| Cholesterol and its esters (RS) | ↑ | ↑↑ | ||||
| S-S bridge (RS) | ↑↑ | ↑ | ||||
| Cytochromes (RS) | ↑ | ↑↑ | ||||
| Carbohydrates (RS) | ↑↑ | ↑ | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kujdowicz, M.; Placha, W.; Mech, B.; Chrabaszcz, K.; Okoń, K.; Malek, K. In Vitro Spectroscopy-Based Profiling of Urothelial Carcinoma: A Fourier Transform Infrared and Raman Imaging Study. Cancers 2021, 13, 123. https://doi.org/10.3390/cancers13010123
Kujdowicz M, Placha W, Mech B, Chrabaszcz K, Okoń K, Malek K. In Vitro Spectroscopy-Based Profiling of Urothelial Carcinoma: A Fourier Transform Infrared and Raman Imaging Study. Cancers. 2021; 13(1):123. https://doi.org/10.3390/cancers13010123
Chicago/Turabian StyleKujdowicz, Monika, Wojciech Placha, Brygida Mech, Karolina Chrabaszcz, Krzysztof Okoń, and Kamilla Malek. 2021. "In Vitro Spectroscopy-Based Profiling of Urothelial Carcinoma: A Fourier Transform Infrared and Raman Imaging Study" Cancers 13, no. 1: 123. https://doi.org/10.3390/cancers13010123
APA StyleKujdowicz, M., Placha, W., Mech, B., Chrabaszcz, K., Okoń, K., & Malek, K. (2021). In Vitro Spectroscopy-Based Profiling of Urothelial Carcinoma: A Fourier Transform Infrared and Raman Imaging Study. Cancers, 13(1), 123. https://doi.org/10.3390/cancers13010123