TGFBR3L—An Uncharacterised Pituitary Specific Membrane Protein Detected in the Gonadotroph Cells in Non-Neoplastic and Tumour Tissue
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
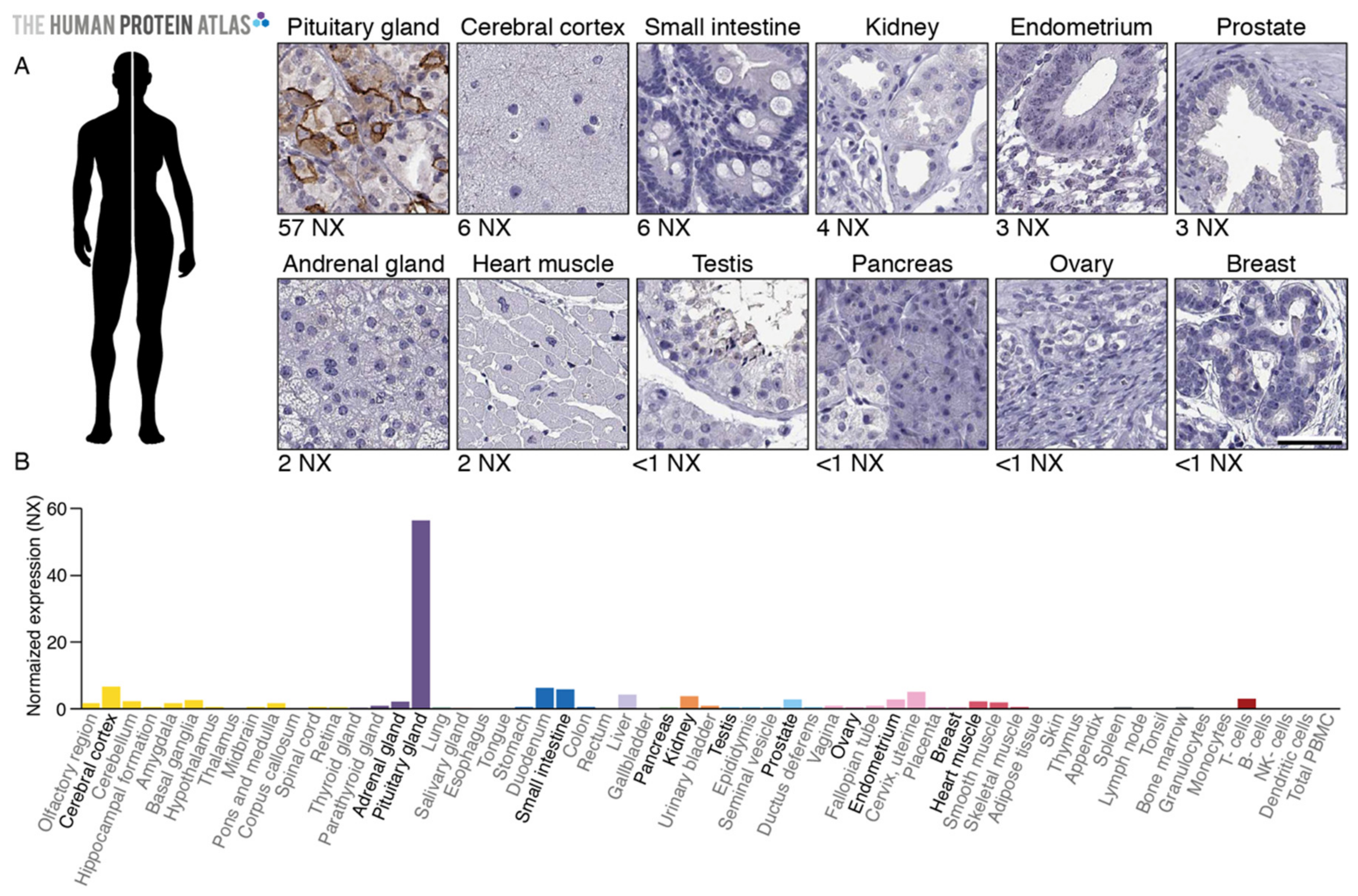
2.1. TGFBR3L Tissue Profiling in the Human Protein Atlas
2.2. TGFBR3L is Selectively Detected in a Subset of Gonadotroph Cells in the Non-Neoplastic Anterior Pituitary Gland
2.3. TGFBR3L is Only Detected in Gonadotroph Tumours
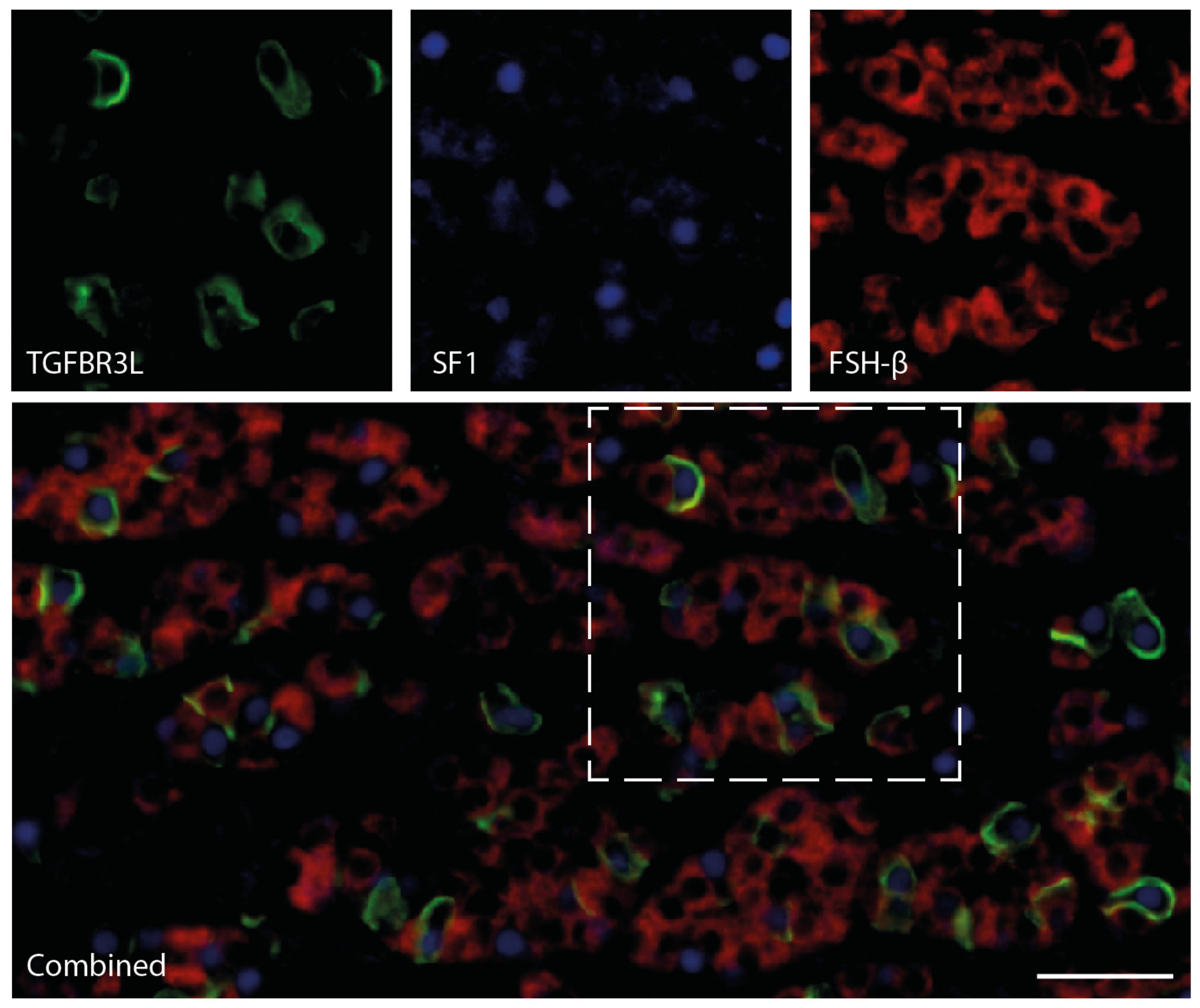
2.4. TGFBR3L and FSH/LH Staining
2.5. TGFBR3L Expression Shows an Inverse Correlation with E-Cadherin and Oestrogen Receptor β
3. Discussion
4. Materials and Methods
4.1. Patient Cohort
4.2. Immunohistochemical Characterisation of the PitNET Cohort
4.3. Whole Tumour Sections and Non-Neoplastic Anterior Pituitary Gland
4.4. TGFBR3L Antibody
4.5. TGFBR3L Chromogen Immunohistochemistry
4.6. TGFBR3L Multiplex Fluorescence Immunohistochemistry
4.7. Real Time-qPCR
4.8. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scully, K.M.; Rosenfeld, M.G. Pituitary development: Regulatory codes in mammalian organogenesis. Science 2002, 295, 2231–2235. [Google Scholar] [CrossRef] [PubMed]
- Asa, S.L.; Casar-Borota, O.; Chanson, P.; Delgrange, E.; Earls, P.; Ezzat, S.; Grossman, A.; Ikeda, H.; Inoshita, N.; Karavitaki, N.; et al. From pituitary adenoma to pituitary neuroendocrine tumor (PitNET): An International Pituitary Pathology Club proposal. Endocr. Relat. Cancer 2017, 24, C5–C8. [Google Scholar] [CrossRef] [PubMed]
- Osamura, R.Y.G.A.; Korbonits, M. Pituitary Adenomas, 4th ed.; IARC Press: Lyon, France, 2017. [Google Scholar]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G.; et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020, 21, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chetty, R.; Serra, S. Nuclear E-cadherin immunoexpression: From biology to potential applications in diagnostic pathology. Adv. Anat. Pathol. 2008, 15, 234–240. [Google Scholar] [CrossRef]
- Elston, M.S.; Gill, A.J.; Conaglen, J.V.; Clarkson, A.; Cook, R.J.; Little, N.S.; Robinson, B.G.; Clifton-Bligh, R.J.; McDonald, K.L. Nuclear accumulation of e-cadherin correlates with loss of cytoplasmic membrane staining and invasion in pituitary adenomas. J. Clin. Endocrinol. Metab. 2009, 94, 1436–1442. [Google Scholar] [CrossRef]
- Fougner, S.L.; Lekva, T.; Borota, O.C.; Hald, J.K.; Bollerslev, J.; Berg, J.P. The expression of E-cadherin in somatotroph pituitary adenomas is related to tumor size, invasiveness, and somatostatin analog response. J. Clin. Endocrinol Metab. 2010, 95, 2334–2342. [Google Scholar] [CrossRef]
- Evang, J.A.; Berg, J.P.; Casar-Borota, O.; Lekva, T.; Kringen, M.K.; Ramm-Pettersen, J.; Bollerslev, J. Reduced levels of E-cadherin correlate with progression of corticotroph pituitary tumours. Clin. Endocrinol. 2011, 75, 811–818. [Google Scholar] [CrossRef]
- Oesterreich, S.; Deng, W.; Jiang, S.; Cui, X.; Ivanova, M.; Schiff, R.; Kang, K.; Hadsell, D.L.; Behrens, J.; Lee, A.V. Estrogen-mediated down-regulation of E-cadherin in breast cancer cells. Cancer Res. 2003, 63, 5203–5208. [Google Scholar] [PubMed]
- Zhou, W.; Song, Y.; Xu, H.; Zhou, K.; Zhang, W.; Chen, J.; Qin, M.; Yi, H.; Gustafsson, J.-A.; Yang, H.; et al. In nonfunctional pituitary adenomas, estrogen receptors and slug contribute to development of invasiveness. J. Clin. Endocrinol. Metab. 2011, 96, E1237–E1245. [Google Scholar] [CrossRef] [PubMed]
- Oystese, K.A.; Casar-Borota, O.; Normann, K.R.; Zucknick, M.; Berg, J.P.; Bollerslev, J. Estrogen Receptor alpha, a Sex-Dependent Predictor of Aggressiveness in Nonfunctioning Pituitary Adenomas: SSTR and Sex Hormone Receptor Distribution in NFPA. J. Clin. Endocrinol. Metab. 2017, 102, 3581–3590. [Google Scholar] [CrossRef] [PubMed]
- Casar-Borota, O.; Heck, A.; Schulz, S.; Nesland, J.M.; Ramm-Pettersen, J.; Lekva, T.; Alafuzoff, I.; Bollerslev, J. Expression of SSTR2a, but not of SSTRs 1, 3, or 5 in somatotroph adenomas assessed by monoclonal antibodies was reduced by octreotide and correlated with the acute and long-term effects of octreotide. J. Clin. Endocrinol. Metab. 2013, 98, E1730–E1739. [Google Scholar] [CrossRef]
- Lee, M.; Lupp, A.; Mendoza, N.; Martin, N.; Beschorner, R.; Honegger, J.; Schlegel, J.; Shively, T.; Pulz, E.; Schulz, S.; et al. SSTR3 is a putative target for the medical treatment of gonadotroph adenomas of the pituitary. Endocr. Relat. Cancer. 2015, 22, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- The Human Protein Atlas. Available online: http://www.proteinatlas.org (accessed on 20 October 2020).
- UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [PubMed]
- Fagerberg, L.; Jonasson, K.; von Heijne, G.; Uhlen, M.; Berglund, L. Prediction of the human membrane proteome. Proteomics 2010, 10, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Gray, K.A.; Yates, B.; Seal, R.L.; Wright, M.W.; Bruford, E.A. Genenames.org: The HGNC resources in 2015. Nucleic Acids Res. 2015, 43, D1079–D1085. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K.A.; Gray, P.C.; Blount, A.L.; MacConell, L.A.; Wiater, E.; Bilezikjian, L.M.; Vale, W. Betaglycan binds inhibin and can mediate functional antagonism of activin signalling. Nature 2000, 404, 411–414. [Google Scholar] [CrossRef]
- Chapman, S.C.; Woodruff, T.K. Betaglycan localization in the female rat pituitary: Implications for the regulation of follicle-stimulating hormone by inhibin. Endocrinology 2003, 144, 5640–5649. [Google Scholar] [CrossRef][Green Version]
- MacConell, L.A.; Leal, A.M.; Vale, W.W. The distribution of betaglycan protein and mRNA in rat brain, pituitary, and gonads: Implications for a role for betaglycan in inhibin-mediated reproductive functions. Endocrinology 2002, 143, 1066–1075. [Google Scholar] [CrossRef][Green Version]
- Uhlen, M.; Bandrowski, A.; Carr, S.; Edwards, A.; Ellenberg, J.; Lundberg, M.U.E.; Rimm, D.L.; Rodriguez, H.; Hiltke, H.R.T.; Snyder, M.; et al. A proposal for validation of antibodies. Nat. Methods 2016, 13, 823–827. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Karlsson, M.J.; Zhong, W.; Tebani, A.; Pou, C.; Mikes, J.; Lakshmikanth, T.; Forsström, B.; Edfors, F.; Odeberg, J.; et al. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science 2019, 366, eaax9198. [Google Scholar] [CrossRef] [PubMed]
- Cheung, L.Y.M.; George, A.S.; McGee, S.R.; Daly, A.Z.; Brinkmeier, M.L.; Ellsworth, B.S.; Camper, S.A. Single-Cell RNA Sequencing Reveals Novel Markers of Male Pituitary Stem Cells and Hormone-Producing Cell Types. Endocrinology 2018, 159, 3910–3924. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, P.A.; Smiljanic, K.; Prévide, R.M.; Iben, J.R.; Li, T.; Rokic, M.B.; Sherman, A.; Coon, S.L.; Stojilkovic, S.S. Cell Type- and Sex-Dependent Transcriptome Profiles of Rat Anterior Pituitary Cells. Front. Endocrinol. (Lausanne) 2019, 10, 623. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cui, Y.; Ma, X.; Yong, J.; Yan, L.; Yang, M.; Ren, J.; Tang, F.; Wen, L.; Qiao, J. Single-cell transcriptomics identifies divergent developmental lineage trajectories during human pituitary development. Nat. Commun. 2020, 11, 5275. [Google Scholar] [CrossRef]
- Sjöstedt, E.; Zhong, W.; Fagerberg, L.; Karlsson, M.J.; Mitsios, N.; Adori, C.; Oksvold, P.; Edfors, F.; Limiszewska, A.; Hikmet, F.; et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 2020, 367, eaay5947. [Google Scholar] [CrossRef]
- Mollard, P.; Hodson, D.J.; Lafont, C.; Rizzoti, K.; Drouin, J. A tridimensional view of pituitary development and function. Trends Endocrinol. Metab. 2012, 23, 261–269. [Google Scholar] [CrossRef]
- Agustsson, T.T.; Baldvinsdottir, T.; Jonasson, J.G.; Olafsdottir, E.; Steinthorsdottir, V.; Sigurdsson, G.; Thorsson, A.V.; Carroll, P.V.; Korbonits, M.; Benediktsson, R. The epidemiology of pituitary adenomas in Iceland, 1955–2012: A nationwide population-based study. Eur. J. Endocrinol. 2015, 173, 655–664. [Google Scholar] [CrossRef]
- Tjörnstrand, A.; Gunnarsson, K.; Evert, M.; Holmberg, E.; Ragnarsson, O.; Rosén, T.; Nyström, H.F. The incidence rate of pituitary adenomas in western Sweden for the period 2001–2011. Eur. J. Endocrinol. 2014, 171, 519–526. [Google Scholar] [CrossRef]
- Manojlovic-Gacic, E.; Engstrom, B.E.; Casar-Borota, O. Histopathological classification of non-functioning pituitary neuroendocrine tumors. Pituitary 2018, 21, 119–129. [Google Scholar] [CrossRef]
- Mete, O.; Kefeli, M.; Caliskan, S.; Asa, S.L. GATA3 immunoreactivity expands the transcription factor profile of pituitary neuroendocrine tumors. Mod. Pathol. 2019, 32, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Nishioka, H.; Inoshita, N.; Mete, O.; Asa, S.L.; Hayashi, K.; Takeshita, A.; Fukuhara, N.; Yamaguchi-Okada, M.; Takeuchi, Y.; Yamada, S. The Complementary Role of Transcription Factors in the Accurate Diagnosis of Clinically Nonfunctioning Pituitary Adenomas. Endocr. Pathol. 2015, 26, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Manojlovic-Gacic, E.; Bollerslev, J.; Casar-Borota, O. Invited Review: Pathology of pituitary neuroendocrine tumours: Present status, modern diagnostic approach, controversies and future perspectives from a neuropathological and clinical standpoint. Neuropathol. Appl. Neurobiol. 2020, 46, 89–110. [Google Scholar] [CrossRef] [PubMed]
- Shaw, N.D.; Histed, S.N.; Srouji, S.S.; Yang, J.; Lee, H.; Hall, J.E. Estrogen negative feedback on gonadotropin secretion: Evidence for a direct pituitary effect in women. J. Clin. Endocrinol. Metab. 2010, 95, 1955–1961. [Google Scholar] [CrossRef]
- Baratta, M.; West, L.A.; Turzillo, A.M.; Nett, T.M. Activin modulates differential effects of estradiol on synthesis and secretion of follicle-stimulating hormone in ovine pituitary cells. Biol. Reprod. 2001, 64, 714–719. [Google Scholar] [CrossRef][Green Version]
- Heaney, A.P.; Fernando, M.; Melmed, S. Functional role of estrogen in pituitary tumor pathogenesis. J. Clin. Investig. 2002, 109, 277–283. [Google Scholar] [CrossRef]
- Haddad, G.; Penabad, J.L.; Bashey, H.M.; Asa, S.L.; Gennarelli, A.T.; Cirullo, R.; Snyder, P.J. Expression of activin/inhibin subunit messenger ribonucleic acids by gonadotroph adenomas. J. Clin. Endocrinol. Metab. 1994, 79, 1399–1403. [Google Scholar]
- Uccella, S.; La Rosa , S.; Genasetti, A. Localization of inhibin/activin subunits in normal pituitary and in pituitary adenomas. Pituitary 2000, 3, 131–139. [Google Scholar] [CrossRef]
- Kolnes, A.J.; Øystese, K.A.B.; Olarescu, N.C.; Ringstad, G.; Berg-Johnsen, J.; Casar-Borota, O.; Bollerslev, J.; Jørgensen, A.P. FSH Levels Are Related to E-cadherin Expression and Subcellular Location in Nonfunctioning Pituitary Tumors. J. Clin. Endocrinol. Metab. 2020, 105, 2587–2594. [Google Scholar] [CrossRef]
- Jin, Y.-Q.; Wang, H.; Han, W.; Lu, J.; Chu, P.; Han, S.; Ni, X.; Ning, B.; Yu, D.; Guo, Y. Single nucleotide polymorphism rs11669203 in TGFBR3L is associated with the risk of neuroblastoma in a Chinese population. Tumour. Biol. 2016, 37, 3739–3747. [Google Scholar] [CrossRef]
- Albihn, A.; Johnsen, J.I.; Henriksson, M.A. MYC in oncogenesis and as a target for cancer therapies. Adv. Cancer Res. 2010, 107, 163–224. [Google Scholar] [PubMed]
- Alaminos, M.; Mora, J.; Cheung, N.-K.V.; Smith, A.; Qin, J.; Chen, L.; Gerald, W.L. Genome-wide analysis of gene expression associated with MYCN in human neuroblastoma. Cancer Res. 2003, 63, 4538–4546. [Google Scholar] [PubMed]
- Dzieran, J.; Rodriguez-Garcia, A.; Westermark, U.K.; Henley, A.B.; Sánchez, E.E.; Träger, C.; Johansson, H.J.; Lehtiö, J.; Arsenian-Henriksson, M. MYCN-amplified neuroblastoma maintains an aggressive and undifferentiated phenotype by deregulation of estrogen and NGF signaling. Proc. Natl. Acad. Sci. USA 2018, 115, E1229–E1238. [Google Scholar] [CrossRef]
- Casar-Borota, O.; Oystese, K.A.; Sundstrom, M.; Melchior, L.; Popovic, V. A high-throughput analysis of the IDH1(R132H) protein expression in pituitary adenomas. Pituitary 2016, 19, 407–414. [Google Scholar] [CrossRef]
- Casar-Borota, O.; Fougner, S.L.; Bollerslev, J.; Nesland, J.M. KIT protein expression and mutational status of KIT gene in pituitary adenomas. Virchows Arch. 2012, 460, 171–181. [Google Scholar] [CrossRef]
- Antibody Details. Available online: https://www.proteinatlas.org/ENSG00000260001-TGFBR3L/antibody (accessed on 20 October 2020).
- Ehrenberg, A.J.; Morales, D.O.; Piergies, A.M.; Li, S.H.; Tejedor, J.S.; Mladinov, M.; Mulder, J.; Grinberg, L.T. A manual multiplex immunofluorescence method for investigating neurodegenerative diseases. J. Neurosci. Methods 2020, 339, 108708. [Google Scholar] [CrossRef] [PubMed]
- Falch, C.M.; Sundaram, A.Y.; Øystese, K.A.; Normann, K.R.; Lekva, T.; Silamikelis, I.; Eieland, A.K.; Andersen, M.S.; Bollerslev, J.; Olarescu, N.C. Gene expression profiling of fast- and slow-growing non-functioning gonadotroph pituitary adenomas. Eur. J. Endocrinol. 2018, 178, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Normann, K.R.; Øystese, K.A.B.; Berg, J.P.; Lekva, T.; Berg-Johnsen, J.; Bollerslev, J.; Olarescu, N.C. Selection and validation of reliable reference genes for RT-qPCR analysis in a large cohort of pituitary adenomas. Mol. Cell Endocrinol. 2016, 437, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Oystese, K.A.B.; Berg, J.P.; Normann, K.R.; Zucknick, M.; Casar-Borota, O.; Bollerslev, J. The role of E and N-cadherin in the postoperative course of gonadotroph pituitary tumours. Endocrine 2018, 62, 351–360. [Google Scholar] [CrossRef]

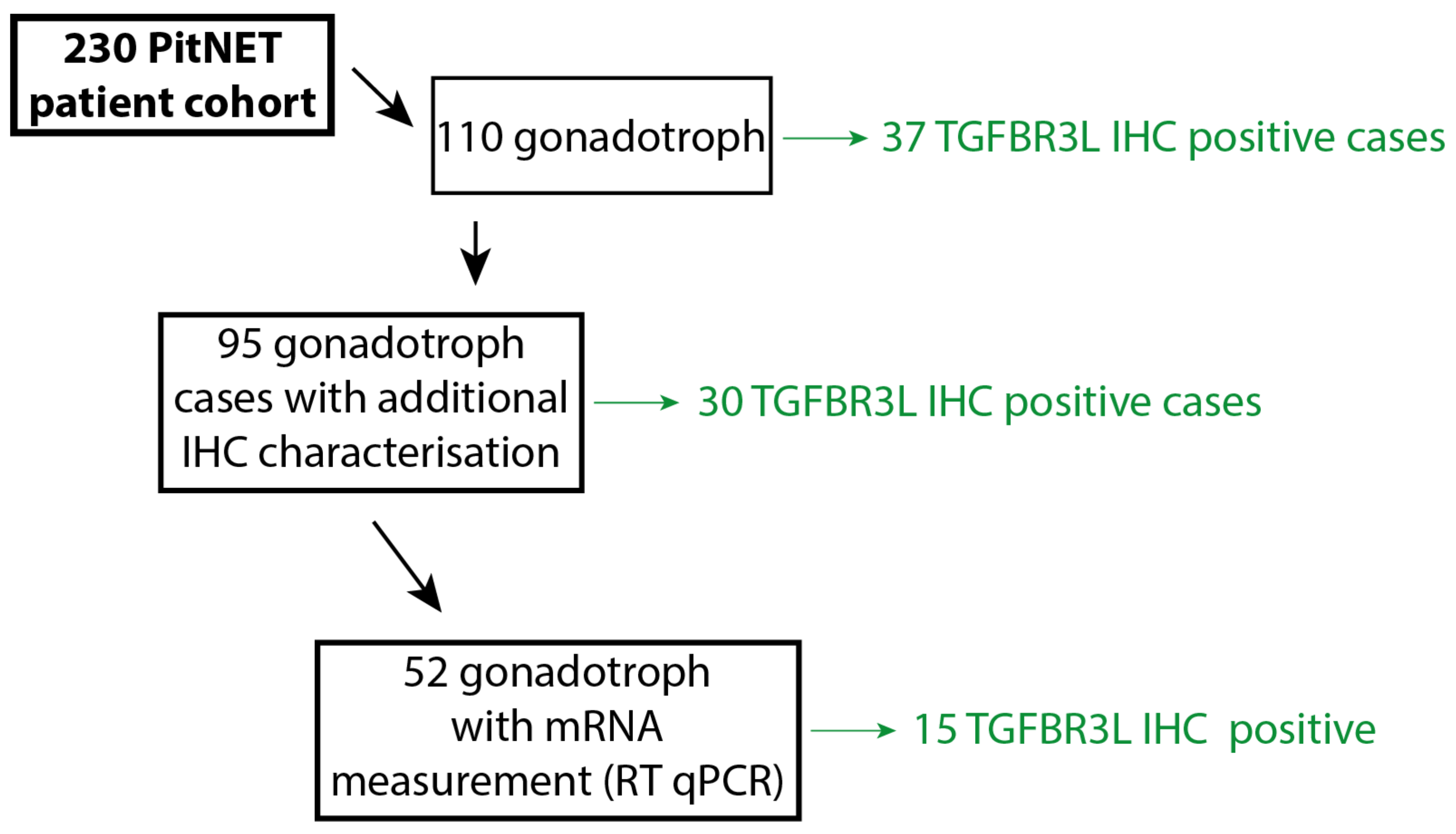
| Tumour Type | Number | IHC Subtype | Clinical Phenotype | TGFBR3L Positive |
|---|---|---|---|---|
| Gonadotroph | 110 | SF1; FSH/LH | NF-PitNET 1 | 37 |
| Corticotroph | 21 | T-Pit; ACTH | Cushing/Nelson | 0 |
| 25 | T-Pit; ACTH | NF-PitNET 1 | 0 | |
| Somatotroph | 24 | Pit-1; GH | Acromegaly | 0 |
| 31 | Pit-1; GH-PRL | Acromegaly | 0 | |
| 2 | Pit-1; GH | NF-PitNET 1 | 0 | |
| Lactotroph | 6 | Pit-1; PRL | Hyperprolactinemia | 0 |
| 2 | Pit-1; PRL | NF-PitNET 1 | 0 | |
| Thyrotroph | 2 | Pit-1; TSH | HyperTSH | 0 |
| Null cell | 7 | TFs 2 neg; Hormone neg. | NF-PitNET 1 | 0 |
| Total | 230 | 37 |
| Staining/mRNA | TGFBR3L Staining | TGFBR3L mRNA |
|---|---|---|
| TGFBR3L mRNA | R = 0.378 | - |
| P = 0.0058 | - | |
| N = 52 | - | |
| FSH staining | R = 0.290 | R = 0.261 |
| P = 0.004 | P = 0.061 | |
| N = 95 | N = 52 | |
| LH staining | R = 0.390 | R = 0.598 |
| P < 0.0001 | P < 0.0001 | |
| N = 95 | N = 52 | |
| E-cadherin IRS 1 (membranous) | R = −0.351 | R = −0.387 |
| P = 0.0005 | P = 0.004 | |
| N = 95 | N = 52 | |
| E-cadherin mRNA | R = −0.086 | R = −0.265 |
| P = 0.482 | P = 0.0598 | |
| N = 70 | N = 51 | |
| ERβ mRNA | R = −0.274 | R = −0.221 |
| P = 0.022 | P = 0.119 | |
| N = 70 | N = 51 | |
| SSTR3 IRS 1 | R = 0.049 | R = −0.218 |
| P = 0.638 | P = 0.061 | |
| N = 95 | N = 52 | |
| SSTR3 mRNA | R = 0.315 | R = 0.818 |
| P = 0.008 | P < 0.0001 | |
| N = 70 | N = 51 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sjöstedt, E.; Kolnes, A.J.; Olarescu, N.C.; Mitsios, N.; Hikmet, F.; Sivertsson, Å.; Lindskog, C.; Øystese, K.A.B.; Jørgensen, A.P.; Bollerslev, J.; et al. TGFBR3L—An Uncharacterised Pituitary Specific Membrane Protein Detected in the Gonadotroph Cells in Non-Neoplastic and Tumour Tissue. Cancers 2021, 13, 114. https://doi.org/10.3390/cancers13010114
Sjöstedt E, Kolnes AJ, Olarescu NC, Mitsios N, Hikmet F, Sivertsson Å, Lindskog C, Øystese KAB, Jørgensen AP, Bollerslev J, et al. TGFBR3L—An Uncharacterised Pituitary Specific Membrane Protein Detected in the Gonadotroph Cells in Non-Neoplastic and Tumour Tissue. Cancers. 2021; 13(1):114. https://doi.org/10.3390/cancers13010114
Chicago/Turabian StyleSjöstedt, Evelina, Anders J. Kolnes, Nicoleta C. Olarescu, Nicholas Mitsios, Feria Hikmet, Åsa Sivertsson, Cecilia Lindskog, Kristin A. B. Øystese, Anders P. Jørgensen, Jens Bollerslev, and et al. 2021. "TGFBR3L—An Uncharacterised Pituitary Specific Membrane Protein Detected in the Gonadotroph Cells in Non-Neoplastic and Tumour Tissue" Cancers 13, no. 1: 114. https://doi.org/10.3390/cancers13010114
APA StyleSjöstedt, E., Kolnes, A. J., Olarescu, N. C., Mitsios, N., Hikmet, F., Sivertsson, Å., Lindskog, C., Øystese, K. A. B., Jørgensen, A. P., Bollerslev, J., & Casar-Borota, O. (2021). TGFBR3L—An Uncharacterised Pituitary Specific Membrane Protein Detected in the Gonadotroph Cells in Non-Neoplastic and Tumour Tissue. Cancers, 13(1), 114. https://doi.org/10.3390/cancers13010114





