Signal Transduction Pathway Activity in High-Grade, Serous Ovarian Carcinoma Reveals a More Favorable Prognosis in Tumors with Low PI3K and High NF-κB Pathway Activity: A Novel Approach to a Long-Standing Enigma
Simple Summary
Abstract
1. Introduction
2. Results
2.1. Datasets and Patient Characteristics
2.1.1. GSE9891
2.1.2. GSE32062
2.2. Clustering of HGSC Samples
2.2.1. GSE9891
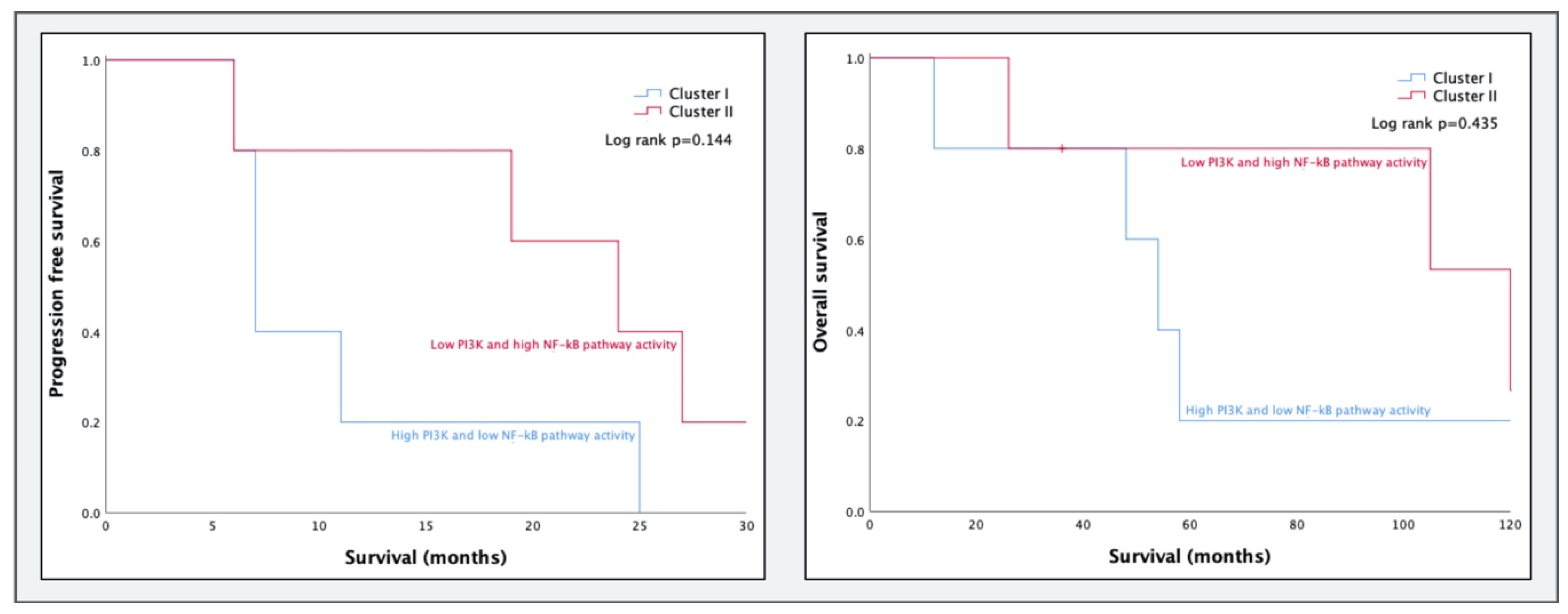
2.2.2. GSE32062
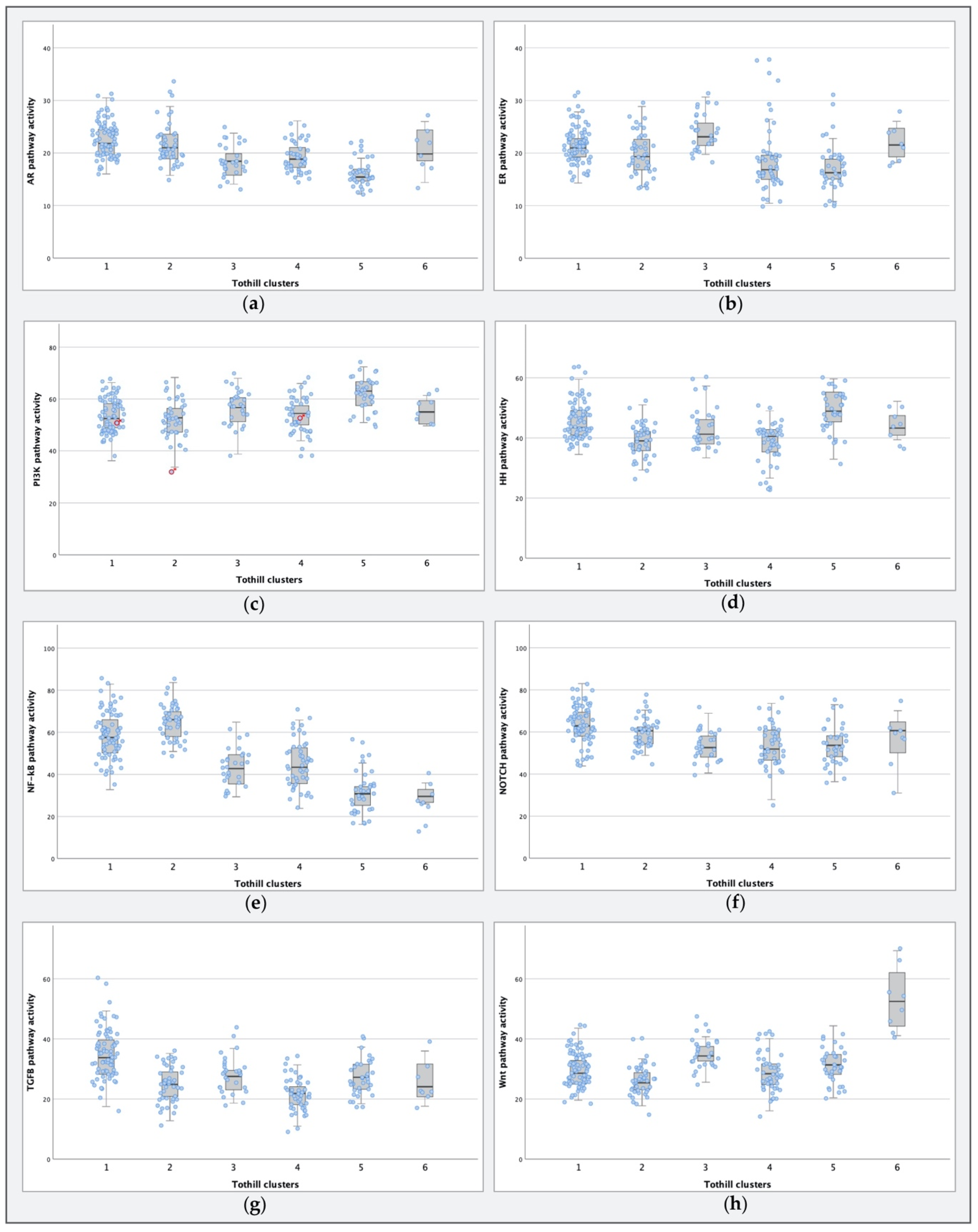
2.3. STA-Analysis of Molecular Subtypes as Defined by Tothill et al.
3. Discussion
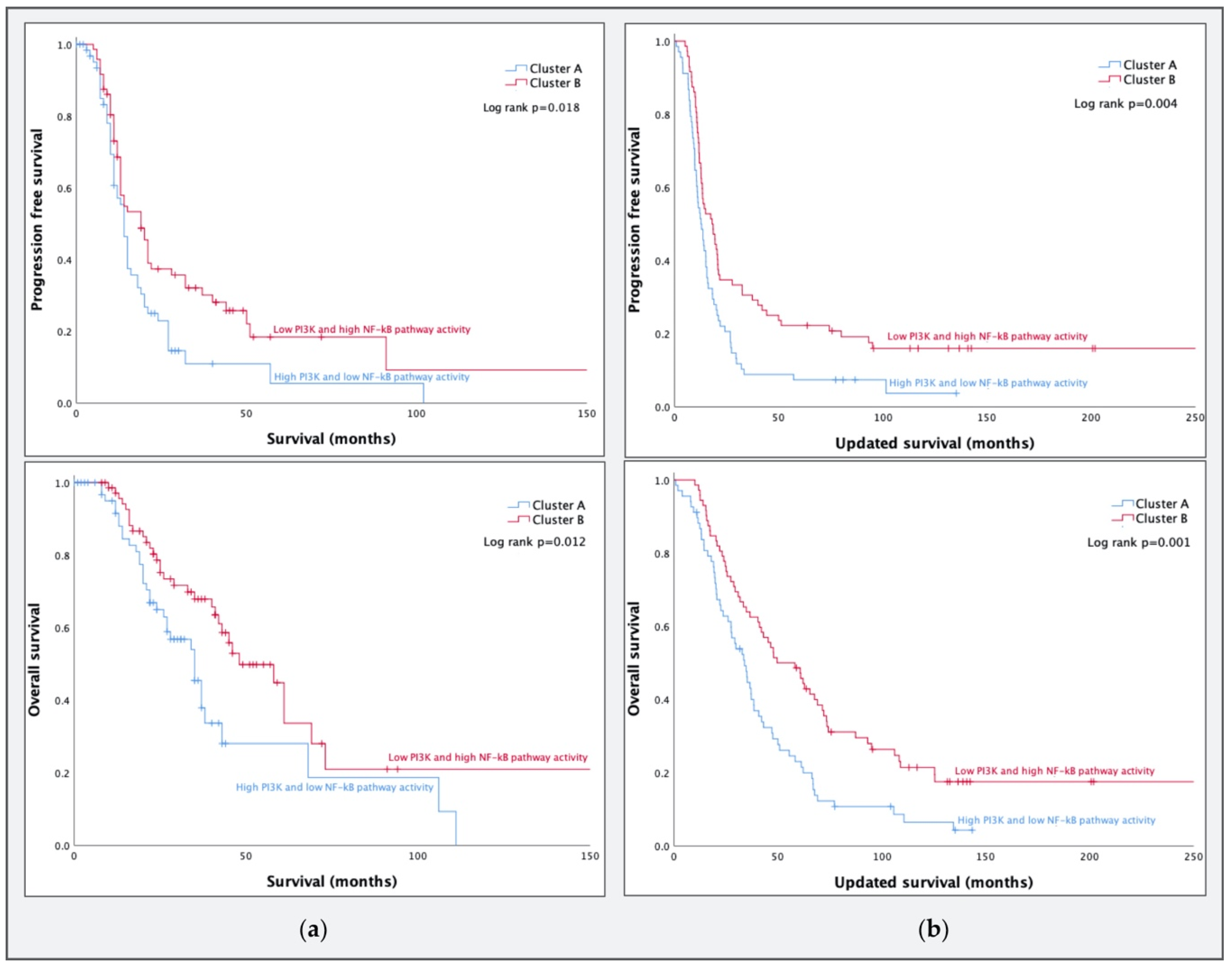
3.1. PI3K and NF-κB Pathway Activity in HGSCs
3.2. Comparing Molecular Subtypes as Formed by Tothill et al. to STA Analysis Results
3.3. Strengths and Limitations
3.4. Future Studies
4. Materials and Methods
4.1. STA Analysis
4.2. Datasets
4.3. Statistical Analysis
4.3.1. Patient Characteristics
4.3.2. Clustering Analysis
4.3.3. STA Analysis of Molecular Subtypes as Defined by Tothill et al.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA. Cancer J. Clin. 2018, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Cantley, L.C.; Hunter, T.; Sever, R.; Thorner, J. Signal Transduction: Principles, Pathways, and Processes; Cold Spring Harbor: New York, NY, USA, 2013. [Google Scholar]
- Hanahan, D.; Weinberg, R.A. Leading Edge Review Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- DeVita, V.T.; Lawrence, T.S.; Rosenberg, S.A. Cancer: Principles and; Practice of Oncology; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 1993; ISBN 9781451192940. [Google Scholar]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A.; Kinzler, K.W. Cancer genome landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Verhaegh, W.; Van Ooijen, H.; Inda, M.A.; Hatzis, P.; Versteeg, R.; Smid, M.; Martens, J.; Foekens, J.; Van De Wiel, P.; Clevers, H.; et al. Selection of personalized patient therapy through the use of knowledge-based computational models that identify tumor-driving signal transduction pathways. Cancer Res. 2014, 74, 2936–2945. [Google Scholar] [CrossRef]
- Massard, C.; Michiels, S.; Ferté, C.; Le Deley, M.-C.; Lacroix, L.; Hollebecque, A.; Verlingue, L.; Ileana, E.; Rosellini, S.; Ammari, S.; et al. High-Throughput Genomics and Clinical Outcome in Hard-to-Treat Advanced Cancers: Results of the MOSCATO 01 Trial. Cancer Discov. 2017, 7, 586–595. [Google Scholar] [CrossRef]
- Verhaegh, W.; van de Stolpe, A. Knowledge-based computational models. Oncotarget 2014, 5, 5196–5197. [Google Scholar] [CrossRef]
- Van de Stolpe, A.; Holtzer, L.; van Ooijen, H.; de Inda, M.A.; Verhaegh, W. Enabling precision medicine by unravelling disease pathophysiology: Quantifying signal transduction pathway activity across cell and tissue types. Sci. Rep. 2019, 9, 1603. [Google Scholar] [CrossRef]
- Van Ooijen, H.; Hornsveld, M.; Dam-de Veen, C.; Velter, R.; Dou, M.; Verhaegh, W.; Burgering, B.; van de Stolpe, A. Assessment of Functional Phosphatidylinositol 3-Kinase Pathway Activity in Cancer Tissue Using Forkhead Box-O Target Gene Expression in a Knowledge-Based Computational Model. Am. J. Pathol. 2018, 188, 1956–1972. [Google Scholar] [CrossRef]
- Van de Stolpe, A.; Verhaegh, W.; van Doorn, A.; Noël, G.; Gu-Trantien, C.; Willard-Gallo, K. Breast cancer induces tolerogenic state of healthy activated CD4+ lymphocytes, characterized by reduced PI3K, NFκB, JAK-STAT, Notch, and increased TGFβpathway activity. Cancer Res. 2019. [Google Scholar] [CrossRef]
- Cante-Barrett, K.; Holtzer, L.; van Ooijen, H.; Hagelaar, R.; Cordo, V.; Verhaegh, W.; van de Stolpe, A.; Meijerink, J. A Test to Quantify NOTCH Pathway Activity in T Cell Acute Lymphoblastic Leukemia Patients. In Proceedings of the American Society of Hematology Annual Meeting. Blood 2019, 134, e4661. [Google Scholar] [CrossRef]
- Inda, M.A.; Blok, E.J.; Kuppen, P.J.; Charehbili, A.; den Biezen-Timmermans, E.C.; Van Brussel, A.; Fruytier, S.E.; Kranenbarg, E.M. Estrogen Receptor pathway activity score to predict clinical response or resistance to neo—Adjuvant endocrine therapy in primary breast cancer. Mol. Cancer Ther. 2019. [Google Scholar] [CrossRef]
- Van Weelden, W.J.; van der Putten, L.J.M.; Inda, M.A.; van Brussel, A.; Snijders, M.P.L.M.; Schriever, L.M.M.; Bulten, J.; Massuger, L.F.A.G.; van de Stolpe, A.; Pijnenborg, J.M.A. Oestrogen Receptor Pathway Activity Is Associated With Outcome in Endometrial Cancer. Br. J. Cancer 2020, 123, 785–792. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef]
- Tothill, R.W.; Tinker, A.V.; George, J.; Brown, R.; Fox, S.B.; Lade, S.; Johnson, D.S.; Trivett, M.K.; Etemadmoghadam, D.; Locandro, B.; et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin. Cancer Res. 2008, 14, 5198–5208. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Tsunoda, T.; Shigemizu, D.; Fujiwara, H.; Hatae, M.; Fujiwara, H.; Masuzaki, H.; Katabuchi, H.; Kawakami, Y.; Okamoto, A.; et al. High-Risk Ovarian Cancer Based on 126-Gene Expression Signature Is Uniquely Characterized by Downregulation of Antigen Presentation Pathway. Progn. Clin. Cancer Res. 2012, 18. [Google Scholar] [CrossRef] [PubMed]
- Prat, J. FIGO Committee on Gynecologic Oncology. Staging classification for cancer of the ovary, fallopian tube, and peritoneum. Int. J. Gynaecol. Obstet. 2014, 124, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Van De Wiel, P.; Helleman, J.; Den Biezen-Timmermans, E.; Wrobel, J.; van Ooijen, H.; Berns, E.; Van de Stolpe, A. Analysis of Active Oncogenic Signal Transduction Pathways in Ovarian Cancer. Association for Molecular Pathology 2017, November 16-18, Salt Lake City, UT. J. Mol. Diagn. 2017, 19, 943–1067. [Google Scholar]
- Silverberg, S.G. Histopathologic grading of ovarian carcinoma: A review and proposal. Int. J. Gynecol. Pathol. 2000, 19, 7–15. [Google Scholar] [CrossRef]
- Yang, J.; Nie, J.; Ma, X.; Wei, Y.; Peng, Y.; Wei, X. Targeting PI3K in cancer: Mechanisms and advances in clinical trials 06 Biological Sciences 0601 Biochemistry and Cell Biology. Mol. Cancer 2019, 18, 26. [Google Scholar] [CrossRef]
- Mabuchi, S.; Kawase, C.; Altomare, D.A.; Morishige, K.; Sawada, K.; Hayashi, M.; Tsujimoto, M.; Yamoto, M.; Klein-Szanto, A.J.; Schilder, R.J.; et al. mTOR Is a Promising Therapeutic Target Both in Cisplatin-Sensitive and Cisplatin-Resistant Clear Cell Carcinoma of the Ovary. Clin. Cancer Res. 2009, 15, 5404–5413. [Google Scholar] [CrossRef] [PubMed]
- Dobbin, Z.; Landen, C. The Importance of the PI3K/AKT/MTOR Pathway in the Progression of Ovarian Cancer. Int. J. Mol. Sci. 2013, 14, 8213–8227. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Wang, J.; Lei, Y.; Cong, C.; Tan, D.; Zhou, X. Research progress on the PI3K/AKT signaling pathway in gynecological cancer (Review). Mol. Med. Rep. 2019, 19, 4529–4535. [Google Scholar] [CrossRef]
- Ghoneum, A.; Abdulfattah, A.Y.; Said, N. Targeting the PI3K/AKT/mTOR/NFκB Axis in Ovarian Cancer. J. Cell. Immunol. 2020, 2, 68. [Google Scholar] [PubMed]
- Harrington, B.S.; Annunziata, C.M. NF-κB Signaling in Ovarian Cancer. Cancers 2019, 11, 1182. [Google Scholar] [CrossRef] [PubMed]
- Annunziata, C.M.; Stavnes, H.T.; Kleinberg, L.; Berner, A.; Hernandez, L.F.; Birrer, M.J.; Steinberg, S.M.; Davidson, B.; Kohn, E.C. Nuclear factor κB transcription factors are coexpressed and convey a poor outcome in ovarian cancer. Cancer 2010, 116, 3276–3284. [Google Scholar] [CrossRef]
- Xiao, X.; Yang, G.; Bai, P.; Gui, S.; Nyuyen, T.M.B.; Mercado-Uribe, I.; Yang, M.; Zou, J.; Li, Q.; Xiao, J.; et al. Inhibition of nuclear factor-kappa B enhances the tumor growth of ovarian cancer cell line derived from a low-grade papillary serous carcinoma in p53-independent pathway. BMC Cancer 2016, 16, 582. [Google Scholar] [CrossRef]
- Hoesel, B.; Schmid, J.A. The complexity of NF-kB signaling in inflammation and cancer. Mol. Cancer 2013, 86. [Google Scholar] [CrossRef]
- Zhang, L.; Conejo-Garcia, J.R.; Katsaros, D.; Gimotty, P.A.; Massobrio, M.; Regnani, G.; Makrigiannakis, A.; Gray, H.; Schlienger, K.; Liebman, M.N.; et al. Intratumoral T Cells, Recurrence, and Survival in Epithelial Ovarian Cancer. N. Engl. J. Med. 2003, 348, 203–213. [Google Scholar] [CrossRef]
- Kurman, R.J.; Shih, I.-M. The Origin and Pathogenesis of Epithelial Ovarian Cancer: A Proposed Unifying Theory. Am. J. Surg. Pathol. 2010, 34, 433–443. [Google Scholar] [CrossRef]
- Emmanuel, C.; Chiew, Y.E.; George, J.; Etemadmoghadam, D.; Anglesio, M.S.; Sharma, R.; Russell, P.; Kennedy, C.; Fereday, S.; Hung, J.; et al. Genomic classification of serous ovarian cancer with adjacent borderline differentiates Ras pathway and TP53-mutant tumors and identifies NRAS as an oncogenic driver. Clin. Cancer Res. 2014, 20, 6618–6630. [Google Scholar] [CrossRef] [PubMed]
- Mariathasan, S.; Turley, S.J.; Nickles, D.; Castiglioni, A.; Yuen, K.; Wang, Y.; Kadel, E.E.; Koeppen, H.; Astarita, J.L.; Cubas, R.; et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018, 554, 544–548. [Google Scholar] [CrossRef]
- Sever, R.; Brugge, J.S. Signal transduction in cancer. Cold Spring Harb. Perspect. Med. 2015, 5. [Google Scholar] [CrossRef]
- Syed, V. TGF-β Signaling in Cancer. J. Cell. Biochem. 2016, 117, 1279–1287. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Azizun-Nisa, B.Y.; Raza, F.; Kayani, N. Comparison of ER, PR & HER-2/neu (C-erb B 2) reactivity pattern with histologic grade, tumor size and lymph node status in breast cancer. Asian Pac. J. Cancer Prev. 2008, 9, 553–556. [Google Scholar] [PubMed]
- Duffy, D.J.; Krstic, A.; Schwarzl, T.; Halasz, M.; Iljin, K.; Fey, D.; Haley, B.; Whilde, J.; Haapa-Paananen, S.; Fey, V.; et al. Wnt signalling is a bi-directional vulnerability of cancer cells. Oncotarget 2016, 7, 60310–60331. [Google Scholar] [CrossRef]
- Hoffmann, K.; Berger, H.; Kulbe, H.; Thillainadarasan, S.; Mollenkopf, H.; Zemojtel, T.; Taube, E.; Darb-esfahani, S.; Mangler, M.; Sehouli, J.; et al. Stable expansion of high-grade serous ovarian cancer organoids requires a low-Wnt environment. Embo J. 2020, 39, e104013. [Google Scholar] [CrossRef]
- Takebe, N.; Miele, L.; Harris, P.J.; Jeong, W.; Bando, H.; Kahn, M.; Yang, S.X.; Ivy, S.P. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: Clinical update. Nat. Rev. Clin. Oncol. 2015, 12, 445–464. [Google Scholar] [CrossRef]
- Najafi, M.; Farhood, B.; Mortezaee, K. Cancer stem cells (CSCs) in cancer progression and therapy. J. Cell. Physiol. 2019, 234, 8381–8395. [Google Scholar] [CrossRef]
- Xia, P.; Xu, X.Y. PI3K/Akt/mTOR signaling pathway in cancer stem cells: From basic research to clinical application. Am. J. Cancer Res. 2015, 5, 1602–1609. [Google Scholar] [PubMed]
- Okkenhaug, K.; Roychoudhuri, R. Oncogenic PI3Kα promotes multipotency in breast epithelial cells. Sci. Signal. 2015, 8, 8–10. [Google Scholar] [CrossRef] [PubMed]
- Van Keymeulen, A.; Lee, M.Y.; Ousset, M.; Brohée, S.; Rorive, S.; Giraddi, R.R.; Wuidart, A.; Bouvencourt, G.; Dubois, C.; Salmon, I.; et al. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature 2015, 525, 119–123. [Google Scholar] [CrossRef]
- Ingram, D.R.; Dillon, L.M.; Lev, D.C.; Lazar, A.; Demicco, E.G.; Eisenberg, B.L.; Miller, T.W. Estrogen receptor alpha and androgen receptor are commonly expressed in well-differentiated liposarcoma. BMC Clin. Pathol. 2014, 14, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mollerup, S.; Jørgensen, K.; Berge, G.; Haugen, A. Expression of estrogen receptors α and β in human lung tissue and cell lines. Lung Cancer 2002, 37, 153–159. [Google Scholar] [CrossRef]
- Park, S.; Koo, J.; Park, H.S.; Kim, J.H.; Choi, S.Y.; Lee, J.H.; Park, B.W.; Lee, K.S. Expression of androgen receptors in primary breast cancer. Ann. Oncol. 2009, 21, 488–492. [Google Scholar] [CrossRef]
- Wu, C.T.; Chang, Y.L.; Lee, Y.C. Expression of the estrogen receptor β in 37 surgically treated pulmonary sclerosing hemangiomas in comparison with non-small cell lung carcinomas. Hum. Pathol. 2005, 36, 1108–1112. [Google Scholar] [CrossRef]
- Thomas, P.D.; Kejariwal, A.; Guo, N.; Mi, H.; Campbell, M.J.; Muruganujan, A.; Lazareva-Ulitsky, B. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006, 34, W645–W650. [Google Scholar] [CrossRef]
- Bhateja, P.; Cherian, M.; Majumder, S.; Ramaswamy, B. The hedgehog signaling pathway: A viable target in breast cancer? Cancers 2019, 11, 1126. [Google Scholar] [CrossRef]
- Giroux-Leprieur, E.; Costantini, A.; Ding, V.W.; He, B. Hedgehog signaling in lung cancer: From oncogenesis to cancer treatment resistance. Int. J. Mol. Sci. 2018, 19, 2835. [Google Scholar] [CrossRef]
- Piek, J.M.J.; van Diest, P.J.; Zweemer, R.P.; Jansen, J.W.; Poort-Keesom, R.J.J.; Menko, F.H.; Gille, J.J.P.; Jongsma, A.P.M.; Pals, G.; Kenemans, P.; et al. Dysplastic changes in prophylactically removed Fallopian tubes of women predisposed to developing ovarian cancer. J. Pathol. 2001, 195, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Soong, T.R.; Howitt, B.E.; Horowitz, N.; Nucci, M.R.; Crum, C.P. The fallopian tube, “precursor escape” and narrowing the knowledge gap to the origins of high-grade serous carcinoma. Gynecol. Oncol. 2019, 152, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Van de Stolpe, A. Quantitative Measurement of Functional Activity of the PI3K Signaling Pathway in Cancer. Cancers 2019, 11, 293. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.L.; Miller, C.J. Simpleaffy: A BioConductor package for Affymetrix Quality Control and data analysis. Bioinform. Appl. Note 2005, 21, 3683–3685. [Google Scholar] [CrossRef]
- Heber, S.; Sick, B. Quality Assessment of Affymetrix GeneChip Data. Omics A J. Integr. Biol. 2006, 10, 358–368. [Google Scholar] [CrossRef]
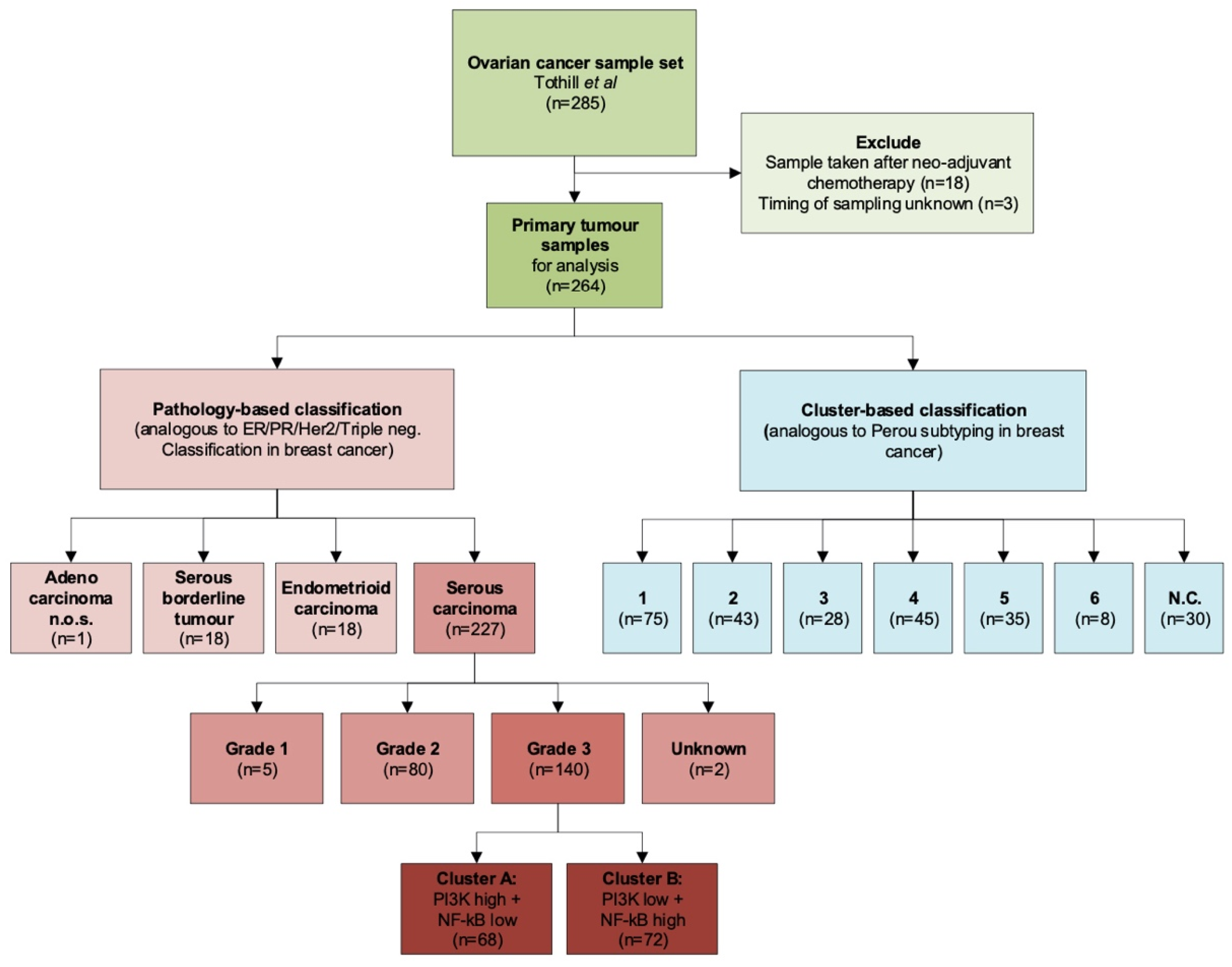
| Feature | Total | Tothill Clusters | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | NC1 | ||
| Number of samples | 264 | 75 (28%) | 43 (16%) | 28 (12%) | 45 (17%) | 35 (13%) | 8 (3%) | 30 (11%) |
| Age at diagnosis (y)2 | 60 (±11) | 59 (±11) | 62 (±10) | 52 (±12) | 60 (±9) | 63 (±10) | 54 (±6) | 61 (±11) |
| Stage at diagnosis | ||||||||
| I | 24 (9%) | 1 (1%) | 5 (12%) | 10 (36%) | 1 (2%) | 2 (6%) | 5 (62%) | |
| II | 18 (7%) | 1 (1%) | 4 (9%) | 4 (14%) | 5 (11%) | 2 (6%) | 1 (13%) | 1 (3%) |
| III | 206 (78%) | 65 (87%) | 31 (72%) | 13 (46%) | 39 (87%) | 30 (85%) | 2 (25%) | 26 (87%) |
| IV | 16 (6%) | 8 (11%) | 3 (7%) | 1 (4%) | 1 (3%) | 3 (10%) | ||
| Histology | ||||||||
| Serous carcinoma | 227 (86%) | 74 (99%) | 40 (93%) | 10 (36%) | 40 (89%) | 34 (97%) | 1 (12%) | 28 (93%) |
| Endometrioid carcinoma | 18 (7%) | 1 (1%) | 2 (5%) | 5 (11%) | 1 (3%) | 7 (88%) | 2 (7%) | |
| Serous borderline tumor | 18 (7%) | 18 (64%) | ||||||
| Adenocarcinoma NOS3 | 1 (0%) | 1 (2%) | ||||||
| Differentiation degree | ||||||||
| 1 | 17 (7%) | 1 (1%) | 2 (5%) | 11 (39%) | 3 (38%) | |||
| 2 | 88 (33%) | 25 (34%) | 13 (30%) | 4 (15%) | 19 (42%) | 14 (40%) | 4 (50%) | 9 (30%) |
| 3 | 156 (59%) | 48 (64%) | 28 (65%) | 11 (39%) | 26 (58%) | 21 (60%) | 1 (12%) | 21 (70%) |
| Unknown | 3 (1%) | 1 (1%) | 2 (7%) | |||||
| Status | ||||||||
| Deceased | 103 (39%) | 44 (59%) | 14 (33%) | 1 (4%) | 16 (36%) | 15 (43%) | 13 (43%) | |
| Deceased, other cause | 2 (1%) | 1 (1%) | 1 (2%) | |||||
| Progressed | 70 (26%) | 19 (25%) | 17 (39%) | 3 (11%) | 14 (31%) | 9 (26%) | 8 (27%) | |
| Progression-free | 89 (34%) | 11 (15%) | 12 (28%) | 24 (85%) | 14 (31%) | 11 (31%) | 8 (100%) | 9 (30%) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Lieshout, L.; van de Stolpe, A.; van der Ploeg, P.; Bowtell, D.; de Hullu, J.; Piek, J. Signal Transduction Pathway Activity in High-Grade, Serous Ovarian Carcinoma Reveals a More Favorable Prognosis in Tumors with Low PI3K and High NF-κB Pathway Activity: A Novel Approach to a Long-Standing Enigma. Cancers 2020, 12, 2660. https://doi.org/10.3390/cancers12092660
van Lieshout L, van de Stolpe A, van der Ploeg P, Bowtell D, de Hullu J, Piek J. Signal Transduction Pathway Activity in High-Grade, Serous Ovarian Carcinoma Reveals a More Favorable Prognosis in Tumors with Low PI3K and High NF-κB Pathway Activity: A Novel Approach to a Long-Standing Enigma. Cancers. 2020; 12(9):2660. https://doi.org/10.3390/cancers12092660
Chicago/Turabian Stylevan Lieshout, Laura, Anja van de Stolpe, Phyllis van der Ploeg, David Bowtell, Joanne de Hullu, and Jurgen Piek. 2020. "Signal Transduction Pathway Activity in High-Grade, Serous Ovarian Carcinoma Reveals a More Favorable Prognosis in Tumors with Low PI3K and High NF-κB Pathway Activity: A Novel Approach to a Long-Standing Enigma" Cancers 12, no. 9: 2660. https://doi.org/10.3390/cancers12092660
APA Stylevan Lieshout, L., van de Stolpe, A., van der Ploeg, P., Bowtell, D., de Hullu, J., & Piek, J. (2020). Signal Transduction Pathway Activity in High-Grade, Serous Ovarian Carcinoma Reveals a More Favorable Prognosis in Tumors with Low PI3K and High NF-κB Pathway Activity: A Novel Approach to a Long-Standing Enigma. Cancers, 12(9), 2660. https://doi.org/10.3390/cancers12092660






