Classifying Lung Neuroendocrine Neoplasms through MicroRNA Sequence Data Mining
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Materials and Study Design
2.2. Total RNA Isolation and Quality Control
2.3. Small RNA Sequencing
2.4. Data Preprocessing
2.5. High Expression and Discovery Analyses
2.6. miRNA-Based Classifier for Discriminating Carcinoids from NECs
2.7. Candidate miRNA Markers for Discriminating Pathological Types
2.8. Statistical Analyses
3. Results
3.1. Clinicopathologic Characteristics of Discovery and Validation Sample Sets
3.2. Barcoded Small RNA Sequencing
3.3. High Expression Analyses
3.4. Discovery Analyses
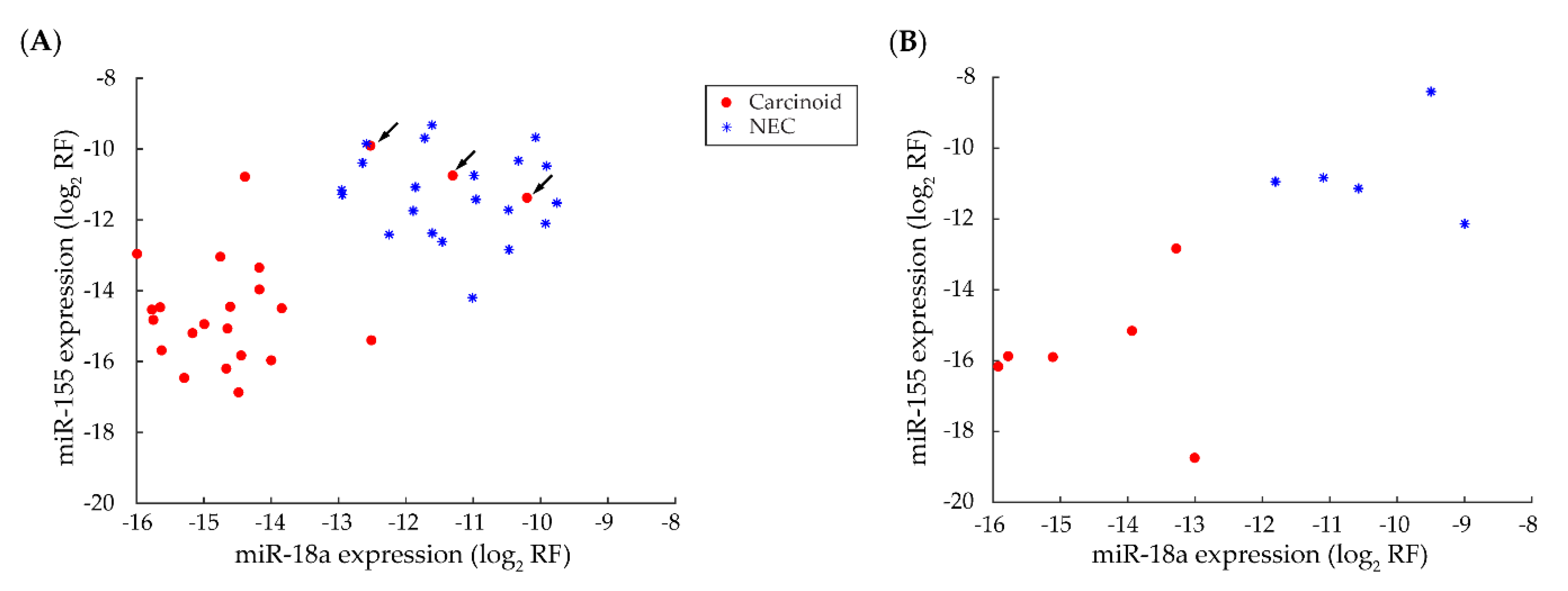
3.5. miRNA-Based Classifier for Discriminating Carcinoids from NECs
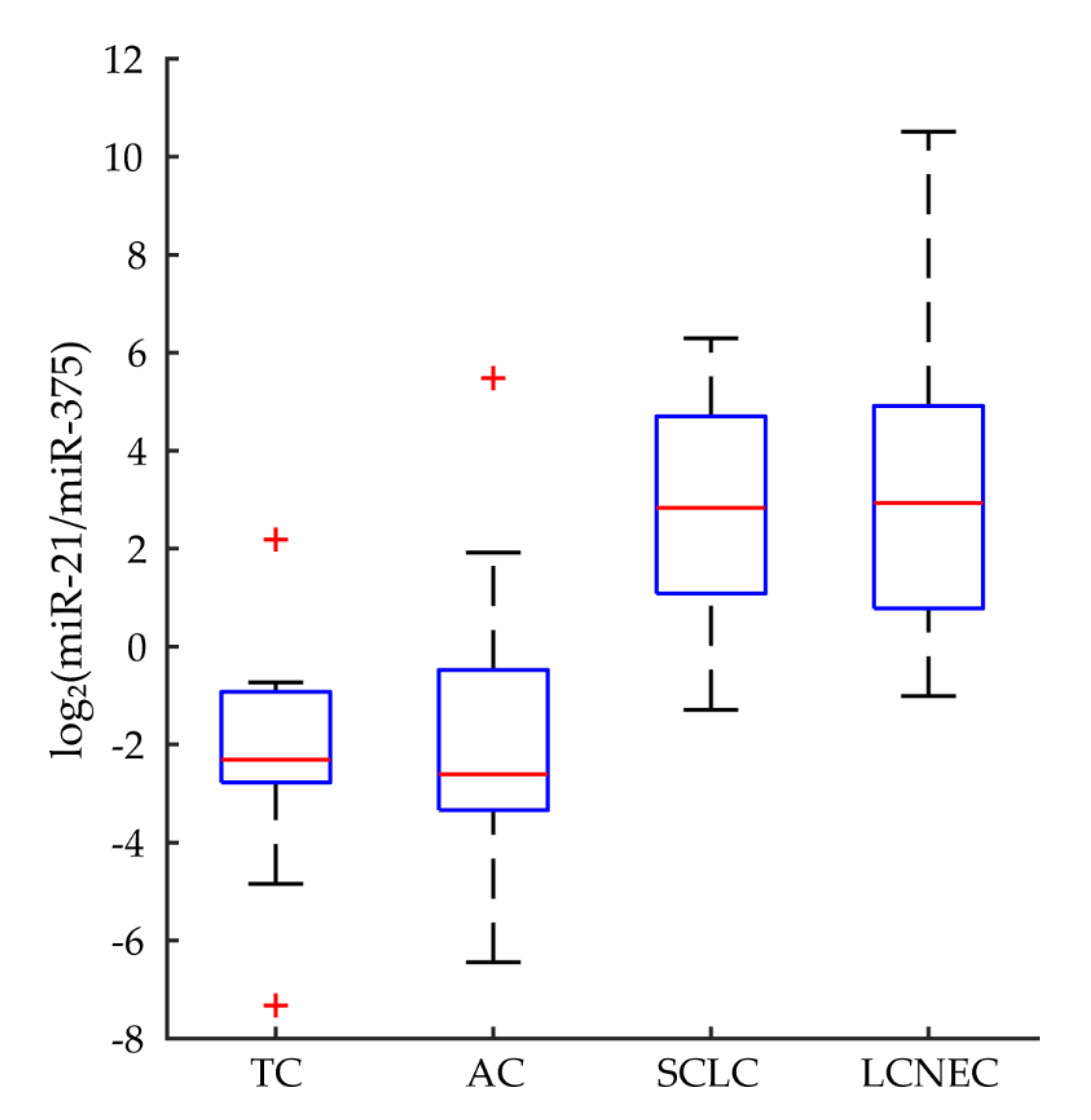
3.6. Candidate miRNA Markers for Discriminating Pathological Types
3.7. Correlation of Candidate miRNA Markers and Pathologic Parameters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Volante, M.; Gatti, G.; Papotti, M. Classification of lung neuroendocrine tumors: Lights and shadows. Endocrine 2015, 50, 315–319. [Google Scholar] [CrossRef]
- Travis, W.D.; Brambilla, E.; Nicholson, A.G.; Yatabe, Y.; Austin, J.H.M.; Beasley, M.B.; Chirieac, L.R.; Dacic, S.; Duhig, E.; Flieder, D.B.; et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J. Thorac. Oncol. 2015, 10, 1243–1260. [Google Scholar] [CrossRef]
- Rindi, G.; Klimstra, D.S.; Abedi-Ardekani, B.; Asa, S.L.; Bosman, F.T.; Brambilla, E.; Busam, K.J.; de Krijger, R.R.; Dietel, M.; El-Naggar, A.K.; et al. A common classification framework for neuroendocrine neoplasms: An International Agency for Research on Cancer (IARC) and World Health Organization (WHO) expert consensus proposal. Mod. Pathol. 2018, 31, 1770–1786. [Google Scholar] [CrossRef] [PubMed]
- Caplin, M.E.; Baudin, E.; Ferolla, P.; Filosso, P.; Garcia-Yuste, M.; Lim, E.; Oberg, K.; Pelosi, G.; Perren, A.; Rossi, R.E.; et al. Pulmonary neuroendocrine (carcinoid) tumors: European Neuroendocrine Tumor Society expert consensus and recommendations for best practice for typical and atypical pulmonary carcinoids. Ann. Oncol. 2015, 26, 1604–1620. [Google Scholar] [CrossRef] [PubMed]
- Marchevsky, A.M.; Gal, A.A.; Shah, S.; Koss, M.N. Morphometry confirms the presence of considerable nuclear size overlap between “small cells” and “large cells” in high-grade pulmonary neuroendocrine neoplasms. Am. J. Clin. Pathol. 2001, 116, 466–472. [Google Scholar] [CrossRef]
- Phan, A.T.; Oberg, K.; Choi, J.; Harrison, L.H., Jr.; Hassan, M.M.; Strosberg, J.R.; Krenning, E.P.; Kocha, W.; Woltering, E.A. NANETS consensus guideline for the diagnosis and management of neuroendocrine tumors: Well-differentiated neuroendocrine tumors of the thorax (includes lung and thymus). Pancreas 2010, 39, 784–798. [Google Scholar] [CrossRef] [PubMed]
- Dasari, A.; Shen, C.; Halperin, D.; Zhao, B.; Zhou, S.; Xu, Y.; Shih, T.; Yao, J.C. Trends in the Incidence, Prevalence, and Survival Outcomes in Patients With Neuroendocrine Tumors in the United States. JAMA Oncol. 2017, 3, 1335–1342. [Google Scholar] [CrossRef]
- Ramirez, R.A.; Chauhan, A.; Gimenez, J.; Thomas, K.E.H.; Kokodis, I.; Voros, B.A. Management of pulmonary neuroendocrine tumors. Rev. Endocr. Metab. Disord. 2017, 18, 433–442. [Google Scholar] [CrossRef]
- Oberg, K.; Hellman, P.; Ferolla, P.; Papotti, M.; Group, E.G.W. Neuroendocrine bronchial and thymic tumors: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2012, 23 (Suppl. 7), vii120–vii123. [Google Scholar] [CrossRef]
- Hendifar, A.E.; Marchevsky, A.M.; Tuli, R. Neuroendocrine Tumors of the Lung: Current Challenges and Advances in the Diagnosis and Management of Well-Differentiated Disease. J. Thorac. Oncol. 2017, 12, 425–436. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, D.; Tyryshkin, K.; Renwick, N. microRNA-guided diagnostics in clinical samples. Best Pract. Res. Clin. Endocrinol. Metab. 2016, 30, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Miko, E.; Czimmerer, Z.; Csanky, E.; Boros, G.; Buslig, J.; Dezso, B.; Scholtz, B. Differentially expressed microRNAs in small cell lung cancer. Exp. Lung Res. 2009, 35, 646–664. [Google Scholar] [CrossRef]
- Gilad, S.; Lithwick-Yanai, G.; Barshack, I.; Benjamin, S.; Krivitsky, I.; Edmonston, T.B.; Bibbo, M.; Thurm, C.; Horowitz, L.; Huang, Y.; et al. Classification of the four main types of lung cancer using a microRNA-based diagnostic assay. J. Mol. Diagn. 2012, 14, 510–517. [Google Scholar] [CrossRef]
- Lee, H.W.; Lee, E.H.; Ha, S.Y.; Lee, C.H.; Chang, H.K.; Chang, S.; Kwon, K.Y.; Hwang, I.S.; Roh, M.S.; Seo, J.W. Altered expression of microRNA miR-21, miR-155, and let-7a and their roles in pulmonary neuroendocrine tumors. Pathol. Int. 2012, 62, 583–591. [Google Scholar] [CrossRef]
- Mairinger, F.D.; Ting, S.; Werner, R.; Walter, R.F.; Hager, T.; Vollbrecht, C.; Christoph, D.; Worm, K.; Mairinger, T.; Sheu-Grabellus, S.Y.; et al. Different micro-RNA expression profiles distinguish subtypes of neuroendocrine tumors of the lung: Results of a profiling study. Mod. Pathol. 2014, 27, 1632–1640. [Google Scholar] [CrossRef]
- Rapa, I.; Votta, A.; Felice, B.; Righi, L.; Giorcelli, J.; Scarpa, A.; Speel, E.J.; Scagliotti, G.V.; Papotti, M.; Volante, M. Identification of MicroRNAs Differentially Expressed in Lung Carcinoid Subtypes and Progression. Neuroendocrinology 2015, 101, 246–255. [Google Scholar] [CrossRef]
- Demes, M.; Aszyk, C.; Bartsch, H.; Schirren, J.; Fisseler-Eckhoff, A. Differential miRNA-Expression as an Adjunctive Diagnostic Tool in Neuroendocrine Tumors of the Lung. Cancers 2016, 8, 38. [Google Scholar] [CrossRef]
- Malczewska, A.; Kidd, M.; Matar, S.; Kos-Kudla, B.; Modlin, I.M. A Comprehensive Assessment of the Role of miRNAs as Biomarkers in Gastroenteropancreatic Neuroendocrine Tumors. Neuroendocrinology 2018, 107, 73–90. [Google Scholar] [CrossRef]
- Loudig, O.; Liu, C.; Rohan, T.; Ben-Dov, I.Z. Retrospective MicroRNA Sequencing: Complementary DNA Library Preparation Protocol Using Formalin-fixed Paraffin-embedded RNA Specimens. J. Vis. Exp. 2018, 5, 57471. [Google Scholar] [CrossRef]
- Hafner, M.; Renwick, N.; Farazi, T.A.; Mihailovic, A.; Pena, J.T.; Tuschl, T. Barcoded cDNA library preparation for small RNA profiling by next-generation sequencing. Methods 2012, 58, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Max, K.E.A.; Bertram, K.; Akat, K.M.; Bogardus, K.A.; Li, J.; Morozov, P.; Ben-Dov, I.Z.; Li, X.; Weiss, Z.R.; Azizian, A.; et al. Human plasma and serum extracellular small RNA reference profiles and their clinical utility. Proc. Natl. Acad. Sci. USA 2018, 115, E5334–E5343. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.; Suryawanshi, H.; Hafner, M.; Farazi, T.A.; Tuschl, T. Mammalian miRNA curation through next-generation sequencing. Front Genet. 2013, 4, 145. [Google Scholar] [CrossRef] [PubMed]
- Panarelli, N.; Tyryshkin, K.; Wong, J.J.M.; Majewski, A.; Yang, X.J.; Scognamiglio, T.; Kim, M.K.; Bogardus, K.; Tuschl, T.; Chen, Y.T.; et al. Evaluating gastroenteropancreatic neuroendocrine tumors through microRNA sequencing. Endocr. Relat. Cancer 2019, 26, 47–57. [Google Scholar] [CrossRef]
- Travis, W.D.; Brambilla, E.; Burke, A.P.; Marx, A.; Nicholson, A.G. WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart, 4th ed.; International Agency for Research on Cancer: Lyon, France, 2015. [Google Scholar]
- Travis, W.D.; Rush, W.; Flieder, D.B.; Falk, R.; Fleming, M.V.; Gal, A.A.; Koss, M.N. Survival Analysis of 200 Pulmonary Neuroendocrine Tumors With Clarification of Criteria for Atypical Carcinoid and Its Separation From Typical Carcinoid. Am. J. Surg. Pathol. 1998, 22, 934–944. [Google Scholar] [CrossRef]
- Rindi, G.; Participants, A.A.O.F.C.C.; Klöppel, G.; Alhman, H.; Caplin, M.; Couvelard, A.; De Herder, W.W.; Erikssson, B.; Falchetti, A.; Falconi, M.; et al. TNM staging of foregut (neuro)endocrine tumors: A consensus proposal including a grading system. Virchows Archiv 2006, 449, 395–401. [Google Scholar] [CrossRef]
- Duda, R.O.; Hart, P.E.; Stork, D.G. Pattern Classification, 2nd ed.; Wiley: New York, NY, USA, 2001; Chapter 9; p. 484. [Google Scholar]
- Hafner, M.; Renwick, N.; Brown, M.; Mihailovic, A.; Holoch, D.; Lin, C.; Pena, J.T.; Nusbaum, J.D.; Morozov, P.; Ludwig, J.; et al. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA 2011, 17, 1697–1712. [Google Scholar] [CrossRef]
- Farazi, T.A.; Brown, M.; Morozov, P.; Ten Hoeve, J.J.; Ben-Dov, I.Z.; Hovestadt, V.; Hafner, M.; Renwick, N.; Mihailovic, A.; Wessels, L.F.; et al. Bioinformatic analysis of barcoded cDNA libraries for small RNA profiling by next-generation sequencing. Methods 2012, 58, 171–187. [Google Scholar] [CrossRef]
- Ren, R.; Tyryshkin, K.; Graham, C.H.; Koti, M.; Siemens, D.R. Comprehensive immune transcriptomic profiling analysis on bladder cancer reveals subtype specific gene expression patterns of prognostic relevance. Oncotarget 2017, 8, 70982–71001. [Google Scholar] [CrossRef]
- Mann, H.B.; Whitney, D.R. On a Test of Whether One of 2 Random Variables Is Stochastically Larger Than the Other. Ann. Math. Statist. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Spearman, C. ‘Footrule’ for Measuring Correlation. Brit. J. Psychol. 1906, 2, 89–108. [Google Scholar] [CrossRef]
- Fisher, R.A. On the interpretation of x(2) from contingency tables, and the calculation of P. J. R Stat. Soc. 1922, 85, 87–94. [Google Scholar] [CrossRef]
- Renwick, N.; Cekan, P.; Masry, P.A.; McGeary, S.E.; Miller, J.B.; Hafner, M.; Li, Z.; Mihailovic, A.; Morozov, P.; Brown, M.; et al. Multicolor microRNA FISH effectively differentiates tumor types. J. Clin. Investig. 2013, 123, 2694–2702. [Google Scholar] [CrossRef]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef]
- Farazi, T.A.; Hoell, J.I.; Morozov, P.; Tuschl, T. MicroRNAs in human cancer. Adv. Exp. Med. Biol. 2013, 774, 1–20. [Google Scholar] [CrossRef]
- Yan, J.-W.; Lin, J.-S.; He, X.-X. The emerging role of miR-375 in cancer. Int. J. Cancer 2014, 135, 1011–1018. [Google Scholar] [CrossRef]
| Features | Discovery Set | Validation Set | ||||||
|---|---|---|---|---|---|---|---|---|
| Carcinoids | NECs | Carcinoids | NECs | |||||
| TC (n = 11) | AC (n = 12) | SCLC (n = 9) | LCNEC (n = 12) | TC (n = 3) | AC (n = 3) | SCLC (n = 2) | LCNEC (n = 3) | |
| Male:female | 1:10 | 2:10 | 4:5 | 4:8 | 0:3 | 1:2 | 1:1 | 1:2 |
| Age avg (min, max) | 64 (41, 85) | 61 (43, 79) | 67 (50, 86) | 69 (45, 85) | 64 (50, 74) | 60 (54, 64) | 69 (65, 73) | 71 (67, 73) |
| Tumor size avg in mm (min, max) | 18 (5, 30) | 25 (12, 66) | 27 (11, 70) | 28 (10, 70) | 12 (3, 22) | 28 (4, 43) | 26 (18, 35) | 26 (15, 32) |
| Ki-67 avg (min, max) | 1 (<1, 3) | 5 (<1, 38) | 61 (33, 73) | 27.5 (7, 51) | <1 (<1, 3) | <1 (<1, 5) | 56 (53, 59) | 69 (62, 75) |
| Mitosis avg (min, max) | 0.3 (0, 1.3) | 3.9 (2, 18) | 88 (49, 183) | 27 (11, 85.3) | 0.3 (0, 1.3) | 2 (2, 3) | 80 (63, 97) | 42.7 (39, 60) |
| Necrosis (yes, no, focal) | 0, 11, 0 | 0, 4, 8 | 9, 0, 0 | 12, 0, 0 | 0, 3, 0 | 0, 3, 0 | 2, 0, 0 | 2, 0, 1 |
| pT category | ||||||||
| 1 | 10 (91%) | 8 (67%) | 6 (67%) | 4 (33%) | 3 (100%) | 1 (33%) | 1 (50%) | 1 (33%) |
| 2 | 0 (0%) | 4 (33%) | 2 (22%) | 8 (67%) | 0 (0%) | 2 (67%) | 1 (50%) | 2 (67%) |
| 3 | 0 (0%) | 0 (0%) | 1 (11%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| 4 | 1 (9%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| pN category | ||||||||
| Unknown | 1 (9%) | 0 (0%) | 1 (11%) | 1 (8%) | 1 (33%) | 0 (0%) | 0 (0%) | 0 (0%) |
| 0 | 8 (73%) | 9 (75%) | 5 (56%) | 5 (42%) | 2 (67%) | 3 (100%) | 2 (100%) | 2 (67%) |
| 1 | 2 (18%) | 1 (8%) | 1 (11%) | 4 (33%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (33%) |
| 2 | 0 (0%) | 2 (17%) | 2 (22%) | 2 (17%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Inter-set comparison | TC | AC | SCLC | LCNEC | ||||
| Gender | χ2 = 0.029, df = 1, p = 0.588 | χ2 = 0.417, df = 1, p = 0.519 | χ2 = 0.020, df = 1, p = 0.887 | χ2 = 0.000, df = 1, p = 1.000 | ||||
| Age | U = 15.0, p = 0.863, r = −0.063 | U = 14.5, p = 0.664, r = −0.131 | U = 8.0, p = 0.909, r = −0.071 | U= 17.5, p = 0.966, r = −0.019 | ||||
| miRNA | Median % of miRNA in all Samples |
|---|---|
| miR-375 | 8.2 |
| miR-21 | 7.9 |
| miR-143 | 4.1 |
| miR-141 | 2.9 |
| let-7a | 2.9 |
| let-7f | 2.5 |
| miR-30d | 2.4 |
| miR-148a | 2.0 |
| miRNA Cistron | Median % of miRNA Cistron in all Samples |
| cluster-mir-98(13) | 10.8 |
| cluster-mir-375(1) | 8.2 |
| cluster-mir-21(1) | 7.9 |
| cluster-mir-143(2) | 4.4 |
| Hierarchical Classifier Designation | Pathologic Diagnosis | |||
|---|---|---|---|---|
| Discovery Set | Validation Set | |||
| Carcinoids | NECs | Carcinoids | NECs | |
| Carcinoids | 20 | 0 | 6 | 0 |
| NECs | 3 | 21 | 0 | 5 |
| Overall accuracy | 41/44 (93%) | 11/11 (100%) | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wong, J.J.M.; Ginter, P.S.; Tyryshkin, K.; Yang, X.; Nanayakkara, J.; Zhou, Z.; Tuschl, T.; Chen, Y.-T.; Renwick, N. Classifying Lung Neuroendocrine Neoplasms through MicroRNA Sequence Data Mining. Cancers 2020, 12, 2653. https://doi.org/10.3390/cancers12092653
Wong JJM, Ginter PS, Tyryshkin K, Yang X, Nanayakkara J, Zhou Z, Tuschl T, Chen Y-T, Renwick N. Classifying Lung Neuroendocrine Neoplasms through MicroRNA Sequence Data Mining. Cancers. 2020; 12(9):2653. https://doi.org/10.3390/cancers12092653
Chicago/Turabian StyleWong, Justin J. M., Paula S. Ginter, Kathrin Tyryshkin, Xiaojing Yang, Jina Nanayakkara, Zier Zhou, Thomas Tuschl, Yao-Tseng Chen, and Neil Renwick. 2020. "Classifying Lung Neuroendocrine Neoplasms through MicroRNA Sequence Data Mining" Cancers 12, no. 9: 2653. https://doi.org/10.3390/cancers12092653
APA StyleWong, J. J. M., Ginter, P. S., Tyryshkin, K., Yang, X., Nanayakkara, J., Zhou, Z., Tuschl, T., Chen, Y.-T., & Renwick, N. (2020). Classifying Lung Neuroendocrine Neoplasms through MicroRNA Sequence Data Mining. Cancers, 12(9), 2653. https://doi.org/10.3390/cancers12092653







