Isolation and Enumeration of CTC in Colorectal Cancer Patients: Introduction of a Novel Cell Imaging Approach and Comparison to Cellular and Molecular Detection Techniques
Simple Summary
Abstract
1. Introduction
2. Results
2.1. Patients and Demographics
2.2. Spiking Experiments and Validation of Cytological and RT-qPCR Detection Techniques
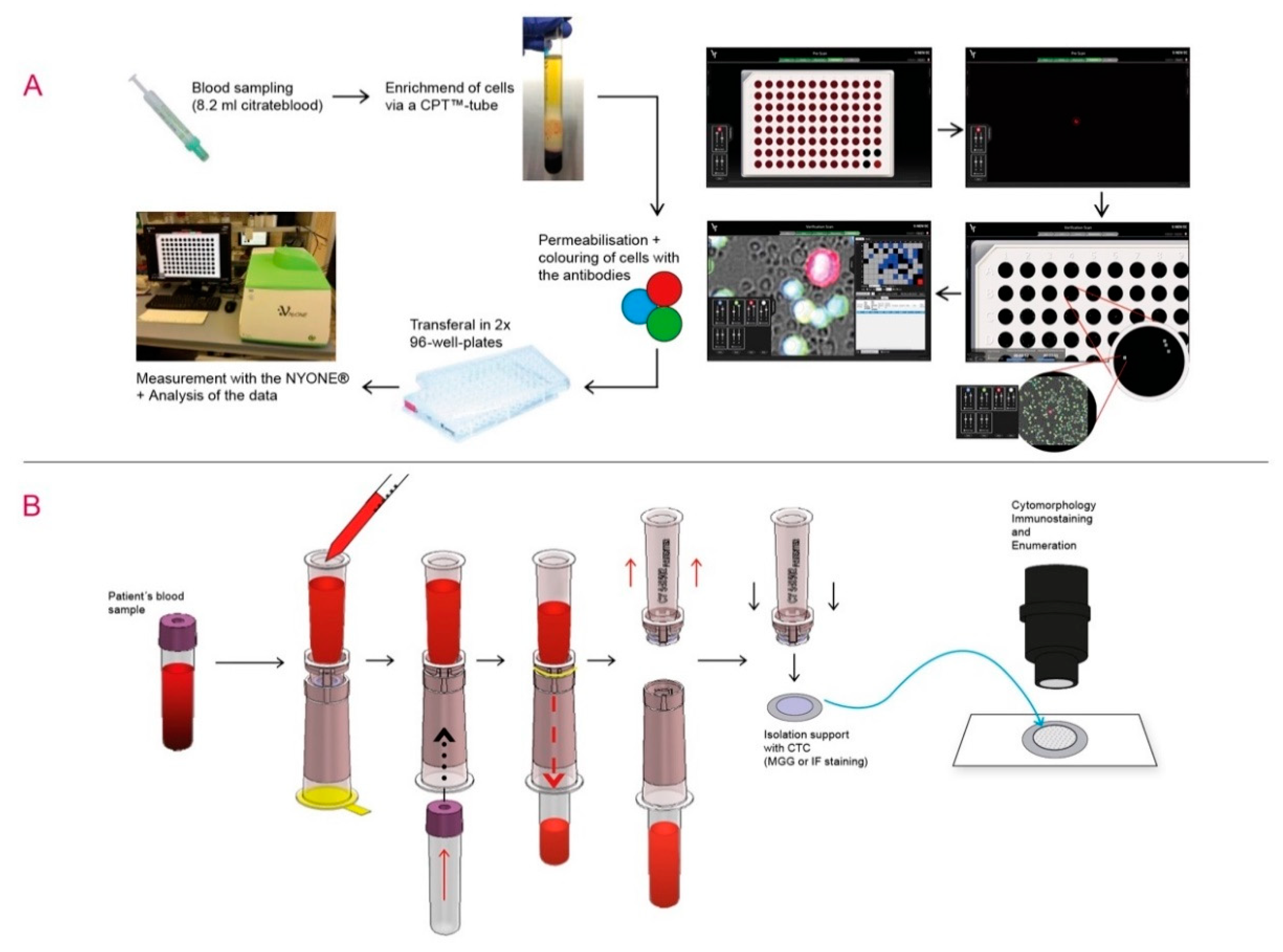
2.3. Detection of CTC by Automated Microscopy with the Cell Imager NYONE®
2.4. Capture and Detection of CTC by ScreenCell® Cyto IS Device
2.5. Relative Quantification of the CTC Load by CK20 RT-qPCR
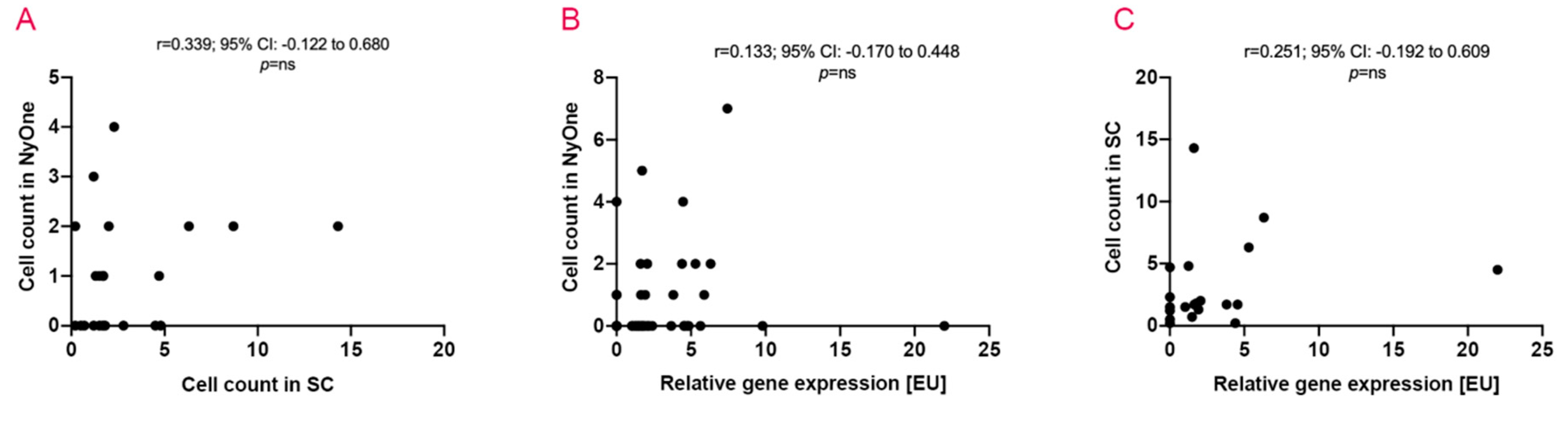
2.6. The Coherence of Applied Detection Methods
3. Discussion
4. Materials and Methods
4.1. Patient/Proband Recruitment and Sample Preparation
4.2. Sample Analysis by IF Staining and Semi-Automated Microscopy—NYONE®
4.3. Sample Analysis by Size-Dependent Filtration and IF Staining—ScreenCell®
4.4. Sample Analysis by Molecular Analysis of mRNA—Semi-Quantitative CK20 RT-qPCR
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CK-20 | Cytokeratin-20 |
| CRC | Colorectal cancer |
| CTC | Circulating tumour cells |
| EGFR | Epithelial Growth Factor Receptor |
| EMT | Epithelial-Mesenchymal-Transition |
| EpCAM | Epithelial adhesion molecule |
| EU | expression units |
| pan-CK | pan-Cytokeratin |
| RT-qPCR | Real Time quantitative Polymerase Chain Reaction |
| UICC | Union internationale contre le cancer |
References
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef]
- Schreuders, E.H.; Ruco, A.; Rabeneck, L.; Schoen, R.E.; Sung, J.J.Y.; Young, G.P.; Kuipers, E.J. Colorectal cancer screening: A global overview of existing programmes. Gut 2015, 64, 1637–1649. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Le Tourneau, C.; Delord, J.-P.; Gonçalves, A.; Gavoille, C.; Dubot, C.; Isambert, N.; Campone, M.; Trédan, O.; Massiani, M.-A.; Mauborgne, C.; et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): A multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015, 16, 1324–1334. [Google Scholar] [CrossRef]
- Sicklick, J.K.; Kato, S.; Okamura, R.; Schwaederle, M.; Hahn, M.E.; Williams, C.B.; De, P.; Krie, A.; Piccioni, D.E.; Miller, V.A.; et al. Molecular profiling of cancer patients enables personalized combination therapy: The I-PREDICT study. Nat. Med. 2019, 25, 744–750. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. Liquid biopsy and minimal residual disease—Latest advances and implications for cure. Nat. Rev. Clin. Oncol 2019, 16, 409–424. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Circulating tumor cells: Liquid biopsy of cancer. Clin. Chem. 2013, 59, 110–118. [Google Scholar] [CrossRef]
- Hendricks, A.; Eggebrecht, G.-L.; Bernsmeier, A.; Geisen, R.; Dall, K.; Trauzold, A.; Becker, T.; Kalthoff, H.; Schafmayer, C.; Röder, C.; et al. Identifying patients with an unfavorable prognosis in early stages of colorectal carcinoma. Oncotarget 2018, 9, 27423–27434. [Google Scholar] [CrossRef]
- Hinz, S.; Röder, C.; Tepel, J.; Hendricks, A.; Schafmayer, C.; Becker, T.; Kalthoff, H. Cytokeratin 20 positive circulating tumor cells are a marker for response after neoadjuvant chemoradiation but not for prognosis in patients with rectal cancer. BMC Cancer 2015, 15, 953. [Google Scholar] [CrossRef]
- Hinz, S.; Hendricks, A.; Wittig, A.; Schafmayer, C.; Tepel, J.; Kalthoff, H.; Becker, T.; Röder, C. Detection of circulating tumor cells with CK20 RT-PCR is an independent negative prognostic marker in colon cancer patients—A prospective study. BMC Cancer 2017, 17, 53. [Google Scholar] [CrossRef]
- Huang, X.; Gao, P.; Song, Y.; Sun, J.; Chen, X.; Zhao, J.; Xu, H.; Wang, Z. Meta-analysis of the prognostic value of circulating tumor cells detected with the CellSearch system in colorectal cancer. BMC Cancer 2015, 15, 202. [Google Scholar] [CrossRef]
- Normanno, N.; Cervantes, A.; Ciardiello, F.; De Luca, A.; Pinto, C. The liquid biopsy in the management of colorectal cancer patients: Current applications and future scenarios. Cancer Treat. Rev. 2018, 70, 1–8. [Google Scholar] [CrossRef]
- Lv, Q.; Gong, L.; Zhang, T.; Ye, J.; Chai, L.; Ni, C.; Mao, Y. Prognostic value of circulating tumor cells in metastatic breast cancer: A systemic review and meta-analysis. Clin. Transl. Oncol. 2016, 18, 322–330. [Google Scholar] [CrossRef]
- Zhou, Y.; Bian, B.; Yuan, X.; Xie, G.; Ma, Y.; Shen, L. Prognostic value of circulating tumor cells in ovarian cancer: A meta-analysis. PLoS ONE 2015, 10, e0130873. [Google Scholar] [CrossRef]
- Ma, X.; Xiao, Z.; Li, X.; Wang, F.; Zhang, J.; Zhou, R.; Wang, J.; Liu, L. Prognostic role of circulating tumor cells and disseminated tumor cells in patients with prostate cancer: A systematic review and meta-analysis. Tumour Biol. 2014, 35, 5551–5560. [Google Scholar] [CrossRef]
- Bork, U.; Rahbari, N.N.; Schölch, S.; Reissfelder, C.; Kahlert, C.; Büchler, M.W.; Weitz, J.; Koch, M. Circulating tumour cells and outcome in non-metastatic colorectal cancer: A prospective study. Br. J. Cancer 2015, 112, 1306–1313. [Google Scholar] [CrossRef]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009, 119, 1420–1428. [Google Scholar] [CrossRef]
- Joosse, S.A.; Hannemann, J.; Spötter, J.; Bauche, A.; Andreas, A.; Müller, V.; Pantel, K. Changes in keratin expression during metastatic progression of breast cancer: Impact on the detection of circulating tumor cells. Clin. Cancer Res. 2012, 18, 993–1003. [Google Scholar] [CrossRef]
- Gorges, T.M.; Tinhofer, I.; Drosch, M.; Röse, L.; Zollner, T.M.; Krahn, T.; von Ahsen, O. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer 2012, 12, 178. [Google Scholar] [CrossRef]
- Zhang, D.; Zhao, L.; Zhou, P.; Ma, H.; Huang, F.; Jin, M.; Dai, X.; Zheng, X.; Huang, S.; Zhang, T. Circulating tumor microemboli (CTM) and vimentin+ circulating tumor cells (CTCs) detected by a size-based platform predict worse prognosis in advanced colorectal cancer patients during chemotherapy. Cancer Cell Int. 2017, 17, 6. [Google Scholar] [CrossRef]
- Tsutsuyama, M.; Nakanishi, H.; Yoshimura, M.; Oshiro, T.; Kinoshita, T.; Komori, K.; Shimizu, Y.; Ichinosawa, Y.; Kinuta, S.; Wajima, K.; et al. Detection of circulating tumor cells in drainage venous blood from colorectal cancer patients using a new filtration and cytology-based automated platform. PLoS ONE 2019, 14, e0212221. [Google Scholar] [CrossRef]
- Tan, K.; Leong, S.M.; Kee, Z.; Caramat, P.V.; Teo, J.; Blanco, M.V.M.; Koay, E.S.C.; Cheong, W.K.; Soh, T.I.-P.; Yong, W.P.; et al. Longitudinal monitoring reveals dynamic changes in circulating tumor cells (CTCs) and CTC-associated miRNAs in response to chemotherapy in metastatic colorectal cancer patients. Cancer Lett. 2018, 423, 1–8. [Google Scholar] [CrossRef]
- Yu, M.; Stott, S.; Toner, M.; Maheswaran, S.; Haber, D.A. Circulating tumor cells: Approaches to isolation and characterization. J. Cell Biol. 2011, 192, 373–382. [Google Scholar] [CrossRef]
- Meng, S.; Tripathy, D.; Frenkel, E.P.; Shete, S.; Naftalis, E.Z.; Huth, J.F.; Beitsch, P.D.; Leitch, M.; Hoover, S.; Euhus, D.; et al. Circulating tumor cells in patients with breast cancer dormancy. Clin. Cancer Res. 2004, 10, 8152–8162. [Google Scholar] [CrossRef]
- Wong, S.C.C.; Chan, C.M.L.; Ma, B.B.Y.; Hui, E.P.; Ng, S.S.M.; Lai, P.B.S.; Cheung, M.T.; Lo, E.S.F.; Chan, A.K.C.; Lam, M.Y.Y.; et al. Clinical significance of cytokeratin 20-positive circulating tumor cells detected by a refined immunomagnetic enrichment assay in colorectal cancer patients. Clin. Cancer Res. 2009, 15, 1005–1012. [Google Scholar] [CrossRef]
- Kamiyama, H.; Noda, H.; Konishi, F.; Rikiyama, T. Molecular biomarkers for the detection of metastatic colorectal cancer cells. World J. Gastroenterol. 2014, 20, 8928–8938. [Google Scholar] [CrossRef]
- Gorges, T.M.; Stein, A.; Quidde, J.; Hauch, S.; Röck, K.; Riethdorf, S.; Joosse, S.A.; Pantel, K. Improved detection of circulating tumor cells in metastatic colorectal cancer by the combination of the CellSearch® system and the AdnaTest®. PLoS ONE 2016, 11, e0155126. [Google Scholar] [CrossRef]
- Van der Toom, E.E.; Verdone, J.E.; Gorin, M.A.; Pienta, K.J. Technical challenges in the isolation and analysis of circulating tumor cells. Oncotarget 2016, 7, 62754–62766. [Google Scholar] [CrossRef]
- Todenhöfer, T.; Hennenlotter, J.; Feyerabend, S.; Aufderklamm, S.; Mischinger, J.; Kühs, U.; Gerber, V.; Fetisch, J.; Schilling, D.; Hauch, S.; et al. Preliminary experience on the use of the Adnatest® system for detection of circulating tumor cells in prostate cancer patients. Anticancer Res. 2012, 32, 3507–3513. [Google Scholar]
- Obermayr, E.; Bednarz-Knoll, N.; Orsetti, B.; Weier, H.-U.; Lambrechts, S.; Castillo-Tong, D.C.; Reinthaller, A.; Braicu, E.I.; Mahner, S.; Sehouli, J.; et al. Circulating tumor cells: Potential markers of minimal residual disease in ovarian cancer? A study of the OVCAD consortium. Oncotarget 2017, 8, 106415–106428. [Google Scholar] [CrossRef]
- Nicolazzo, C.; Raimondi, C.; Francescangeli, F.; Ceccarelli, S.; Trenta, P.; Magri, V.; Marchese, C.; Zeuner, A.; Gradilone, A.; Gazzaniga, P. EpCAM-expressing circulating tumor cells in colorectal cancer. Int. J. Biol. Markers 2017, 32, e415–e420. [Google Scholar] [CrossRef]
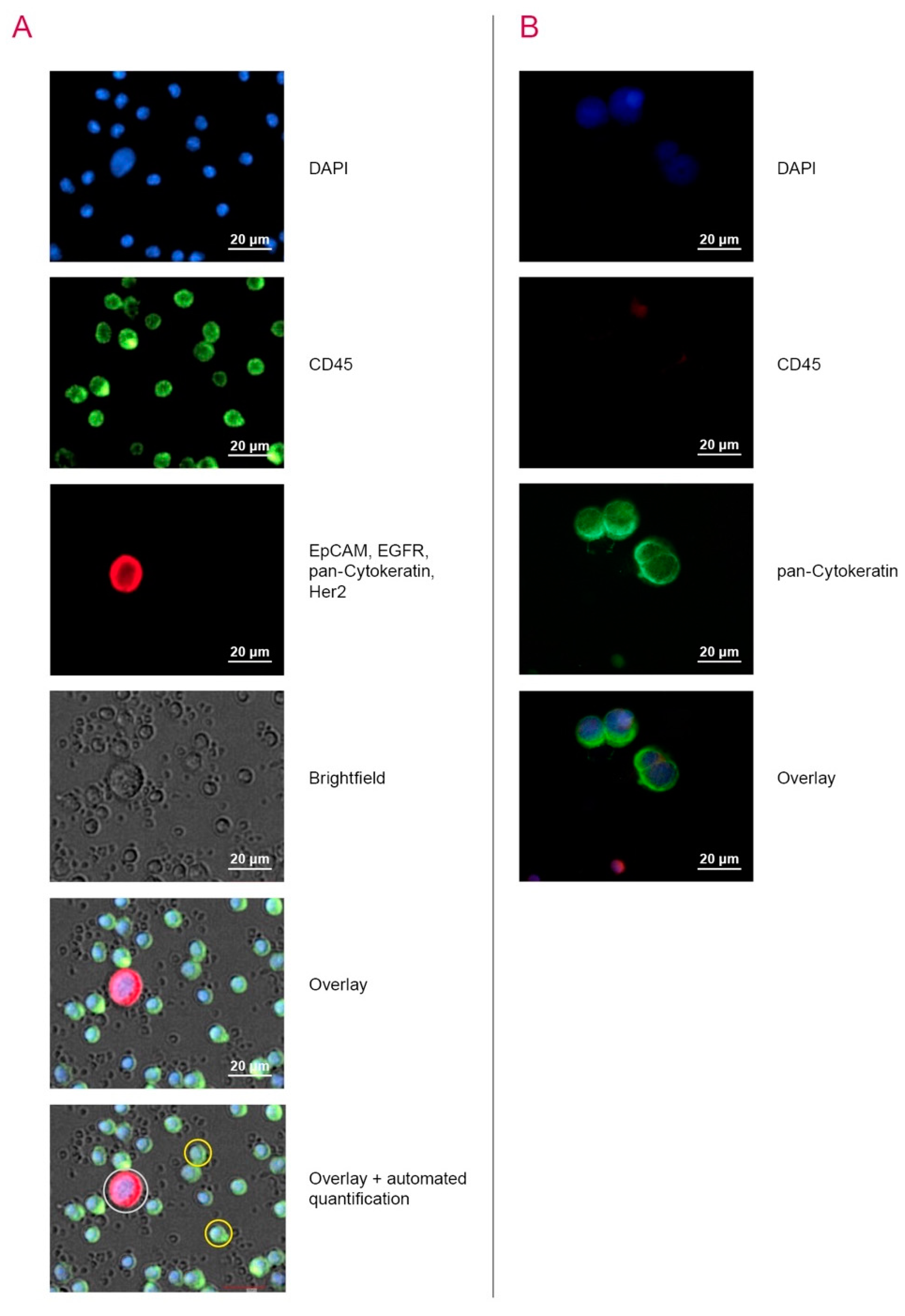
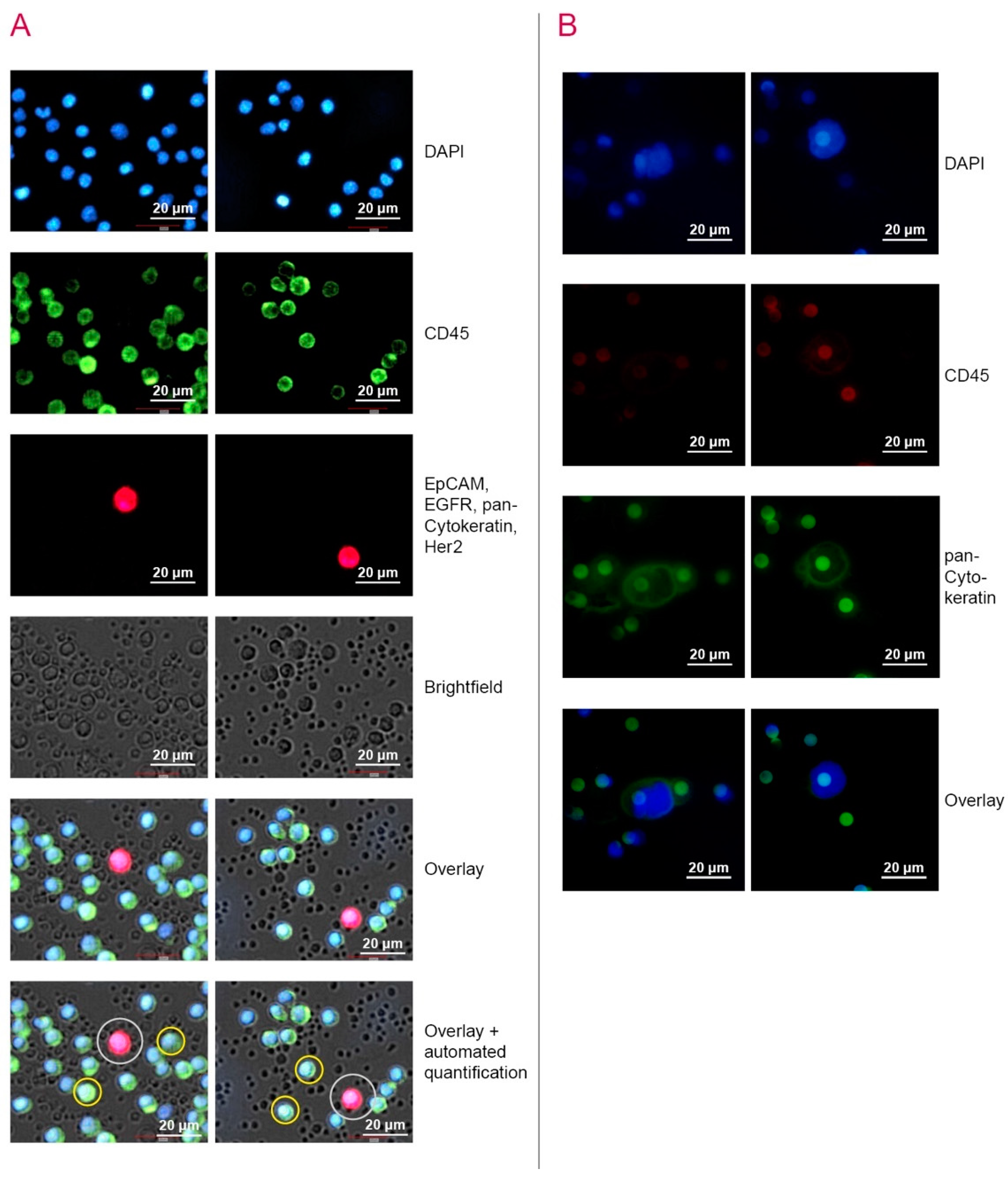
| Parameters | Total N (%) | NYONE® N (%) | ScreenCell® N (%) | CK20 RT-qPCR N (%) |
|---|---|---|---|---|
| 57 (100) | 44 (100) | 31 (100) | 41 (100) | |
| Gender | ||||
| Male | 36 (63.2) | 26 (59.1) | 21 (67.7) | 24 (58.5) |
| Female | 21 (36.8) | 18 (40.9) | 10 (32.3) | 17 (41.5) |
| Age | ||||
| Median (range) | 66 (42–89) | 66 (45–89) | 66 (42–89) | 68 (45–89) |
| <65 | 27 (47.4) | 18 (40.9) | 15 (48.4) | 17 (41.5) |
| ≥65 | 30 (52.6) | 26 (59.1) | 16 (51.6) | 24 (58.5) |
| Tumour site | ||||
| colon | 38 (66.7) | 32 (72.7) | 22 (71.0) | 29 (70.7) |
| right | 17 (44.7) | 16 (50.0) | 9 (40.9) | 15 (51.7) |
| left | 21 (55.3) | 16 (50.0) | 13 (59.1) | 14 (48.3) |
| Rectum | 19 (33.3) | 12 (27.3) | 9 (29.0) | 12 (29.3) |
| UICC stage | ||||
| I | 15 (26.3) | 12 (27.3) | 9 (29.0) | 12 (29.3) |
| II | 10 (17.5) | 9 (20.5) | 6 (19.4) | 8 (19.5) |
| III | 24 (42.1) | 18 (40.9) | 12 (38.7) | 16 (39.0) |
| IV | 8 (14.0) | 5 (11.4) | 4 (12.9) | 5 (12.2) |
| Parameters | NYONE® | ScreenCell® | CK20 RT-qPCR | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Positive N (%) | Mean (SD) | p | Positive N (%) | Mean (SD) | p | Positive N (%) | Mean (SD) | p | |
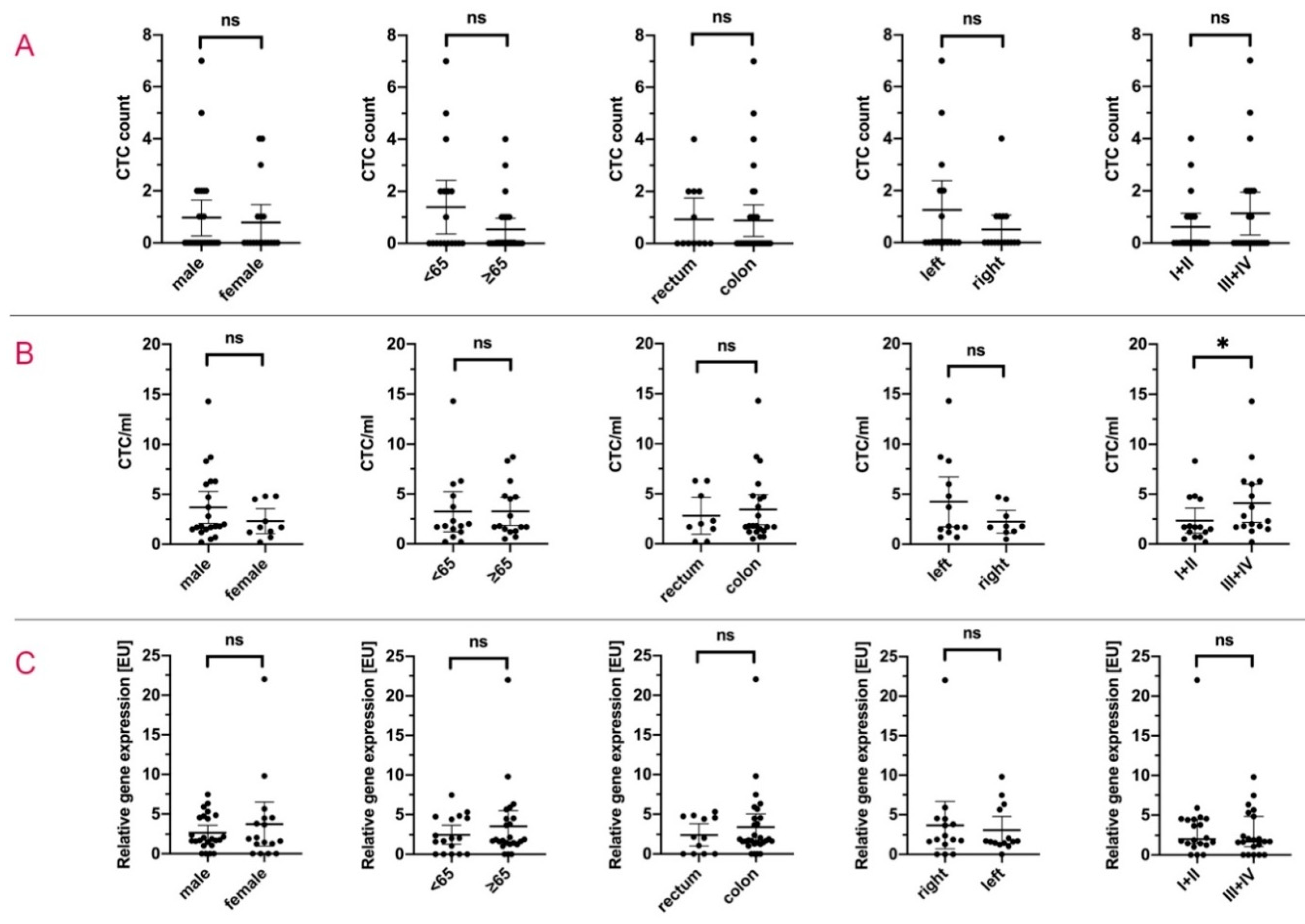
| Total | 16/44 (36.4) | 0.89 (1.57) | 31/31 (100) | 3.25 (3.10) | 33/41 (80.5) | 3.11 (3.81) | |||
| Gender | |||||||||
| Male | 10/26 (38.5) | 0.96 (1.71) | 0.708 | 21/21 (100) | 3.69 (3.52) | 0.257 | 20/24 (83.3) | 2.67 (2.19) | 0.383 |
| Female | 6/18 (33.3) | 0.78 (1.40) | 10/10 (100) | 2.32 (1.74) | 13/17 (76.5) | 3.74 (5.35) | |||
| Age | |||||||||
| <65 | 8/18 (44.4) | 1.39 (2.06) | 0.078 | 15/15 (100) | 3.23 (3.62) | 0.980 | 12/17 (70.6) | 2.48 (2.31) | 0.379 |
| ≥65 | 8/26 (30.8) | 0.54 (1.03) | 16/16 (100) | 3.26 (2.65) | 21/24 (87.5) | 3.56 (4.58) | |||
| Tumour site | |||||||||
| Rectum | 5/12 (41.7) | 0.92 (1.31) | 0.939 | 9/9 (100) | 2.81 (2.39) | 0.624 | 8/12 (66.7) | 2.43 (2.20) | 0.466 |
| colon | 11/32 (34.4) | 0.88 (1.68) | 22/22 (100) | 3.43 (3.38) | 25/29 (86.2) | 3.40 (4.30) | |||
| right | 5/16 (31.3) | 0.50 (1.03) | 0.212 | 9/9 (100) | 2.26 (1.46) | 0.182 | 12/15 (80.0) | 3.69 (5.37) | 0.713 |
| left | 6/16 (37.5) | 1.25 (2.11) | 13/13 (100) | 4.24 (4.10) | 13/14 (92.9) | 3.08 (2.94) | |||
| UICC stage | |||||||||
| I + II | 3/12 (25.0) | 0.58 (1.24) | 0.503 | 15/15 (100) | 2.35 (2.24) | 0.039 | 11/12 (91.7) | 3.54 (4.70) | 0.491 |
| III + IV | 6/18 (33.3) | 1.06 (1.89) | 16/16 (100) | 4.09 (3.60) | 13/16 (81.3) | 2.71 (2.76) | |||
| Parameters | NYONE® | ScreenCell® | CK20 RT-qPCR | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Positive N (%) | Mean (SD) | p | Positive N (%) | Mean (SD) | p | Positive N (%) | Mean (SD) | p | |
| Total | 11/21 (52.4) | 0.90 (1.03) | 21/21 (100) | 3.01 (3.16) | 15/21 (71.4) | 2.90 (3.81) | |||
| Gender | |||||||||
| Male | 7/14 (50.0) | 0.86 (0.95) | 0.785 | 14/14 (100) | 3.34 (3.97) | 0.541 | 10/14 (71.4) | 2.16 (2.13) | 0.328 |
| Female | 4/7 (57.1) | 1.00 (1.41) | 7/7 (100) | 2.36 (1.69) | 5/7 (71.4) | 4.37 (7.88) | |||
| Age | |||||||||
| <65 | 6/9 (66.7) | 1.44 (1.33) | 0.047 | 9/9 (100) | 3.30 (4.50) | 0.746 | 5/9 (55.6) | 1.99 (2.22) | 0.464 |
| ≥65 | 5/12 (41.7) | 0.50 (0.67) | 12/12 (100) | 2.80 (2.40) | 10/12 (83.3) | 3.58 (6.04) | |||
| Tumour site | |||||||||
| Rectum | 5/7 (71.4) | 1.57 (1.40) | 0.045 | 7/7 (100) | 2.03 (2.06) | 0.356 | 4/7 (57.1) | 2.33 (2.39) | 0.709 |
| colon | 6/14 (42.9) | 0.57 (0.76) | 14/14 (100) | 3.51 (3.84) | 11/14 (78.6) | 3.18 (5.66) | |||
| right | 3/7 (42.9) | 0.43 (0.54) | 0.502 | 7/7 (100) | 2.24 (1.67) | 0.834 | 4/7 (57.1) | 4.22 (7.96) | 0.517 |
| left | 3/7 (42.9) | 0.71 (0.95) | 7/7 (100) | 2.03 (2.06) | 7/7 (100) | 2.15 (1.85) | |||
| UICC stage | |||||||||
| I + II | 4/12 (33.3) | 0.42 (0.67) | 0.017 | 8/8 (100) | 2.08 (1.64) | 0.148 | 7/8 (87.5) | 3.50 (6.05) | 0.521 |
| III + IV | 7/9 (77.8) | 1.56 (1.24) | 6/6 (100) | 4.26 (4.65) | 4/6 (66.7) | 2.10 (2.28) | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hendricks, A.; Brandt, B.; Geisen, R.; Dall, K.; Röder, C.; Schafmayer, C.; Becker, T.; Hinz, S.; Sebens, S. Isolation and Enumeration of CTC in Colorectal Cancer Patients: Introduction of a Novel Cell Imaging Approach and Comparison to Cellular and Molecular Detection Techniques. Cancers 2020, 12, 2643. https://doi.org/10.3390/cancers12092643
Hendricks A, Brandt B, Geisen R, Dall K, Röder C, Schafmayer C, Becker T, Hinz S, Sebens S. Isolation and Enumeration of CTC in Colorectal Cancer Patients: Introduction of a Novel Cell Imaging Approach and Comparison to Cellular and Molecular Detection Techniques. Cancers. 2020; 12(9):2643. https://doi.org/10.3390/cancers12092643
Chicago/Turabian StyleHendricks, Alexander, Burkhard Brandt, Reinhild Geisen, Katharina Dall, Christian Röder, Clemens Schafmayer, Thomas Becker, Sebastian Hinz, and Susanne Sebens. 2020. "Isolation and Enumeration of CTC in Colorectal Cancer Patients: Introduction of a Novel Cell Imaging Approach and Comparison to Cellular and Molecular Detection Techniques" Cancers 12, no. 9: 2643. https://doi.org/10.3390/cancers12092643
APA StyleHendricks, A., Brandt, B., Geisen, R., Dall, K., Röder, C., Schafmayer, C., Becker, T., Hinz, S., & Sebens, S. (2020). Isolation and Enumeration of CTC in Colorectal Cancer Patients: Introduction of a Novel Cell Imaging Approach and Comparison to Cellular and Molecular Detection Techniques. Cancers, 12(9), 2643. https://doi.org/10.3390/cancers12092643







