Therapeutically Significant MicroRNAs in Primary and Metastatic Brain Malignancies
Simple Summary
Abstract
1. Introduction
2. Brain Cancer
2.1. Primary Brain Cancers
2.2. Brain Metastasis
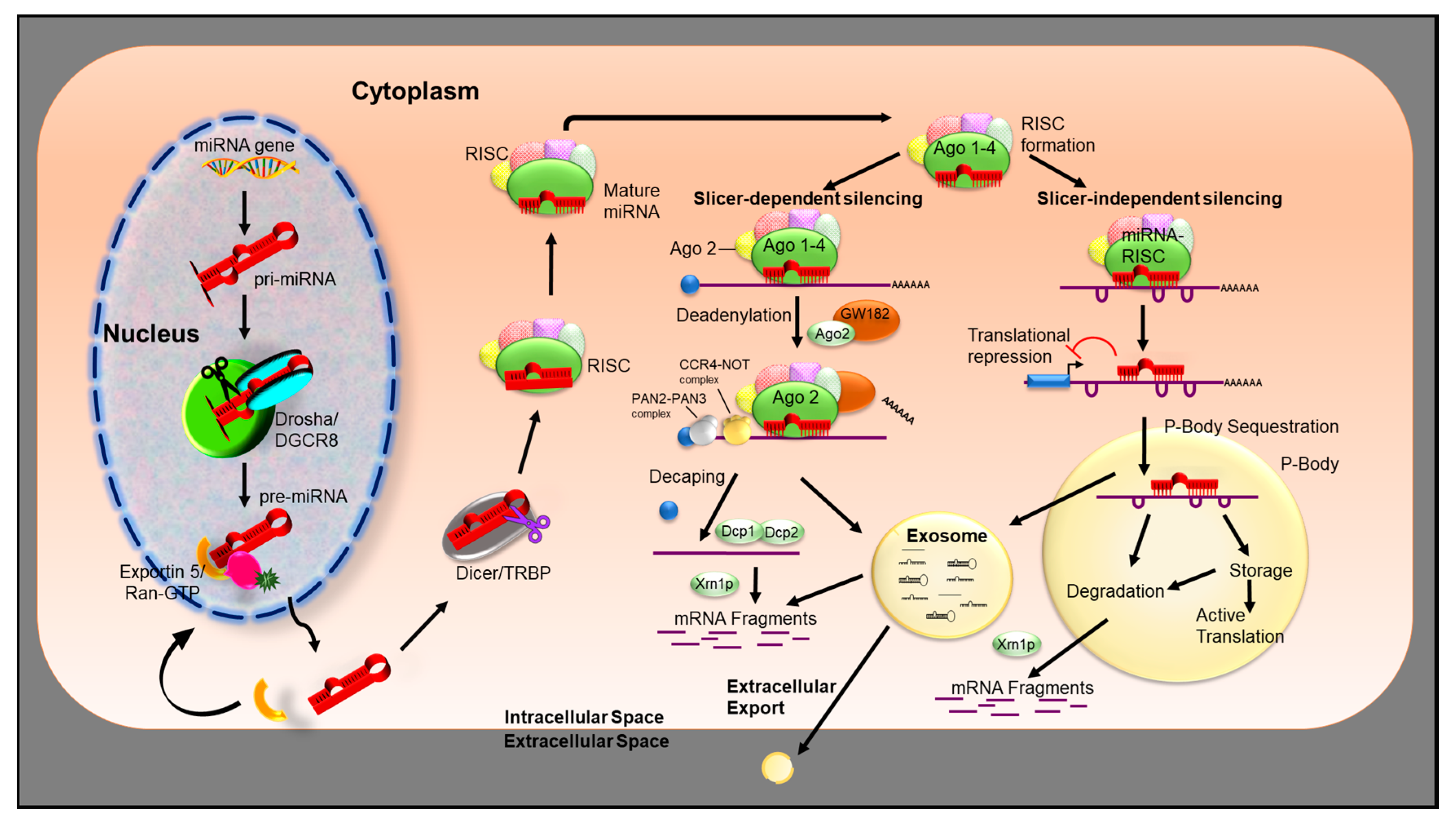
3. miRNA Biogenesis
miRNA Mechanism of Action
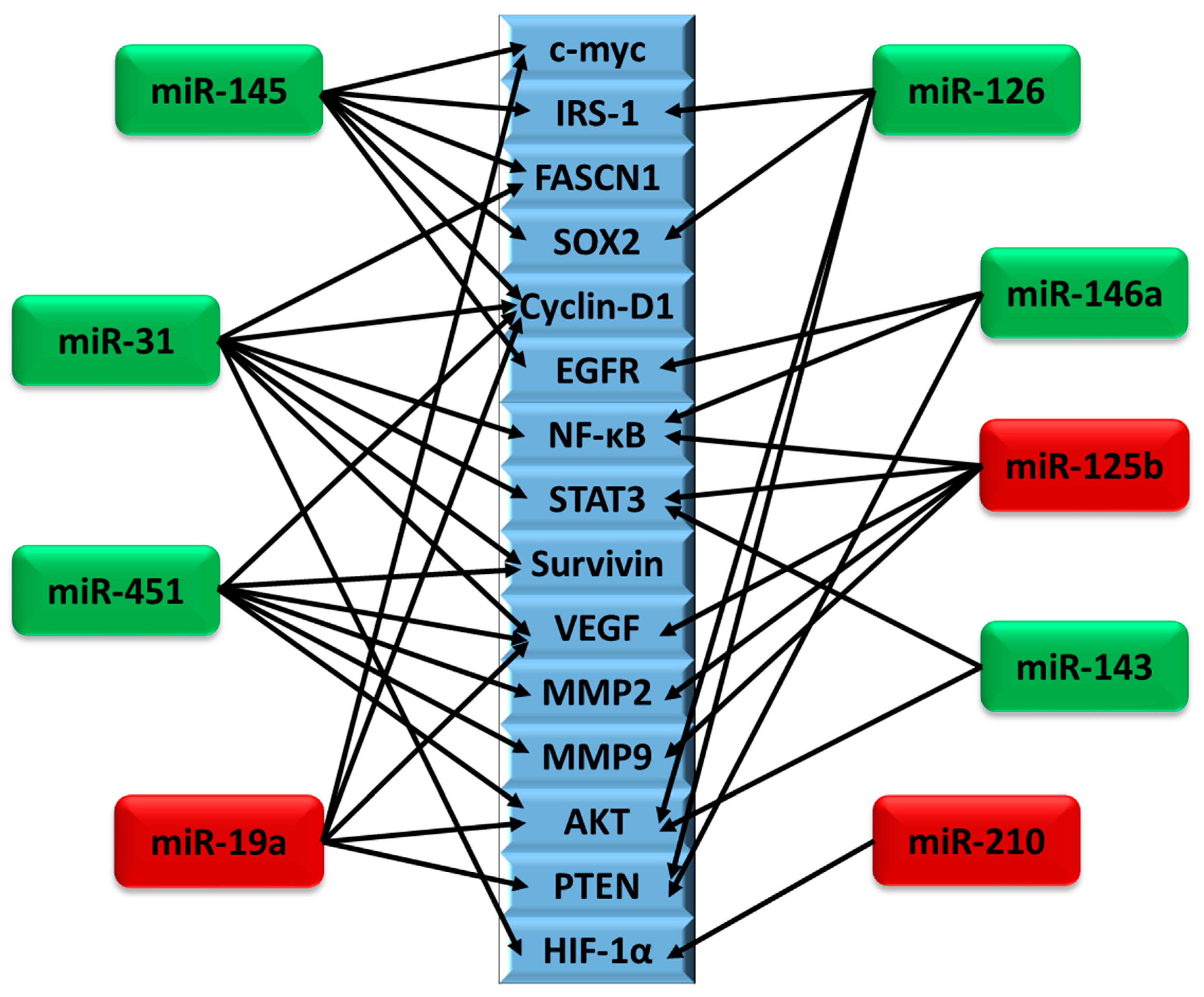
4. miRNAs Differentially Regulated in Brain Cancer
4.1. miR-145
4.2. miR-31
4.3. miR-451
4.4. miR-19a
4.5. miR-143
4.6. miR-125b
4.7. miR-328
4.8. miR-210
4.9. miR-146a
4.10. miR-126
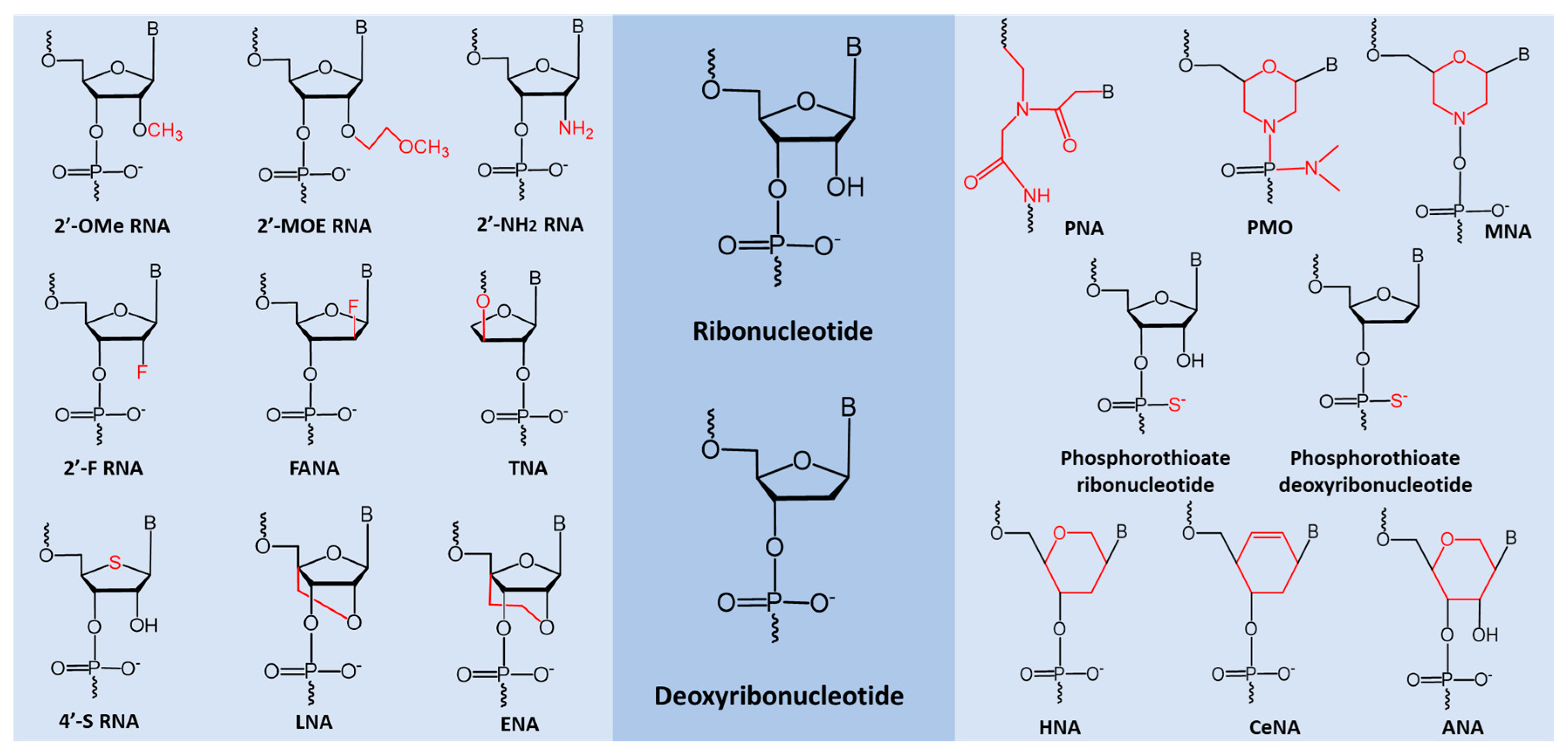
5. miRNA Based Therapeutic Oligonucleotides
6. Conclusions and Prospects
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McNeill, K.A. Epidemiology of brain tumors. Neurol. Clin. 2016, 34, 981–998. [Google Scholar] [CrossRef] [PubMed]
- Shah, V.; Kochar, P. Brain cancer: Implication to disease, therapeutic strategies and tumor targeted drug delivery approaches. Recent Pat. Anticancer Drug Discov. 2018, 13, 70–85. [Google Scholar] [CrossRef] [PubMed]
- Franchino, F.; Ruda, R.; Soffietti, R. Mechanisms and therapy for cancer metastasis to the brain. Front. Oncol. 2018, 8, 161. [Google Scholar] [CrossRef]
- Lapointe, S.; Perry, A.; Butowski, N.A. Primary brain tumours in adults. Lancet 2018, 392, 432–446. [Google Scholar] [CrossRef]
- Lukas, R.V.; Wainwright, D.A.; Ladomersky, E.; Sachdev, S.; Sonabend, A.M.; Stupp, R. Newly diagnosed glioblastoma: A review on clinical management. Oncology 2019, 33, 91–100. [Google Scholar]
- Ohgaki, H.; Kleihues, P. The definition of primary and secondary glioblastoma. Clin. Cancer Res. 2013, 19, 764–772. [Google Scholar] [CrossRef]
- Achrol, A.S.; Rennert, R.C.; Anders, C.; Soffietti, R.; Ahluwalia, M.S.; Nayak, L.; Peters, S.; Arvold, N.D.; Harsh, G.R.; Steeg, P.S.; et al. Brain metastases. Nat. Rev. Dis. Primers 2019, 5, 5. [Google Scholar] [CrossRef]
- Fidler, I.J. The biology of brain metastasis: Challenges for therapy. Cancer J. 2015, 21, 284–293. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S. The nuclear rnase iii drosha initiates microrna processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: Microrna biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. Microrna genes are transcribed by rna polymerase ii. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef] [PubMed]
- Borchert, G.M.; Lanier, W.; Davidson, B.L. Rna polymerase iii transcribes human micrornas. Nat. Struct. Mol. Biol. 2006, 13, 1097–1101. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, Y.; Yeom, K.-H.; Nam, J.-W.; Heo, I.; Rhee, J.-K.; Sohn, S.Y.; Cho, Y.; Zhang, B.-T.; Kim, V.N. Molecular basis for the recognition of primary micrornas by the drosha-dgcr8 complex. Cell 2006, 125, 887–901. [Google Scholar] [CrossRef] [PubMed]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary micrornas by the microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Gregory, R.I.; Yan, K.-p.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The microprocessor complex mediates the genesis of micrornas. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, Y.; Yeom, K.-H.; Kim, Y.-K.; Jin, H.; Kim, V.N. The drosha-dgcr8 complex in primary microrna processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-micrornas and short hairpin rnas. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef]
- Bohnsack, M.T.; Czaplinski, K.; GÖRLICH, D. Exportin 5 is a rangtp-dependent dsrna-binding protein that mediates nuclear export of pre-mirnas. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef]
- Lund, E.; Dahlberg, J. Substrate selectivity of exportin 5 and dicer in the biogenesis of micrornas. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 59–66. [Google Scholar] [CrossRef]
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A high-resolution structure of the pre-microrna nuclear export machinery. Science 2009, 326, 1275–1279. [Google Scholar] [CrossRef]
- Zeng, Y.; Cullen, B.R. Structural requirements for pre-microrna binding and nuclear export by exportin 5. Nucleic Acids Res. 2004, 32, 4776–4785. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Zhou, K.; Smith, A.M.; Noland, C.L.; Doudna, J.A. Differential roles of human dicer-binding proteins trbp and pact in small rna processing. Nucleic Acids Res. 2013, 41, 6568–6576. [Google Scholar] [CrossRef] [PubMed]
- Kawamata, T.; Yoda, M.; Tomari, Y. Multilayer checkpoints for microrna authenticity during risc assembly. EMBO Rep. 2011, 12, 944–949. [Google Scholar] [CrossRef] [PubMed]
- Vella, M.C.; Choi, E.-Y.; Lin, S.-Y.; Reinert, K.; Slack, F.J. The c. Elegans microrna let-7 binds to imperfect let-7 complementary sites from the lin-41 3′ utr. Genes Dev. 2004, 18, 132–137. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in c. Elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Brennecke, J.; Stark, A.; Russell, R.B.; Cohen, S.M. Principles of microrna–target recognition. PLoS Biol. 2005, 3, e85. [Google Scholar] [CrossRef]
- Doench, J.G.; Sharp, P.A. Specificity of microrna target selection in translational repression. Genes Dev. 2004, 18, 504–511. [Google Scholar] [CrossRef]
- Krek, A.; Grün, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; Da Piedade, I.; Gunsalus, K.C.; Stoffel, M. Combinatorial microrna target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef]
- Lewis, B.P.; Shih, I.-h.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of mammalian microrna targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef]
- Jeffries, C.D.; Fried, H.M.; Perkins, D.O. Nuclear and cytoplasmic localization of neural stem cell micrornas. RNA 2011, 17, 675–686. [Google Scholar] [CrossRef]
- Miyoshi, K.; Miyoshi, T.; Siomi, H. Many ways to generate microrna-like small rnas: Non-canonical pathways for microrna production. Mol. Genet Genom. 2010, 284, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Okamura, K.; Hagen, J.W.; Duan, H.; Tyler, D.M.; Lai, E.C. The mirtron pathway generates microrna-class regulatory rnas in drosophila. Cell 2007, 130, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microrna precursors that bypass drosha processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Ender, C.; Krek, A.; Friedlander, M.R.; Beitzinger, M.; Weinmann, L.; Chen, W.; Pfeffer, S.; Rajewsky, N.; Meister, G. A human snorna with microrna-like functions. Mol. Cell 2008, 32, 519–528. [Google Scholar] [CrossRef]
- Babiarz, J.E.; Ruby, J.G.; Wang, Y.; Bartel, D.P.; Blelloch, R. Mouse es cells express endogenous shrnas, sirnas, and other microprocessor-independent, dicer-dependent small rnas. Genes Dev. 2008, 22, 2773–2785. [Google Scholar] [CrossRef]
- Cole, C.; Sobala, A.; Lu, C.; Thatcher, S.R.; Bowman, A.; Brown, J.W.; Green, P.J.; Barton, G.J.; Hutvagner, G. Filtering of deep sequencing data reveals the existence of abundant dicer-dependent small rnas derived from trnas. RNA 2009, 15, 2147–2160. [Google Scholar] [CrossRef]
- Lee, Y.S.; Shibata, Y.; Malhotra, A.; Dutta, A. A novel class of small rnas: Trna-derived rna fragments (trfs). Genes Dev. 2009, 23, 2639–2649. [Google Scholar] [CrossRef]
- Miyoshi, K.; Miyoshi, T.; Hartig, J.V.; Siomi, H.; Siomi, M.C. Molecular mechanisms that funnel rna precursors into endogenous small-interfering rna and microrna biogenesis pathways in drosophila. RNA 2010, 16, 506–515. [Google Scholar] [CrossRef]
- Bogerd, H.P.; Karnowski, H.W.; Cai, X.; Shin, J.; Pohlers, M.; Cullen, B.R. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral micrornas. Mol. Cell 2010, 37, 135–142. [Google Scholar] [CrossRef]
- Cheloufi, S.; Dos Santos, C.O.; Chong, M.M.; Hannon, G.J. A dicer-independent mirna biogenesis pathway that requires ago catalysis. Nature 2010, 465, 584–589. [Google Scholar] [CrossRef]
- Cifuentes, D.; Xue, H.; Taylor, D.W.; Patnode, H.; Mishima, Y.; Cheloufi, S.; Ma, E.; Mane, S.; Hannon, G.J.; Lawson, N.D.; et al. A novel mirna processing pathway independent of dicer requires argonaute2 catalytic activity. Science 2010, 328, 1694. [Google Scholar] [CrossRef] [PubMed]
- Havens, M.A.; Reich, A.A.; Duelli, D.M.; Hastings, M.L. Biogenesis of mammalian micrornas by a non-canonical processing pathway. Nucleic Acids Res. 2012, 40, 4626–4640. [Google Scholar] [CrossRef] [PubMed]
- Westholm, J.O.; Lai, E.C. Mirtrons: Microrna biogenesis via splicing. Biochimie 2011, 93, 1897–1904. [Google Scholar] [CrossRef]
- Yang, J.S.; Lai, E.C. Alternative mirna biogenesis pathways and the interpretation of core mirna pathway mutants. Mol. Cell 2011, 43, 892–903. [Google Scholar] [CrossRef] [PubMed]
- Abdelfattah, A.M.; Park, C.; Choi, M.Y. Update on non-canonical micrornas. Biomol. Concepts 2014, 5, 275–287. [Google Scholar] [CrossRef]
- Stavast, C.J.; Erkeland, S.J. The non-canonical aspects of micrornas: Many roads to gene regulation. Cells 2019, 8, 1465. [Google Scholar] [CrossRef]
- Bartel, D.P. Micrornas: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microrna target sites in mammalian mrnas. Elife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Agrawal, R.; Pandey, P.; Jha, P.; Dwivedi, V.; Sarkar, C.; Kulshreshtha, R. Hypoxic signature of micrornas in glioblastoma: Insights from small rna deep sequencing. BMC Genom. 2014, 15, 686. [Google Scholar] [CrossRef]
- Schirle, N.T.; Sheu-Gruttadauria, J.; MacRae, I.J. Structural basis for microrna targeting. Science 2014, 346, 608–613. [Google Scholar] [CrossRef]
- Schirle, N.T.; Sheu-Gruttadauria, J.; Chandradoss, S.D.; Joo, C.; MacRae, I.J. Water-mediated recognition of t1-adenosine anchors argonaute2 to microrna targets. Elife 2015, 4, e07646. [Google Scholar] [CrossRef] [PubMed]
- Grimson, A.; Farh, K.K.-H.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. Microrna targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Valencia-Sanchez, M.A.; Liu, J.; Hannon, G.J.; Parker, R. Control of translation and mrna degradation by mirnas and sirnas. Genes Dev. 2006, 20, 515–524. [Google Scholar] [CrossRef]
- Mallory, A.C.; Reinhart, B.J.; Jones-Rhoades, M.W.; Tang, G.; Zamore, P.D.; Barton, M.K.; Bartel, D.P. Microrna control of phabulosa in leaf development: Importance of pairing to the microrna 5′ region. EMBO J. 2004, 23, 3356–3364. [Google Scholar] [CrossRef] [PubMed]
- Mallory, A.C.; Vaucheret, H. Micrornas: Something important between the genes. Curr. Opin. Plant Biol. 2004, 7, 120–125. [Google Scholar] [CrossRef]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human argonaute2 mediates rna cleavage targeted by mirnas and sirnas. Mol. Cell 2004, 15, 185–197. [Google Scholar] [CrossRef]
- Yekta, S.; Shih, I.-h.; Bartel, D.P. Microrna-directed cleavage of hoxb8 mrna. Science 2004, 304, 594–596. [Google Scholar] [CrossRef]
- Liu, J.; Carmell, M.A.; Rivas, F.V.; Marsden, C.G.; Thomson, J.M.; Song, J.-J.; Hammond, S.M.; Joshua-Tor, L.; Hannon, G.J. Argonaute2 is the catalytic engine of mammalian rnai. Science 2004, 305, 1437–1441. [Google Scholar] [CrossRef]
- Diederichs, S.; Haber, D.A. Dual role for argonautes in microrna processing and posttranscriptional regulation of microrna expression. Cell 2007, 131, 1097–1108. [Google Scholar] [CrossRef]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microrna-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421. [Google Scholar] [CrossRef]
- Christie, M.; Boland, A.; Huntzinger, E.; Weichenrieder, O.; Izaurralde, E. Structure of the pan3 pseudokinase reveals the basis for interactions with the pan2 deadenylase and the gw182 proteins. Mol. Cell 2013, 51, 360–373. [Google Scholar] [CrossRef]
- Braun, J.E.; Huntzinger, E.; Fauser, M.; Izaurralde, E. Gw182 proteins directly recruit cytoplasmic deadenylase complexes to mirna targets. Mol. Cell 2011, 44, 120–133. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-Y.A.; Zheng, D.; Xia, Z.; Shyu, A.-B. Ago–tnrc6 triggers microrna-mediated decay by promoting two deadenylation steps. Nat. Struct. Mol. Biol. 2009, 16, 1160. [Google Scholar] [CrossRef] [PubMed]
- Behm-Ansmant, I.; Rehwinkel, J.; Doerks, T.; Stark, A.; Bork, P.; Izaurralde, E. Mrna degradation by mirnas and gw182 requires both ccr4: Not deadenylase and dcp1: Dcp2 decapping complexes. Genes Dev. 2006, 20, 1885–1898. [Google Scholar] [CrossRef] [PubMed]
- Chekulaeva, M.; Mathys, H.; Zipprich, J.T.; Attig, J.; Colic, M.; Parker, R.; Filipowicz, W. Mirna repression involves gw182-mediated recruitment of ccr4–not through conserved w-containing motifs. Nat. Struct. Mol. Biol. 2011, 18, 1218–1226. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; Cieplak, M.K.; Frank, F.; Morita, M.; Green, J.; Srikumar, T.; Nagar, B.; Yamamoto, T.; Raught, B.; Duchaine, T.F. Mirna-mediated deadenylation is orchestrated by gw182 through two conserved motifs that interact with ccr4–not. Nat. Struct. Mol. Biol. 2011, 18, 1211. [Google Scholar] [CrossRef]
- Parker, R.; Song, H. The enzymes and control of eukaryotic mrna turnover. Nat. Struct. Mol. Biol. 2004, 11, 121–127. [Google Scholar] [CrossRef]
- Coller, J.; Parker, R. Eukaryotic mrna decapping. Annu. Rev. Biochem. 2004, 73, 861–890. [Google Scholar] [CrossRef]
- Braun, J.E.; Truffault, V.; Boland, A.; Huntzinger, E.; Chang, C.-T.; Haas, G.; Weichenrieder, O.; Coles, M.; Izaurralde, E. A direct interaction between dcp1 and xrn1 couples mrna decapping to 5′ exonucleolytic degradation. Nat. Struct. Mol. Biol. 2012, 19, 1324. [Google Scholar] [CrossRef]
- Krützfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of micrornas in vivo with ‘antagomirs’. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef]
- Ameres, S.L.; Horwich, M.D.; Hung, J.-H.; Xu, J.; Ghildiyal, M.; Weng, Z.; Zamore, P.D. Target rna–directed trimming and tailing of small silencing rnas. Science 2010, 328, 1534–1539. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.A.; Shyu, A.B. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip. Rev. RNA 2011, 2, 167–183. [Google Scholar] [CrossRef] [PubMed]
- Pillai, R.S. Microrna function: Multiple mechanisms for a tiny rna? RNA 2005, 11, 1753–1761. [Google Scholar] [CrossRef] [PubMed]
- Pillai, R.S.; Bhattacharyya, S.N.; Artus, C.G.; Zoller, T.; Cougot, N.; Basyuk, E.; Bertrand, E.; Filipowicz, W. Inhibition of translational initiation by let-7 microrna in human cells. Science 2005, 309, 1573–1576. [Google Scholar] [CrossRef]
- Kong, Y.W.; Cannell, I.G.; de Moor, C.H.; Hill, K.; Garside, P.G.; Hamilton, T.L.; Meijer, H.A.; Dobbyn, H.C.; Stoneley, M.; Spriggs, K.A. The mechanism of micro-rna-mediated translation repression is determined by the promoter of the target gene. Proc. Natl. Acad. Sci. USA 2008, 105, 8866–8871. [Google Scholar] [CrossRef]
- Meijer, H.; Kong, Y.; Lu, W.; Wilczynska, A.; Spriggs, R.; Robinson, S.; Godfrey, J.; Willis, A.; Bushell, M. Translational repression and eif4a2 activity are critical for microrna-mediated gene regulation. Science 2013, 340, 82–85. [Google Scholar] [CrossRef]
- Fukaya, T.; Iwakawa, H.-o.; Tomari, Y. Micrornas block assembly of eif4f translation initiation complex in drosophila. Mol. Cell 2014, 56, 67–78. [Google Scholar] [CrossRef]
- Fukao, A.; Mishima, Y.; Takizawa, N.; Oka, S.; Imataka, H.; Pelletier, J.; Sonenberg, N.; Thoma, C.; Fujiwara, T. Micrornas trigger dissociation of eif4ai and eif4aii from target mrnas in humans. Mol. Cell 2014, 56, 79–89. [Google Scholar] [CrossRef]
- Xu, W.; San Lucas, A.; Wang, Z.; Liu, Y. Identifying microrna targets in different gene regions. BMC Bioinform. 2014, 15, S4. [Google Scholar] [CrossRef]
- Ellwanger, D.C.; Büttner, F.A.; Mewes, H.-W.; Stümpflen, V. The sufficient minimal set of mirna seed types. Bioinformatics 2011, 27, 1346–1350. [Google Scholar] [CrossRef]
- Brengues, M.; Teixeira, D.; Parker, R. Movement of eukaryotic mrnas between polysomes and cytoplasmic processing bodies. Science 2005, 310, 486–489. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Valencia-Sanchez, M.A.; Hannon, G.J.; Parker, R. Microrna-dependent localization of targeted mrnas to mammalian p-bodies. Nat. Cell Biol. 2005, 7, 719–723. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Rivas, F.V.; Wohlschlegel, J.; Yates, J.R.; Parker, R.; Hannon, G.J. A role for the p-body component gw182 in microrna function. Nat. Cell Biol. 2005, 7, 1261–1266. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.; Stoecklin, G.; Ayodele, M.; Yacono, P.; Lykke-Andersen, J.; Fritzler, M.J.; Scheuner, D.; Kaufman, R.J.; Golan, D.E.; Anderson, P. Stress granules and processing bodies are dynamically linked sites of mrnp remodeling. J. Cell Biol. 2005, 169, 871–884. [Google Scholar] [CrossRef]
- Andrei, M.A.; Ingelfinger, D.; Heintzmann, R.; Achsel, T.; Rivera-Pomar, R.; Lührmann, R. A role for eif4e and eif4e-transporter in targeting mrnps to mammalian processing bodies. RNA 2005, 11, 717–727. [Google Scholar] [CrossRef]
- Anderson, P.; Kedersha, N. Rna granules. J. Cell Biol. 2006, 172, 803–808. [Google Scholar] [CrossRef]
- Teixeira, D.; Sheth, U.; Valencia-Sanchez, M.A.; Brengues, M.; Parker, R. Processing bodies require rna for assembly and contain nontranslating mrnas. RNA 2005, 11, 371–382. [Google Scholar] [CrossRef]
- Sato-Kuwabara, Y.; Melo, S.A.; Soares, F.A.; Calin, G.A. The fusion of two worlds: Non-coding rnas and extracellular vesicles-diagnostic and therapeutic implications. Int. J. Oncol. 2015, 46, 17–27. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Cosmopoulos, K.; Thorley-Lawson, D.A.; van Eijndhoven, M.A.; Hopmans, E.S.; Lindenberg, J.L.; de Gruijl, T.D.; Würdinger, T.; Middeldorp, J.M. Functional delivery of viral mirnas via exosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 6328–6333. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mrnas and micrornas is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Goldie, B.J.; Dun, M.D.; Lin, M.; Smith, N.D.; Verrills, N.M.; Dayas, C.V.; Cairns, M.J. Activity-associated mirna are packaged in map1b-enriched exosomes released from depolarized neurons. Nucleic Acids Res. 2014, 42, 9195–9208. [Google Scholar] [CrossRef] [PubMed]
- Guduric-Fuchs, J.; O’Connor, A.; Camp, B.; O’Neill, C.L.; Medina, R.J.; Simpson, D.A. Selective extracellular vesicle-mediated export of an overlapping set of micrornas from multiple cell types. BMC Genom. 2012, 13, 357. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, K.; Inoue, K.; Fujiwara, A.; Hatakeyama, K.; Kanto, K.; Watanabe, Y.; Muramatsu, K.; Fukuda, Y.; Ogura, S.-i.; Yamaguchi, K. Let-7 microrna family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS ONE 2010, 5, e13247. [Google Scholar] [CrossRef] [PubMed]
- Skog, J.; Würdinger, T.; Van Rijn, S.; Meijer, D.H.; Gainche, L.; Curry, W.T.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport rna and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; García, V.; Zaballos, A.; Provencio, M.; Lombardía, L.; Almonacid, L.; García, J.M.; Domínguez, G.; Peña, C.; Diaz, R. Vesicle-related micrornas in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur. Respir. J. 2011, 37, 617–623. [Google Scholar] [CrossRef]
- Taylor, D.D.; Gercel-Taylor, C. Microrna signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.F.; Chin, A.R. Cancer-secreted mir-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef]
- Van Balkom, B.W.; De Jong, O.G.; Smits, M.; Brummelman, J.; den Ouden, K.; de Bree, P.M.; van Eijndhoven, M.A.; Pegtel, D.M.; Stoorvogel, W.; Würdinger, T. Endothelial cells require mir-214 to secrete exosomes that suppress senescence and induce angiogenesis in human and mouse endothelial cells. BloodJ. Am. Soc. Hematol. 2013, 121, 3997–4006. [Google Scholar] [CrossRef]
- Umezu, T.; Ohyashiki, K.; Kuroda, M.; Ohyashiki, J. Leukemia cell to endothelial cell communication via exosomal mirnas. Oncogene 2013, 32, 2747–2755. [Google Scholar] [CrossRef]
- Iftikhar, H.; Carney, G.E. Evidence and potential in vivo functions for biofluid mirnas: From expression profiling to functional testing: Potential roles of extracellular mirnas as indicators of physiological change and as agents of intercellular information exchange. Bioessays 2016, 38, 367–378. [Google Scholar] [CrossRef]
- Gallo, A.; Tandon, M.; Alevizos, I.; Illei, G.G. The majority of micrornas detectable in serum and saliva is concentrated in exosomes. PLoS ONE 2012, 7, e30679. [Google Scholar] [CrossRef] [PubMed]
- Tabet, F.; Vickers, K.C.; Torres, L.F.C.; Wiese, C.B.; Shoucri, B.M.; Lambert, G.; Catherinet, C.; Prado-Lourenco, L.; Levin, M.G.; Thacker, S. Hdl-transferred microrna-223 regulates icam-1 expression in endothelial cells. Nat. Commun. 2014, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. Micrornas are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, S.; Weber, J.; Baxter, D.; Galas, D.J. Export of micrornas and microrna-protective protein by mammalian cells. Nucleic Acids Res. 2010, 38, 7248–7259. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L. Argonaute2 complexes carry a population of circulating micrornas independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Weiz, L.; Burwinkel, B. Extracellular mirnas: The mystery of their origin and function. Trends Biochem. Sci. 2012, 37, 460–465. [Google Scholar] [CrossRef]
- Calin, G.A.; Ferracin, M.; Cimmino, A.; Di Leva, G.; Shimizu, M.; Wojcik, S.E.; Iorio, M.V.; Visone, R.; Sever, N.I.; Fabbri, M. A microrna signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005, 353, 1793–1801. [Google Scholar] [CrossRef]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M. Human microrna genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef]
- Bonci, D.; Coppola, V.; Musumeci, M.; Addario, A.; Giuffrida, R.; Memeo, L.; D’urso, L.; Pagliuca, A.; Biffoni, M.; Labbaye, C. The mir-15a–mir-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat. Med. 2008, 14, 1271–1277. [Google Scholar] [CrossRef]
- Creighton, C.J.; Hernandez-Herrera, A.; Jacobsen, A.; Levine, D.A.; Mankoo, P.; Schultz, N.; Du, Y.; Zhang, Y.; Larsson, E.; Sheridan, R. Integrated analyses of micrornas demonstrate their widespread influence on gene expression in high-grade serous ovarian carcinoma. PLoS ONE 2012, 7, e34546. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of tissue-specific micrornas from mouse. Curr. Biol. 2002, 12, 735–739. [Google Scholar] [CrossRef]
- Svoronos, A.A.; Engelman, D.M.; Slack, F.J. Oncomir or tumor suppressor? The duplicity of micrornas in cancer. Cancer Res. 2016, 76, 3666–3670. [Google Scholar] [CrossRef] [PubMed]
- Bartels, C.L.; Tsongalis, G.J. Micrornas: Novel biomarkers for human cancer. Clin. Chem. 2009, 55, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Hosseinahli, N.; Aghapour, M.; Duijf, P.H.G.; Baradaran, B. Treating cancer with microrna replacement therapy: A literature review. J. Cell Physiol. 2018, 233, 5574–5588. [Google Scholar] [CrossRef]
- Allen, K.E.; Weiss, G.J. Resistance may not be futile: Microrna biomarkers for chemoresistance and potential therapeutics. Mol. Cancer 2010, 9, 3126–3136. [Google Scholar] [CrossRef]
- Meijer, L.L.; Puik, J.R.; Vivaldi, C.; Capula, M.; Vasile, E.; Kazemier, G.; Giovannetti, E. Chapter 15 modification of drug response in cancer by micrornas. In Micrornas in Diseases and Disorders: Emerging Therapeutic Targets; The Royal Society of Chemistry: London, UK, 2019; pp. 416–451. [Google Scholar]
- Pang, J.C.; Kwok, W.K.; Chen, Z.; Ng, H.K. Oncogenic role of micrornas in brain tumors. Acta Neuropathol. 2009, 117, 599–611. [Google Scholar] [CrossRef]
- Areeb, Z.; Stylli, S.S.; Koldej, R.; Ritchie, D.S.; Siegal, T.; Morokoff, A.P.; Kaye, A.H.; Luwor, R.B. Microrna as potential biomarkers in glioblastoma. J. Neurooncol. 2015, 125, 237–248. [Google Scholar] [CrossRef]
- Bronisz, A.; Godlewski, J.; Chiocca, E.A. Extracellular vesicles and micrornas: Their role in tumorigenicity and therapy for brain tumors. Cell Mol. Neurobiol. 2016, 36, 361–376. [Google Scholar] [CrossRef]
- Saadatpour, L.; Fadaee, E.; Fadaei, S.; Nassiri Mansour, R.; Mohammadi, M.; Mousavi, S.M.; Goodarzi, M.; Verdi, J.; Mirzaei, H. Glioblastoma: Exosome and microrna as novel diagnosis biomarkers. Cancer Gene 2016, 23, 415–418. [Google Scholar] [CrossRef]
- Shea, A.; Harish, V.; Afzal, Z.; Chijioke, J.; Kedir, H.; Dusmatova, S.; Roy, A.; Ramalinga, M.; Harris, B.; Blancato, J.; et al. Micrornas in glioblastoma multiforme pathogenesis and therapeutics. Cancer Med. 2016, 5, 1917–1946. [Google Scholar] [CrossRef]
- Ahir, B.K.; Ozer, H.; Engelhard, H.H.; Lakka, S.S. Micrornas in glioblastoma pathogenesis and therapy: A comprehensive review. Crit. Rev. Oncol. Hematol. 2017, 120, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Beyer, S.; Fleming, J.; Meng, W.; Singh, R.; Haque, S.J.; Chakravarti, A. The role of mirnas in angiogenesis, invasion and metabolism and their therapeutic implications in gliomas. Cancers 2017, 9, 85. [Google Scholar] [CrossRef] [PubMed]
- Mercatelli, N.; Galardi, S.; Ciafre, S.A. Micrornas as multifaceted players in glioblastoma multiforme. Int. Rev. Cell Mol. Biol. 2017, 333, 269–323. [Google Scholar] [PubMed]
- Anthiya, S.; Griveau, A.; Loussouarn, C.; Baril, P.; Garnett, M.; Issartel, J.P.; Garcion, E. Microrna-based drugs for brain tumors. Trends Cancer 2018, 4, 222–238. [Google Scholar] [CrossRef] [PubMed]
- Godlewski, J.; Lenart, J.; Salinska, E. Microrna in brain pathology: Neurodegeneration the other side of the brain cancer. Noncoding RNA 2019, 5, 20. [Google Scholar] [CrossRef]
- Krichevsky, A.M.; Uhlmann, E.J. Oligonucleotide therapeutics as a new class of drugs for malignant brain tumors: Targeting mrnas, regulatory rnas, mutations, combinations, and beyond. Neurotherapeutics 2019, 16, 319–347. [Google Scholar] [CrossRef]
- Macharia, L.W.; Wanjiru, C.M.; Mureithi, M.W.; Pereira, C.M.; Ferrer, V.P.; Moura-Neto, V. Micrornas, hypoxia and the stem-like state as contributors to cancer aggressiveness. Front. Genet 2019, 10, 125. [Google Scholar] [CrossRef]
- Paulmurugan, R.; Malhotra, M.; Massoud, T.F. The protean world of non-coding rnas in glioblastoma. J. Mol. Med. 2019, 97, 909–925. [Google Scholar] [CrossRef]
- Baffa, R.; Fassan, M.; Volinia, S.; O’Hara, B.; Liu, C.G.; Palazzo, J.P.; Gardiman, M.; Rugge, M.; Gomella, L.G.; Croce, C.M.; et al. Microrna expression profiling of human metastatic cancers identifies cancer gene targets. J. Pathol. 2009, 219, 214–221. [Google Scholar] [CrossRef]
- Aigner, A. Micrornas (mirnas) in cancer invasion and metastasis: Therapeutic approaches based on metastasis-related mirnas. J. Mol. Med. 2011, 89, 445–457. [Google Scholar] [CrossRef]
- Li, Z.; Gu, X.; Fang, Y.; Xiang, J.; Chen, Z. Microrna expression profiles in human colorectal cancers with brain metastases. Oncol Lett. 2012, 3, 346–350. [Google Scholar] [CrossRef]
- Alsidawi, S.; Malek, E.; Driscoll, J.J. Micrornas in brain metastases: Potential role as diagnostics and therapeutics. Int. J. Mol. Sci. 2014, 15, 10508–10526. [Google Scholar] [CrossRef]
- Pastorkova, Z.; Skarda, J.; Andel, J. The role of microrna in metastatic processes of non-small cell lung carcinoma. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc. Czech Repub. 2016, 160, 343–357. [Google Scholar] [CrossRef]
- Wu, K.; Sharma, S.; Venkat, S.; Liu, K.; Zhou, X.; Watabe, K. Non-coding rnas in cancer brain metastasis. Front. Biosci. (Sch. Ed.) 2016, 8, 187–202. [Google Scholar]
- Yousefi, M.; Bahrami, T.; Salmaninejad, A.; Nosrati, R.; Ghaffari, P.; Ghaffari, S.H. Lung cancer-associated brain metastasis: Molecular mechanisms and therapeutic options. Cell Oncol 2017, 40, 419–441. [Google Scholar] [CrossRef] [PubMed]
- Fassan, M.; Sachsenmeir, K.; Rugge, M.; Baffa, R. Role of mirna in distinguishing primary brain tumors from secondary tumors metastatic to the brain. Front. Biosci. (Sch. Ed.) 2011, 3, 970–979. [Google Scholar] [CrossRef] [PubMed]
- Donzelli, S.; Mori, F.; Bellissimo, T.; Sacconi, A.; Casini, B.; Frixa, T.; Roscilli, G.; Aurisicchio, L.; Facciolo, F.; Pompili, A.; et al. Epigenetic silencing of mir-145-5p contributes to brain metastasis. Oncotarget 2015, 6, 35183–35201. [Google Scholar] [CrossRef] [PubMed]
- Anandappa, G.; Lampis, A.; Cunningham, D.; Khan, K.H.; Kouvelakis, K.; Vlachogiannis, G.; Hedayat, S.; Tunariu, N.; Rao, S.; Watkins, D.; et al. Mir-31-3p expression and benefit from anti-egfr inhibitors in metastatic colorectal cancer patients enrolled in the prospective phase ii prospect-c trial. Clin. Cancer Res. 2019, 25, 3830–3838. [Google Scholar] [CrossRef]
- Gong, J.; Maia, M.C.; Dizman, N.; Govindarajan, A.; Pal, S.K. Metastasis in renal cell carcinoma: Biology and implications for therapy. Asian J. Urol. 2016, 3, 286–292. [Google Scholar] [CrossRef]
- Wa, Q.; Li, L.; Lin, H.; Peng, X.; Ren, D.; Huang, Y.; He, P.; Huang, S. Downregulation of mir19a3p promotes invasion, migration and bone metastasis via activating tgfbeta signaling in prostate cancer. Oncol. Rep. 2018, 39, 81–90. [Google Scholar]
- Chen, L.J.; Li, X.Y.; Zhao, Y.Q.; Liu, W.J.; Wu, H.J.; Liu, J.; Mu, X.Q.; Wu, H.B. Down-regulated microrna-375 expression as a predictive biomarker in non-small cell lung cancer brain metastasis and its prognostic significance. Pathol. Res. Pr. 2017, 213, 882–888. [Google Scholar] [CrossRef] [PubMed]
- Daugaard, I.; Veno, M.T.; Yan, Y.; Kjeldsen, T.E.; Lamy, P.; Hager, H.; Kjems, J.; Hansen, L.L. Small rna sequencing reveals metastasis-related micrornas in lung adenocarcinoma. Oncotarget 2017, 8, 27047–27061. [Google Scholar] [CrossRef] [PubMed]
- Heinzelmann, J.; Unrein, A.; Wickmann, U.; Baumgart, S.; Stapf, M.; Szendroi, A.; Grimm, M.O.; Gajda, M.R.; Wunderlich, H.; Junker, K. Micrornas with prognostic potential for metastasis in clear cell renal cell carcinoma: A comparison of primary tumors and distant metastases. Ann. Surg. Oncol. 2014, 21, 1046–1054. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Y.; Lang, Z.Q.; Ren, C.; Yang, P.; Zhang, B. Mir143 acts as a novel big mitogenactivated protein kinase 1 suppressor and may inhibit invasion of glioma. Oncol. Rep. 2019, 42, 1194–1204. [Google Scholar]
- Wu, Z.; Sun, L.; Wang, H.; Yao, J.; Jiang, C.; Xu, W.; Yang, Z. Mir-328 expression is decreased in high-grade gliomas and is associated with worse survival in primary glioblastoma. PLoS ONE 2012, 7, e47270. [Google Scholar] [CrossRef]
- Chen, S.R.; Cai, W.P.; Dai, X.J.; Guo, A.S.; Chen, H.P.; Lin, G.S.; Lin, R.S. Research on mir-126 in glioma targeted regulation of pten/pi3k/akt and mdm2-p53 pathways. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3461–3470. [Google Scholar]
- Godlewski, J.; Nowicki, M.O.; Bronisz, A.; Nuovo, G.; Palatini, J.; De Lay, M.; Van Brocklyn, J.; Ostrowski, M.C.; Chiocca, E.A.; Lawler, S.E. Microrna-451 regulates lkb1/ampk signaling and allows adaptation to metabolic stress in glioma cells. Mol. Cell 2010, 37, 620–632. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, A.; Hao, Y.; Wang, G.; Jia, Z. The emerging role of mir-19 in glioma. J. Cell Mol. Med. 2018, 22, 4611–4616. [Google Scholar] [CrossRef]
- Cosset, E.; Petty, T.; Dutoit, V.; Tirefort, D.; Otten-Hernandez, P.; Farinelli, L.; Dietrich, P.Y.; Preynat-Seauve, O. Human tissue engineering allows the identification of active mirna regulators of glioblastoma aggressiveness. Biomaterials 2016, 107, 74–87. [Google Scholar] [CrossRef]
- GlioVis. Available online: Http://gliovis.Bioinfo.Cnio.Es/ (accessed on 24 August 2020).
- Zhao, H.; Shen, J.; Hodges, T.R.; Song, R.; Fuller, G.N.; Heimberger, A.B. Serum microrna profiling in patients with glioblastoma: A survival analysis. Mol. Cancer 2017, 16, 59. [Google Scholar] [CrossRef]
- Srinivasan, S.; Patric, I.R.; Somasundaram, K. A ten-microrna expression signature predicts survival in glioblastoma. PLoS ONE 2011, 6, e17438. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Jiao, B.; Geng, S.; Song, J.; Liang, Z.; Lu, S. Concomitant microrna-31 downregulation and radixin upregulation predicts advanced tumor progression and unfavorable prognosis in patients with gliomas. J. Neurol. Sci. 2014, 338, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Y.; Ho, H.L.; Lin, S.C.; Ho, T.D.; Hsu, C.Y. Upregulation of mir-125b, mir-181d, and mir-221 predicts poor prognosis in mgmt promoter-unmethylated glioblastoma patients. Am. J. Clin. Pathol. 2018, 149, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Henriksen, M.; Johnsen, K.B.; Olesen, P.; Pilgaard, L.; Duroux, M. Microrna expression signatures and their correlation with clinicopathological features in glioblastoma multiforme. Neuromolecular Med. 2014, 16, 565–577. [Google Scholar] [CrossRef]
- Permuth-Wey, J.; Thompson, R.C.; Burton Nabors, L.; Olson, J.J.; Browning, J.E.; Madden, M.H.; Ann Chen, Y.; Egan, K.M. A functional polymorphism in the pre-mir-146a gene is associated with risk and prognosis in adult glioma. J. Neurooncol. 2011, 105, 639–646. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, P.; Sun, T.; Li, D.; Xu, X.; Rui, Y.; Li, C.; Chong, M.; Ibrahim, T.; Mercatali, L.; et al. Mir-126 and mir-126* repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat. Cell Biol. 2013, 15, 284–294. [Google Scholar] [CrossRef]
- Sun, D.; Li, X.; Ma, M.; Liu, J.; Xu, Y.; Ye, L.; Hou, H.; Wang, C.; Li, X.; Jiang, Y. The predictive value and potential mechanisms of mirna-328 and mirna-378 for brain metastases in operable and advanced non-small-cell lung cancer. Jpn. J. Clin. Oncol. 2015, 45, 464–473. [Google Scholar] [CrossRef]
- Zeinali, T.; Mansoori, B.; Mohammadi, A.; Baradaran, B. Regulatory mechanisms of mir-145 expression and the importance of its function in cancer metastasis. Biomed. Pharm. 2019, 109, 195–207. [Google Scholar] [CrossRef]
- Sachdeva, M.; Mo, Y.Y. Mir-145-mediated suppression of cell growth, invasion and metastasis. Am. J. Transl. Res. 2010, 2, 170–180. [Google Scholar]
- Speranza, M.C.; Frattini, V.; Pisati, F.; Kapetis, D.; Porrati, P.; Eoli, M.; Pellegatta, S.; Finocchiaro, G. Nedd9, a novel target of mir-145, increases the invasiveness of glioblastoma. Oncotarget 2012, 3, 723–734. [Google Scholar] [CrossRef]
- Haapa-Paananen, S.; Chen, P.; Hellstrom, K.; Kohonen, P.; Hautaniemi, S.; Kallioniemi, O.; Perala, M. Functional profiling of precursor micrornas identifies micrornas essential for glioma proliferation. PLoS ONE 2013, 8, e60930. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Kim, S.J.; Seo, H.H.; Shin, S.P.; Kim, D.; Park, C.S.; Kim, K.T.; Kim, Y.H.; Jeong, J.S.; Kim, I.H. Over-expression of mir-145 enhances the effectiveness of hsvtk gene therapy for malignant glioma. Cancer Lett. 2012, 320, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.P.; Chien, Y.; Chiou, G.Y.; Cherng, J.Y.; Wang, M.L.; Lo, W.L.; Chang, Y.L.; Huang, P.I.; Chen, Y.W.; Shih, Y.H.; et al. Inhibition of cancer stem cell-like properties and reduced chemoradioresistance of glioblastoma using microrna145 with cationic polyurethane-short branch pei. Biomaterials 2012, 33, 1462–1476. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Bier, A.; Cazacu, S.; Finniss, S.; Xiang, C.; Twito, H.; Poisson, L.M.; Mikkelsen, T.; Slavin, S.; Jacoby, E.; et al. Microrna-145 is downregulated in glial tumors and regulates glioma cell migration by targeting connective tissue growth factor. PLoS ONE 2013, 8, e54652. [Google Scholar] [CrossRef]
- Liu, H.; Liu, Z.; Jiang, B.; Huo, L.; Liu, J.; Lu, J. Synthetic mir-145 mimic enhances the cytotoxic effect of the antiangiogenic drug sunitinib in glioblastoma. Cell Biochem. Biophys. 2015, 72, 551–557. [Google Scholar] [CrossRef]
- Xu, Z.; Zeng, X.; Xu, J.; Xu, D.; Li, J.; Jin, H.; Jiang, G.; Han, X.; Huang, C. Isorhapontigenin suppresses growth of patient-derived glioblastoma spheres through regulating mir-145/sox2/cyclin d1 axis. Neuro Oncol. 2016, 18, 830–839. [Google Scholar] [CrossRef]
- Kurogi, R.; Nakamizo, A.; Suzuki, S.O.; Mizoguchi, M.; Yoshimoto, K.; Amano, T.; Amemiya, T.; Takagishi, S.; Iihara, K. Inhibition of glioblastoma cell invasion by hsa-mir-145-5p and hsa-mir-31-5p co-overexpression in human mesenchymal stem cells. J. Neurosurg. 2018, 130, 44–55. [Google Scholar] [CrossRef]
- Koo, S.; Martin, G.S.; Schulz, K.J.; Ronck, M.; Toussaint, L.G. Serial selection for invasiveness increases expression of mir-143/mir-145 in glioblastoma cell lines. BMC Cancer 2012, 12, 143. [Google Scholar] [CrossRef]
- Koo, S.; Martin, G.; Toussaint, L.G. Microrna-145 promotes the phenotype of human glioblastoma cells selected for invasion. Anticancer Res. 2015, 35, 3209–3215. [Google Scholar]
- Wu, N.; Lin, X.; Zhao, X.; Zheng, L.; Xiao, L.; Liu, J.; Ge, L.; Cao, S. Mir-125b acts as an oncogene in glioblastoma cells and inhibits cell apoptosis through p53 and p38mapk-independent pathways. Br. J. Cancer 2013, 109, 2853. [Google Scholar] [CrossRef]
- Hua, D.; Ding, D.; Han, X.; Zhang, W.; Zhao, N.; Foltz, G.; Lan, Q.; Huang, Q.; Lin, B. Human mir-31 targets radixin and inhibits migration and invasion of glioma cells. Oncol. Rep. 2012, 27, 700–706. [Google Scholar] [PubMed]
- Rajbhandari, R.; McFarland, B.C.; Patel, A.; Gerigk, M.; Gray, G.K.; Fehling, S.C.; Bredel, M.; Berbari, N.F.; Kim, H.; Marks, M.P.; et al. Loss of tumor suppressive microrna-31 enhances tradd/nf-κb signaling in glioblastoma. Oncotarget 2015, 6, 17805–17816. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.J.; Xu, X.Y.; Liu, B.X.; Dai, W.Z.; Cai, M.Q.; Bai, C.F.; Zhang, X.F.; Wang, L.M.; Lin, L.; Jia, S.Z.; et al. Growth-inhibitory and chemosensitizing effects of microrna-31 in human glioblastoma multiforme cells. Int. J. Mol. Med. 2015, 36, 1159–1164. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, H.; Yin, C.; Sun, X.; Zheng, S.; Zhang, C.; Shi, L.; Liu, Y.; Lu, S. Dock1 promotes the mesenchymal transition of glioma and is modulated by mir-31. Neuropathol. Appl. Neurobiol. 2017, 43, 419–432. [Google Scholar] [CrossRef]
- Wang, J.; Li, B.; Wang, C.; Luo, Y.; Zhao, M.; Chen, P. Long noncoding rna foxd2-as1 promotes glioma cell cycle progression and proliferation through the foxd2-as1/mir-31/cdk1 pathway. J. Cell. Biochem. 2019, 120, 19784–19795. [Google Scholar] [CrossRef]
- Ye, J.; Zhu, J.; Chen, H.; Qian, J.; Zhang, L.; Wan, Z.; Chen, F.; Sun, S.; Li, W.; Luo, C. A novel lncrna-linc01116 regulates tumorigenesis of glioma by targeting vegfa. Int. J. Cancer 2020, 146, 248–261. [Google Scholar] [CrossRef]
- Wong, H.A.; Fatimy, R.E.; Onodera, C.; Wei, Z.; Yi, M.; Mohan, A.; Gowrisankaran, S.; Karmali, P.; Marcusson, E.; Wakimoto, H.; et al. The cancer genome atlas analysis predicts microrna for targeting cancer growth and vascularization in glioblastoma. Mol. Ther. 2015, 23, 1234–1247. [Google Scholar] [CrossRef]
- Pan, X.; Wang, R.; Wang, Z.X. The potential role of mir-451 in cancer diagnosis, prognosis, and therapy. Mol. Cancer 2013, 12, 1153–1162. [Google Scholar] [CrossRef]
- Nan, Y.; Han, L.; Zhang, A.; Wang, G.; Jia, Z.; Yang, Y.; Yue, X.; Pu, P.; Zhong, Y.; Kang, C. Mirna-451 plays a role as tumor suppressor in human glioma cells. Brain Res. 2010, 1359, 14–21. [Google Scholar] [CrossRef]
- Gal, H.; Pandi, G.; Kanner, A.A.; Ram, Z.; Lithwick-Yanai, G.; Amariglio, N.; Rechavi, G.; Givol, D. Mir-451 and imatinib mesylate inhibit tumor growth of glioblastoma stem cells. Biochem. Biophys. Res. Commun. 2008, 376, 86–90. [Google Scholar] [CrossRef]
- Alural, B.; Ayyildiz, Z.O.; Tufekci, K.U.; Genc, S.; Genc, K. Erythropoietin promotes glioblastoma via mir-451 suppression. Vitam. Horm. 2017, 105, 249–271. [Google Scholar] [PubMed]
- Ogawa, D.; Ansari, K.; Nowicki, M.O.; Salinska, E.; Bronisz, A.; Godlewski, J. Microrna-451 inhibits migration of glioblastoma while making it more susceptible to conventional therapy. Noncoding RNA 2019, 5, 25. [Google Scholar] [CrossRef] [PubMed]
- Godlewski, J.; Bronisz, A.; Nowicki, M.O.; Chiocca, E.A.; Lawler, S. Microrna-451: A conditional switch controlling glioma cell proliferation and migration. Cell Cycle 2010, 9, 2814–2820. [Google Scholar] [CrossRef] [PubMed]
- Malzkorn, B.; Wolter, M.; Liesenberg, F.; Grzendowski, M.; Stuhler, K.; Meyer, H.E.; Reifenberger, G. Identification and functional characterization of micrornas involved in the malignant progression of gliomas. Brain Pathol. 2010, 20, 539–550. [Google Scholar] [CrossRef]
- Chen, Q.; Guo, W.; Zhang, Y.; Wu, Y.; Xiang, J. Mir-19a promotes cell proliferation and invasion by targeting rhob in human glioma cells. Neurosci Lett. 2016, 628, 161–166. [Google Scholar] [CrossRef]
- Sun, J.; Jia, Z.; Li, B.; Zhang, A.; Wang, G.; Pu, P.; Chen, Z.; Wang, Z.; Yang, W. Mir-19 regulates the proliferation and invasion of glioma by runx3 via β-catenin/tcf-4 signaling. Oncotarget 2017, 8, 110785–110796. [Google Scholar] [CrossRef]
- Jia, Z.; Wang, K.; Zhang, A.; Wang, G.; Kang, C.; Han, L.; Pu, P. Mir-19a and mir-19b overexpression in gliomas. Pathol. Oncol. Res. 2013, 19, 847–853. [Google Scholar] [CrossRef]
- Tokudome, T.; Sasaki, A.; Tsuji, M.; Udaka, Y.; Oyamada, H.; Tsuchiya, H.; Oguchi, K. Reduced pten expression and overexpression of mir-17-5p, -19a-3p, -19b-3p, -21-5p, -130b-3p, -221-3p and -222-3p by glioblastoma stem-like cells following irradiation. Oncol. Lett. 2015, 10, 2269–2272. [Google Scholar] [CrossRef]
- Qin, N.; Tong, G.F.; Sun, L.W.; Xu, X.L. Long noncoding rna meg3 suppresses glioma cell proliferation, migration, and invasion by acting as a competing endogenous rna of mir-19a. Oncol. Res. 2017, 25, 1471–1478. [Google Scholar] [CrossRef]
- Ren, S.; Xu, Y. Ac016405.3, a novel long noncoding rna, acts as a tumor suppressor through modulation of tet2 by microrna-19a-5p sponging in glioblastoma. Cancer Sci. 2019, 110, 1621–1632. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, S.; Yao, J.; Lowery, F.J.; Zhang, Q.; Huang, W.C.; Li, P.; Li, M.; Wang, X.; Zhang, C.; et al. Microenvironment-induced pten loss by exosomal microrna primes brain metastasis outgrowth. Nature 2015, 527, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.J.; Zhou, Z.G.; Wang, L.; Yang, L.; Zhou, B.; Gu, J.; Chen, H.Y.; Sun, X.F. Clinicopathological significance of microrna-31, -143 and -145 expression in colorectal cancer. Dis. Markers 2009, 26, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.G.; Wang, L.; Li, W.; Li, J.Z.; Li, J. Mir-143 inhibits oncogenic traits by degrading nuak2 in glioblastoma. Int. J. Mol. Med. 2016, 37, 1627–1635. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Liu, H.; Liu, Y.; Wu, J.; Wang, C.; Hou, X.; Chen, X.; Yang, G.; Zhao, L.; Che, H.; et al. Mir-143 inhibits glycolysis and depletes stemness of glioblastoma stem-like cells. Cancer Lett. 2013, 333, 253–260. [Google Scholar] [CrossRef]
- Wang, L.; Shi, Z.M.; Jiang, C.F.; Liu, X.; Chen, Q.D.; Qian, X.; Li, D.M.; Ge, X.; Wang, X.F.; Liu, L.Z.; et al. Mir-143 acts as a tumor suppressor by targeting n-ras and enhances temozolomide-induced apoptosis in glioma. Oncotarget 2014, 5, 5416–5427. [Google Scholar] [CrossRef]
- Liu, J.; Qu, C.B.; Xue, Y.X.; Li, Z.; Wang, P.; Liu, Y.H. Mir-143 enhances the antitumor activity of shikonin by targeting bag3 expression in human glioblastoma stem cells. Biochem. Biophys. Res. Commun. 2015, 468, 105–112. [Google Scholar] [CrossRef]
- Lozada-Delgado, E.L.; Grafals-Ruiz, N.; Miranda-Roman, M.A.; Santana-Rivera, Y.; Valiyeva, F.; Rivera-Diaz, M.; Marcos-Martinez, M.J.; Vivas-Mejia, P.E. Targeting microrna-143 leads to inhibition of glioblastoma tumor progression. Cancers 2018, 10, 382. [Google Scholar] [CrossRef]
- Slaby, O.; Lakomy, R.; Fadrus, P.; Hrstka, R.; Kren, L.; Lzicarova, E.; Smrcka, M.; Svoboda, M.; Dolezalova, H.; Novakova, J.; et al. Microrna-181 family predicts response to concomitant chemoradiotherapy with temozolomide in glioblastoma patients. Neoplasma 2010, 57, 264–269. [Google Scholar] [CrossRef]
- Xia, H.F.; He, T.Z.; Liu, C.M.; Cui, Y.; Song, P.P.; Jin, X.H.; Ma, X. Mir-125b expression affects the proliferation and apoptosis of human glioma cells by targeting bmf. Cell. Physiol. Biochem. 2009, 23, 347–358. [Google Scholar] [CrossRef]
- Shi, L.; Zhang, S.; Feng, K.; Wu, F.; Wan, Y.; Wang, Z.; Zhang, J.; Wang, Y.; Yan, W.; Fu, Z.; et al. Microrna-125b-2 confers human glioblastoma stem cells resistance to temozolomide through the mitochondrial pathway of apoptosis. Int. J. Oncol. 2012, 40, 119–129. [Google Scholar] [CrossRef]
- Shi, L.; Fei, X.; Wang, Z.; You, Y. Pi3k inhibitor combined with mir-125b inhibitor sensitize tmz-induced anti-glioma stem cancer effects through inactivation of wnt/beta-catenin signaling pathway. In Vitro Cell Dev. Biol. Anim. 2015, 51, 1047–1055. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Xu, S.; Yu, H.; Yang, B.; Zhao, H.; Zhao, G. Mir-125b inhibits connexin43 and promotes glioma growth. Cell Mol. Neurobiol. 2013, 33, 1143–1148. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Fu, X.; Wan, Y.; Wang, Z.; Jiang, D.; Shi, L. Mir-125b inhibitor enhance the chemosensitivity of glioblastoma stem cells to temozolomide by targeting bak1. Tumour. Biol. 2014, 35, 6293–6302. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Wan, Y.; Sun, G.; Zhang, S.; Wang, Z.; Zeng, Y. Mir-125b inhibitor may enhance the invasion-prevention activity of temozolomide in glioblastoma stem cells by targeting pias3. BioDrugs 2014, 28, 41–54. [Google Scholar] [CrossRef]
- Haemmig, S.; Baumgartner, U.; Gluck, A.; Zbinden, S.; Tschan, M.P.; Kappeler, A.; Mariani, L.; Vajtai, I.; Vassella, E. Mir-125b controls apoptosis and temozolomide resistance by targeting tnfaip3 and nkiras2 in glioblastomas. Cell Death Dis. 2014, 5, e1279. [Google Scholar] [CrossRef]
- Shi, L.; Wan, Y.; Sun, G.; Gu, X.; Qian, C.; Yan, W.; Zhang, S.; Pan, T.; Wang, Z.; You, Y. Functional differences of mir-125b on the invasion of primary glioblastoma cd133-negative cells and cd133-positive cells. Neuromolecular Med. 2012, 14, 303–316. [Google Scholar] [CrossRef]
- Wan, Y.; Fei, X.F.; Wang, Z.M.; Jiang, D.Y.; Chen, H.C.; Yang, J.; Shi, L.; Huang, Q. Expression of mir-125b in the new, highly invasive glioma stem cell and progenitor cell line su3. Chin. J. Cancer 2012, 31, 207–214. [Google Scholar] [CrossRef]
- Shi, L.; Zhang, J.; Pan, T.; Zhou, J.; Gong, W.; Liu, N.; Fu, Z.; You, Y. Mir-125b is critical for the suppression of human u251 glioma stem cell proliferation. Brain Res. 2010, 1312, 120–126. [Google Scholar] [CrossRef]
- Wu, N.; Xiao, L.; Zhao, X.; Zhao, J.; Wang, J.; Wang, F.; Cao, S.; Lin, X. Mir-125b regulates the proliferation of glioblastoma stem cells by targeting e2f2. FEBS Lett. 2012, 586, 3831–3839. [Google Scholar] [CrossRef]
- Wan, Y.; Sun, G.; Wang, Z.; Guo, J.; Shi, L. Mir-125b promotes cell proliferation by directly targeting lin28 in glioblastoma stem cells with low expression levels of mir-125b. Neuroreport 2014, 25, 289–296. [Google Scholar] [CrossRef]
- Smits, M.; Wurdinger, T.; van het Hof, B.; Drexhage, J.A.; Geerts, D.; Wesseling, P.; Noske, D.P.; Vandertop, W.P.; de Vries, H.E.; Reijerkerk, A. Myc-associated zinc finger protein (maz) is regulated by mir-125b and mediates vegf-induced angiogenesis in glioblastoma. Faseb J. 2012, 26, 2639–2647. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zheng, J.; Chen, L.; Diao, H.; Liu, Y. Predictive and prognostic roles of abnormal expression of tissue mir-125b, mir-221, and mir-222 in glioma. Mol. Neurobiol. 2016, 53, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Jesionek-Kupnicka, D.; Braun, M.; Trabska-Kluch, B.; Czech, J.; Szybka, M.; Szymanska, B.; Kulczycka-Wojdala, D.; Bienkowski, M.; Kordek, R.; Zawlik, I. Mir-21, mir-34a, mir-125b, mir-181d and mir-648 levels inversely correlate with mgmt and tp53 expression in primary glioblastoma patients. Arch. Med. Sci. 2019, 15, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Drusco, A.; Bottoni, A.; Lagana, A.; Acunzo, M.; Fassan, M.; Cascione, L.; Antenucci, A.; Kumchala, P.; Vicentini, C.; Gardiman, M.P.; et al. A differentially expressed set of micrornas in cerebro-spinal fluid (csf) can diagnose cns malignancies. Oncotarget 2015, 6, 20829–20839. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.Z.; Morris, M.E.; Yu, A.M. Microrna-328 negatively regulates the expression of breast cancer resistance protein (bcrp/abcg2) in human cancer cells. Mol Pharm. 2009, 75, 1374–1379. [Google Scholar] [CrossRef] [PubMed]
- Delic, S.; Lottmann, N.; Stelzl, A.; Liesenberg, F.; Wolter, M.; Gotze, S.; Zapatka, M.; Shiio, Y.; Sabel, M.C.; Felsberg, J.; et al. Mir-328 promotes glioma cell invasion via sfrp1-dependent wnt-signaling activation. Neuro Oncol. 2014, 16, 179–190. [Google Scholar] [CrossRef]
- Arora, S.; Ranade, A.R.; Tran, N.L.; Nasser, S.; Sridhar, S.; Korn, R.L.; Ross, J.T.; Dhruv, H.; Foss, K.M.; Sibenaller, Z.; et al. Microrna-328 is associated with (non-small) cell lung cancer (nsclc) brain metastasis and mediates nsclc migration. Int. J. Cancer 2011, 129, 2621–2631. [Google Scholar] [CrossRef]
- Chan, Y.C.; Banerjee, J.; Choi, S.Y.; Sen, C.K. Mir-210: The master hypoxamir. Microcirculation 2012, 19, 215–223. [Google Scholar] [CrossRef]
- Lai, N.S.; Dong, Q.S.; Ding, H.; Miao, Z.L.; Lin, Y.C. Microrna-210 overexpression predicts poorer prognosis in glioma patients. J. Clin. Neurosci. 2014, 21, 755–760. [Google Scholar] [CrossRef]
- Rosenberg, T.; Thomassen, M.; Jensen, S.S.; Larsen, M.J.; Sorensen, K.P.; Hermansen, S.K.; Kruse, T.A.; Kristensen, B.W. Acute hypoxia induces upregulation of microrna-210 expression in glioblastoma spheroids. CNS Oncol. 2015, 4, 25–35. [Google Scholar] [CrossRef]
- Zhang, S.; Lai, N.; Liao, K.; Sun, J.; Lin, Y. Microrna-210 regulates cell proliferation and apoptosis by targeting regulator of differentiation 1 in glioblastoma cells. Folia Neuropathol. 2015, 53, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Shang, C.; Hong, Y.; Guo, Y.; Liu, Y.H.; Xue, Y.X. Mir-210 up-regulation inhibits proliferation and induces apoptosis in glioma cells by targeting sin3a. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2014, 20, 2571–2577. [Google Scholar]
- Lee, D.; Sun, S.; Zhang, X.Q.; Zhang, P.D.; Ho, A.S.; Kiang, K.M.; Fung, C.F.; Lui, W.M.; Leung, G.K. Microrna-210 and endoplasmic reticulum chaperones in the regulation of chemoresistance in glioblastoma. J. Cancer 2015, 6, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Jiang, T.; Zhong, Y.; Yu, Y. Mir-210 inhibits cell migration and invasion by targeting the brain-derived neurotrophic factor in glioblastoma. J. Cell. Biochem. 2019, 120, 11375–11382. [Google Scholar] [CrossRef]
- Paterson, M.R.; Kriegel, A.J. Mir-146a/b: A family with shared seeds and different roots. Physiol. Genom. 2017, 49, 243–252. [Google Scholar] [CrossRef]
- Iacona, J.R.; Lutz, C.S. Mir-146a-5p: Expression, regulation, and functions in cancer. Wiley Interdiscip Rev. RNA 2019, 10, e1533. [Google Scholar] [CrossRef]
- Mei, J.; Bachoo, R.; Zhang, C.L. Microrna-146a inhibits glioma development by targeting notch1. Mol. Cell Biol. 2011, 31, 3584–3592. [Google Scholar] [CrossRef]
- Liu, R.; Li, W.; Wu, C. A functional polymorphism in the premir146a gene influences the prognosis of glioblastoma multiforme by interfering with the balance between notch1 and notch2. Mol. Med. Rep. 2015, 12, 5475–5481. [Google Scholar] [CrossRef]
- Hu, H.Q.; Sun, L.G.; Guo, W.J. Decreased mirna-146a in glioblastoma multiforme and regulation of cell proliferation and apoptosis by target notch1. Int. J. Biol. Markers 2016, 31, e270–e275. [Google Scholar] [CrossRef]
- Wu, H.; Liu, Q.; Cai, T.; Chen, Y.D.; Wang, Z.F. Induction of microrna-146a is involved in curcumin-mediated enhancement of temozolomide cytotoxicity against human glioblastoma. Mol. Med. Rep. 2015, 12, 5461–5466. [Google Scholar] [CrossRef]
- Hwang, S.J.; Seol, H.J.; Park, Y.M.; Kim, K.H.; Gorospe, M.; Nam, D.H.; Kim, H.H. Microrna-146a suppresses metastatic activity in brain metastasis. Mol. Cells 2012, 34, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Crawford, M.; Brawner, E.; Batte, K.; Yu, L.; Hunter, M.G.; Otterson, G.A.; Nuovo, G.; Marsh, C.B.; Nana-Sinkam, S.P. Microrna-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem. Biophys. Res. Commun. 2008, 373, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Han, I.B.; Kim, M.; Lee, S.H.; Kim, J.K.; Kim, S.H.; Chang, J.H.; Teng, Y.D. Down-regulation of microrna-126 in glioblastoma and its correlation with patient prognosis: A pilot study. Anticancer Res. 2016, 36, 6691–6697. [Google Scholar] [CrossRef]
- Cui, H.; Mu, Y.; Yu, L.; Xi, Y.G.; Matthiesen, R.; Su, X.; Sun, W. Methylation of the mir-126 gene associated with glioma progression. Fam. Cancer 2016, 15, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Luan, Y.; Zuo, L.; Zhang, S.; Wang, G.; Peng, T. Microrna-126 acts as a tumor suppressor in glioma cells by targeting insulin receptor substrate 1 (irs-1). Int. J. Clin. Exp. Pathol. 2015, 8, 10345–10354. [Google Scholar]
- Li, Y.; Li, Y.; Ge, P.; Ma, C. Mir-126 regulates the erk pathway via targeting kras to inhibit the glioma cell proliferation and invasion. Mol. Neurobiol. 2017, 54, 137–145. [Google Scholar] [CrossRef]
- Xu, Y.; Xu, W.; Lu, T.; Dai, Y.; Liang, W. Mir-126 affects the invasion and migration of glioma cells through gata4. Artif. Cells Nanomed. Biotechnol. 2017, 45, 1–7. [Google Scholar] [CrossRef]
- Luo, W.; Yan, D.; Song, Z.; Zhu, X.; Liu, X.; Li, X.; Zhao, S. Mir-126-3p sensitizes glioblastoma cells to temozolomide by inactivating wnt/beta-catenin signaling via targeting sox2. Life Sci. 2019, 226, 98–106. [Google Scholar] [CrossRef]
- Tavazoie, S.F.; Alarcon, C.; Oskarsson, T.; Padua, D.; Wang, Q.; Bos, P.D.; Gerald, W.L.; Massague, J. Endogenous human micrornas that suppress breast cancer metastasis. Nature 2008, 451, 147–152. [Google Scholar] [CrossRef]
- Lundin, K.E.; Gissberg, O.; Smith, C.I. Oligonucleotide therapies: The past and the present. Hum. Gene Ther. 2015, 26, 475–485. [Google Scholar] [CrossRef]
- Ling, H.; Fabbri, M.; Calin, G.A. Micrornas and other non-coding rnas as targets for anticancer drug development. Nat. Rev. Drug Discov. 2013, 12, 847–865. [Google Scholar] [CrossRef] [PubMed]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase i study of mrx34, a liposomal mir-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Van Zandwijk, N.; Pavlakis, N.; Kao, S.C.; Linton, A.; Boyer, M.J.; Clarke, S.; Huynh, Y.; Chrzanowska, A.; Fulham, M.J.; Bailey, D.L.; et al. Safety and activity of microrna-loaded minicells in patients with recurrent malignant pleural mesothelioma: A first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017, 18, 1386–1396. [Google Scholar] [CrossRef]
- Lennox, K.A.; Behlke, M.A. Chemical modification and design of anti-mirna oligonucleotides. Gene Ther. 2011, 18, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Piva, R.; Spandidos, D.A.; Gambari, R. From microrna functions to microrna therapeutics: Novel targets and novel drugs in breast cancer research and treatment (review). Int. J. Oncol. 2013, 43, 985–994. [Google Scholar] [CrossRef] [PubMed]
- Poltronieri, P.; D’Urso, P.I.; Mezzolla, V.; D’Urso, O.F. Potential of anti-cancer therapy based on anti-mir-155 oligonucleotides in glioma and brain tumours. Chem. Biol. Drug Des. 2013, 81, 79–84. [Google Scholar] [CrossRef]
- Lipi, F.; Chen, S.; Chakravarthy, M.; Rakesh, S.; Veedu, R.N. In vitro evolution of chemically-modified nucleic acid aptamers: Pros and cons, and comprehensive selection strategies. RNA Biol. 2016, 13, 1232–1245. [Google Scholar] [CrossRef]
- Majlessi, M.; Nelson, N.C.; Becker, M.M. Advantages of 2′-o-methyl oligoribonucleotide probes for detecting rna targets. Nucleic Acids Res. 1998, 26, 2224–2229. [Google Scholar] [CrossRef]
- Miroshnichenko, S.K.; Amirloo, B.; Bichenkova, E.V.; Vlassov, V.V.; Zenkova, M.A.; Patutina, O.A. 2′ome modification of anti-mirna-21 oligonucleotide–peptide conjugate improves its hybridization properties and catalytic activity. Russ. J. Bioorg. Chem. 2019, 45, 803–812. [Google Scholar] [CrossRef]
- Geary, R.S.; Watanabe, T.A.; Truong, L.; Freier, S.; Lesnik, E.A.; Sioufi, N.B.; Sasmor, H.; Manoharan, M.; Levin, A.A. Pharmacokinetic properties of 2′-o-(2-methoxyethyl)-modified oligonucleotide analogs in rats. J. Pharmacol. Exp. Ther. 2001, 296, 890–897. [Google Scholar]
- Lin, Y.; Qiu, Q.; Gill, S.C.; Jayasena, S.D. Modified rna sequence pools for in vitro selection. Nucleic Acids Res. 1994, 22, 5229–5234. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Le, B.T.; Chakravarthy, M.; Kosbar, T.R.; Veedu, R.N. Systematic evaluation of 2′-fluoro modified chimeric antisense oligonucleotide-mediated exon skipping in vitro. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Alves Ferreira-Bravo, I.; Cozens, C.; Holliger, P.; DeStefano, J.J. Selection of 2′-deoxy-2′-fluoroarabinonucleotide (fana) aptamers that bind hiv-1 reverse transcriptase with picomolar affinity. Nucleic Acids Res. 2015, 43, 9587–9599. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhang, S.; Chaput, J.C. Darwinian evolution of an alternative genetic system provides support for tna as an rna progenitor. Nat. Chem. 2012, 4, 183–187. [Google Scholar] [CrossRef]
- Kato, Y.; Minakawa, N.; Komatsu, Y.; Kamiya, H.; Ogawa, N.; Harashima, H.; Matsuda, A. New ntp analogs: The synthesis of 4′-thioutp and 4′-thioctp and their utility for selex. Nucleic Acids Res. 2005, 33, 2942–2951. [Google Scholar] [CrossRef]
- Veedu, R.N.; Wengel, J. Locked nucleic acids: Promising nucleic acid analogs for therapeutic applications. Chem. Biodivers. 2010, 7, 536–542. [Google Scholar] [CrossRef]
- Veedu, R.N.; Wengel, J. Locked nucleic acid as a novel class of therapeutic agents. RNA Biol. 2009, 6, 321–323. [Google Scholar] [CrossRef]
- Le, B.T.; Adams, A.M.; Fletcher, S.; Wilton, S.D.; Veedu, R.N. Rational design of short locked nucleic acid-modified 2′-o-methyl antisense oligonucleotides for efficient exon-skipping in vitro. Mol. Ther. Nucleic Acids 2017, 9, 155–161. [Google Scholar] [CrossRef]
- Koizumi, M.; Takagi-Sato, M.; Okuyama, R.; Araki, K.; Sun, W.; Nakai, D.; Tsutsumi, S.; Kawai, K. Direct comparison of in vivo antisense activity of ena oligonucleotides targeting ptp1b mrna with that of 2′-o-(2-methoxy)ethyl-modified oligonucleotides. Oligonucleotides 2006, 16, 253–262. [Google Scholar] [CrossRef]
- Hyrup, B.; Nielsen, P.E. Peptide nucleic acids (pna): Synthesis, properties and potential applications. Bioorg. Med. Chem. 1996, 4, 5–23. [Google Scholar] [CrossRef]
- Summerton, J.; Weller, D. Morpholino antisense oligomers: Design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 1997, 7, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Le, B.T.; Rahimizadeh, K.; Shaikh, K.; Mohal, N.; Veedu, R.N. Synthesis of a morpholino nucleic acid (mna)-uridine phosphoramidite, and exon skipping using mna/2′-o-methyl mixmer antisense oligonucleotide. Molecules 2016, 21, 1582. [Google Scholar] [CrossRef] [PubMed]
- Eckstein, F. Phosphorothioate oligodeoxynucleotides: What is their origin and what is unique about them? Antisense Nucleic Acid Drug Dev. 2000, 10, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Le, B.T.; Chen, S.; Abramov, M.; Herdewijn, P.; Veedu, R.N. Evaluation of anhydrohexitol nucleic acid, cyclohexenyl nucleic acid and d-altritol nucleic acid-modified 2′-o-methyl rna mixmer antisense oligonucleotides for exon skipping in vitro. Chem. Commun. 2016, 52, 13467–13470. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E.; Kauppinen, S. Development of micro rna therapeutics is coming of age. EMBO Mol. Med. 2014, 6, 851–864. [Google Scholar] [CrossRef] [PubMed]
- Lindow, M.; Kauppinen, S. Discovering the first microrna-targeted drug. J. Cell Biol. 2012, 199, 407–412. [Google Scholar] [CrossRef]
- Titze-de-Almeida, R.; David, C.; Titze-de-Almeida, S.S. The race of 10 synthetic rnai-based drugs to the pharmaceutical market. Pharm. Res. 2017, 34, 1339–1363. [Google Scholar] [CrossRef]
- Hanna, J.; Hossain, G.S.; Kocerha, J. The potential for microrna therapeutics and clinical research. Front. Genet 2019, 10, 478. [Google Scholar] [CrossRef]

| Names | Total | miRNAs |
|---|---|---|
| BrM-down BrM-up GBM-down GBM-up | 1 | miR-145 |
| BrM-down GBM-down GBM-up | 2 | miR-31 miR-451 |
| BrM-up GBM-down GBM-up | 5 | miR-19a miR-143 miR-125b miR-328 miR-210 |
| BrM-down BrM-up GBM-down | 2 | miR-146a miR-126 |
| GBM-down GBM-up | 31 | miR-221/222 miR-200b miR-132 miR-185 miR-100 miR-135b miR-329 miR-106a miR-873 miR-205 miR-23a miR-323 miR-26b miR-200a miR-30bc miR-330 miR-146 miR-29b miR-9 miR-16 miR-15b miR-195 miR-130a miR-25 miR-23b miR-296 miR-141 miR-29a miR-27a miR-26a miR-107 |
| BrM-down GBM-down | 1 | miR-7 |
| BrM-up GBM-down | 5 | miR-197 miR-133a miR-378 miR-184 miR-133b |
| BrM-down GBM-up | 1 | miR-95 |
| BrM-up GBM-up | 2 | miR-21 miR-10b |
| GBM-down | 61 | miR-497 miR-429 miR-137 miR-181d miR-129 miR-153 miR-487b miR-340 miR-150 miR-320 miR-339-5p miR-34a miR-491 miR-520b miR-331 miR-101 miR-410 miR-432 miR-181b miR-149 miR-377 miR-190 miR-422a miR-203 miR-124 miR-299 miR-136 miR-485 miR-610 miR-152 miR-146b-5p miR-634 miR-139 miR-219 miR-297 miR-513 LET-7 miR-365a miR-548b miR-181a miR-511-1 miR-218 miR-32 miR-128a miR-885 miR-154 miR-326 miR-433 miR-187 miR-379 miR-519a miR-483 miR-181 miR-874 miR-181c miR-138 miR-663 miR-182 miR-125a miR-148 miR-128 |
| GBM-up | 51 | miR-142 miR-204 miR-92a miR-155 miR-20a miR-425 miR-519d miR-28 miR-182/183 miR-10a miR-193 miR-23 miR-93 miR-595 miR-24 miR-140 miR-30a miR-196 miR-148a miR-15a miR-215 miR-96 miR-655 miR-455 miR-196ab miR-130b miR-371-373 miR-18a miR-19b miR-92 miR-486 miR-27b miR-200c miR-301a miR-363 miR-383 miR-134 miR-92b miR-106b miR-123 miR-381 miR-17 miR-27 miR-367-302 miR-339 miR-603 miR-335 miR-372 miR-516-3p miR-17-92 cluster miR-582-5p |
| BrM-down | 6 | HS-170 miR-509 miR-1258 miR-30c miR-768-3p miR-29c |
| BrM-up | 9 | miR-576-5p miR-1 miR-199b HS-287 miR-200 miR-330-3p miR-28-5p miR-22 miR-199a |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balachandran, A.A.; Larcher, L.M.; Chen, S.; Veedu, R.N. Therapeutically Significant MicroRNAs in Primary and Metastatic Brain Malignancies. Cancers 2020, 12, 2534. https://doi.org/10.3390/cancers12092534
Balachandran AA, Larcher LM, Chen S, Veedu RN. Therapeutically Significant MicroRNAs in Primary and Metastatic Brain Malignancies. Cancers. 2020; 12(9):2534. https://doi.org/10.3390/cancers12092534
Chicago/Turabian StyleBalachandran, Akilandeswari A., Leon M. Larcher, Suxiang Chen, and Rakesh N. Veedu. 2020. "Therapeutically Significant MicroRNAs in Primary and Metastatic Brain Malignancies" Cancers 12, no. 9: 2534. https://doi.org/10.3390/cancers12092534
APA StyleBalachandran, A. A., Larcher, L. M., Chen, S., & Veedu, R. N. (2020). Therapeutically Significant MicroRNAs in Primary and Metastatic Brain Malignancies. Cancers, 12(9), 2534. https://doi.org/10.3390/cancers12092534





