Critical Role for Cold Shock Protein YB-1 in Cytokinesis
Simple Summary
Abstract
1. Introduction
2. Results
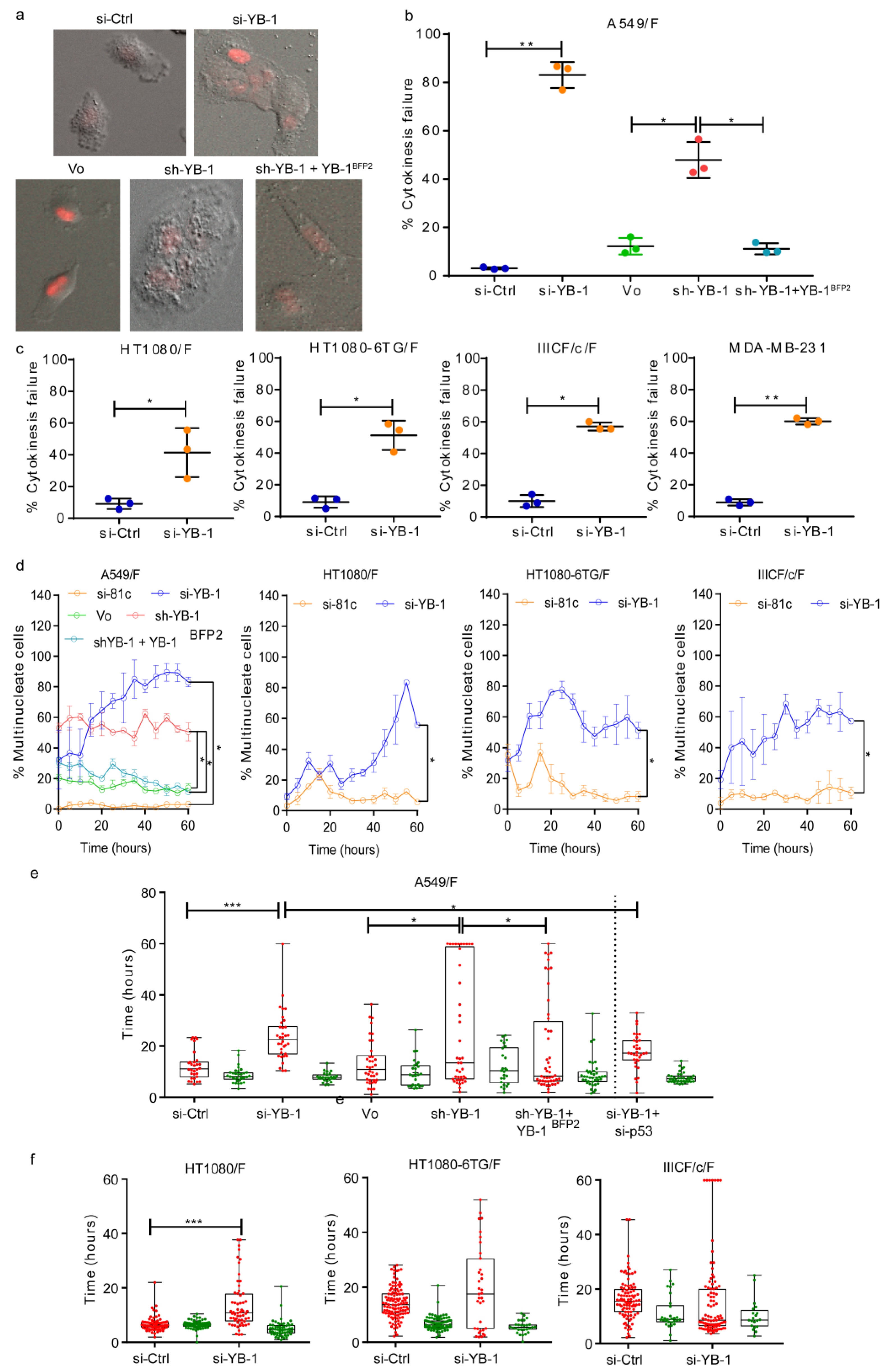
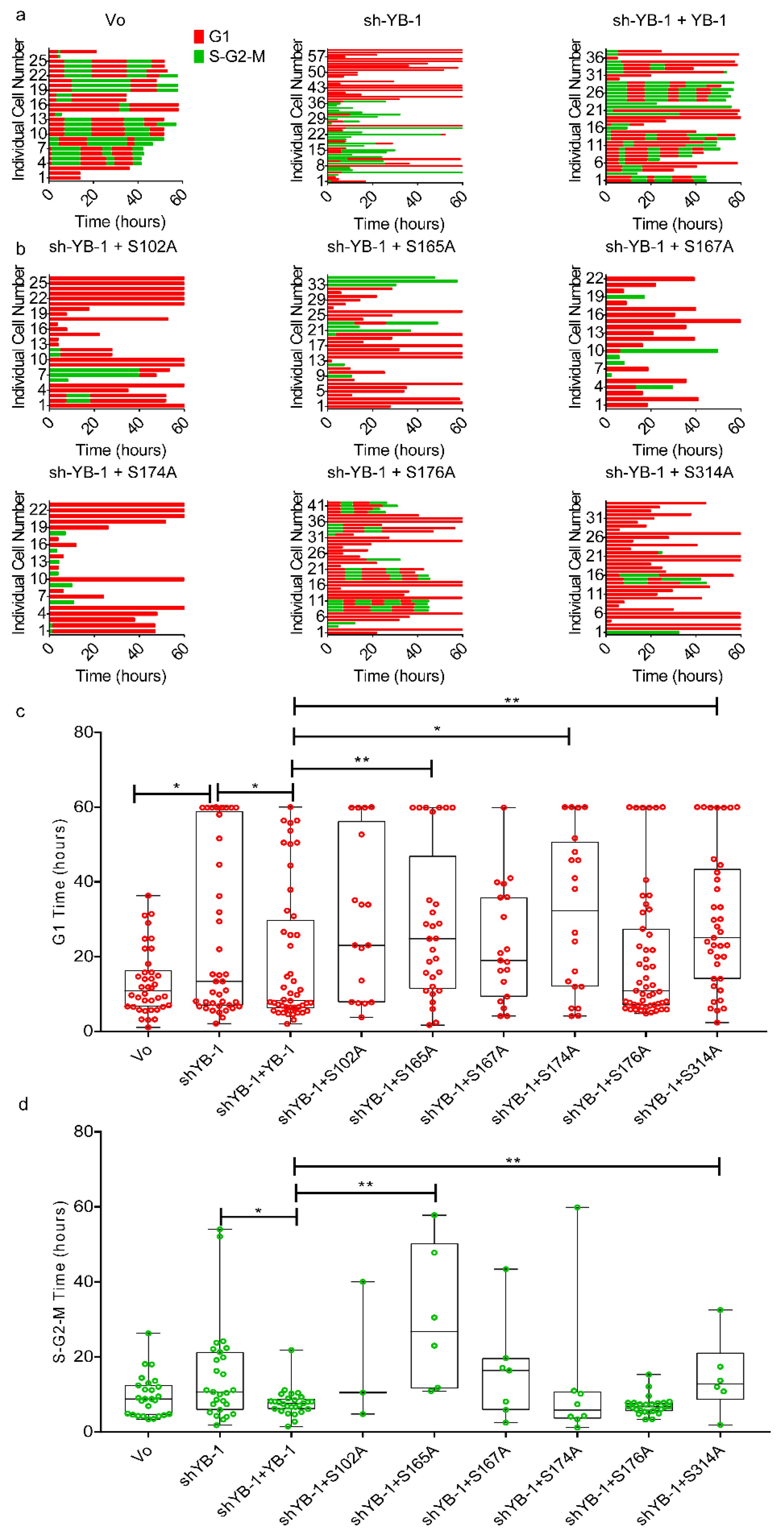
2.1. YB-1 Depletion Causes Cytokinesis Failure, Multinucleation and Accumulation in G1
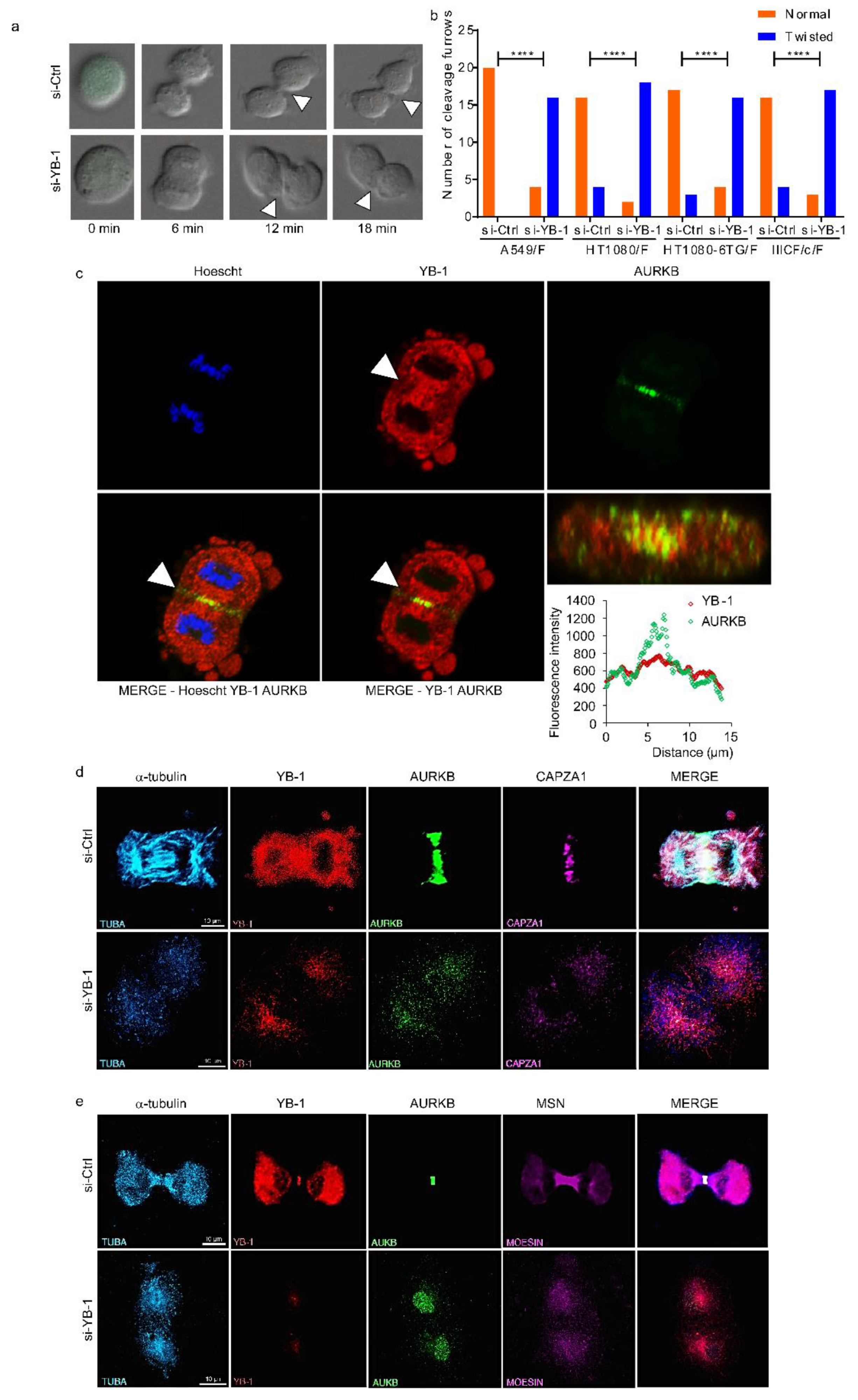
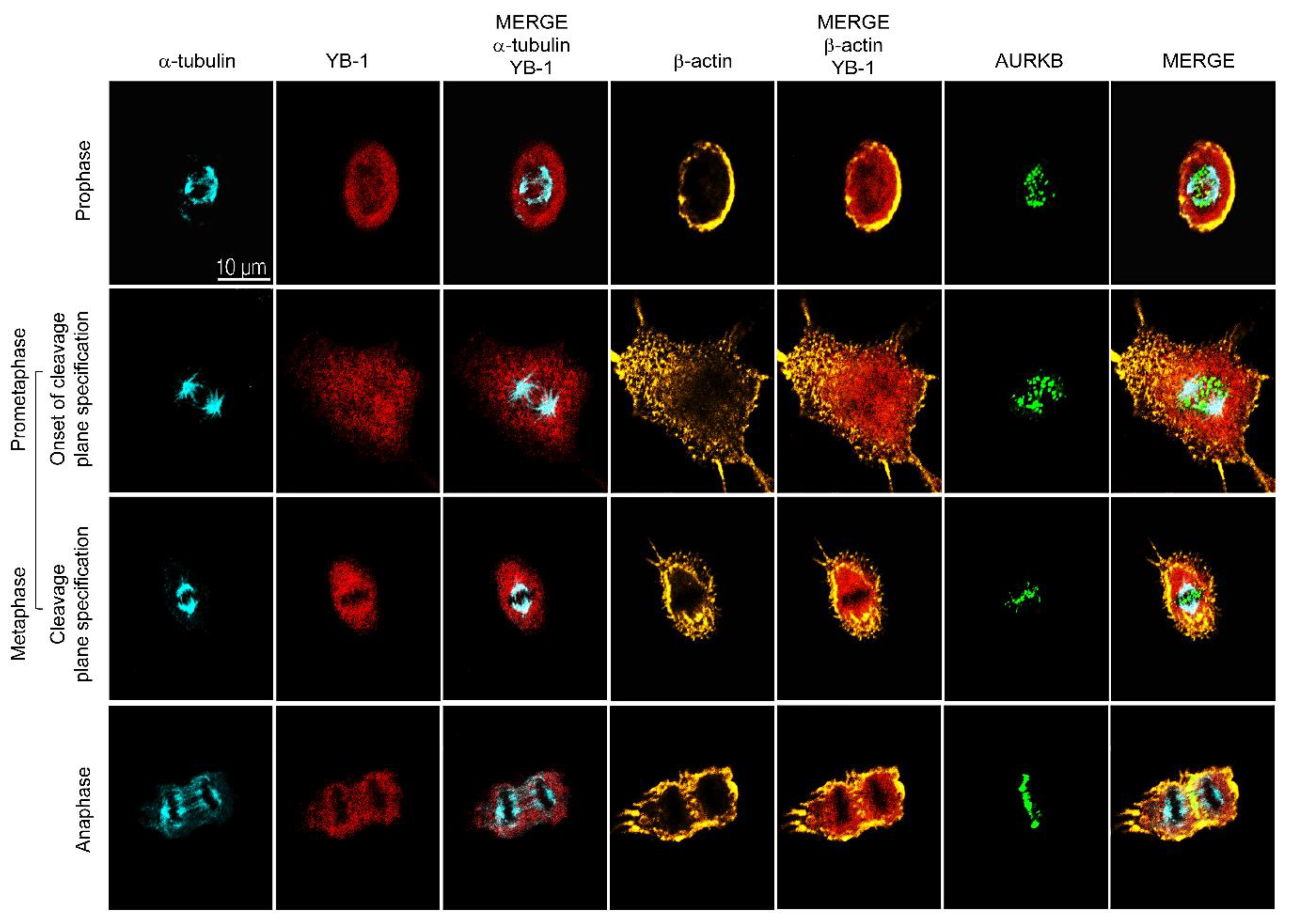
2.2. YB-1 Is Required for Initiation of Cleavage Furrow Formation
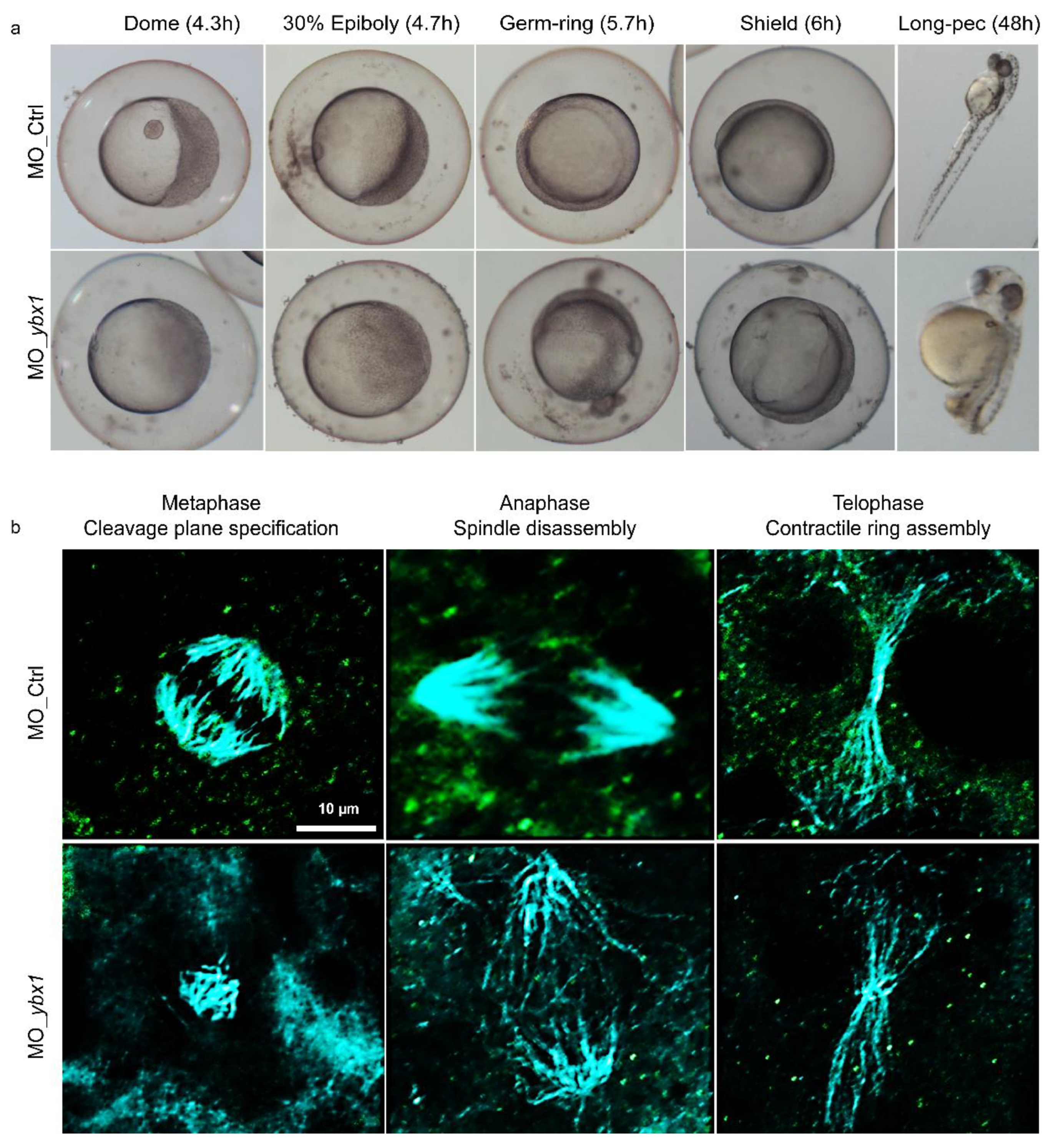
2.3. YB-1 Depletion Causes Cytokinesis Failure in Zebrafish Embryos Resulting in Developmental Defects
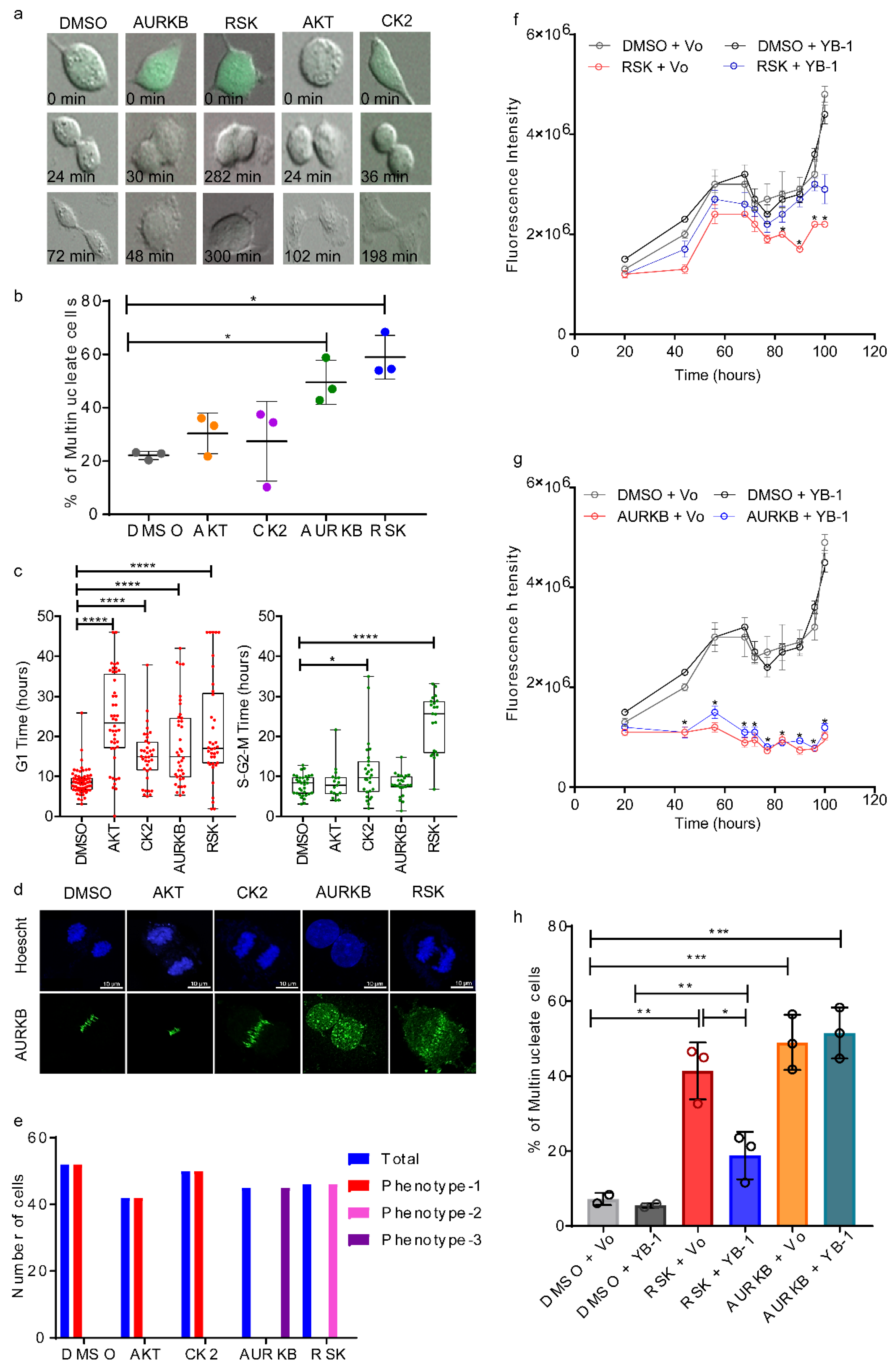
2.4. p90 Ribosomal-S6 Kinase (RSK) Phosphorylates YB-1 and Is Required for Cytokinesis.
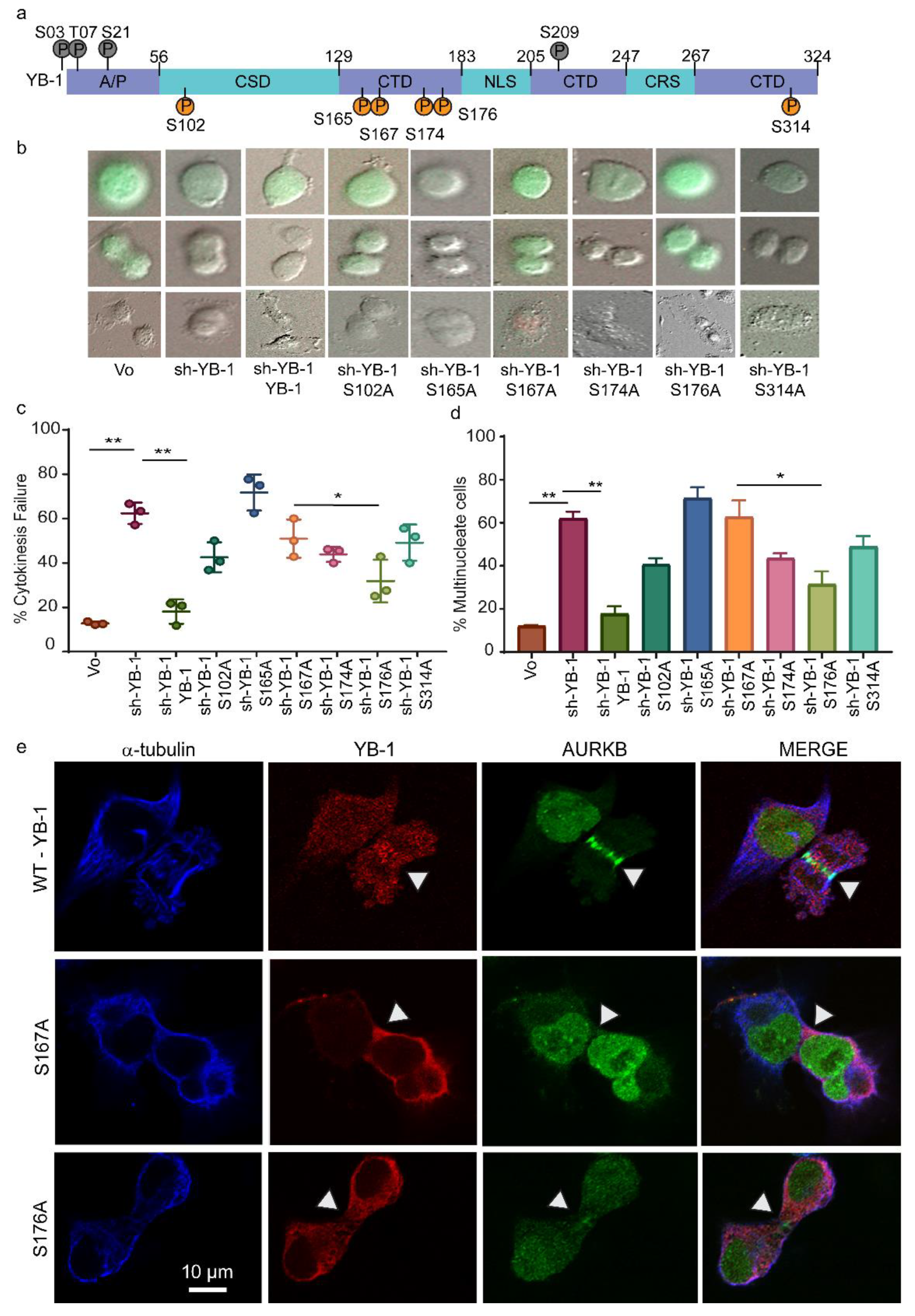
2.5. Phosphorylation Defective YB-1 Mutants Fail to Complete Cytokinesis.
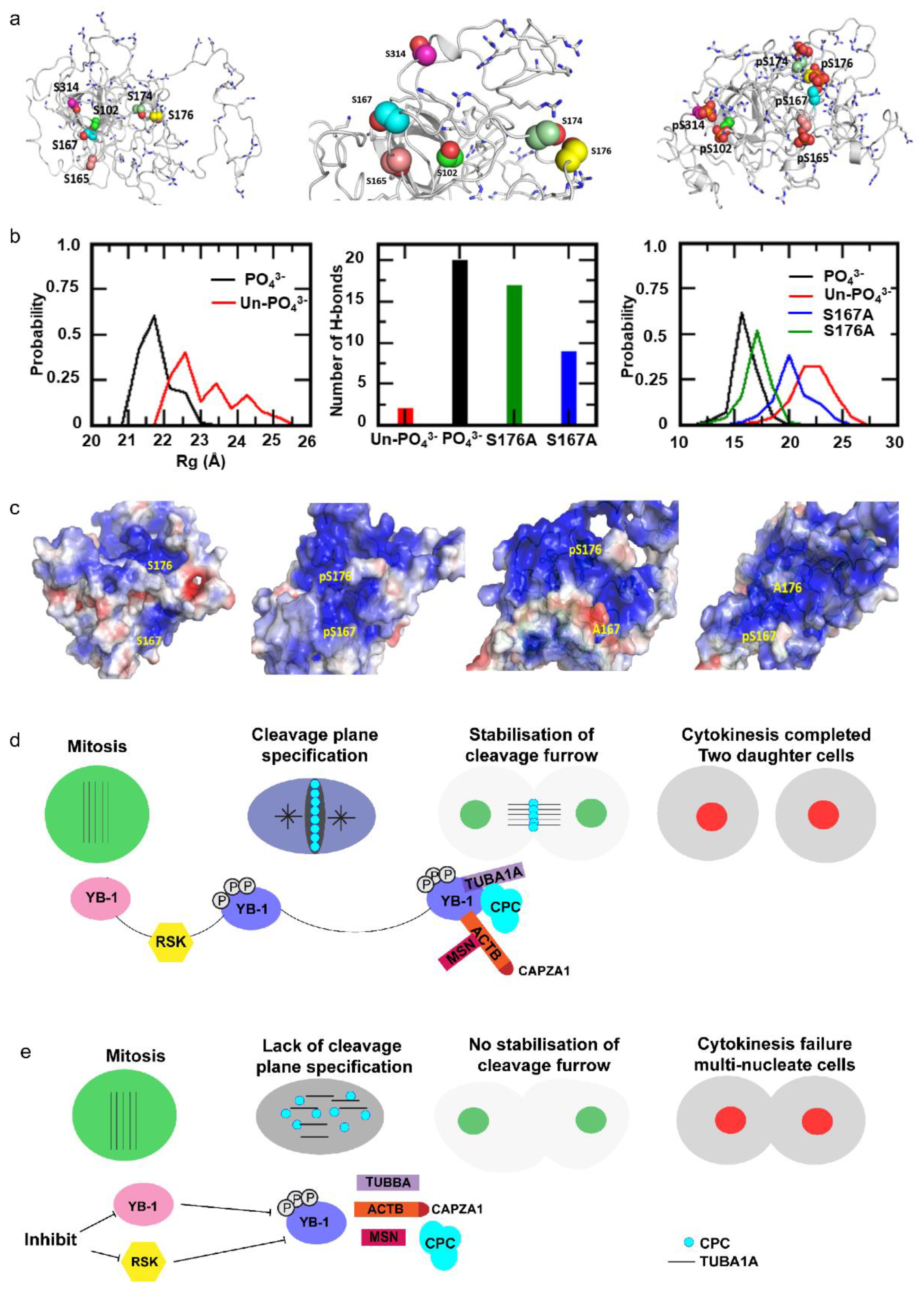
2.6. Cooperative YB-1 Phosphorylation on Multiple residues Alters Protein Folding.
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Generation of FUCCI Cell Lines
4.3. siRNA Transfection
4.4. Generation of YB-1:EBFP2 Expression Constructs
4.5. Plasmid Transfection
4.6. Kinase Inhibitor Treatment and Cell Cycle
4.7. Cell Extracts and Western Blot
4.8. Live Imaging and Processing of Time-lapse Data
4.9. Cell Synchronization
4.10. Cell Proliferation Assays
4.11. Immunofluorescence
4.12. Zebrafish Lines
4.13. Microinjection of Zebrafish Embryos
4.14. Whole-Mount Immunofluorescence of Zebrafish Embryos
4.15. Subcellular Fractionation of Cultured Cells
4.16. Immunopurification of YB-1
4.17. Immunopurification of Cross-Linked YB-1
4.18. Mass Spectrometry
4.19. Modeling the Structure of YB-1 Protein
4.20. Molecular Dynamics Simulations
4.21. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lasham, A.; Woolley, A.G.; Dunn, S.E.; Braithwaite, A.W. Yb-1: Oncoprotein, prognostic marker and therapeutic target? Biochem. J. 2013, 449, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Eliseeva, I.; Kim, E.; Guryanov, S.; Ovchinnikov, L.; Lyabin, D. Y-box-binding protein 1 (yb-1) and its functions. Biochemistry 2011, 76, 1402–1433. [Google Scholar] [CrossRef] [PubMed]
- Habibi, G.; Leung, S.; Law, J.H.; Gelmon, K.; Masoudi, H.; Turbin, D.; Pollak, M.; Nielsen, T.O.; Huntsman, D.; Dunn, S.E. Redefining prognostic factors for breast cancer: Yb-1 is a stronger predictor of relapse and disease-specific survival than estrogen receptor or her-2 across all tumor subtypes. Breast Cancer Res. 2008, 10, R86. [Google Scholar] [CrossRef] [PubMed]
- Lasham, A.; Samuel, W.; Cao, H.; Patel, R.; Mehta, R.; Stern, J.L.; Reid, G.; Woolley, A.G.; Miller, L.D.; Black, M.A.; et al. Yb-1, the e2f pathway, and regulation of tumor cell growth. J. Natl. Cancer Inst. 2011, 104, 133–146. [Google Scholar] [CrossRef]
- Woolley, A.G.; Algie, M.; Samuel, W.; Harfoot, R.; Wiles, A.; Hung, N.A.; Tan, P.-H.; Hains, P.; Valova, V.A.; Huschtscha, L. Prognostic association of yb-1 expression in breast cancers: A matter of antibody. PLoS ONE 2011, 6, e20603. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Bergmann, S.; Royer-Pokora, B.; Fietze, E.; Jürchott, K.; Hildebrandt, B.; Trost, D.; Leenders, F.; Claude, J.-C.; Theuring, F.; Bargou, R. Yb-1 provokes breast cancer through the induction of chromosomal instability that emerges from mitotic failure and centrosome amplification. Cancer Res. 2005, 65, 4078–4087. [Google Scholar] [CrossRef]
- Gao, Y.; Fotovati, A.; Lee, C.; Wang, M.; Cote, G.; Guns, E.; Toyota, B.; Faury, D.; Jabado, N.; Dunn, S.E. Inhibition of y-box binding protein-1 slows the growth of glioblastoma multiforme and sensitizes to temozolomide independent o6-methylguanine-DNA methyltransferase. Mol. Cancer Ther. 2009, 8, 3276–3284. [Google Scholar] [CrossRef]
- Lee, C.; Dhillon, J.; Wang, M.Y.; Gao, Y.; Hu, K.; Park, E.; Astanehe, A.; Hung, M.-C.; Eirew, P.; Eaves, C.J. Targeting yb-1 in her-2 overexpressing breast cancer cells induces apoptosis via the mtor/stat3 pathway and suppresses tumor growth in mice. Cancer Res. 2008, 68, 8661–8666. [Google Scholar] [CrossRef]
- Miao, X.; Wu, Y.; Wang, Y.; Zhu, X.; Yin, H.; He, Y.; Li, C.; Liu, Y.; Lu, X.; Chen, Y. Y-box-binding protein-1 (yb-1) promotes cell proliferation, adhesion and drug resistance in diffuse large b-cell lymphoma. Exp. Cell Res. 2016, 346, 157–166. [Google Scholar] [CrossRef]
- Guo, T.; Zhao, S.; Wang, P.; Xue, X.; Zhang, Y.; Yang, M.; Li, N.; Li, Z.; Xu, L.; Jiang, L. Yb-1 regulates tumor growth by promoting macc1/c-met pathway in human lung adenocarcinoma. Oncotarget 2017, 8, 48110. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sun, R.; Chi, Z.; Li, S.; Hao, L. Silencing of y-box binding protein-1 by rna interference inhibits proliferation, invasion, and metastasis, and enhances sensitivity to cisplatin through nf-κb signaling pathway in human neuroblastoma sh-sy5y cells. Mol. Cell. Biochem. 2017, 433, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chao, H.-M.; Huang, H.-X.; Chang, P.-H.; Tseng, K.-C.; Miyajima, A.; Chern, E. Y-box binding protein-1 promotes hepatocellular carcinoma-initiating cell progression and tumorigenesis via wnt/β-catenin pathway. Oncotarget 2017, 8, 2604. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Gao, S.; Yu, C.; Li, M.; Liu, P.; Zhang, G.; Song, J.; Zheng, J. Effect and mechanism of yb-1 knockdown on glioma cell growth, migration, and apoptosis. Acta Biochim. Et Biophys. Sin. 2020, 52, 168–179. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Xu, L.; Lu, X.; Fu, D.; Su, J.; Geng, H.; Qin, G.; Chen, R.; Quan, C. Cd4+ t cells promote renal cell carcinoma proliferation via modulating ybx1. Exp. Cell Res. 2018, 363, 95–101. [Google Scholar] [CrossRef]
- Schittek, B.; Psenner, K.; Sauer, B.; Meier, F.; Iftner, T.; Garbe, C. The increased expression of y box-binding protein 1 in melanoma stimulates proliferation and tumor invasion, antagonizes apoptosis and enhances chemoresistance. Int. J. Cancer 2007, 120, 2110–2118. [Google Scholar] [CrossRef]
- Johnson, T.G.; Schelch, K.; Mehta, S.; Burgess, A.; Reid, G. Why be one protein when you can affect many? The multiple roles of yb-1 in lung cancer and mesothelioma. Front. Cell Dev. Biol. 2019, 7, 221. [Google Scholar] [CrossRef]
- Lim, W.K.; Lyashenko, E.; Califano, A. Master regulators used as breast cancer metastasis classifier. Pac. Symp. Biocomput. 2009, 504–515. [Google Scholar]
- Homer, C.; Knight, D.A.; Hananeia, L.; Sheard, P.; Risk, J.; Lasham, A.; Royds, J.A.; Braithwaite, A.W. Y-box factor yb1 controls p53 apoptotic function. Oncogene 2005, 24, 8314–8325. [Google Scholar] [CrossRef]
- Davies, A.H.; Barrett, I.; Pambid, M.R.; Hu, K.; Stratford, A.L.; Freeman, S.; Berquin, I.M.; Pelech, S.; Hieter, P.; Maxwell, C.; et al. Yb-1 evokes susceptibility to cancer through cytokinesis failure, mitotic dysfunction and her2 amplification. Oncogene 2011, 30, 3649–3660. [Google Scholar] [CrossRef]
- Bargou, R.C.; Jürchott, K.; Wagener, C.; Bergmann, S.; Metzner, S.; Bommert, K.; Mapara, M.Y.; Winzer, K.-J.; Dietel, M.; Dörken, B. Nuclear localization and increased levels of transcription factor yb-1 in primary human breast cancers are associated with intrinsic mdr1 gene expression. Nat. Med. 1997, 3, 447. [Google Scholar] [CrossRef] [PubMed]
- Lasham, A.; Mehta, S.Y.; Fitzgerald, S.J.; Woolley, A.G.; Hearn, J.I.; Hurley, D.G.; Ruza, I.; Algie, M.; Shelling, A.N.; Braithwaite, A.W.; et al. A novel egr-1 dependent mechanism for yb-1 modulation of paclitaxel response in a triple negative breast cancer cell line. Int. J. Cancer 2016, 139, 1157–1170. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Zhao, K.; Zhang, J.; Zhu, J.; Xiao, J. Yb-1 modulates the drug resistance of glioma cells by activation of mdm2/p53 pathway. Drug Des. Dev. Ther. 2019, 13, 317. [Google Scholar] [CrossRef] [PubMed]
- Uchiumi, T.; Fotovati, A.; Sasaguri, T.; Shibahara, K.; Shimada, T.; Fukuda, T.; Nakamura, T.; Izumi, H.; Tsuzuki, T.; Kuwano, M.; et al. Yb-1 is important for an early stage embryonic development: Neural tube formation and cell proliferation. J. Biol. Chem. 2006, 281, 40440–40449. [Google Scholar] [CrossRef]
- Lu, Z.H.; Books, J.T.; Ley, T.J. Yb-1 is important for late-stage embryonic development, optimal cellular stress responses, and the prevention of premature senescence. Mol. Cell. Biol. 2005, 25, 4625–4637. [Google Scholar] [CrossRef]
- Raffetseder, U.; Rauen, T.; Boor, P.; Ostendorf, T.; Hanssen, L.; Floege, J.; En-Nia, A.; Djudjaj, S.; Frye, B.C.; Mertens, P.R. Extracellular yb-1 blockade in experimental nephritis upregulates notch-3 receptor expression and signaling. Nephron. Exp. Nephrol. 2011, 118, e100–e108. [Google Scholar] [CrossRef]
- Sun, J.; Yan, L.; Shen, W.; Meng, A. Maternal ybx1 safeguards zebrafish oocyte maturation and maternal-to-zygotic transition by repressing global translation. Development 2018, 145, dev166587. [Google Scholar] [CrossRef]
- Jürchott, K.; Bergmann, S.; Stein, U.; Walther, W.; Janz, M.; Manni, I.; Piaggio, G.; Fietze, E.; Dietel, M.; Royer, H.-D. Yb-1 as a cell cycle regulated transcription factor facilitating cyclin a and cyclin b1 gene expression. J. Biol. Chem. 2003. [Google Scholar] [CrossRef]
- Basaki, Y.; Taguchi, K.-I.; Izumi, H.; Murakami, Y.; Kubo, T.; Hosoi, F.; Watari, K.; Nakano, K.; Kawaguchi, H.; Ohno, S.; et al. Y-box binding protein-1 (yb-1) promotes cell cycle progression through cdc6-dependent pathway in human cancer cells. Eur. J. Cancer 2010, 46, 954–965. [Google Scholar] [CrossRef]
- Khandelwal, P.; Padala, M.K.; Cox, J.; Guntaka, R.V. The n-terminal domain of y-box binding protein-1 induces cell cycle arrest in g2/m phase by binding to cyclin d1. Int. J. Cell. Biol. 2009, 2009. [Google Scholar] [CrossRef]
- Guarino, A.M.; Troiano, A.; Pizzo, E.; Bosso, A.; Vivo, M.; Pinto, G.; Amoresano, A.; Pollice, A.; La Mantia, G.; Calabrò, V. Oxidative stress causes enhanced secretion of yb-1 protein that restrains proliferation of receiving cells. Genes 2018, 9, 513. [Google Scholar] [CrossRef] [PubMed]
- Kotake, Y.; Arikawa, N.; Tahara, K.; Maru, H.; Naemura, M. Y-box binding protein 1 is involved in regulating the g2/m phase of the cell cycle. Anticancer Res. 2017, 37, 1603–1608. [Google Scholar] [CrossRef] [PubMed]
- Zielke, N.; Edgar, B. Fucci sensors: Powerful new tools for analysis of cell proliferation. Wiley. Interdiscip. Rev. Dev. Biol. 2015, 4, 469–487. [Google Scholar] [CrossRef]
- Lehman, T.A.; Bennett, W.P.; Metcalf, R.A.; Welsh, J.A.; Ecker, J.; Modali, R.V.; Ullrich, S.; Romano, J.W.; Appella, E.; Testa, J.R. P53 mutations, ras mutations, and p53-heat shock 70 protein complexes in human lung carcinoma cell lines. Cancer. Res. 1991, 51, 4090–4096. [Google Scholar] [PubMed]
- Pellegata, N.S.; Antoniono, R.J.; Redpath, J.L.; Stanbridge, E.J. DNA damage and p53-mediated cell cycle arrest: A reevaluation. Proc. Natl. Acad. Sci. USA 1996, 93, 15209–15214. [Google Scholar] [CrossRef] [PubMed]
- Scott, A.F.; Phillips, J.A.; Migeon, B.R. DNA restriction endonuclease analysis for localization of human beta-and delta-globin genes on chromosome 11. Proc. Natl. Acad. Sci. USA 1979, 76, 4563–4565. [Google Scholar] [CrossRef] [PubMed]
- Whyte, P.; Buchkovich, K.J.; Horowitz, J.M.; Friend, S.H.; Raybuck, M.; Weinberg, R.A.; Harlow, E. Association between an oncogene and an anti-oncogene: The adenovirus e1a proteins bind to the retinoblastoma gene product. Nature 1988, 334, 124. [Google Scholar] [CrossRef] [PubMed]
- D’Avino, P.P.; Giansanti, M.G.; Petronczki, M. Cytokinesis in animal cells. Cold. Spring. Harb. Perspect. Biol. 2015, 7, a015834. [Google Scholar] [CrossRef]
- Carmena, M.; Wheelock, M.; Funabiki, H.; Earnshaw, W.C. The chromosomal passenger complex (cpc): From easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell. Biol. 2012, 13, 789. [Google Scholar] [CrossRef]
- Landino, J.; Norris, S.R.; Li, M.; Ballister, E.R.; Lampson, M.A.; Ohi, R. Two mechanisms coordinate the recruitment of the chromosomal passenger complex to the plane of cell division. Mol. Biol. Cell 2017, 28, 3634–3646. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.B.; Ma, W.; Valova, V.A.; Algie, M.; Harfoot, R.; Woolley, A.G.; Robinson, P.J.; Braithwaite, A.W. Genotoxic stress-induced nuclear localization of oncoprotein yb-1 in the absence of proteolytic processing. Oncogene 2009, 29, 403. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.; McKinney, C.; Algie, M.; Verma, C.S.; Kannan, S.; Harfoot, R.; Bartolec, T.K.; Bhatia, P.; Fisher, A.J.; Gould, M.L.; et al. Dephosphorylation of yb-1 is required for nuclear localisation during g2 phase of the cell cycle. Cancers 2020, 12, 315. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. Panther version 11: Expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids. Res. 2016, 45, D183–D189. [Google Scholar] [CrossRef] [PubMed]
- Chernov, K.G.; Mechulam, A.; Popova, N.V.; Pastre, D.; Nadezhdina, E.S.; Skabkina, O.V.; Shanina, N.A.; Vasiliev, V.D.; Tarrade, A.; Melki, J.; et al. Yb-1 promotes microtubule assembly in vitro through interaction with tubulin and microtubules. BMC Biochem. 2008, 9, 23. [Google Scholar] [CrossRef] [PubMed]
- Kumari, P.; Gilligan, P.C.; Lim, S.; Tran, L.D.; Winkler, S.; Philp, R.; Sampath, K. An essential role for maternal control of nodal signaling. Elife 2013, 2, e00683. [Google Scholar] [CrossRef] [PubMed]
- Keene, J.D. Rna regulons: Coordination of post-transcriptional events. Nat. Rev. Genet. 2007, 8, 533. [Google Scholar] [CrossRef]
- Zaucker, A.; Nagorska, A.; Kumari, P.; Hecker, N.; Wang, Y.; Huang, S.; Cooper, L.; Sivashanmugam, L.; VijayKumar, S.; Brosens, J. Translational co-regulation of a ligand and inhibitor by a conserved rna element. Nucleic Acids. Res. 2017, 46, 104–119. [Google Scholar] [CrossRef]
- Sutherland, B.W.; Kucab, J.; Wu, J.; Lee, C.; Cheang, M.C.; Yorida, E.; Turbin, D.; Dedhar, S.; Nelson, C.; Pollak, M. Akt phosphorylates the y-box binding protein 1 at ser102 located in the cold shock domain and affects the anchorage-independent growth of breast cancer cells. Oncogene 2005, 24, 4281. [Google Scholar] [CrossRef]
- Sechi, M.; Lall, R.K.; Afolabi, S.O.; Singh, A.; Joshi, D.C.; Chiu, S.-Y.; Mukhtar, H.; Syed, D.N. Fisetin targets yb-1/rsk axis independent of its effect on erk signaling: Insights from in vitro and in vivo melanoma models. Sci. Rep. 2018, 8, 15726. [Google Scholar] [CrossRef]
- Stratford, A.L.; Habibi, G.; Astanehe, A.; Jiang, H.; Hu, K.; Park, E.; Shadeo, A.; Buys, T.P.; Lam, W.; Pugh, T. Epidermal growth factor receptor (egfr) is transcriptionally induced by the y-box binding protein-1 (yb-1) and can be inhibited with iressa in basal-like breast cancer, providing a potential target for therapy. Breast. Cancer Res. 2007, 9, R61. [Google Scholar] [CrossRef]
- Astanehe, A.; Finkbeiner, M.; Hojabrpour, P.; To, K.; Fotovati, A.; Shadeo, A.; Stratford, A.; Lam, W.; Berquin, I.; Duronio, V. The transcriptional induction of pik3ca in tumor cells is dependent on the oncoprotein y-box binding protein-1. Oncogene 2009, 28, 2406. [Google Scholar] [CrossRef] [PubMed]
- Reipas, K.M.; Law, J.H.; Couto, N.; Islam, S.; Li, Y.; Li, H.; Cherkasov, A.; Jung, K.; Cheema, A.S.; Jones, S.J.M.; et al. Luteolin is a novel p90 ribosomal s6 kinase (rsk) inhibitor that suppresses notch4 signaling by blocking the activation of y-box binding protein-1 (yb-1). Oncotarget 2013, 4, 329–345. [Google Scholar] [CrossRef] [PubMed]
- Stratford, A.L.; Fry, C.J.; Desilets, C.; Davies, A.H.; Cho, Y.Y.; Li, Y.; Dong, Z.; Berquin, I.M.; Roux, P.P.; Dunn, S.E. Y-box binding protein-1 serine 102 is a downstream target of p90 ribosomal s6 kinase in basal-like breast cancer cells. Breast. Cancer Res. 2008, 10, R99. [Google Scholar] [CrossRef] [PubMed]
- Donaubauer, E.M.; Hunzicker-Dunn, M.E. Extracellular signal-regulated kinase (erk)-dependent phosphorylation of y-box-binding protein 1 (yb-1) enhances gene expression in granulosa cells in response to follicle-stimulating hormone (fsh). J. Biol. Chem. 2016, 291, 12145–12160. [Google Scholar] [CrossRef]
- Tiwari, A.; Rebholz, S.; Maier, E.; Dehghan Harati, M.; Zips, D.; Sers, C.; Rodemann, H.P.; Toulany, M. Stress-induced phosphorylation of nuclear yb-1 depends on nuclear trafficking of p90 ribosomal s6 kinase. Int. J. Mol. Sci. 2018, 19, 2441. [Google Scholar] [CrossRef]
- Shiota, M.; Yokomizo, A.; Takeuchi, A.; Itsumi, M.; Imada, K.; Kashiwagi, E.; Inokuchi, J.; Tatsugami, K.; Uchiumi, T.; Naito, S. Inhibition of rsk/yb-1 signaling enhances the anti-cancer effect of enzalutamide in prostate cancer. Prostate 2014, 74, 959–969. [Google Scholar] [CrossRef]
- Xue, Y.; Liu, Z.; Cao, J.; Ma, Q.; Gao, X.; Wang, Q.; Jin, C.; Zhou, Y.; Wen, L.; Ren, J. Gps 2.1: Enhanced prediction of kinase-specific phosphorylation sites with an algorithm of motif length selection. Protein Eng. Des. Sel. 2010, 24, 255–260. [Google Scholar] [CrossRef]
- Evdokimova, V.; Ruzanov, P.; Anglesio, M.S.; Sorokin, A.V.; Ovchinnikov, L.P.; Buckley, J.; Triche, T.J.; Sonenberg, N.; Sorensen, P.H. Akt-mediated yb-1 phosphorylation activates translation of silent mrna species. Mol. Cell. Biol. 2006, 26, 277–292. [Google Scholar] [CrossRef]
- Yokoyama, T.; Goto, H.; Izawa, I.; Mizutani, H.; Inagaki, M. Aurora-b and rho-kinase/rock, the two cleavage furrow kinases, independently regulate the progression of cytokinesis: Possible existence of a novel cleavage furrow kinase phosphorylates ezrin/radixin/moesin (erm). Genes Cells 2005, 10, 127–137. [Google Scholar] [CrossRef]
- Mathew, S.S.; Nieves, B.; Sequeira, S.; Sambandamoorthy, S.; Pumiglia, K.; Larsen, M.; LaFlamme, S.E. Integrins promote cytokinesis through the rsk signaling axis. J. Cell Sci. 2014, 127, 534–545. [Google Scholar] [CrossRef]
- Martin, M.; Hua, L.; Wang, B.; Wei, H.; Prabhu, L.; Hartley, A.-V.; Jiang, G.; Liu, Y.; Lu, T. Novel serine 176 phosphorylation of ybx1 activates nf-κb in colon cancer. J. Biol. Chem. 2017, 292, 3433–3444. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, L.; Mundade, R.; Wang, B.; Wei, H.; Hartley, A.-V.; Martin, M.; McElyea, K.; Temm, C.J.; Sandusky, G.; Liu, Y. Critical role of phosphorylation of serine 165 of ybx1 on the activation of nf-κb in colon cancer. Oncotarget 2015, 6, 29396. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M.; Network, C.G.A.R. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013, 45, 1113. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E. Integrative analysis of complex cancer genomics and clinical profiles using the cbioportal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. I-tasser server for protein 3d structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef]
- Kloks, C.P.A.M.; Spronk, C.A.E.M.; Lasonder, E.; Hoffmann, A.; Vuister, G.W.; Grzesiek, S.; Hilbers, C.W. The solution structure and DNA-binding properties of the cold-shock domain of the human y-box protein yb-111edited by p. E. Wright. J. Mol. Biol. 2002, 316, 317–326. [Google Scholar] [CrossRef]
- Nguyen, P.A.; Groen, A.C.; Loose, M.; Ishihara, K.; Wühr, M.; Field, C.M.; Mitchison, T.J. Spatial organization of cytokinesis signaling reconstituted in a cell-free system. Science 2014, 346, 244–247. [Google Scholar] [CrossRef]
- Priel, A.; Tuszynski, J.A.; Woolf, N.J. Transitions in microtubule c-termini conformations as a possible dendritic signaling phenomenon. Eur. Biophys. J. EBJ 2005, 35, 40–52. [Google Scholar] [CrossRef]
- Tang, J.X.; Janmey, P.A. The polyelectrolyte nature of f-actin and the mechanism of actin bundle formation. J. Biol. Chem. 1996, 271, 8556–8563. [Google Scholar] [CrossRef]
- Fava, L.L.; Schuler, F.; Sladky, V.; Haschka, M.D.; Soratroi, C.; Eiterer, L.; Demetz, E.; Weiss, G.; Geley, S.; Nigg, E.A.; et al. The piddosome activates p53 in response to supernumerary centrosomes. Genes Dev. 2017, 31, 34–45. [Google Scholar] [CrossRef]
- Ganem, N.J.; Cornils, H.; Chiu, S.-Y.; O’Rourke, K.P.; Arnaud, J.; Yimlamai, D.; Théry, M.; Camargo, F.D.; Pellman, D. Cytokinesis failure triggers hippo tumor suppressor pathway activation. Cell 2014, 158, 833–848. [Google Scholar] [CrossRef] [PubMed]
- Lasham, A.; Moloney, S.; Hale, T.; Homer, C.; Zhang, Y.F.; Murison, J.G.; Braithwaite, A.W.; Watson, J. The y-box-binding protein, yb1, is a potential negative regulator of the p53 tumor suppressor. J. Biol. Chem. 2003, 278, 35516–35523. [Google Scholar] [CrossRef] [PubMed]
- Sakaue-Sawano, A.; Kurokawa, H.; Morimura, T.; Hanyu, A.; Hama, H.; Osawa, H.; Kashiwagi, S.; Fukami, K.; Miyata, T.; Miyoshi, H.; et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 2008, 132, 487–498. [Google Scholar] [CrossRef]
- Reid, G.; Wallant, N.C.; Patel, R.; Antonic, A.; Saxon-Aliifaalogo, F.; Cao, H.; Webster, G.; Watson, J.D. Potent subunit-specific effects on cell growth and drug sensitivity from optimised sirna-mediated silencing of ribonucleotide reductase. J. RNAi Gene Silenc. 2009, 5, 321–330. [Google Scholar] [PubMed]
- Westerfield, M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Brachydanio Rerio); University of Oregon Press: Eugene, OR, USA, 1995. [Google Scholar]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Zhu, G.; Wang, Y.; Sun, J.; Liu, X.; Chen, Y.-G.; Meng, A. Maternal eomesodermin regulates zygotic nodal gene expression for mesendoderm induction in zebrafish embryos. J. Mol. Cell Biol. 2014, 6, 272–285. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef]
- Thingholm, T.E.; Jørgensen, T.J.; Jensen, O.N.; Larsen, M.R. Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat. Protoc. 2006, 1, 1929. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Käll, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 2007, 4, 923. [Google Scholar] [CrossRef]
- Pearlman, D.A.; Case, D.A.; Caldwell, J.W.; Ross, W.S.; Cheatham, T.E.; DeBolt, S.; Ferguson, D.; Seibel, G.; Kollman, P. Amber, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995, 91, 1–41. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. Ff14sb: Improving the accuracy of protein side chain and backbone parameters from ff99sb. J. Chem. Theory. Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Homeyer, N.; Horn, A.H.; Lanig, H.; Sticht, H. Amber force-field parameters for phosphorylated amino acids in different protonation states: Phosphoserine, phosphothreonine, phosphotyrosine, and phosphohistidine. J. Mol. Model. 2006, 12, 281–289. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh ewald: An n log (n) method for ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Miyamoto, S.; Kollman, P.A. Settle: An analytical version of the shake and rattle algorithm for rigid water models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. Vmd: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- DeLano, W.L. The Pymol Molecular Graphics System. 2002. Available online: http://www.pymol.org (accessed on 5 March 2018).
| Amino Acid | MDA-MB231 | A549 | T47D | |||
|---|---|---|---|---|---|---|
| Nucleus | Cytoplasm | Nucleus | Cytoplasm | Nucleus | Cytoplasm | |
| S102 | 3/8 | 3/8 | ||||
| S165 | 2/2 | 2/2 | 5/8 | 5/8 | 1/1 | 1/1 |
| S167 | 1/2 | 3/8 | 3/8 | 1/1 | 1/1 | |
| S174 | 1/2 | 3/8 | 4/8 | 1/1 | 1/1 | |
| S176 | 1/2 | 4/8 | 4/8 | 1/1 | 1/1 | |
| S314 | 1/2 | 2/2 | 5/8 | 5/8 | 1/1 | 1/1 |
| S03 | 1/8 | 2/8 | ||||
| T07 | 1/8 | 1/8 | ||||
| S21 | 3/8 | |||||
| S209 | 4/8 | 3/8 | ||||
| T271 | 2/8 | |||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mehta, S.; Algie, M.; Al-Jabry, T.; McKinney, C.; Kannan, S.; Verma, C.S.; Ma, W.; Zhang, J.; Bartolec, T.K.; Masamsetti, V.P.; et al. Critical Role for Cold Shock Protein YB-1 in Cytokinesis. Cancers 2020, 12, 2473. https://doi.org/10.3390/cancers12092473
Mehta S, Algie M, Al-Jabry T, McKinney C, Kannan S, Verma CS, Ma W, Zhang J, Bartolec TK, Masamsetti VP, et al. Critical Role for Cold Shock Protein YB-1 in Cytokinesis. Cancers. 2020; 12(9):2473. https://doi.org/10.3390/cancers12092473
Chicago/Turabian StyleMehta, Sunali, Michael Algie, Tariq Al-Jabry, Cushla McKinney, Srinivasaraghavan Kannan, Chandra S Verma, Weini Ma, Jessie Zhang, Tara K. Bartolec, V. Pragathi Masamsetti, and et al. 2020. "Critical Role for Cold Shock Protein YB-1 in Cytokinesis" Cancers 12, no. 9: 2473. https://doi.org/10.3390/cancers12092473
APA StyleMehta, S., Algie, M., Al-Jabry, T., McKinney, C., Kannan, S., Verma, C. S., Ma, W., Zhang, J., Bartolec, T. K., Masamsetti, V. P., Parker, K., Henderson, L., Gould, M. L., Bhatia, P., Harfoot, R., Chircop, M., Kleffmann, T., Cohen, S. B., Woolley, A. G., ... Braithwaite, A. (2020). Critical Role for Cold Shock Protein YB-1 in Cytokinesis. Cancers, 12(9), 2473. https://doi.org/10.3390/cancers12092473






