Detection of Germline Mutations in a Cohort of 139 Patients with Bilateral Breast Cancer by Multi-Gene Panel Testing: Impact of Pathogenic Variants in Other Genes beyond BRCA1/2
Simple Summary
Abstract
1. Introduction
2. Results
2.1. Clinical Features of Bilateral Breast Cancer Patients
2.2. Detection of Germline Pathogenic Variants in Cancer Susceptibility Genes by Multi-Gene Panel Testing
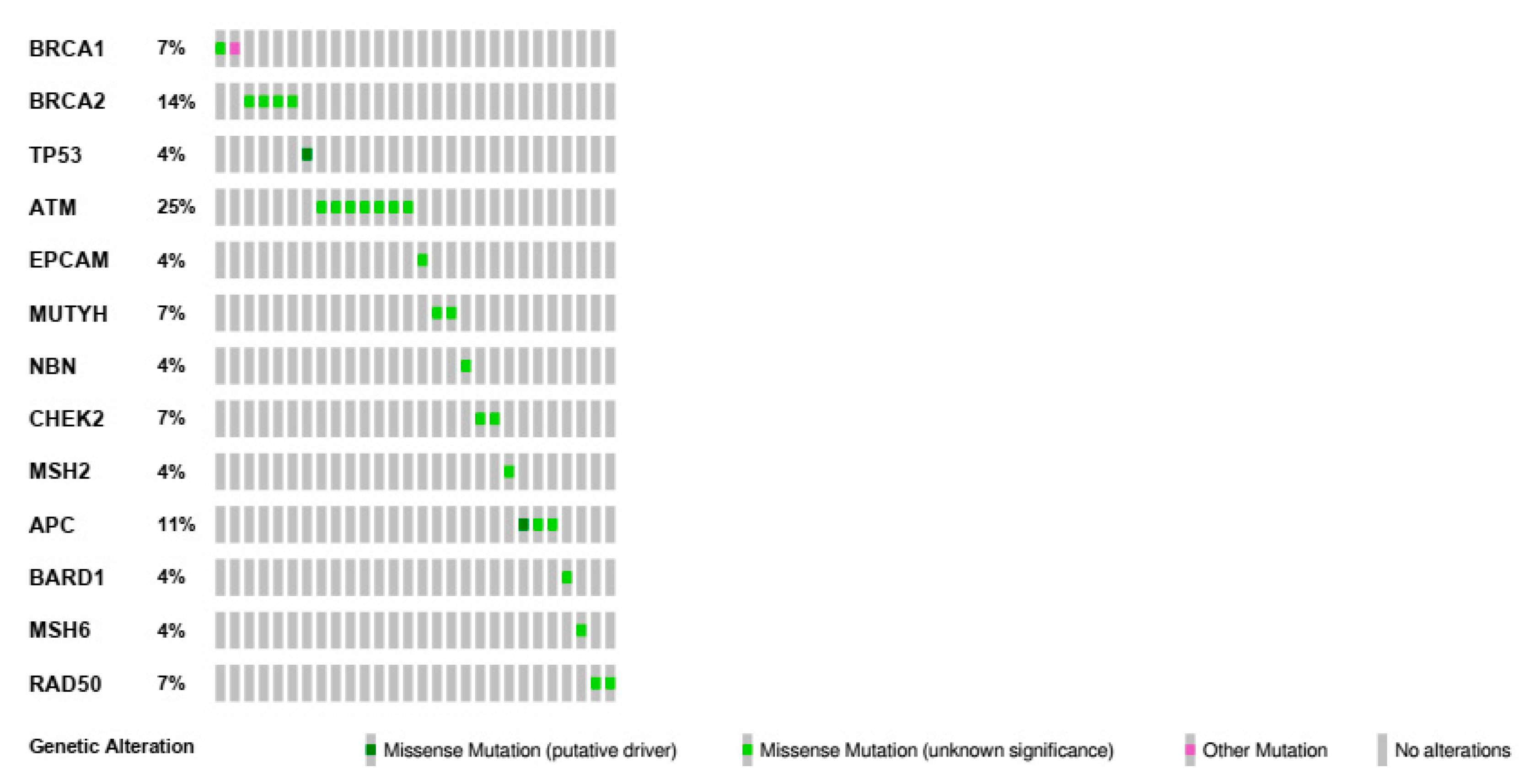
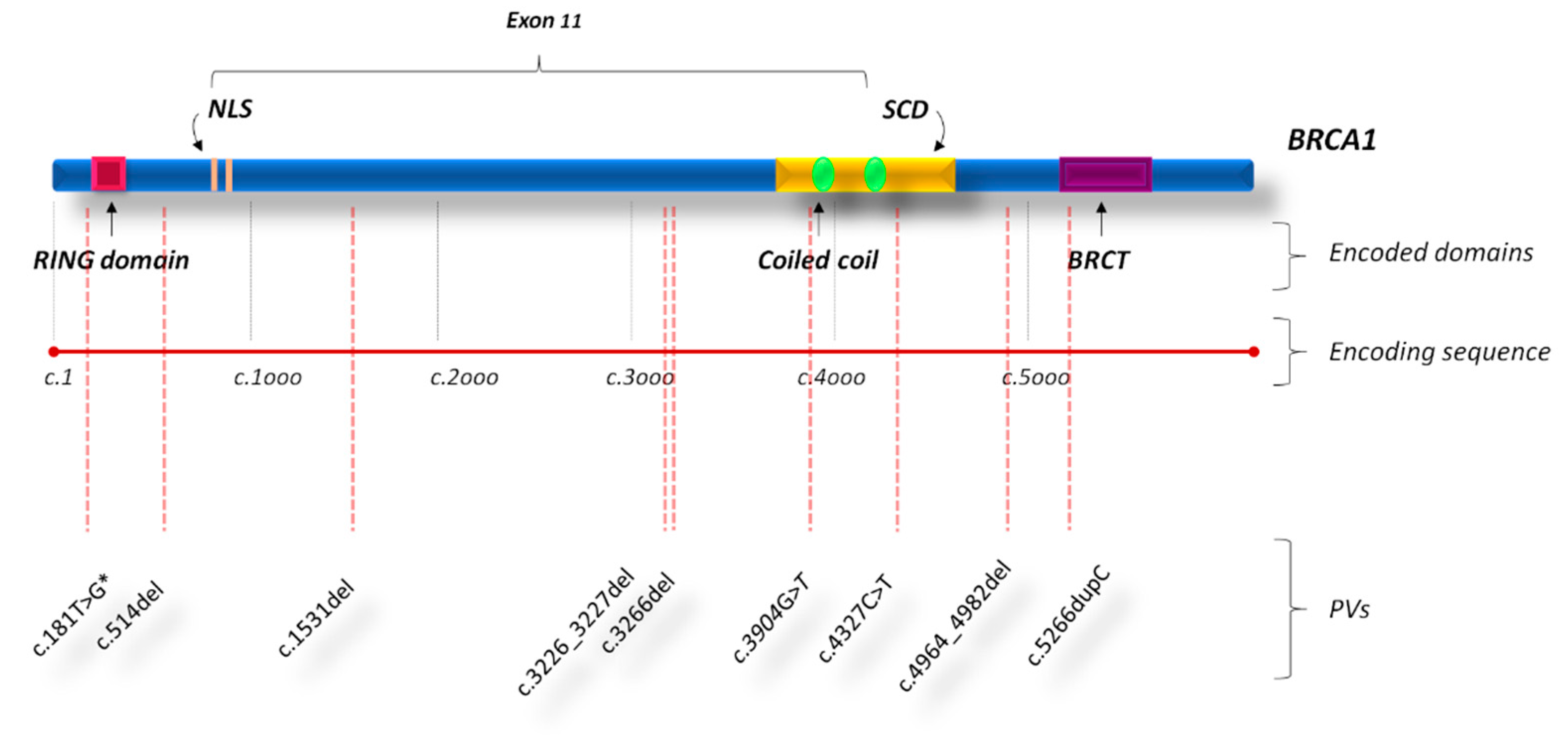
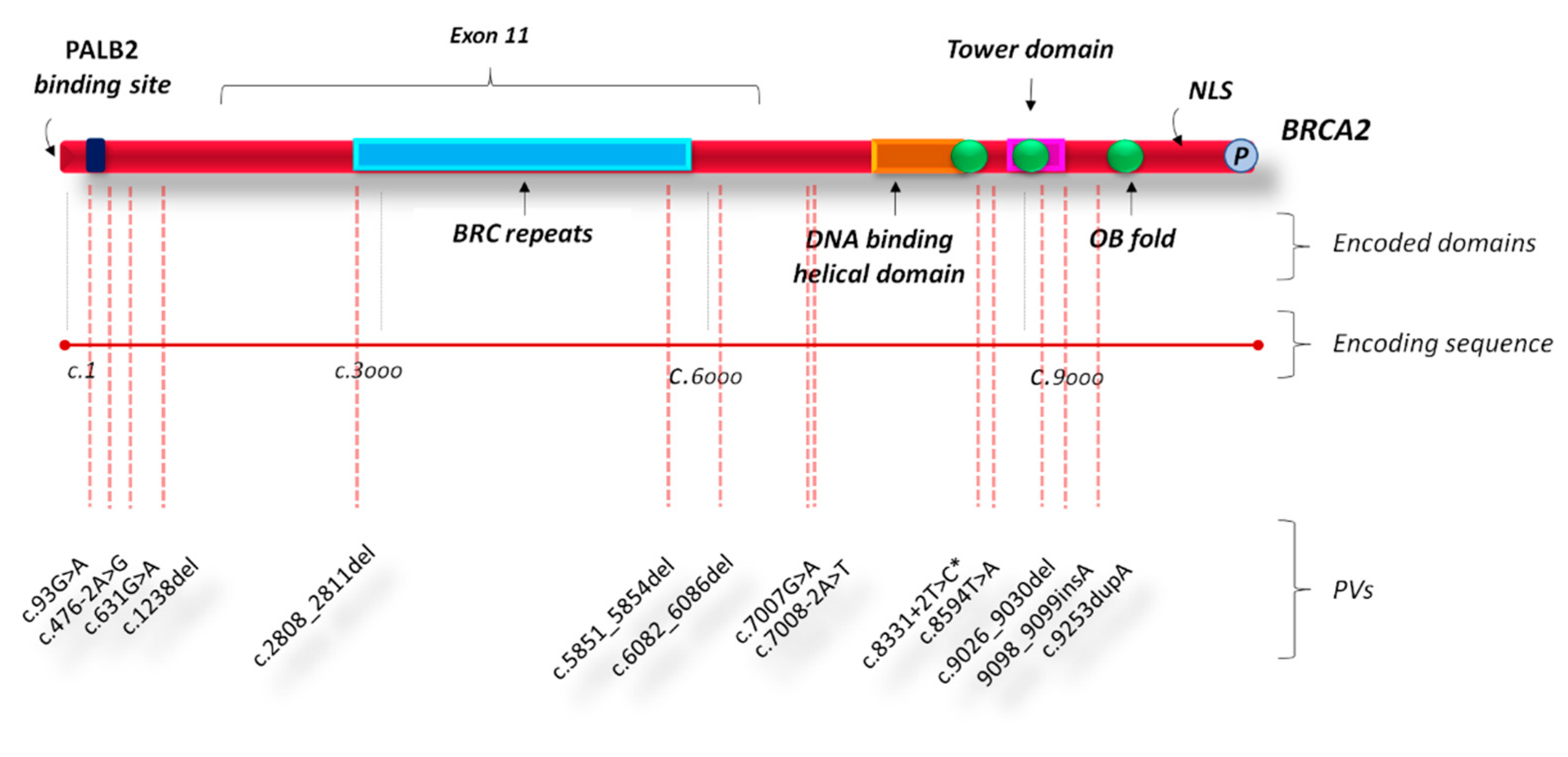
2.3. Type and Gene Location of Genetic Variants in Bilateral Breast Cancers
3. Discussion
4. Patients and Methods
4.1. Study Population
4.2. Sample Collection and Next-Generation Sequencing Analysis by Multi-Gene Panel
4.3. Sanger Sequencing
4.4. Genetic Variant Classification
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pan, B.; Xu, Y.; Zhou, Y.D.; Yao, R.; Wu, H.W.; Zhu, Q.L.; Wang, C.J.; Mao, F.; Lin, Y.; Shen, S.J.; et al. The Prognostic Comparison Among Unilateral, Bilateral, Synchronous Bilateral, and Metachronous Bilateral Breast Cancer: A Meta-Analysis of Studies from Recent Decade (2008–2018). Cancer Med. 2019. [Google Scholar] [CrossRef]
- Sim, Y.; Tan, V.K.M.; Sidek, N.A.B.; Chia, D.K.A.; Tan, B.K.T.; Madhukumar, P.; Yong, W.S.; Wong, C.Y.; Ong, K.W. Bilateral Breast Cancers in An Asian Population, and A Comparison Between Synchronous and Metachronous Tumours. ANZ J. Surg. 2018, 88, 982–987. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Schaapveld, M.; Jansen, L.; Bagherzadegan, E.; Sahinovic, M.M.; Baas, P.C.; Hanssen, L.M.H.C.; van Der Mijle, H.C.J.; Brandenburg, J.D.; Wiggers, T.; et al. The Value of Surveillance Mammography of the Contralateral Breast in Patients with a History of Breast Cancer. Eur. J. Cancer 2009, 45, 3000–3007. [Google Scholar] [CrossRef] [PubMed]
- Hartman, M.; Czene, K.; Reilly, M.; Adolfsson, J.; Bergh, J.; Adami, H.-O.; Dickman, P.W.; Hall, P. Incidence and Prognosis of Synchronous and Metachronous Bilateral Breast Cancer. J. Clin. Oncol. 2007, 25, 4210–4216. [Google Scholar] [CrossRef] [PubMed]
- Corredor, J.; Woodson, A.H.; Gutierrez Barrera, A.; Arun, B. Multigene Panel Testing Results in Patients with Multiple Breast Cancer Primaries. Breast J. 2020. [Google Scholar] [CrossRef]
- Apostolou, P.; Fostira, F. Hereditary Breast Cancer: The Era of New Susceptibility Genes. Biomed Res. Int. 2013, 2013, 1–11. [Google Scholar] [CrossRef]
- Valencia, O.M.; Samuel, S.E.; Viscusi, R.K.; Riall, T.S.; Neumayer, L.A.; Aziz, H. The Role of Genetic Testing in Patients with Breast Cancer. JAMA Surg. 2017, 152, 589. [Google Scholar] [CrossRef]
- Kotsopoulos, J.; Huzarski, T.; Gronwald, J.; Singer, C.F.; Moller, P.; Lynch, H.T.; Armel, S.; Karlan, B.; Foulkes, W.D.; Neuhausen, S.L.; et al. Bilateral Oophorectomy and Breast Cancer Risk Inbrca1andbrca2mutation Carriers. JNCI J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef]
- Miki, Y.; Swensen, J.; Shattuck-Eidens, D.; Futreal, P.; Harshman, K.; Tavtigian, S.; Liu, Q.; Cochran, C.; Bennett, L.; Ding, W.; et al. A Strong Candidate for the Breast and Ovarian Cancer Susceptibility Gene Brca1. Science 1994, 266, 66–71. [Google Scholar] [CrossRef]
- Wooster, R.; Bignell, G.; Lancaster, J.; Swift, S.; Seal, S.; Mangion, J.; Collins, N.; Gregory, S.; Gumbs, C.; Micklem, G.; et al. Identification of the Breast Cancer Susceptibility Gene Brca2. Nature 1995, 378, 789–792. [Google Scholar] [CrossRef]
- Kuchenbaecker, K.B.; Hopper, J.L.; Barnes, D.R.; Phillips, K.-A.; Mooij, T.M.; Roos-Blom, M.-J.; Jervis, S.; van Leeuwen, F.E.; Milne, R.L.; Andrieu, N.; et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for Brca1 and Brca2 Mutation Carriers. JAMA 2017, 317, 2402. [Google Scholar] [CrossRef] [PubMed]
- Metcalfe, K.; Gershman, S.; Lynch, H.T.; Ghadirian, P.; Tung, N.; Kim-Sing, C.; Olopade, O.I.; Domchek, S.; Mclennan, J.; Eisen, A.; et al. Predictors of Contralateral Breast Cancer in Brca1 and Brca2 Mutation Carriers. Br. J. Cancer 2011, 104, 1384–1392. [Google Scholar] [CrossRef] [PubMed]
- Graeser, M.K.; Engel, C.; Rhiem, K.; Gadzicki, D.; Bick, U.; Kast, K.; Froster, U.G.; Schlehe, B.; Bechtold, A.; Arnold, N.; et al. Contralateral Breast Cancer Risk in Brca1 and Brca2 Mutation Carriers. J. Clin. Oncol. 2009, 27, 5887–5892. [Google Scholar] [CrossRef] [PubMed]
- Incorvaia, L.; Fanale, D.; Badalamenti, G.; Bono, M.; Calò, V.; Cancelliere, D.; Castiglia, M.; Fiorino, A.; Pivetti, A.; Barraco, N.; et al. Hereditary Breast and Ovarian Cancer in Families from Southern Italy (Sicily)—Prevalence and Geographic Distribution of Pathogenic Variants in Brca1/2 Genes. Cancers 2020, 12, 1158. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D. Cancer Incidence in Brca1 Mutation Carriers. J. Natl. Cancer Inst. 2002, 94, 1358–1365. [Google Scholar] [CrossRef] [PubMed]
- Breast Cancer Linkage Consortium. Cancer Risks in Brca2 Mutation Carriers. J. Natl. Cancer Inst. 1999, 91, 1310–1316. [Google Scholar] [CrossRef]
- Bevers, T.B.; Helvie, M.; Bonaccio, E.; Calhoun, K.E.; Daly, M.B.; Farrar, W.B.; Garber, J.E.; Gray, R.; Greenberg, C.C.; Greenup, R.; et al. Breast Cancer Screening and Diagnosis, Version 3.2018, Nccn Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2018, 16, 1362–1389. [Google Scholar] [CrossRef]
- Senkus, E.; Kyriakides, S.; Ohno, S.; Penault-Llorca, F.; Poortmans, P.; Rutgers, E.; Zackrisson, S.; Cardoso, F. Primary Breast Cancer: Esmo Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2015, 26, 8–30. [Google Scholar] [CrossRef]
- Tedaldi, G.; Tebaldi, M.; Zampiga, V.; Danesi, R.; Arcangeli, V.; Ravegnani, M.; Cangini, I.; Pirini, F.; Petracci, E.; Rocca, A.; et al. Multiple-Gene Panel Analysis in A Case Series of 255 Women with Hereditary Breast and Ovarian Cancer. Oncotarget 2017, 8, 47064–47075. [Google Scholar] [CrossRef]
- Slavin, T.P.; Maxwell, K.N.; Lilyquist, J.; Vijai, J.; Neuhausen, S.L.; Hart, S.N.; Ravichandran, V.; Thomas, T.; Maria, A.; Villano, D.; et al. The Contribution of Pathogenic Variants in Breast Cancer Susceptibility Genes to Familial Breast Cancer Risk. NPJ Breast Cancer 2017, 3. [Google Scholar] [CrossRef]
- Economopoulou, P.; Dimitriadis, G.; Psyrri, A. Beyond Brca: New Hereditary Breast Cancer Susceptibility Genes. Cancer Treat. Rev. 2015, 41, 1–8. [Google Scholar] [CrossRef]
- Singer, C.F.; Balmaña, J.; Bürki, N.; Delaloge, S.; Filieri, M.E.; Gerdes, A.-M.; Grindedal, E.M.; Han, S.; Johansson, O.; Kaufman, B.; et al. Genetic Counselling and Testing of Susceptibility Genes for Therapeutic Decision-Making in Breast Cancer—An European Consensus Statement and Expert Recommendations. Eur. J. Cancer 2019, 106, 54–60. [Google Scholar] [CrossRef]
- Narod, S.A. Bilateral Breast Cancers. Nat. Rev. Clin. Oncol. 2014, 11, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Phelan, C.M.; Zhang, P.; Rousseau, F.; Ghadirian, P.; Robidoux, A.; Foulkes, W.; Hamel, N.; Mccready, D.; Trudeau, M.; et al. Frequency of the Chek2 1100delc Mutation Among Women with Breast Cancer: An international Study. Cancer Res. 2008, 68, 2154–2157. [Google Scholar] [CrossRef] [PubMed]
- Broeks, A.; De Witte, L.; Nooijen, A.; Huseinovic, A.; Klijn, J.G.M.; van Leeuwen, F.E.; Russell, N.S.; van’t Veer, L.J. Excess Risk for Contralateral Breast Cancer in Chek2*1100delc Germline Mutation Carriers. Breast Cancer Res. Treat. 2004, 83, 91–93. [Google Scholar] [CrossRef] [PubMed]
- Plon, S.E.; Eccles, D.M.; Easton, D.; Foulkes, W.D.; Genuardi, M.; Greenblatt, M.S.; Hogervorst, F.B.L.; Hoogerbrugge, N.; Spurdle, A.B.; Tavtigian, S.V. Sequence Variant Classification and Reporting: Recommendations for Improving the Interpretation of Cancer Susceptibility Genetic Test Results. Hum. Mutat. 2008, 29, 1282–1291. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.; Calò, V.; Bruno, L.; Schirò, V.; Agnese, V.; Cascio, S.; Foddai, E.; Fanale, D.; Rizzo, S.; Di Gaudio, F.; et al. Is Brca1-5083del19, Identified in Breast Cancer Patients of Sicilian Origin, a Calabrian Founder Mutation? Breast Cancer Res. Treat. 2008, 113, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Janavičius, R. Founder Brca1/2 Mutations in the Europe: Implications for Hereditary Breast-Ovarian Cancer Prevention and Control. EPMA J. 2010, 1, 397–412. [Google Scholar] [CrossRef]
- Ding, D.; Zhang, Y.; He, X.; Meng, W.; Ma, W.; Zheng, W. Frequency of the Chek2 1100delc Mutation among Women with Early-Onset and Bilateral Breast Cancer. Breast Cancer Res. 2012, 14. [Google Scholar] [CrossRef]
- Weischer, M.; Nordestgaard, B.G.; Pharoah, P.; Bolla, M.K.; Nevanlinna, H.; van’t Veer, L.J.; Garcia-Closas, M.; Hopper, J.L.; Hall, P.; Andrulis, I.L.; et al. Chek2*1100delc Heterozygosity in Women with Breast Cancer Associated with Early Death, Breast Cancer–Specific Death, and Increased Risk of a Second Breast Cancer. J. Clin. Oncol. 2012, 30, 4308–4316. [Google Scholar] [CrossRef]
- Stjepanovic, N.; Moreira, L.; Carneiro, F.; Balaguer, F.; Cervantes, A.; Balmaña, J.; Martinelli, E. Hereditary Gastrointestinal Cancers: Esmo Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2019, 30, 1558–1571. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, N. Synchronous Bilateral Breast Cancers. J. Clin. Diagn. Res. 2015. [Google Scholar] [CrossRef] [PubMed]
- Huber, A.; Seidler, S.J.; Huber, D.E. Clinicopathological Characteristics, Treatment and Outcome of 123 Patients with Synchronous or Metachronous Bilateral Breast Cancer in a Swiss Institutional Retrospective Series. Eur. J. Breast Health 2020, 16, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Desmond, A.; Kurian, A.W.; Gabree, M.; Mills, M.A.; Anderson, M.J.; Kobayashi, Y.; Horick, N.; Yang, S.; Shannon, K.M.; Tung, N.; et al. Clinical Actionability of Multigene Panel Testing for Hereditary Breast and Ovarian Cancer Risk Assessment. JAMA Oncol. 2015, 1, 943. [Google Scholar] [CrossRef]
- Tung, N.; Battelli, C.; Allen, B.; Kaldate, R.; Bhatnagar, S.; Bowles, K.; Timms, K.; Garber, J.E.; Herold, C.; Ellisen, L.; et al. Frequency of Mutations in Individuals with Breast Cancer Referred Forbrca1 and brca2 testing Using Next-Generation Sequencing with a 25-Gene Panel. Cancer 2015, 121, 25–33. [Google Scholar] [CrossRef]
- Easton, D.F.; Pharoah, P.D.P.; Antoniou, A.C.; Tischkowitz, M.; Tavtigian, S.V.; Nathanson, K.L.; Devilee, P.; Meindl, A.; Couch, F.J.; Southey, M.; et al. Gene-Panel Sequencing and the Prediction of Breast-Cancer Risk. N. Engl. J. Med. 2015, 372, 2243–2257. [Google Scholar] [CrossRef]
- Shin, H.-C.; Lee, H.-B.; Yoo, T.-K.; Lee, E.-S.; Kim, R.N.; Park, B.; Yoon, K.-A.; Park, C.; Lee, E.S.; Moon, H.-G.; et al. Detection of Germline Mutations in Breast Cancer Patients with Clinical Features of Hereditary Cancer Syndrome Using A Multi-Gene Panel Test. Cancer Res. Treat. 2020, 52, 697–713. [Google Scholar] [CrossRef]
- Crawford, B.; Adams, S.B.; Sittler, T.; van den Akker, J.; Chan, S.; Leitner, O.; Ryan, L.; Gil, E.; van ’t Veer, L. Multi-Gene Panel Testing for Hereditary Cancer Predisposition in Unsolved High-Risk Breast and Ovarian Cancer Patients. Breast Cancer Res. Treat. 2017, 163, 383–390. [Google Scholar] [CrossRef]
- Kurian, A.W.; Hare, E.E.; Mills, M.A.; Kingham, K.E.; Mcpherson, L.; Whittemore, A.S.; Mcguire, V.; Ladabaum, U.; Kobayashi, Y.; Lincoln, S.E.; et al. Clinical Evaluation of a Multiple-Gene Sequencing Panel for Hereditary Cancer Risk Assessment. J. Clin. Oncol. 2014, 32, 2001–2009. [Google Scholar] [CrossRef]
- Laduca, H.; Stuenkel, A.J.; Dolinsky, J.S.; Keiles, S.; Tandy, S.; Pesaran, T.; Chen, E.; Gau, C.-L.; Palmaer, E.; Shoaepour, K.; et al. Utilization of Multigene Panels in Hereditary Cancer Predisposition Testing: Analysis of More Than 2000 Patients. Genet. Med. 2014, 16, 830–837. [Google Scholar] [CrossRef]
- Hoheisel, J.D.; Simbolo, M.; Gottardi, M.; Corbo, V.; Fassan, M.; Mafficini, A.; Malpeli, G.; Lawlor, R.T.; Scarpa, A. DNA Qualification Workflow for Next Generation Sequencing of Histopathological Samples. PLoS ONE 2013, 8, e62692. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the Cbioportal. Sci. Signal. 2013, 6, Pl1. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The Cbio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data: Figure 1. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Den Dunnen, J.T.; Dalgleish, R.; Maglott, D.R.; Hart, R.K.; Greenblatt, M.S.; Mcgowan-Jordan, J.; Roux, A.-F.; Smith, T.; Antonarakis, S.E.; Taschner, P.E.M. Hgvs Recommendations for the Description of Sequence Variants: 2016 Update. Hum. Mutat. 2016, 37, 564–569. [Google Scholar] [CrossRef] [PubMed]
| BBC (n = 139) | No. Patients (%) |
|---|---|
| Tumor description | |
| Metachronous | 93 (66.9%) |
| Synchronous | 46 (33.1%) |
| Sex | |
| Female | 137 |
| Male | 2 |
| Age at diagnosis (years) | |
| Primary tumor | |
| Median (range) | 45 (21–77) |
| ≤40 | 41 (29.5%) |
| 41–50 | 53 (38.12%) |
| 51–60 | 29 (20.9%) |
| ≥60 | 16 (11.5%) |
| Secondary tumor | |
| Median (range) | 51 (21–80) |
| ≤40 | 21 (15.1%) |
| 41–50 | 48 (34.5%) |
| 51–60 | 40 (28.8%) |
| ≥60 | 30 (21.6%) |
| Time between 1st and 2nd tumors (years) | |
| Median ± DS | 4 ± 6.08 |
| ≤5 | 88 (63.3%) |
| 6–10 | 27 (19.4%) |
| >10 | 24 (17.3%) |
| Histology 1st breast cancer | |
| Ductal in situ | 96 (69%) |
| Ductal invasive | 13 (9.4%) |
| Lobular invasive | 12 (8.6%) |
| Others | 18 (13%) |
| Histology 2nd breast cancer | |
| Ductal in situ | 16 (11.5%) |
| Ductal invasive | 92 (66.2%) |
| Lobular invasive | 15 (10.8%) |
| Others | 16 (11.5%) |
| Molecular phenotype 1st breast cancer | |
| Luminal A | 28 (20.1%) |
| Luminal B/HER2 - | 49 (35.3%) |
| Luminal B/HER2 + | 7 (5%) |
| HER2 + (non-luminal) | 6 (4.3%) |
| Triple negative | 12 (8.6%) |
| Unknown | 37 (26.7%) |
| Molecular phenotype 2nd breast cancer | |
| Luminal A | 35 (25.2%) |
| Luminal B/HER2 - | 35 (25.2%) |
| Luminal B/HER2 + | 10 (7.2%) |
| HER2 + (non-luminal) | 5 (3.6%) |
| Triple negative | 17 (12.2%) |
| Unknown | 37 (26.6%) |
| Genetic testing results | |
| Pathogenic variant BRCA1 | 13 (9.3%) |
| Pathogenic variant BRCA2 | 19 (13.7%) |
| Pathogenic variant BRCA1/BRCA2 | 1 (0.7%) |
| Pathogenic variant CHEK2 | 5 (3.6%) |
| Pathogenic variant PALB2 | 8 (5.8 %) |
| Pathogenic variant ATM | 3 (2.2%) |
| Pathogenic variant PTEN | 1 (0.7%) |
| Pathogenic variant RAD51C | 2 (1.4%) |
| Pathogenic variant MUTHY | 1 (0.7%) |
| Variant of uncertain significance | 27 (19.4%) |
| Negative | 59 (42.5%) |
| Type of surgery 1st breast cancer | |
| Mastectomy | 50 (36.2%) |
| Breast conserving therapy | 74 (53.2%) |
| Unknown | 15 (10.9%) |
| Type of surgery 2nd breast cancer | |
| Mastectomy | 48 (34.5%) |
| Breast conserving therapy | 55 (39.9%) |
| Unknown | 35 (25.3%) |
| Gene | Variant Type | HGVS Nomenclature | Protein Change | No. Patients | Allele Frequency (ExAC */GnomAD **) | Allele Frequency (Study Cohort) |
|---|---|---|---|---|---|---|
| BRCA1 | fs | c.4964_4982del | p.Ser1655fs | 4 (7.4%) | ExAC 0.000008 | 0.0.14 |
| BRCA1 | fs | c.3226_3227AG [1] | p.Gly1077fs | 2 (3.8%) | GnomAD 0.000004 | 0.0072 |
| BRCA1 | NS | c.3904G>T | p.Glu1302Ter | 2 (3.8%) | / | 0.0072 |
| BRCA1 | fs | c.1531del | p.Gly512Terfs | 1 (1.9%) | / | 0.0036 |
| BRCA1 | fs | c.514del | p.Gln172fs | 1 (1.9%) | ExAC 0.000008 GnomAD 0.000004 | 0.0036 |
| BRCA1 | fs | c.5266dupC | p.Gln1756Profs | 1 (1.9%) | ExAC 0.000156 GnomAD 0.00018 | 0.0036 |
| BRCA1 | fs | c.3266del | p.Leu1089fs | 1 (1.9%) | / | 0.0036 |
| BRCA1 | NS | c.4327C > T | p.Arg1443Ter | 1 (1.9%) | GnomAD 0.000024 | 0.0036 |
| BRCA2 | fs | c.1238del | p.Leu413fs | 4 (7.4%) | GnomAD 0.000004 | 0.0.14 |
| BRCA2 | fs | c.9026_9030del | p.Tyr3009fs | 2 (3.8%) | GnomAD 0.000004 | 0.0072 |
| BRCA2 | fs | c.9253dupA | p.Thr3085Asnfs | 2 (3.8%) | / | 0.0072 |
| BRCA2 | fs | c.6082_6086del | p.Glu2028fs | 2 (3.8%) | ExAC 0.000008 GnomAD 0.000004 | 0.0072 |
| BRCA2 | fs | c.9098_9099insA | p.Gln3034fs | 1 (1.9%) | / | 0.0036 |
| BRCA2 | NS | c.8594T > A | p.Leu2865Ter | 1 (1.9%) | / | 0.0036 |
| BRCA2 | M | c.631G > A | p.Val211Ile | 1 (1.9%) | / | 0.0036 |
| BRCA2 | fs | c.5851_5854del | p.Ser1951fs | 1 (1.9%) | / | 0.0036 |
| BRCA2 | fs | c.2808_2811del | p.Ala938Profs | 1 (1.9%) | ExAC 0.000017 GnomAD 0.000008 | 0.0036 |
| BRCA2 | IVS | c.7008-2A > T | / | 1 (1.9%) | / | 0.0036 |
| BRCA2 | IVS | c.476-2A > G | / | 1 (1.9%) | / | 0.0036 |
| BRCA2 | M | c.7007G > A | p.Arg2336His | 1 (1.9%) | / | 0.0036 |
| BRCA2 | NS | c.93G > A | p.Trp31Ter | 1 (1.9%) | GnomAD 0.000004 | 0.0036 |
| BRCA2/BRCA1 | IVS/M | c.8331 + 2T > C/c.181T > G | /p.Cys61Gly | 1 (1.9%) | /-ExAC 0.000067 GnomAD 0.000032 | 0.0036 |
| Gene | Variant Type | HGVS Nomenclature | Protein Change | No. Patients | Allele Frequency (ExAC */GnomAD **) | Allele Frequency (Study Cohort) |
|---|---|---|---|---|---|---|
| CHEK2 | fs | c.1100del | p.Thr367fs | 5 (9.2%) | gnomAD 0.00204 ExAC 0.00182 | 0.018 |
| RAD51C | NS | c.224dup | p.Tyr75Ter | 1 (1.9%) | gnomAD 0.00001 | 0.0036 |
| ATM | NS | c.8818_8821dup | p.Ser2941Ter | 1 (1.9%) | / | 0.0036 |
| PALB2 | fs | c.758dup | p.Ser254fs | 2 (3.8%) | gnomAD 0.00002 ExAC 0.00003 | 0.0072 |
| PALB2 | NS | c.2566C > T | p.Gln856Ter | 2 (3.8%) | gnomAD 0.00000 ExAC 0.00001 | 0.0072 |
| PALB2 | fs | c.1050_1053del | p.Thr351fs | 2 (3.8%) | / | 0.0072 |
| PALB2 | NS | c.2257C > T | p.Arg753Ter | 2 (3.8%) | gnomAD 0.00002 ExAC 0.00003 | 0.0072 |
| MUTYH | M | c.1103G > A | p.Gly368Asp | 1 (1.9%) | gnomAD 0.00303 ExAC 0.00280 | 0.0036 |
| PTEN | M | c.284C > A | p.Pro95Gln | 1 (1.9%) | / | 0.0036 |
| ATM | M | c.8147T > C | p.Val2716Ala | 1 (1.9%) | gnomAD 0.00003 ExAC 0.00004 | 0.0036 |
| RAD51C | IVS | c.1026 + 5_1026 + 7del | / | 1 (1.9%) | / | 0.0036 |
| ATM | LGR | Exon 57–61del | / | 1 (1.9%) | / | 0.0036 |
| Gene | Variant Type | HGVS Nomenclature | Protein Change | Variant Interpretation | No. Patients | Allele Frequency (ExAC a/GnomAD b) | Allele Frequency (Cohort Study) |
|---|---|---|---|---|---|---|---|
| BRCA1 | M | c.4054G > A | p.Glu1352Lys | VUS | 1 (3.70%) | gnomAD 0.00002 ExAC 0.00004 | 0.0036 |
| BRCA1 | IVS | c.670 + 31A > C | / | VUS | 1 (3.70%) | / | 0.0036 |
| BRCA2 | M | c.5267T > A | p.Val1756Glu | VUS | 1 (3.70%) | gnomAD 0.00000 ExAC 0.00001 | 0.0036 |
| BRCA2 | M | c.8299C > T | p.Pro2767Ser | VUS | 1 (3.70%) | / | 0.0036 |
| BRCA2 | M | c.1769T > G | p.Phe590Cys | CIP | 1 (3.70%) | gnomAD 0.00001 ExAC 0.00002 | 0.0036 |
| BRCA2 | M | c.9581C > A | p.Pro3194Gln | CIP | 1 (3.70%) | gnomAD 0.00001 ExAC 0.00001 | 0.0036 |
| TP53 | M | c.446C > T * | p.Ser149Phe | VUS | 1 (3.70%) | / | 0.0036 |
| ATM | M | c.1229T > C * | p.Val410Ala | CIP | gnomAD 0.00222 ExAC 0.00217 | 0.0036 | |
| EPCAM | M | c.334G > A | p.Gly112Ser | VUS | 1 (3.70%) | / | 0.0036 |
| MUTYH | M | c.1378 C > T ** | p.Arg460Cys | VUS | 1 (3.70%) | gnomAD 0.00005 ExAC 0.00005 | 0.0036 |
| NBN | M | c.283G > A ** | p.Asp95Asn | CIP | gnomAD 0.00173 ExAC 0.00186 | 0.0036 | |
| ATM | M | c.6407G > C | p.Arg2136Thr | VUS | 1 (3.70%) | / | 0.0036 |
| MUTYH | M | c.202T > C | p.Ser68Pro | VUS | 1 (3.70%) | gnomAD 0.00001 ExAC 0.00001 | 0.0036 |
| CHEK2 | M | c.1388G > A | p.Cys463Tyr | VUS | 2 (7.4%) | gnomAD 0.00000 | 0.0072 |
| MSH2 | M | c.1111G > C | p.Glu371Gln | VUS | 1 (3.70%) | / | 0.0036 |
| APC | M | c.3920T > A | p.Ile1307Lys | CIP, Risk Factor | 1 (3.70%) | gnomAD 0.00201 ExAC 0.00169 | 0.0036 |
| ATM | M | c.4060C > A *** | p.Pro1354Thr | CIP | 1 (3.70%) | gnomAD 0.00019 ExAC 0.00021 | 0.0036 |
| CHEK2 | M | c.1441 G > T *** | p.Asp481Tyr | CIP | 2 (7.4%) | gnomAD 0.00039 ExAC 0.00028 | 0.0072 |
| ATM | M | c.6983C > T | p.Pro2328Leu | VUS | 1 (3.70%) | gnomAD 0.00000 | 0.0036 |
| APC | M | c.3949G > C **** | p.Glu1317Gln | CIP | 1 (3.70%) | gnomAD 0.00438 ExAC 0.00413 | 0.0036 |
| ATM | M | c.4258C > T **** | p.Leu1420Phe | CIP | gnomAD 0.01104 ExAC 0.01271 | 0.0036 | |
| BARD1 | M | c.1793C > T | p.Thr598Ile | VUS | 1 (3.70%) | gnomAD 0.000008 | 0.0036 |
| ATM | M | c.6067G > A | p.Gly2023Arg | CIP | 1 (3.70%) | gnomAD 0.00143 ExAC 0.00157 | 0.0036 |
| MSH6 | M | c.663A > C | p.Glu221Asp | CIP | 2 (7.4%) | gnomAD 0.00071 ExAC 0.00063 | 0.0072 |
| ATM | M | c.6293T > C | p.Leu2098Pro | VUS | 1 (3.70%) | gnomAD 0.00000 ExAC 0.00001 | 0.0036 |
| RAD50 | M | c.1277A > G | p.Gln426Arg | CIP | 1 (3.70%) | gnomAD 0.00014 ExAC 0.00015 | 0.0036 |
| APC | M | c.5338C > T | p.Pro1780Ser | VUS | 1 (3.70%) | / | 0.0036 |
| RAD50 | M | c.1094G > A | p.Arg365Gln | CIP | 1 (3.70%) | gnomAD 0.00046 ExAC 0.00040 | 0.0036 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fanale, D.; Incorvaia, L.; Filorizzo, C.; Bono, M.; Fiorino, A.; Calò, V.; Brando, C.; Corsini, L.R.; Barraco, N.; Badalamenti, G.; et al. Detection of Germline Mutations in a Cohort of 139 Patients with Bilateral Breast Cancer by Multi-Gene Panel Testing: Impact of Pathogenic Variants in Other Genes beyond BRCA1/2. Cancers 2020, 12, 2415. https://doi.org/10.3390/cancers12092415
Fanale D, Incorvaia L, Filorizzo C, Bono M, Fiorino A, Calò V, Brando C, Corsini LR, Barraco N, Badalamenti G, et al. Detection of Germline Mutations in a Cohort of 139 Patients with Bilateral Breast Cancer by Multi-Gene Panel Testing: Impact of Pathogenic Variants in Other Genes beyond BRCA1/2. Cancers. 2020; 12(9):2415. https://doi.org/10.3390/cancers12092415
Chicago/Turabian StyleFanale, Daniele, Lorena Incorvaia, Clarissa Filorizzo, Marco Bono, Alessia Fiorino, Valentina Calò, Chiara Brando, Lidia Rita Corsini, Nadia Barraco, Giuseppe Badalamenti, and et al. 2020. "Detection of Germline Mutations in a Cohort of 139 Patients with Bilateral Breast Cancer by Multi-Gene Panel Testing: Impact of Pathogenic Variants in Other Genes beyond BRCA1/2" Cancers 12, no. 9: 2415. https://doi.org/10.3390/cancers12092415
APA StyleFanale, D., Incorvaia, L., Filorizzo, C., Bono, M., Fiorino, A., Calò, V., Brando, C., Corsini, L. R., Barraco, N., Badalamenti, G., Russo, A., & Bazan, V. (2020). Detection of Germline Mutations in a Cohort of 139 Patients with Bilateral Breast Cancer by Multi-Gene Panel Testing: Impact of Pathogenic Variants in Other Genes beyond BRCA1/2. Cancers, 12(9), 2415. https://doi.org/10.3390/cancers12092415







