Interphase Cytogenetic Analysis of G0 Lymphocytes Exposed to α-Particles, C-Ions, and Protons Reveals their Enhanced Effectiveness for Localized Chromosome Shattering—A Critical Risk for Chromothripsis
Abstract
1. Introduction
2. Results
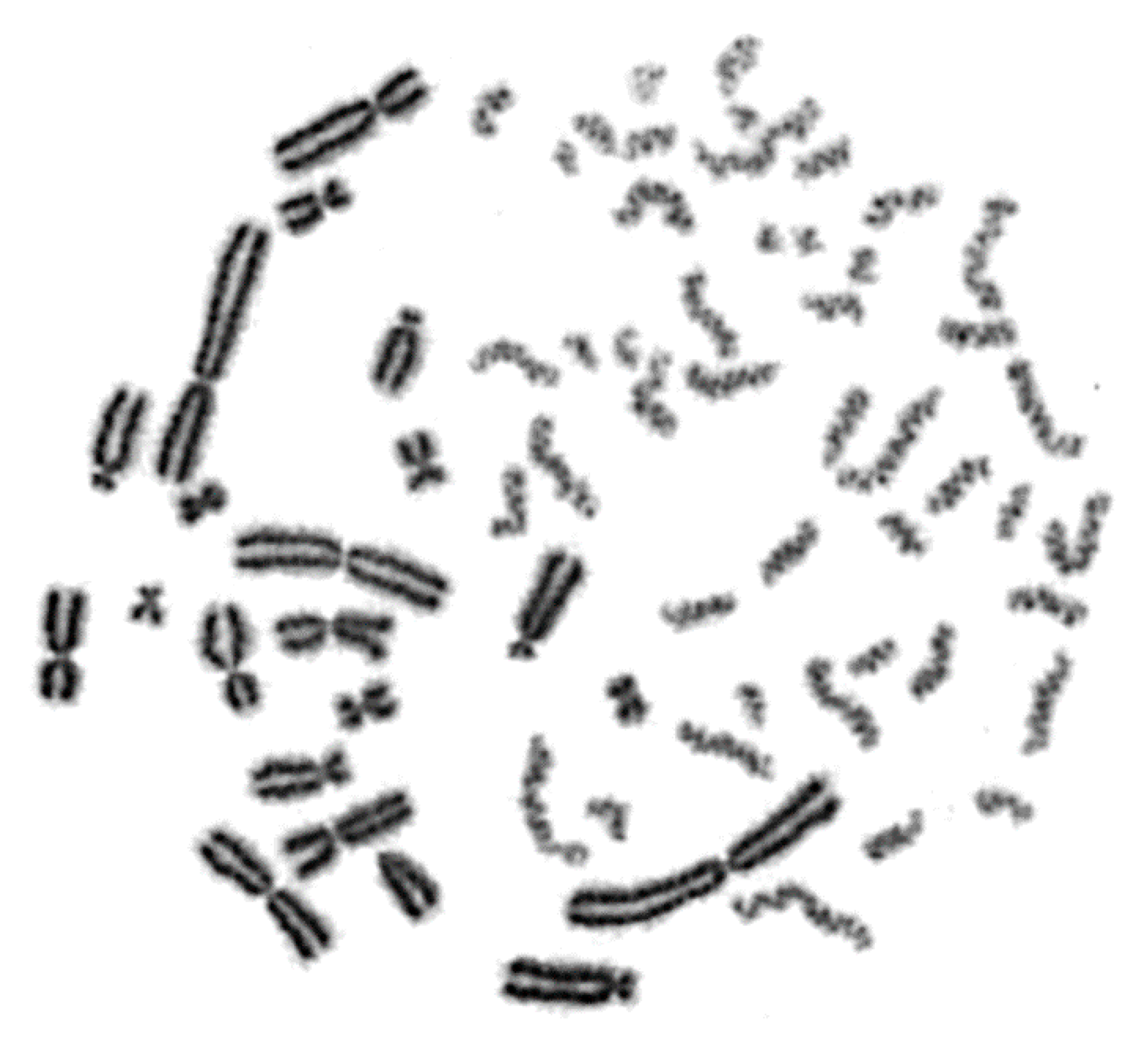
2.1. Interphase Cytogenetic Analysis of G0 Lymphocytes by Means of the PCC Assay is a Promising Tool to Study the Mechanisms Underlying the Biological Effectiveness of Particle Irradiation
2.2. RBE Values for Different Radiation Qualities Can Be Obtained Using Chromosome Fragmentation Analysis Directly in Interphase G0 Lymphocyte PCCs
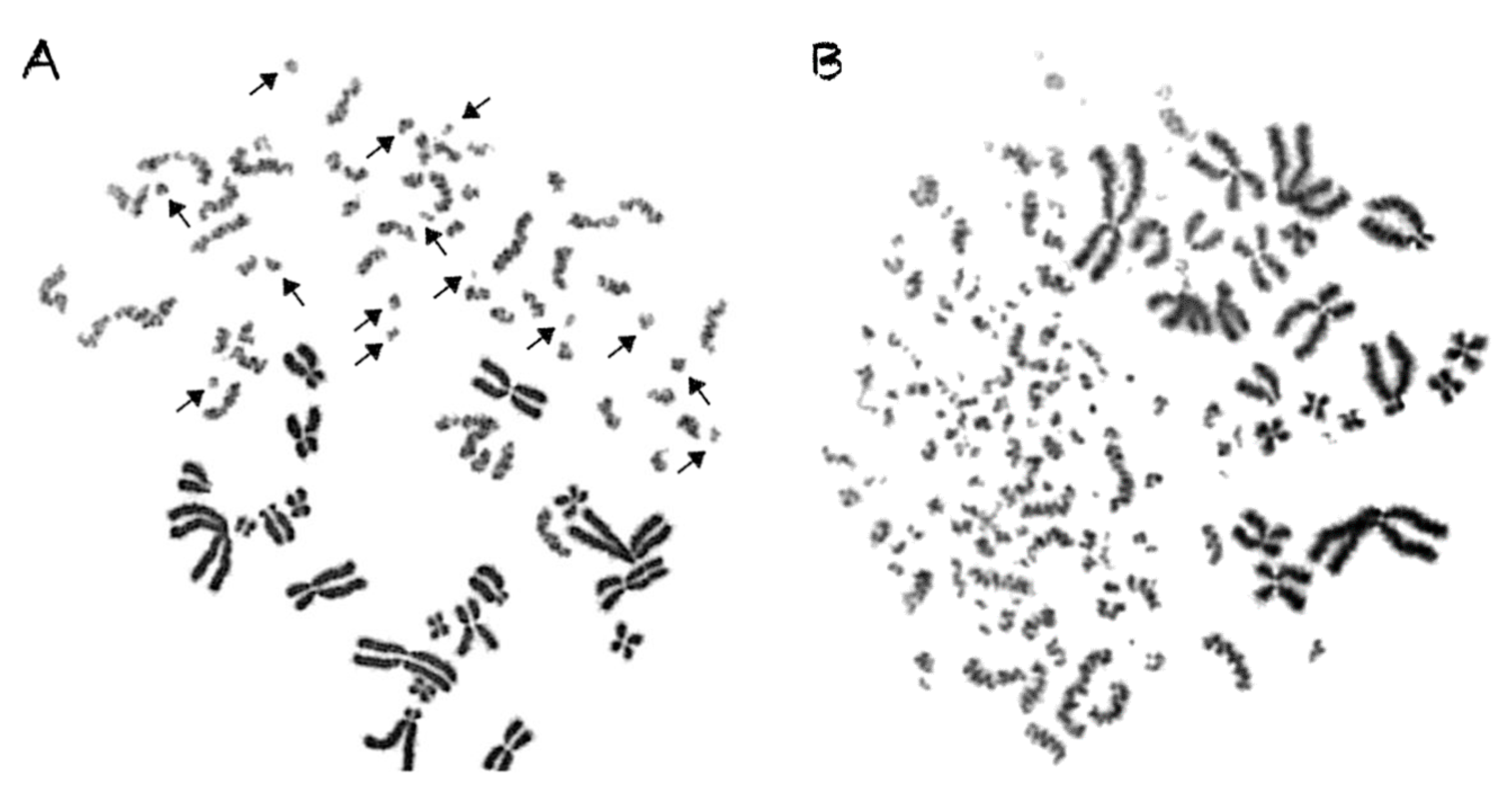
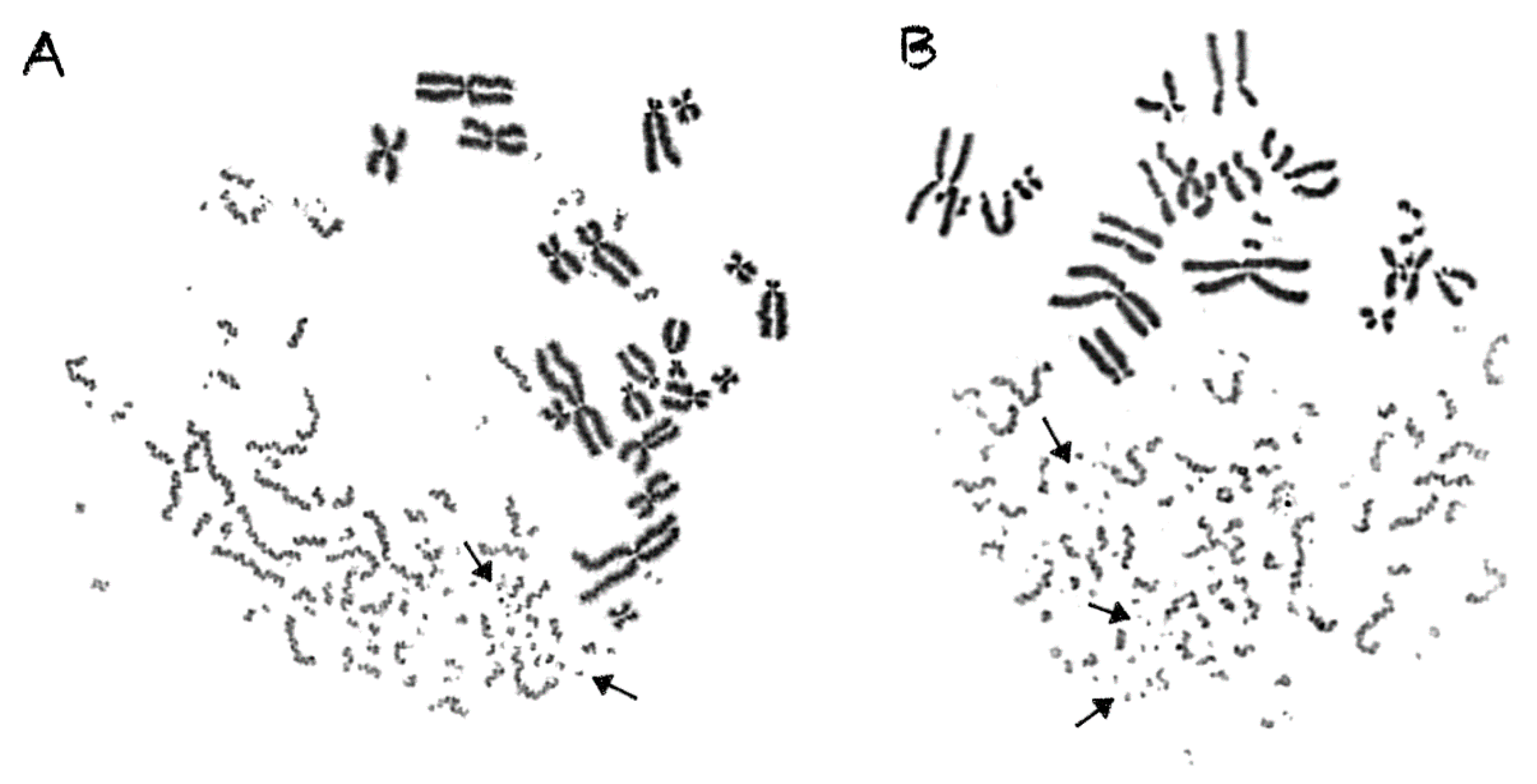
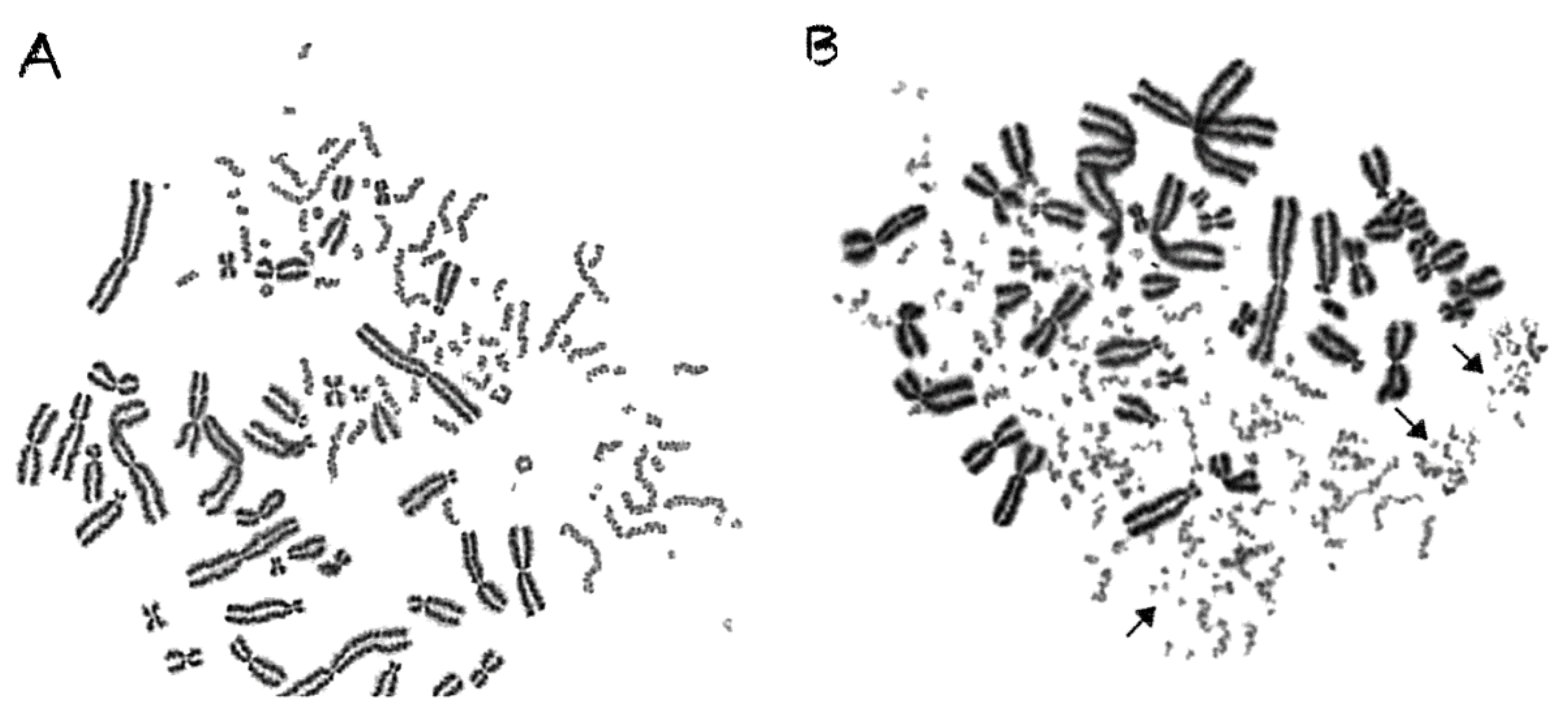
2.3. Shattered Chromosome Domains are a Fingerprint of Exposure to High-LET Particle Radiation and Their Yield Depends on Dose and Radiation Quality
2.4. Persistent Shattered Chromosome Domains May Explain Differences in Biological Effectiveness among Different Radiation Qualities and the Induction of Chromothripsis-Like Rearrangements
3. Discussion
3.1. Can Clustered DNA Lesions Alone Account for the Formation of Complex Chromosomal Aberrations and the Increased Relative Biological Effectiveness of Particle Irradiation?
3.2. Chromosome Aberration Analysis of G0 Lymphocyte PCCs Enables the Assessment of DNA Damage without the Requirement of Irradiated Cells Entering into Metaphase
3.3. Reliable RBE Values for Particle Radiations Can Be Established Using Fragmentation of Interphase Chromosomes as a Biological Endpoint
3.4. Localized Chromosome Shattering Induced by Energetic Nuclei May be Used as a Fingerprint of Exposure
3.5. Our Model: Clustered DNA Lesions are Transformed through Chromatin Condensation into Localized Chromosome Shattering and, via Random Rejoining, Evolve into Chromothripsis-Like Rearrangements
4. Materials and Methods
4.1. Cell Cultures and Preparation of PCC-Inducer Mitotic CHO Cells
4.2. Lymphocyte Isolation from Human Peripheral Blood
4.3. Irradiation and Sample Preparation
4.4. Cell Fusion-Mediated Induction of Premature Chromosome Condensation in Lymphocytes
4.5. Cytogenetic Analysis, Scoring Criteria, Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Li, Y.R.; Halliwill, K.D.; Adams, C.J.; Iyer, V.; Riva, L.; Mamunur, R.; Jen, K.-Y.; del Rosario, R.; Fredlund, E.; Hirst, G.; et al. Mutational signatures in tumours induced by high and low energy radiation in Trp53 deficient mice. Nat. Commun. 2020, 11, 394. [Google Scholar]
- Morishita, M.; Muramatsu, T.; Suto, Y.; Hirai, M.; Konishi, T.; Hayashi, S.; Shigemizu, D.; Tsunoda, T.; Moriyama, K.; Inazawa, J. Chromothripsis-like chromosomal rearrangements induced by ionizing radiation using proton microbeam Irradiation system. Oncotarget 2016, 7, 10182–10192. [Google Scholar] [CrossRef] [PubMed]
- Stephens, P.J.; Greenman, C.D.; Fu, B.; Yang, F.; Bignell, G.R.; Mudie, L.J.; Pleasance, E.D.; Lau, K.W.; Beare, D.; Stebbings, L.A.; et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 2011, 144, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.-Z.; Spektor, A.; Cornils, H.; Francis, J.M.; Jackson, E.K.; Liu, S.; Meyerson, M.; Pellman, D. Chromothripsis from DNA damage in micronuclei. Nature 2015, 522, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Korbel, J.O.; Campbell, P.J. Criteria for Inference of Chromothripsis in Cancer Genomes. Cell 2013, 152, 1226–1236. [Google Scholar] [CrossRef]
- Leibowitz, M.L.; Zhang, C.-Z.; Pellman, D. Chromothripsis: A New Mechanism for Rapid Karyotype Evolution. Annu. Rev. Genet. 2015, 49, 183–211. [Google Scholar] [CrossRef]
- Aziz, K.; Nowsheen, S.; Pantelias, G.; Iliakis, G.; Gorgoulis, V.G.; Georgakilas, A.G. Targeting DNA damage and repair: Embracing the pharmacological era for successful cancer therapy. Pharmacol. Ther. 2012, 133, 334–350. [Google Scholar] [CrossRef]
- Forment, J.V.; Kaidi, A.; Jackson, S.P. Chromothripsis and cancer: Causes and consequences of chromosome shattering. Nat. Rev. Cancer 2012, 12, 663–670. [Google Scholar] [CrossRef]
- Kloosterman, W.P.; Tavakoli-Yaraki, M.; Van Roosmalen, M.J.; Van Binsbergen, E.; Renkens, I.; Duran, K.; Ballarati, L.; Vergult, S.; Giardino, D.; Hansson, K.; et al. Constitutional Chromothripsis Rearrangements Involve Clustered Double-Stranded DNA Breaks and Nonhomologous Repair Mechanisms. Cell Rep. 2012, 1, 648–655. [Google Scholar] [CrossRef]
- Molenaar, J.J.; Koster, J.; Zwijnenburg, D.A.; Van Sluis, P.; Valentijn, L.J.; Van Der Ploeg, I.; Hamdi, M.; Van Nes, J.; Westerman, B.A.; Van Arkel, J.; et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 2012, 483, 589–593. [Google Scholar] [CrossRef]
- Iliakis, G.; Murmann, T.; Soni, A. Alternative end-joining repair pathways are the ultimate backup for abrogated classical non-homologous end-joining and homologous recombination repair: Implications for the formation of chromosome translocations. Mutat. Res.-Genet. Toxicol. Environ. Mutagen. 2015, 793, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Schipler, A.; Mladenova, V.; Soni, A.; Nikolov, V.; Saha, J.; Mladenov, E.; Iliakis, G. Chromosome thripsis by DNA double strand break clusters causes enhanced cell lethality, chromosomal translocations and 53BP1-recruitment. Nucleic Acids Res. 2016, 44, 7673–7690. [Google Scholar] [CrossRef] [PubMed]
- Hair, J.M.; Terzoudi, G.I.; Hatzi, V.I.; Lehockey, K.A.; Srivastava, D.; Wang, W.; Pantelias, G.E.; Georgakilas, A.G. BRCA1 role in the mitigation of radiotoxicity and chromosomal instability through repair of clustered DNA lesions. Chem. Biol. Interact. 2010, 188, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Pantelias, A.; Karachristou, I.; Georgakilas, A.G.; Terzoudi, G.I. Interphase Cytogenetic Analysis of Micronucleated and Multinucleated Cells Supports the Premature Chromosome Condensation Hypothesis as the Mechanistic Origin of Chromothripsis. Cancers 2019, 11, 1123. [Google Scholar] [CrossRef] [PubMed]
- Ritter, S.; Durante, M. Heavy-ion induced chromosomal aberrations: A review. Mutat. Res.-Toxicol. Environ. Mutagen. 2010, 701, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Durante, M.; Cucinotta, F.A. Heavy ion carcinogenesis and human space exploration. Nat. Rev. Cancer 2008, 8, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Cremer, T.; Cremer, M.; Dietzel, S.; Müller, S.; Solovei, I.; Fakan, S. Chromosome territories—A functional nuclear landscape. Curr. Opin. Cell Biol. 2006, 18, 307–316. [Google Scholar] [CrossRef] [PubMed]
- McCord, R.P.; Balajee, A. 3D Genome Organization Influences the Chromosome Translocation Pattern. Adv. Exp. Med. Biol. 2018, 1044, 113–133. [Google Scholar]
- Zheng, H.; Xie, W. The role of 3D genome organization in development and cell differentiation. Nat. Rev. Mol. Cell Biol. 2019, 20, 535–550. [Google Scholar] [CrossRef]
- Shibata, A.; Jeggo, P.A. DNA Double-strand Break Repair in a Cellular Context. Clin. Oncol. 2014, 26, 243–249. [Google Scholar] [CrossRef]
- Schipler, A.; Iliakis, G. DNA double-strand-break complexity levels and their possible contributions to the probability for error-prone processing and repair pathway choice. Nucleic Acids Res. 2013, 41, 7589–7605. [Google Scholar] [CrossRef]
- Soni, A.; Murmann-Konda, T.; Siemann-Loekes, M.; Pantelias, G.E.; Iliakis, G. Chromosome breaks generated by low doses of ionizing radiation in G2-phase are processed exclusively by gene conversion. DNA Repair 2020, 89, 102828. [Google Scholar] [CrossRef]
- Iliakis, G.; Mladenov, E.; Mladenova, V. Necessities in the Processing of DNA Double Strand Breaks and Their Effects on Genomic Instability and Cancer. Cancers 2019, 11, 1671. [Google Scholar] [CrossRef] [PubMed]
- Hada, M.; Georgakilas, A.G. Formation of clustered DNA damage after high-LET irradiation: A review. J. Radiat. Res. 2008, 49, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Georgakilas, A.G.; O’Neill, P.; Stewart, R.D. Induction and Repair of Clustered DNA Lesions: What Do We Know So Far? Radiat. Res. 2013, 180, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Stewart, R.D.; Yu, V.K.; Georgakilas, A.G.; Koumenis, C.; Park, J.H.; Carlson, D.J. Effects of Radiation Quality and Oxygen on Clustered DNA Lesions and Cell Death. Radiat. Res. 2011, 176, 587–602. [Google Scholar] [CrossRef] [PubMed]
- Nikitaki, Z.; Nikolov, V.; Mavragani, I.V.; Mladenov, E.; Mangelis, A.; Laskaratou, D.A.; Fragkoulis, G.I.; Hellweg, C.E.; Martin, O.A.; Emfietzoglou, D.; et al. Measurement of complex DNA damage induction and repair in human cellular systems after exposure to ionizing radiations of varying linear energy transfer (LET). Free Radic. Res. 2016, 50, S64–S78. [Google Scholar] [CrossRef]
- Mavragani, I.V.; Nikitaki, Z.; Kalospyros, S.A.; Georgakilas, A.G. Ionizing Radiation and Complex DNA Damage: From Prediction to Detection Challenges and Biological Significance. Cancers 2019, 11, 1789. [Google Scholar] [CrossRef]
- Tsao, D.; Kalogerinis, P.; Tabrizi, I.; Dingfelder, M.; Stewart, R.D.; Georgakilas, A.G. Induction and Processing of Oxidative Clustered DNA Lesions in 56 Fe-Ion-Irradiated Human Monocytes. Radiat. Res. 2007, 168, 87–97. [Google Scholar] [CrossRef]
- Saha, J.; Wilson, P.; Thieberger, P.; Lowenstein, D.; Wang, M.; Cucinotta, F.A. Biological Characterization of Low-Energy Ions with High-Energy Deposition on Human Cells. Radiat. Res. 2014, 182, 282. [Google Scholar] [CrossRef]
- Mavragani, I.; Nikitaki, Z.; Souli, M.; Aziz, A.; Nowsheen, S.; Aziz, K.; Rogakou, E.; Georgakilas, A. Complex DNA Damage: A Route to Radiation-Induced Genomic Instability and Carcinogenesis. Cancers 2017, 9, 91. [Google Scholar] [CrossRef] [PubMed]
- Mladenov, E.; Saha, J.; Iliakis, G. Processing-Challenges Generated by Clusters of DNA Double-Strand Breaks Underpin Increased Effectiveness of High-LET Radiation and Chromothripsis. Adv. Exp. Med. Biol. 2018, 1044, 149–168. [Google Scholar] [PubMed]
- Nasonova, E.; Fussel, K.; Berger, S.; Gudowska-Nowak, E.; Ritter, S. Cell cycle arrest and aberration yield in normal human fibroblasts. I. Effects of X-rays and 195 MeV u(−1) C ions. Int. J. Radiat. Biol. 2004, 80, 621–634. [Google Scholar] [CrossRef] [PubMed]
- Tenhumberg, S.; Gudowska-Nowak, E.; Nasonova, E.; Ritter, S. Cell cycle arrest and aberration yield in normal human fibroblasts. II: Effects of 11 MeV u −1 C ions and 9.9 MeV u −1 Ni ions. Int. J. Radiat. Biol. 2007, 83, 501–513. [Google Scholar] [CrossRef]
- Lücke-Huhle, C.; Blakely, E.A.; Chang, P.Y.; Tobias, C.A. Drastic G2 arrest in mammalian cells after irradiation with heavy-ion beams. Radiat. Res. 1979, 79, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Ochab-Marcinek, A.; Gudowska-Nowak, E.; Nasonova, E.; Ritter, S. Modeling radiation-induced cell cycle delays. Radiat. Environ. Biophys. 2009, 48, 361–370. [Google Scholar] [CrossRef]
- Terzoudi, G.I.; Pantelias, G.; Darroudi, F.; Barszczewska, K.; Buraczewska, I.; Depuydt, J.; Georgieva, D.; Hadjidekova, V.; Hatzi, V.I.; Karachristou, I.; et al. Dose assessment intercomparisons within the RENEB network using G0-lymphocyte prematurely condensed chromosomes (PCC assay). Int. J. Radiat. Biol. 2017, 93, 48–57. [Google Scholar] [CrossRef]
- Pantelias, A.; Terzoudi, G.I. Development of an automatable micro-PCC biodosimetry assay for rapid individualized risk assessment in large-scale radiological emergencies. Mutat. Res.-Genet. Toxicol. Environ. Mutagen. 2018, 836, 65–71. [Google Scholar] [CrossRef]
- Hagiwara, Y.; Oike, T.; Niimi, A.; Yamauchi, M.; Sato, H.; Limsirichaikul, S.; Held, K.D.; Nakano, T.; Shibata, A. Clustered DNA double-strand break formation and the repair pathway following heavy-ion irradiation. J. Radiat. Res. 2019, 60, 69–79. [Google Scholar] [CrossRef]
- Hagiwara, Y.; Niimi, A.; Isono, M.; Yamauchi, M.; Yasuhara, T.; Limsirichaikul, S.; Oike, T.; Sato, H.; Held, K.D.; Nakano, T.; et al. 3D-structured illumination microscopy reveals clustered DNA double-strand break formation in widespread γH2AX foci after high LET heavy-ion particle radiation. Oncotarget 2017, 8, 109370–109381. [Google Scholar] [CrossRef]
- Pantelias, G.E.; Terzoudi, G.I. Functional cell-cycle chromatin conformation changes in the presence of DNA damage result into chromatid breaks: A new insight in the formation of radiation-induced chromosomal aberrations based on the direct observation of interphase chromatin. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2010, 701, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Terzoudi, G.I.; Karakosta, M.; Pantelias, A.; Hatzi, V.I.; Karachristou, I.; Pantelias, G. Stress induced by premature chromatin condensation triggers chromosome shattering and chromothripsis at DNA sites still replicating in micronuclei or multinucleate cells when primary nuclei enter mitosis. Mutat. Res.-Genet. Toxicol. Environ. Mutagen. 2015, 793, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Timm, S.; Lorat, Y.; Jakob, B.; Taucher-Scholz, G.; Rübe, C.E. Clustered DNA damage concentrated in particle trajectories causes persistent large-scale rearrangements in chromatin architecture. Radiother. Oncol. 2018, 129, 600–610. [Google Scholar] [CrossRef] [PubMed]
- Ryan, T.L.; Pantelias, A.G.; Terzoudi, G.I.; Pantelias, G.E.; Balajee, A.S. Use of human lymphocyte G0 PCCs to detect intra- and inter-chromosomal aberrations for early radiation biodosimetry and retrospective assessment of radiation-induced effects. PLoS ONE 2019, 14, e0216081. [Google Scholar] [CrossRef]
- Bedford, J.S.; Goodhead, D.T. Breakage of human interphase chromosomes by alpha particles and x-rays. Int. J. Radiat. Biol. 1989, 55, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Loucas, B.D.; Geard, C.R. Initial damage in human interphase chromosomes from alpha particles with linear energy transfers relevant to radon exposure. Radiat. Res. 1994, 139, 9–14. [Google Scholar] [CrossRef]
- Goodwin, E.H.; Blakely, E.A.; Tobias, C.A. Chromosomal damage and repair in G1-phase Chinese hamster ovary cells exposed to charged-particle beams. Radiat. Res. 1994, 138, 343–351. [Google Scholar] [CrossRef]
- Suzuki, M.; Kase, Y.; Kanai, T.; Yatagai, F.; Watanabe, M. LET dependence of cell death and chromatin-break induction in normal human cells irradiated by neon-ion beams. Int. J. Radiat. Biol. 1997, 72, 497–503. [Google Scholar] [CrossRef]
- Durante, M.; Furusawa, Y.; George, K.; Gialanella, G.; Greco, O.; Grossi, G.; Matsufuji, N.; Pugliese, M.; Yang, T.C. Rejoining and misrejoining of radiation-induced chromatin breaks. IV. Charged particles. Radiat. Res. 1998, 149, 446–454. [Google Scholar] [CrossRef]
- Nasonova, E.; Gudowska-Nowak, E.; Ritter, S.; Kraft, G. Analysis of Ar-ion and X-ray-induced chromatin breakage and repair in V79 plateau-phase cells by the premature chromosome condensation technique. Int. J. Radiat. Biol. 2001, 77, 59–70. [Google Scholar] [CrossRef]
- Gudowska-Nowak, E.; Nasonova, E.; Ritter, S.; Scholz, M. Chromosome fragmentation after irradiation with C ions. Radiother. Oncol. 2004, 73, S123–S126. [Google Scholar] [CrossRef]
- Tsuruoka, C.; Suzuki, M.; Hande, M.P.; Furusawa, Y.; Anzai, K.; Okayasu, R. The Difference in LET and Ion Species Dependence for Induction of Initially Measured and Non-rejoined Chromatin Breaks in Normal Human Fibroblasts. Radiat. Res. 2008, 170, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Sekine, E.; Okada, M.; Matsufuji, N.; Yu, D.; Furusawa, Y.; Okayasu, R. High LET heavy ion radiation induces lower numbers of initial chromosome breaks with minimal repair than low LET radiation in normal human cells. Mutat. Res.-Genet. Toxicol. Environ. Mutagen. 2008, 652, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Vitti, E.T.; Parsons, J.L. The Radiobiological Effects of Proton Beam Therapy: Impact on DNA Damage and Repair. Cancers 2019, 11, 946. [Google Scholar] [CrossRef] [PubMed]
- Badie, C.; Iliakis, G.; Foray, N.; Alsbeih, G.; Cedervall, B.; Chavaudra, N.; Pantelias, G.; Arlett, C.; Malaise, E.P. Induction and rejoining of DNA double-strand breaks and interphase chromosome breaks after exposure to X rays in one normal and two hypersensitive human fibroblast cell lines. Radiat. Res. 1995, 144, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Kodym, R.; Hoerth, E. Determination of the radiation sensitivity of the stromal cells in the murine long-term bone marrow culture by measuring the induction and rejoining of interphase chromosome breaks. Int. J. Radiat. Oncol. Biol. Phys. 1993, 25, 829–833. [Google Scholar] [CrossRef]
- Johannes, C.; Chudoba, I.; Obe, G. Analysis of X-ray-induced aberrations in human chromosome 5 using high-resolution multicolour banding FISH (mBAND). Chromosom. Res. 1999, 7, 625–633. [Google Scholar] [CrossRef]
- Hada, M.; Wu, H.; Cucinotta, F.A. mBAND analysis for high- and low-LET radiation-induced chromosome aberrations: A review. Mutat. Res. Mol. Mech. Mutagen. 2011, 711, 187–192. [Google Scholar] [CrossRef]
- Chatzipapas, K.P.; Papadimitroulas, P.; Emfietzoglou, D.; Kalospyros, S.A.; Hada, M.; Georgakilas, A.G.; Kagadis, G.C. Ionizing Radiation and Complex DNA Damage: Quantifying the Radiobiological Damage Using Monte Carlo Simulations. Cancers 2020, 12, 799. [Google Scholar] [CrossRef]
- Cornforth, M.N. Occam’s broom and the dirty DSB: Cytogenetic perspectives on cellular response to changes in track structure and ionization density. Int. J. Radiat. Biol. 2020, 1–10. [Google Scholar] [CrossRef]
- Jakob, B.; Dubiak-Szepietowska, M.; Janiel, E.; Schmidt, A.; Durante, M.; Taucher-Scholz, G. Differential Repair Protein Recruitment at Sites of Clustered and Isolated DNA Double-Strand Breaks Produced by High-Energy Heavy Ions. Sci. Rep. 2020, 10, 1443. [Google Scholar] [CrossRef] [PubMed]
- Sage, E.; Shikazono, N. Radiation-induced clustered DNA lesions: Repair and mutagenesis. Free Radic. Biol. Med. 2017, 107, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Terzoudi, G.I.; Pantelias, G.E. Conversion of DNA damage into chromosome damage in response to cell cycle regulation of chromatin condensation after irradiation. Mutagenesis 1997, 12, 271–276. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Crasta, K.; Ganem, N.J.; Dagher, R.; Lantermann, A.B.; Ivanova, E.V.; Pan, Y.; Nezi, L.; Protopopov, A.; Chowdhury, D.; Pellman, D. DNA breaks and chromosome pulverization from errors in mitosis. Nature 2012, 482, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Belli, M.; Cherubini, R.; Galeazzi, G.; Mazzucato, S.; Moschini, G.; Sapora, O.; Simone, G.; Tabocchini, M.A. Proton irradiation facility for radiobiological studies at a 7 MV Van de Graaff accelerator. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrom. Detect. Assoc. Equip. 1987, 256, 576–580. [Google Scholar] [CrossRef]
- Cera, F.; Cherubini, R.; Dalla Vecchia, M.; Galeazzi, G.; Haque, A.M.I.; Moschini, G.; Tiveron, P. A radiobiological facility set up at LNL XTU-Tandem accelerator for irradiation in air of cultured mammalian cells with heavy ion beams. In INFN-LNL Annual Report 1993 - LNL-INFN(Rep)- 081/94; INFN-LNL: Legnaro, Italy, 1993; pp. 128–129. [Google Scholar]
- Pantelias, G.E.; Maillie, H.D. A simple method for premature chromosome condensation induction in primary human and rodent cells using polyethylene glycol. Somat. Cell Genet. 1983, 9, 533–547. [Google Scholar] [CrossRef]
- Karachristou, I.; Karakosta, M.; Pantelias, A.; Hatzi, V.I.; Karaiskos, P.; Dimitriou, P.; Pantelias, G.; Terzoudi, G.I. Triage biodosimetry using centromeric/telomeric PNA probes and Giemsa staining to score dicentrics or excess fragments in non-stimulated lymphocyte prematurely condensed chromosomes. Mutat. Res.-Genet. Toxicol. Environ. Mutagen. 2015, 793, 107–114. [Google Scholar] [CrossRef]
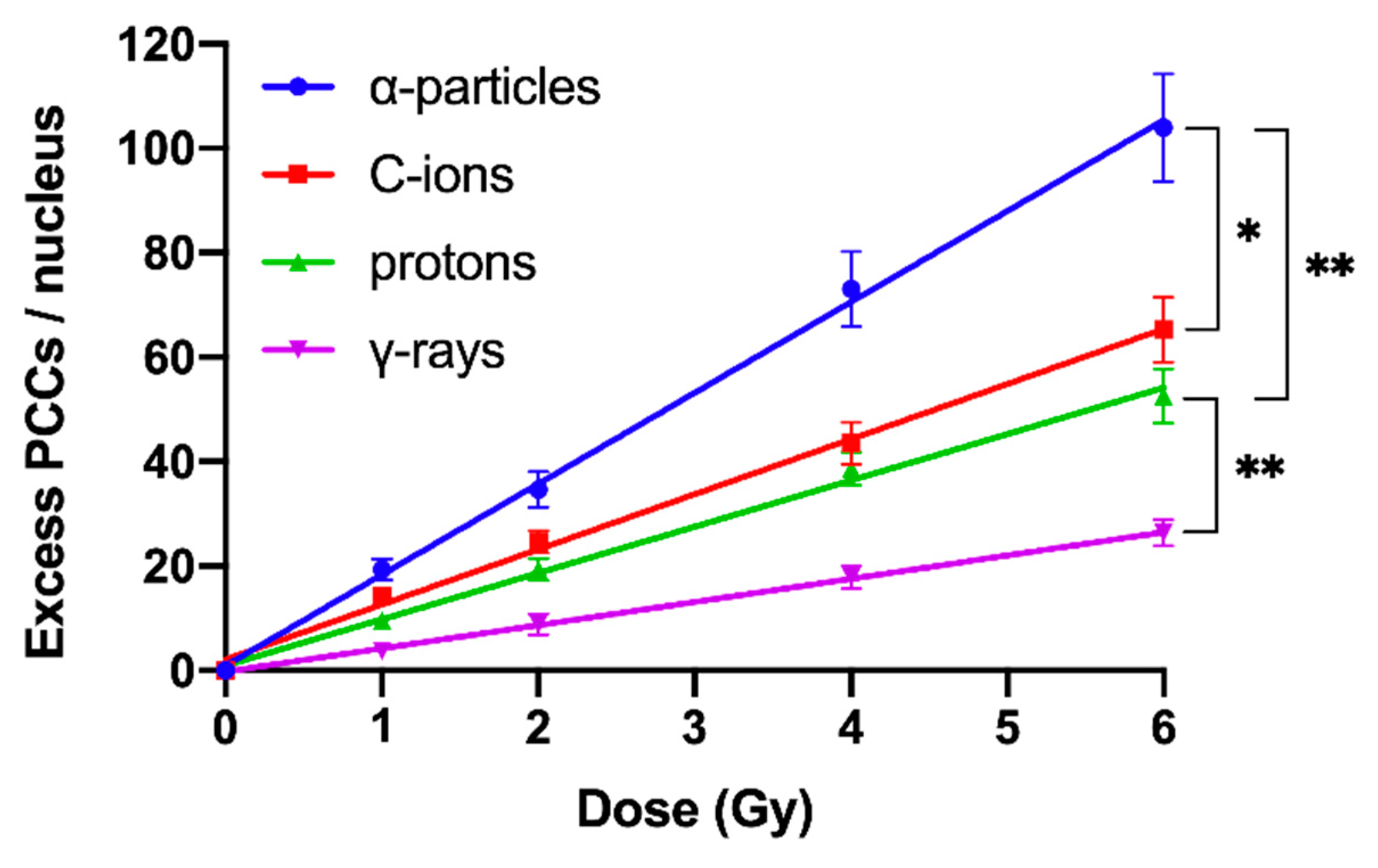
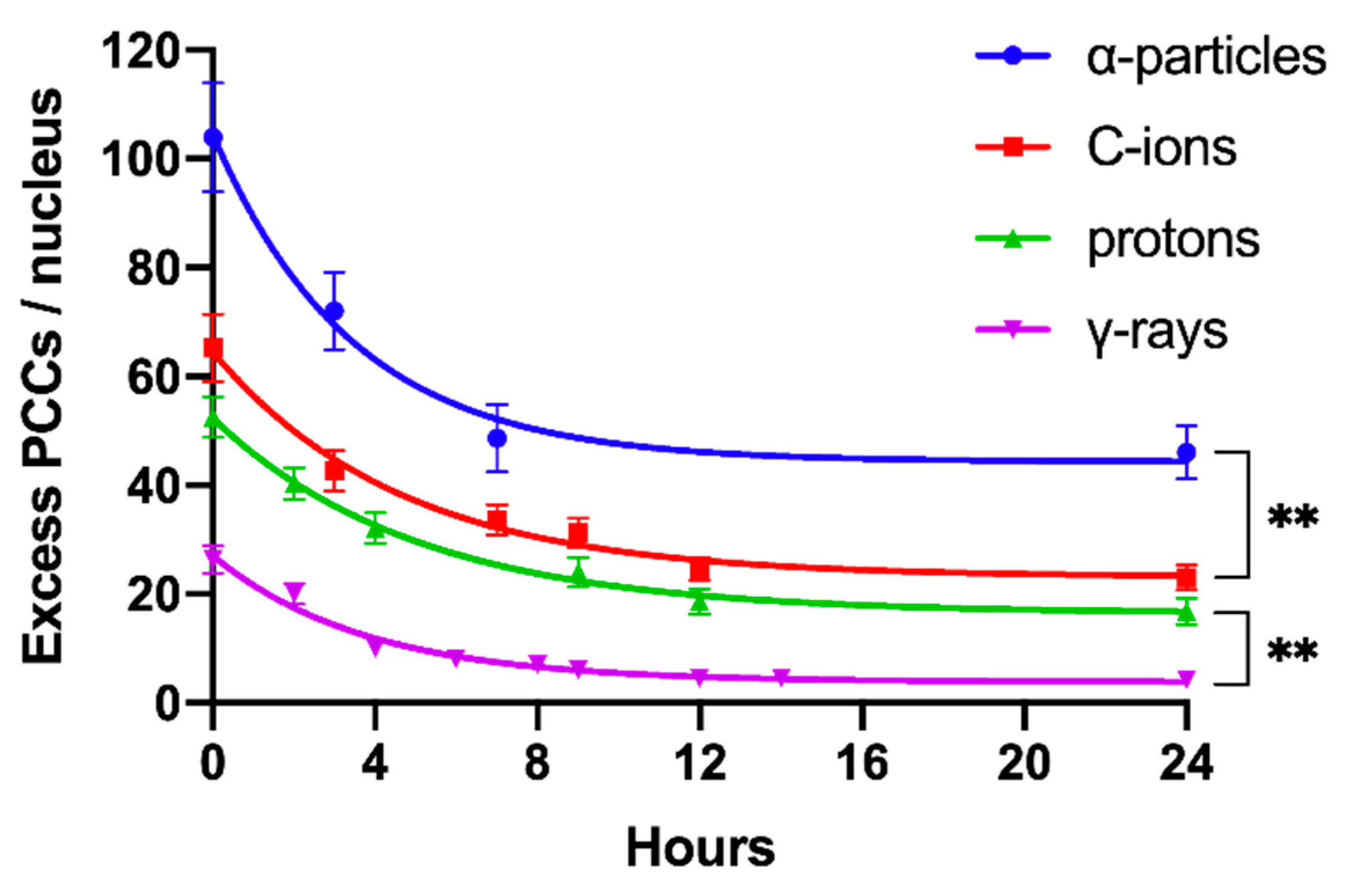

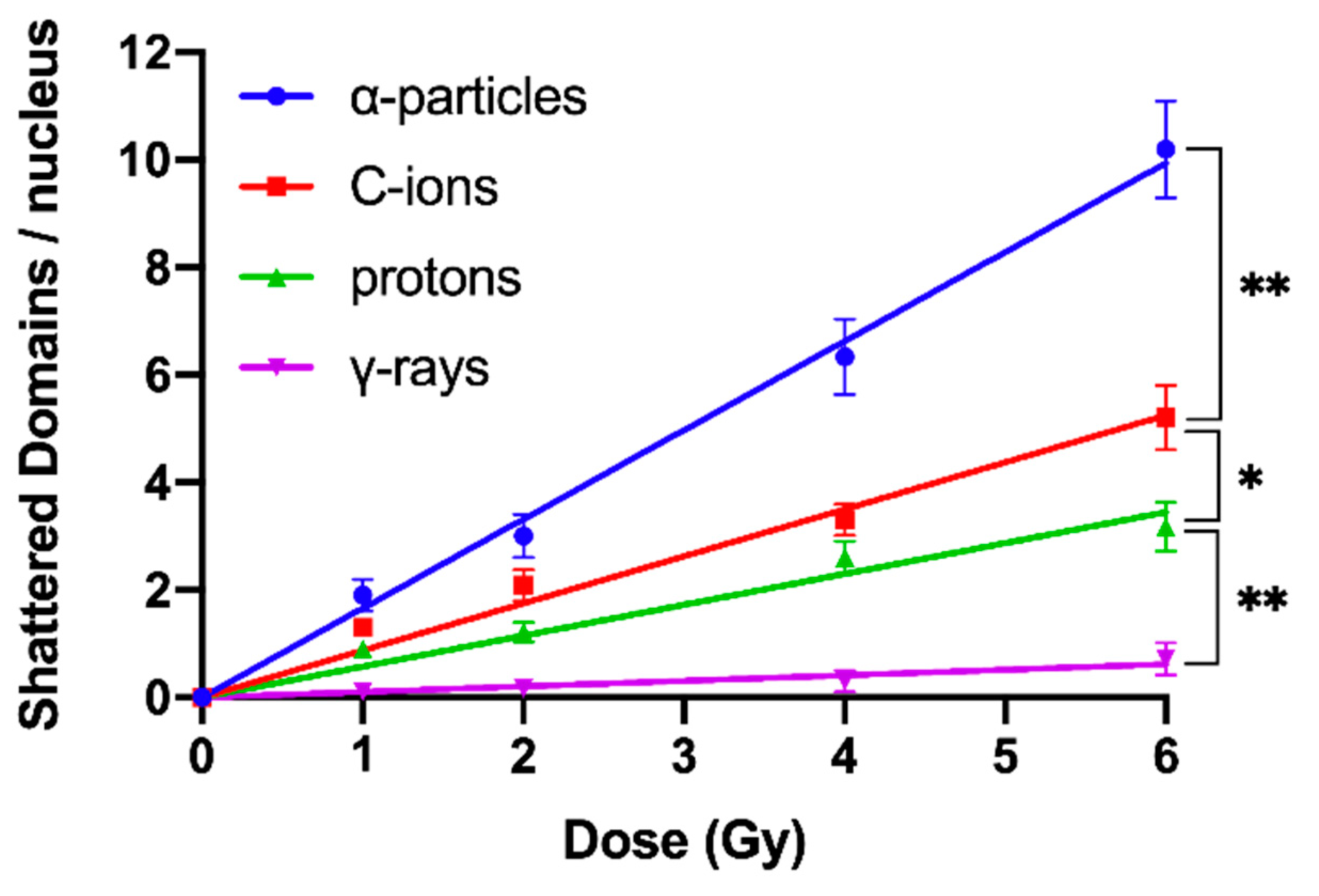
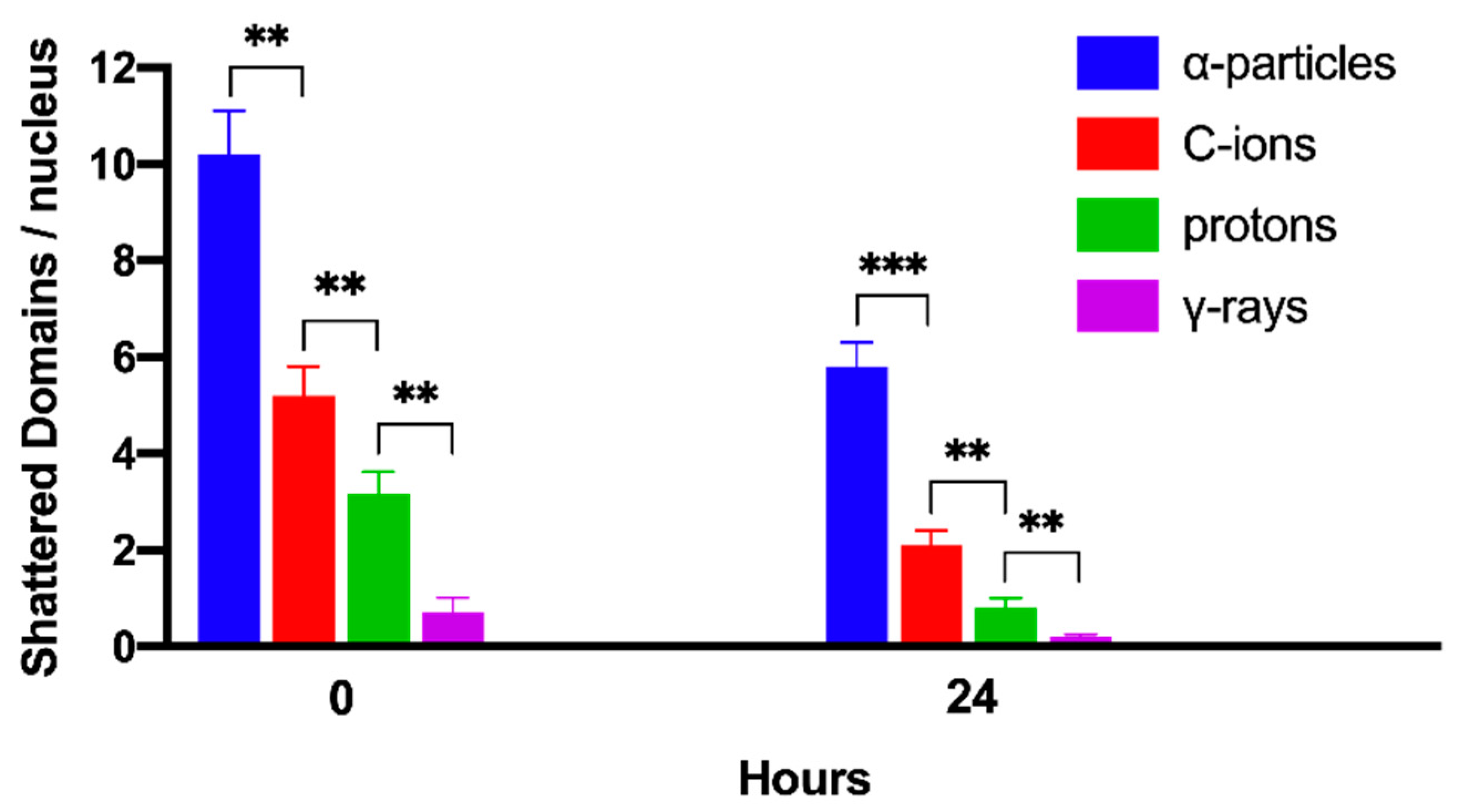
| Radiation Quality | RBE Excess PCCs/Nucleus | RBE Shattered Domains/Nucleus | ||
|---|---|---|---|---|
| 0 h | 24 h | 0 h | 24 h | |
| α-particles | 4.1 | 10.7 | 14.3 | 28.6 |
| C-ions | 2.6 | 5.4 | 7.5 | 10.5 |
| protons | 2.1 | 3.9 | 4.9 | 4 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pantelias, A.; Zafiropoulos, D.; Cherubini, R.; Sarchiapone, L.; De Nadal, V.; Pantelias, G.E.; Georgakilas, A.G.; Terzoudi, G.I. Interphase Cytogenetic Analysis of G0 Lymphocytes Exposed to α-Particles, C-Ions, and Protons Reveals their Enhanced Effectiveness for Localized Chromosome Shattering—A Critical Risk for Chromothripsis. Cancers 2020, 12, 2336. https://doi.org/10.3390/cancers12092336
Pantelias A, Zafiropoulos D, Cherubini R, Sarchiapone L, De Nadal V, Pantelias GE, Georgakilas AG, Terzoudi GI. Interphase Cytogenetic Analysis of G0 Lymphocytes Exposed to α-Particles, C-Ions, and Protons Reveals their Enhanced Effectiveness for Localized Chromosome Shattering—A Critical Risk for Chromothripsis. Cancers. 2020; 12(9):2336. https://doi.org/10.3390/cancers12092336
Chicago/Turabian StylePantelias, Antonio, Demetre Zafiropoulos, Roberto Cherubini, Lucia Sarchiapone, Viviana De Nadal, Gabriel E. Pantelias, Alexandros G. Georgakilas, and Georgia I. Terzoudi. 2020. "Interphase Cytogenetic Analysis of G0 Lymphocytes Exposed to α-Particles, C-Ions, and Protons Reveals their Enhanced Effectiveness for Localized Chromosome Shattering—A Critical Risk for Chromothripsis" Cancers 12, no. 9: 2336. https://doi.org/10.3390/cancers12092336
APA StylePantelias, A., Zafiropoulos, D., Cherubini, R., Sarchiapone, L., De Nadal, V., Pantelias, G. E., Georgakilas, A. G., & Terzoudi, G. I. (2020). Interphase Cytogenetic Analysis of G0 Lymphocytes Exposed to α-Particles, C-Ions, and Protons Reveals their Enhanced Effectiveness for Localized Chromosome Shattering—A Critical Risk for Chromothripsis. Cancers, 12(9), 2336. https://doi.org/10.3390/cancers12092336






