Cell Motility and Cancer
Abstract
1. Introduction
2. Simple Organism Models Are Necessary to Understand Human Cell Behavior
3. The Locomotion System in Unicellular Eukaryotic Organisms and Human Cells
4. External Stimuli, Migration and Cancer
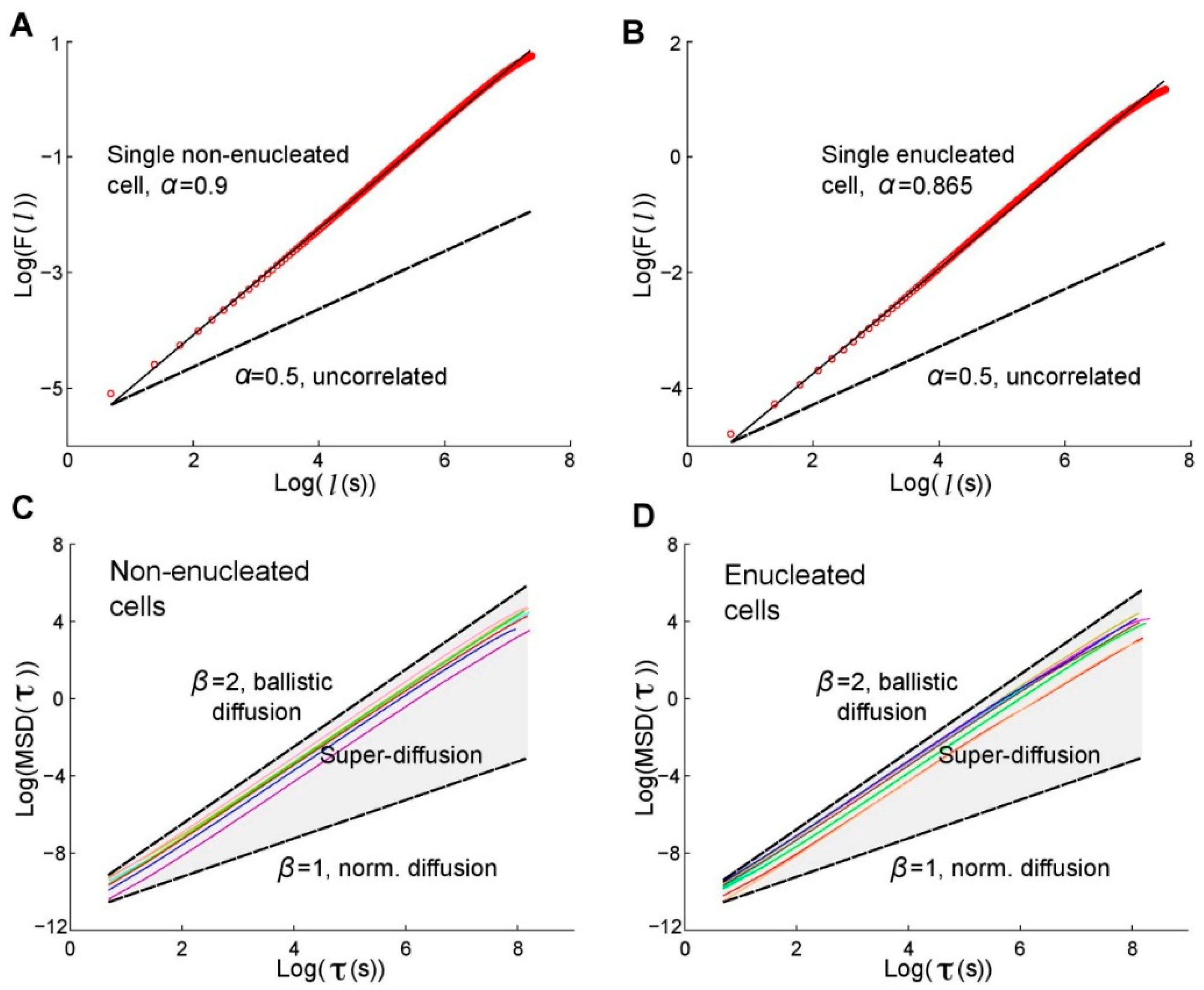
5. The Role of the Nucleus in Cell Migration
6. Conditioned Behavior in Single Cells
- (1)
- Cell locomotion in the absence of stimuli exhibited a random directional distribution in which amoebae and metamoebae explored practically all the directions of the experimental chamber (Figure 2a),
- (2)
- Amoebae and metamoebae showed an unequivocal systemic response consisting in the migration to the cathode when they were exposed to a strong direct electric field of about 300–600 mV/mm (galvanotaxis, Figure 2b),
- (3)
- The response of both organisms was studied during biochemical guidance by exposing them to an nFMLP peptide gradient placed in the anode of a specific set-up. In these experimental conditions, most of the exposed cells migrated towards the peptide in the anode showing stochastic movements with robust directionality (chemotaxis, Figure 2c),
- (4)
- Cells were exposed simultaneously to the galvanotactic and chemotactic stimuli for 30 min (induction process). For such a purpose, the cathode was placed on the right of the set-up and the anode with the nFMLP peptide solution on the left (Figure 2d). The results showed that roughly half of the amoebae and metamoebae migrated towards the anode where the peptide was placed, while the reminders did it to the cathode,
- (5)
- To verify if the cells that moved to the anode during the induction process (Figure 2d) exhibited some degree of persistence in their migratory behavior, those cells that had previously migrated to the anode-peptide in the fourth step were exposed a second time (30 min) to a single electric field without the peptide. Under these experimental conditions, the analysis of the individual trajectories showed that most cells did migrate to the anode where the peptide was absent (Figure 2e). This evidence corroborated that a new locomotion pattern had appeared in amoebae and metamoebae (Figure 2d) (note that without the induction process practically all the cells migrated to the cathode, (Figure 2c) and after the induction process the cells modified their behavior going to the anode instead to the cathode).
7. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Stuelten, C.H.; Parent, C.A.; Montell, D.J. Cell motility in cancer invasion and metastasis: Insights from simple model organisms. Nat. Rev. Cancer 2018, 18, 296–312. [Google Scholar] [CrossRef]
- Titus, M.A.; Goodson, H.V. An evolutionary perspective on cell migration: Digging for the roots of amoeboid motility. J. Cell Biol. 2017, 216, 1509–1511. [Google Scholar] [CrossRef]
- Fritz-Laylin, L.K.; Lord, S.J.; Mullins, R.D. WASP and SCAR are evolutionarily conserved in actin-filled pseudopod-based motility. J. Cell Biol. 2017, 216, 1673–1688. [Google Scholar] [CrossRef] [PubMed]
- Fritz-Laylin, L.K.; Riel-Mehan, M.; Chen, B.; Lord, S.J.; Goddard, T.D.; Ferrin, T.E.; Nicholson-Dykstra, S.; Higgs, H.; Johnson, G.T.; Betzig, E.; et al. Actin-based protrusions of migrating neutrophils are intrinsically lamellar and facilitate direction changes. eLife 2017, 6, e26990. [Google Scholar] [CrossRef] [PubMed]
- Artemenko, Y.; Lampert, T.J.; Devreotes, P.N. Moving towards a paradigm: Common mechanisms of chemotactic signaling in Dictylostelium and mammalian leukocytes. Cell Mol. Life Sci. 2014, 71, 3711–3747. [Google Scholar] [CrossRef] [PubMed]
- Batsios, P.; Ishikawa-Ankerhold, H.; Roth, H.; Schleicher, M.; Wong, C.; Müller-Taubenberger, A. Ate1-mediated posttranslational arginylation affects substrate adhesion and cell migration in Dictyostelium discoideum. Mol. Biol. Cell 2019, 30, 453–456. [Google Scholar] [CrossRef]
- Gilsbach, B.K.; Ho, F.Y.; Vetter, I.R.; Van Haastert, P.J.; Wittinghofer, A.; Kortholt, A. Roco kinase structures give insights into the mechanism of Parkinson disease-related leucine-rich-repeat kinase 2 mutations. Proc. Natl. Acad. Sci. USA 2012, 109, 10322–10327. [Google Scholar] [CrossRef]
- Chida, J.; Araki, H.; Maeda, Y. Specific growth suppression of human cancer cells by targeted delivery of Dictyostelium mitochondrial ribosomal protein S4. Cancer Cell Int. 2014, 14, 56. [Google Scholar] [CrossRef]
- Sherwood, D.R. Cell invasion through basement membranes: An anchor of understanding. Trends Cell. Biol. 2006, 16, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Ellis, H.M.; Horvitz, H.R. Genetic control of programmed cell death in the nematode C. elegans. Cell 1986, 44, 817–829. [Google Scholar] [CrossRef]
- Pinkston-Gosse, J.; Kenyon, C. DAF-16/FOXO targets genes that regulate tumor growth in Caenorhabditis elegans. Nat. Genet. 2007, 39, 1403–1409. [Google Scholar] [CrossRef] [PubMed]
- Nüsslein-Volhard, C.; Wieschaus, E. Mutations affecting segment number and polarity in Drosophila. Nature 1980, 287, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Martorell, O.; Merlos-Suarez, A.; Campbell, K.; Barriga, F.M.; Christov, C.P.; Miguel-Aliaga, I.; Batlle, E.; Casanova, J.; Casali, A. Conserved mechanisms of tumorigenesis in the Drosophila adult midgut. PLoS ONE 2014, 9, e88413. [Google Scholar] [CrossRef] [PubMed]
- Campbell, K.; Rossi, F.; Adams, J.; Pitsidianaki, I.; Barriga, F.M.; García-Gerique, L.; Batlle, E.; Casanova, J.; Casali, A. Collective cell migration and metastases induced by an epithelial-to-mesenchymal transition in Drosophila intestinal tumors. Nat. Commun. 2019, 10, 2311. [Google Scholar] [CrossRef] [PubMed]
- Caussinus, E.; Gonzalez, C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nature Genet. 2005, 37, 1125–1129. [Google Scholar] [CrossRef] [PubMed]
- Sonoshita, M.; Cagan, R.L. Modeling human cancers in Drosophila. Curr. Top. Dev. Biol. 2017, 121, 287–309. [Google Scholar]
- Lammermann, T.; Sixt, M. Mechanical modes of ‘amoeboid’ cell migration. Curr. Opin. Cell Biol. 2009, 21, 636–644. [Google Scholar] [CrossRef]
- Kumar, S.; Xu, J.; Perkins, C.; Guo, F.; Snapper, S.; Finkelman, F.D.; Zheng, Y.; Filippi, M.D. Cdc42 regulates neutrophil migration via crosstalk between WASp, CD11b, and microtubules. Blood 2012, 120, 3563–3574. [Google Scholar] [CrossRef]
- Stengel, K.; Zheng, Y. Cdc42 in oncogenic transformation, invasion, and tumorigenesis. Cell. Signal. 2011, 23, 1415–1423. [Google Scholar] [CrossRef]
- Creed, S.J.; Desouza, M.; Bamburg, J.R.; Gunning, P.; Stehn, J. Tropomyosin isoforms 3 promotes the formation of filopodia by regulating the recruitment of actin-binding proteins to actin filaments. Exp. Cell Res. 2011, 317, 249–261. [Google Scholar] [CrossRef]
- Bornens, M. Cell Polarity: Having and Making Sense of Direction-On the Evolutionary Significance of the Primary Cilium/Centrosome Organ in Metazoa. Open Biol. 2018, 8, 180052. [Google Scholar] [CrossRef] [PubMed]
- Viola, A.; Luster, A.D. Chemokines and their receptors: Drug targets in immunity and inflammation. Annu. Rev. Pharmacol. Toxicol. 2008, 48, 171–197. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, A.T.; Chun, C.; Takeda, K.; Firtel, R.A. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J. Cell Biol. 2004, 167, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Bos, J.L. ras oncogenes in human cancer: A review. Cancer Res. 1989, 49, 4682–4689. [Google Scholar]
- Kumar, A.; Molli, P.R.; Pakala, S.B.; Bui Nguyen, T.M.; Rayala, S.K.; Kumar, R. PAK thread from amoeba to mammals. J. Cell. Biochem. 2009, 107, 579–585. [Google Scholar] [CrossRef]
- Eswaran, J.; Soundararajan, M.; Knapp, S. Targeting group II PAKs in cancer and metastasis. Cancer Metastasis Rev. 2009, 28, 209–217. [Google Scholar] [CrossRef]
- Chan, P.M.; Manser, E. PAKs in human disease. Prog. Mol. Biol. Transl. Sci. 2012, 106, 171–187. [Google Scholar]
- Jaśkiewicz, A.; Pająk, B.; Orzechowski, A. The Many Faces of Rap1 GTPase. Int. J. Mol. Sci. 2018, 19, 2848. [Google Scholar] [CrossRef]
- Takai, Y.; Sasaki, T.; Matozaki, T. Small GTP-binding proteins. Physiol. Rev. 2001, 81, 153–208. [Google Scholar] [CrossRef]
- Jeon, T.J.; Lee, D.J.; Merlot, S.; Weeks, G.; Firtel, R.A. Rap1 controls cell adhesion and cell motility through the regulation of myosin II. J. Cell. Biol. 2007, 176, 1021–1033. [Google Scholar] [CrossRef]
- Sawant, K.; Chen, Y.; Kotian, N.; Preuss, K.M.; McDonald, J.A. Rap1 GTPase promotes coordinated collective cell migration in vivo. Mol. Biol. Cell. 2018, 29, 2656–2673. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.L.; Wang, R.C.; Cheng, K.; Ring, B.Z.; Su, L. Roles of Rap1 signaling in tumor cell migration and invasion. Cancer Biol. Med. 2017, 14, 90–99. [Google Scholar] [PubMed]
- Dale, H.H. Galvanotaxis and chemotaxis of ciliate infusoria: Part 1. J. Physiol 1901, 25, 291–361. [Google Scholar] [CrossRef] [PubMed]
- Roy, J.; Mazzaferri, J.; Filep, J.G.; Costantino, S. A haptotaxis assay for neutrophils using optical patterning and a high-content approach. Sci. Rep. 2017, 7, 2869. [Google Scholar] [CrossRef] [PubMed]
- Oudin, M.J.; Jonas, O.; Kosciuk, T.; Broye, L.C.; Guido, B.C.; Wyckoft, J.; Riquelme, D.; Lamar, J.M.; Asokan, S.B.; Whittaker, C.; et al. Tumor cell-driven extracellular matrix remodeling drives haptotaxis during metastatic progression. Cancer Discov. 2016, 6, 516–531. [Google Scholar] [CrossRef]
- Prentice-Mott, H.V.; Chang, C.H.; Mahadevan, L.; Mitchison, T.J.; Irimia, D.; Shah, J.V. Biased migration of confined neutrophil-like cells in asymmetric hydraulic environments. PNAS 2013, 110, 21006–21011. [Google Scholar] [CrossRef]
- Moreau, H.D.; Blanch-Mercader, C.; Attia, R.; Maurin, M.; Alraies, Z.; Sanséau, D.; Malbec, O.; Delgado, M.G.; Bousso, P.; Joanny, J.F.; et al. Macropinocytosis overcomes directional bias in dendritic cells due to hydraulic resistance and facilitates space exploration. Dev. Cell 2019, 49, 171–188. [Google Scholar] [CrossRef]
- Lo, C.M.; Wang, H.B.; Dembo, M.; Wang, Y.L. Cell movement is guided by the rigidity of the substrate. Biophys. J. 2000, 79, 144–152. [Google Scholar] [CrossRef]
- Feng, J.; Levine, H.; Mao, X.; Sander, L.M. Cell motility, contact guidance, and durotaxis. Soft Matter 2019, 15, 4856–4864. [Google Scholar] [CrossRef]
- Buenemann, M.; Levine, H.; Rappel, W.J.; Sander, L.M. The role of cell contraction and adhesion in Dictyostelium motility. Biophys. J. 2010, 99, 50–58. [Google Scholar] [CrossRef]
- Parida, L.; Padmanabhan, V. Durotaxis in nematode Caenorhabditis elegans. Biophys. J. 2016, 111, 666–674. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Parida, L.; Neogi, S.; Padmanabhan, V. Effect of temperature pre-exposure on the locomotion and chemotaxis of C. elegans. PLoS ONE 2014, 9, e111342. [Google Scholar] [CrossRef] [PubMed]
- Isenberg, B.C.; DiMilla, P.A.; Walker, M.; Kim, S.; Womg, J.Y. Vascular smooth muscle cell durotaxis depends on substrate stiffness gradient strength. Biophys. J. 2009, 97, 1313–1322. [Google Scholar] [CrossRef] [PubMed]
- Vincent, L.G.; Choi, I.S.; Alonso-Latorre, B.; Del Alamo, J.C.; Engler, A.J. Mesenchymal stem cell durotaxis depends on substrate stiffness gradient strength. Biotechnol. J. 2013, 8, 472–484. [Google Scholar] [CrossRef]
- DuChez, B.J.; Doyle, A.D.; Dimitriadis, E.K.; Yamada, K.M. Durotaxis by human cancer cells. Biophys. J. 2019, 116, 670–683. [Google Scholar] [CrossRef]
- Liu, C.; Pei, H.; Tan, F. Matrix stiffness and colorectal cancer. OncoTargets Ther. 2020, 13, 2747–2755. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Kim, D.H.; Levchenko, A. Topotaxis: A new mechanism of directed cell migration in topographic ECM gradients. Biophys. J. 2018, 114, 1257–1263. [Google Scholar] [CrossRef]
- Kim, D.H.; Han, K.; Gupta, K.; Kwon, K.W.; Suh, K.Y.; Levchenko, A. Mechanosensitivity of fibroblast cell shape and movement to anisotropic substratum topography gradients. Biomaterials 2009, 30, 5433–5444. [Google Scholar] [CrossRef]
- Trepat, X.; Fredberg, J.J. Plithotaxis and emergent dynamics in collective cellular migration. Trends Cell Biol. 2011, 21, 638–646. [Google Scholar] [CrossRef]
- Kippenberger, S.; Bernd, A.; Loitsch, S.; Guschel, M.; Muller, J.; Bereiter-Hahn, J.; Kaufmann, R. Signaling of mechanical stretch in human keratinocytes via MAP kinases. J. Investig. Dermatol. 2000, 114, 408–412. [Google Scholar] [CrossRef]
- Nabeshima, K.; Inoue, T.; Shimao, Y.; Kataoka, H.; Koono, M. Cohort migration of carcinoma cells: Differentiated colorectal carcinoma cells move as coherent cell clusters or sheets. Histol. Histopathol. 1999, 14, 1183–1197. [Google Scholar] [PubMed]
- Jeon, K.W. The large, free-living amoebae: Wonderful cells for biological studies. J. Eukaryot. Microbiol. 1995, 42, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Korohoda, W.; Mycielska, M.; Janda, E.; Madeja, Z. Immediate and long-term galvanotactic responses of Amoeba proteus to electric fields. Cell Motil. Cytoskeleton. 2000, 45, 10–26. [Google Scholar] [CrossRef]
- Mycielska, M.E.; Djamgoz, M.B.A. Cellular mechanisms of direct-current electric field affects: Galvanotaxis and metastatic disease. J. Cell Sci. 2004, 117, 1631–1639. [Google Scholar] [CrossRef]
- Borys, P. The role of passive calcium influx through the cell membrane in galvanotaxis. Cell. Mol. Biol. Lett. 2013, 18, 187–199. [Google Scholar] [CrossRef]
- Pollard, T.D. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 451–477. [Google Scholar] [CrossRef]
- Wang, E.; Zhan, M.; Forrester, J.V.; McCraig, C.D. Reorientation and faster directed migration of lens epithelial cells in a physiological electric field. Exp. Eye Res. 2000, 71, 91–98. [Google Scholar] [CrossRef]
- Frazer, S.P.; Diss, J.K.J.; Mycielska, M.E.; Coombes, R.C.; Djamgoz, M.B.A. Voltage-gated sodium channel expression in human breast cancer: Possible functional role in metastasis. Breast Cancer Res. Treat. 2002, 76, S142. [Google Scholar]
- Cahalan, M.D.; Wulff, H.; Chandy, K.G. Molecular properties and physiological roles of ion channels in the immune system. J. Clin. Immunol. 2001, 21, 235–252. [Google Scholar] [CrossRef]
- Shu, L.; Zhang, B.; Queller, D.C.; Strassmann, J.E. Burkholderia bacteria use chemotaxis to find social amoeba Dictyostelium discoideum hosts. ISME J. 2018, 12, 1977–1993. [Google Scholar] [CrossRef]
- Hirata, E.; Sahai, E. Tumor microenvironment and differential responses to therapy. Cold Spring Harb. Perspect. Med. 2017, 7, a026781. [Google Scholar] [CrossRef] [PubMed]
- Marques, P.; Barry, S.; Carlsen, E.; Collier, D.; Ronaldson, A.; Awad, S.; Dorward, N.; Grieve, J.; Mendoza, N.; Muquit, S.; et al. Chemokines modulate the tumour microenvironment in pituitary neuroendocrine tumours. Acta Neuropathol. Commun. 2019, 7, 172. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.; Larrinaga, G.; Errarte, P.; Martín, A.M.; Dotor, A.; Esquinas, C.; Nunes-Xavier, C.E.; Pulido, R.; López, J.I.; Angulo, J.C. The coexpression of fibroblast activation protein (FAP) and basal-type markers (CK5/6 and CD44) predicts prognosis in high-grade invasive urothelial carcinoma of the bladder. Hum. Pathol. 2019, 91, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Errarte, P.; Larrinaga, G.; López, J.I. The role of cancer-associated fibroblasts in renal cell carcinoma. An example of tumor modulation through tumor/non-tumor cell interactions. J. Adv. Res. 2019, 21, 103–108. [Google Scholar] [CrossRef]
- Xiang, H.; Ramil, C.P.; Hai, J.; Zhang, C.; Wang, H.; Watkins, A.A.; Afshar, R.; Georgiev, P.; Sze, M.A.; Song, X.S.; et al. Cancer-associated fibroblasts promote immunosuppression by inducing ROS-generating monocytic MDSCs in lung squamous cell carcinoma. Cancer Immunol. Res. 2020. [Google Scholar] [CrossRef]
- Angulo, J.C.; Shapiro, O. The changing therapeutic landscape of metastatic renal cancer. Cancers 2019, 11, E1227. [Google Scholar] [CrossRef]
- Nunes-Xavier, C.E.; Angulo, J.C.; Pulido, R.; López, J.I. A critical insight into the clinical translation of PD-1/PD-L1 blockade therapy in clear cell renal cell carcinoma. Curr. Urol. Rep. 2019, 20, 1. [Google Scholar] [CrossRef]
- Xinran, W.; Liu, Y. PD-L1 expression in tumor infiltrated lymphocytes predicts survival in triple-negative breast cancer. Pathol. Res. Pract. 2020, 216, 152802. [Google Scholar]
- Evans, M.; O’Sullivan, B.; Hughes, F.; Mullis, T.; Smith, M.; Trim, N.; Taniere, P. The clinicopathological and molecular associations of PD-L1 expression in non-small cell lung cancer: Analysis of a series of 10,005 cases tested with the 22C3 assay. Pathol. Oncol. Res. 2018. [Google Scholar] [CrossRef]
- Morsch, R.; Rose, M.; Maurer, A.; Cassataro, M.A.; Braunschweig, T.; Knüchel, R.; Vögeli, T.A.; Ecke, T.; Eckstein, M.; Weyerer, V.; et al. Therapeutic implications of PD-L1 expression in bladder cancer with squamous differentiation. BMC Cancer 2020, 20, 230. [Google Scholar] [CrossRef]
- Wu, H.; Xia, L.; Jia, D.; Zou, H.; Jin, G.; Qian, W.; Xu, H.; Li, T. PD-L1+ regulatory B cells act as a T-cell suppressor in a PD-L1-dependent manner in melanoma patients with bone metastasis. Mol. Immunol. 2020, 119, 83–91. [Google Scholar] [CrossRef]
- Graham, D.M.; Andersen, T.; Sharek, L.; Uzer, G.; Rothenberg, K.; Hoffman, B.D.; Rubin, J.; Balland, M.; Bear, J.E.; Burridge, K. Enucleated cells reveal differential roles of the nucleus in cell migration, polarity, and mechanotransduction. J. Cell Biol. 2018, 217, 895–914. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, R.J. Do migrating cells need a nucleus? J. Cell Biol. 2018, 217, 799–801. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente, I.M.; Bringas, C.; Malaina, I.; Regner, B.; Pérez-Samartin, A.; Boyano, M.D.; Fedetz, M.; López, J.I.; Pérez-Yarza, G.; Cortés, J.M.; et al. The nucleus does not significantly affect the migratory trajectories of amoeba in two-dimensional environments. Sci. Rep. 2019, 9, 16369. [Google Scholar] [CrossRef] [PubMed]
- Bringas, C.; Malaina, I.; Pérez-Samartín, A.; Boyano, M.D.; Fedetz, M.; Pérez-Yarza, G.; Cortés, J.M.; de la Fuente, I.M. Long-term memory in the migration movements of enucleated Amoeba Proteus. BioRxiv 2017. [Google Scholar] [CrossRef]
- Viswanathan, G.M.; Afanasyev, V.; Buldyrev, S.V.; Murphy, E.J.; Prince, P.A.; Stanley, H.E. Lévy flight search patterns of wandering albatrosses. Nature 1996, 381, 413–415. [Google Scholar] [CrossRef]
- Ivanov, P.C.; Amaral, L.A.; Goldberger, A.L.; Havlin, S.; Rosenblum, M.G.; Strzuik, Z.R.; Stanley, H.E. Multifractality in human heartbeat dynamics. Nature 1999, 399, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Einstein, A. Über die von der molekularkinetischen Theorie der wärme geforderte Bewegung von in ruhenden Flüssigkeiten suspendierten Teilchen. Ann. Phys. 1905, 322, 549–560. [Google Scholar] [CrossRef]
- Wilson, K. Renormalization group and critical phenomena II: Phase space cell analysis of critical behavior. Phys. Rev. B 1971, 4, 3184. [Google Scholar] [CrossRef]
- De la Fuente, I.M. Elements of the cellular metabolic structure. Front. Mol. Biosci. 2015, 2, 16. [Google Scholar] [CrossRef]
- Senoo, H.; Cai, H.; Wang, Y.; Sesaki, H.; Iijima, M. The novel RacE-binding protein GflB sharpens Ras activity at the leading edge of migrating cells. Mol. Biol. Cell 2016, 27, 1596–1605. [Google Scholar] [CrossRef] [PubMed]
- Pavlov, I.P. Conditioned Reflexes: An Investigation of the Physiological Activity of the Cerebral Cortex; Oxford University Press: Oxford, UK, 1927. [Google Scholar]
- De la Fuente, I.M.; Bringas, C.; Malaina, I.; Fedetz, M.; Carrasco-Pujante, J.; Morales, M.; Knafo, S.; Martinez, L.; Pérez-Samartín, A.; López, J.I.; et al. Evidence of conditioned behavior in amoebae. Nat. Commun. 2019, 10, 3690. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente, I.M.; Cortés, J.M.; Pelta, D.A.; Veguillas, J. Attractor metabolic networks. PLoS ONE 2013, 8, e58284. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De la Fuente, I.M.; López, J.I. Cell Motility and Cancer. Cancers 2020, 12, 2177. https://doi.org/10.3390/cancers12082177
De la Fuente IM, López JI. Cell Motility and Cancer. Cancers. 2020; 12(8):2177. https://doi.org/10.3390/cancers12082177
Chicago/Turabian StyleDe la Fuente, Ildefonso M., and José I. López. 2020. "Cell Motility and Cancer" Cancers 12, no. 8: 2177. https://doi.org/10.3390/cancers12082177
APA StyleDe la Fuente, I. M., & López, J. I. (2020). Cell Motility and Cancer. Cancers, 12(8), 2177. https://doi.org/10.3390/cancers12082177





