Mathematical Modeling of MPNs Offers Understanding and Decision Support for Personalized Treatment
Abstract
1. Introduction
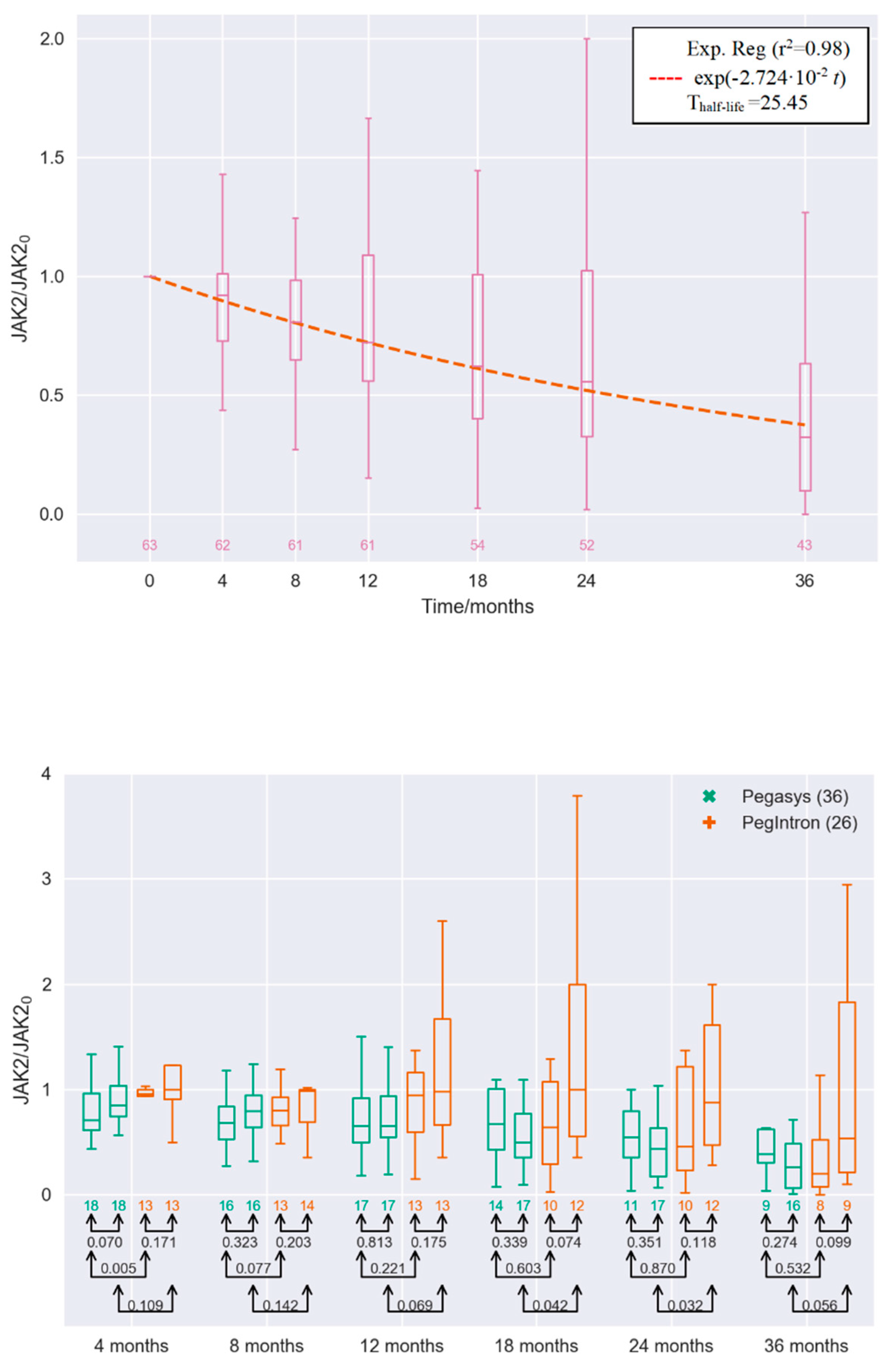
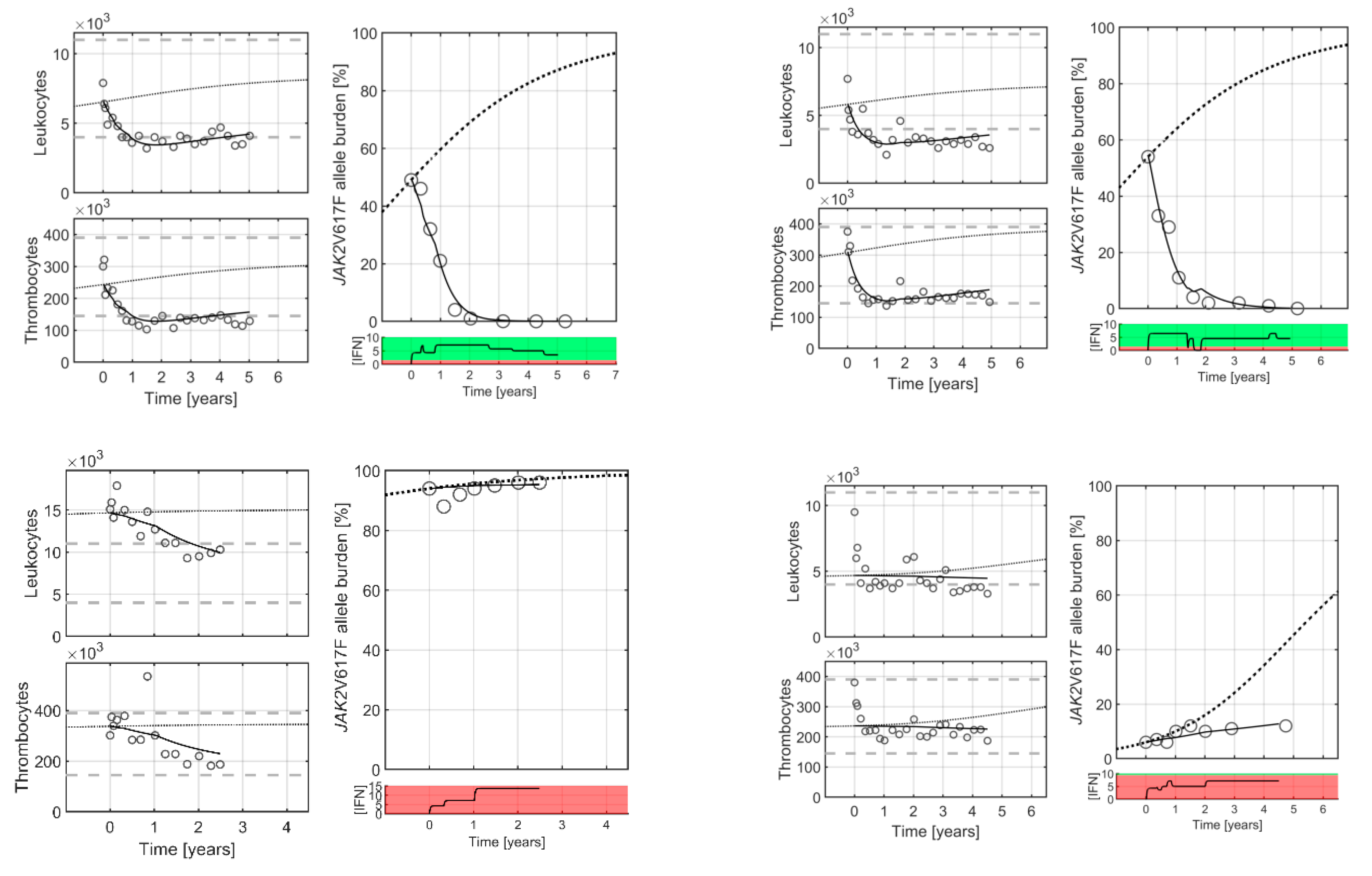
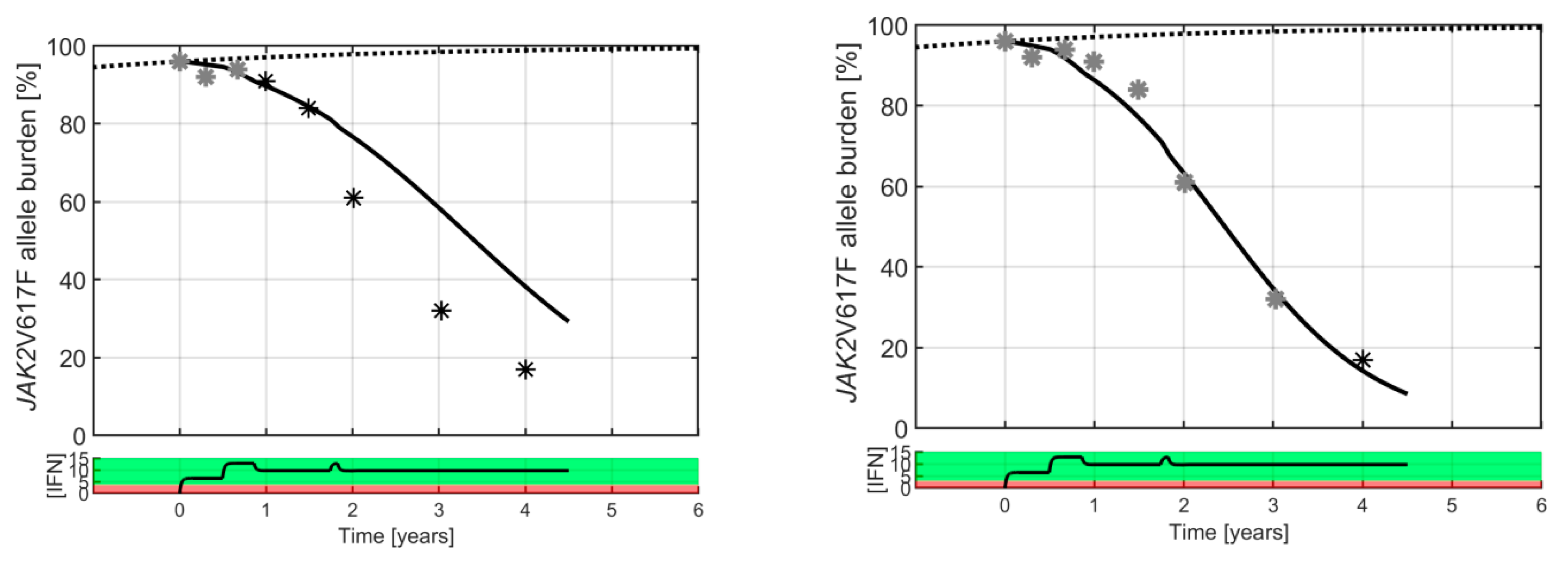
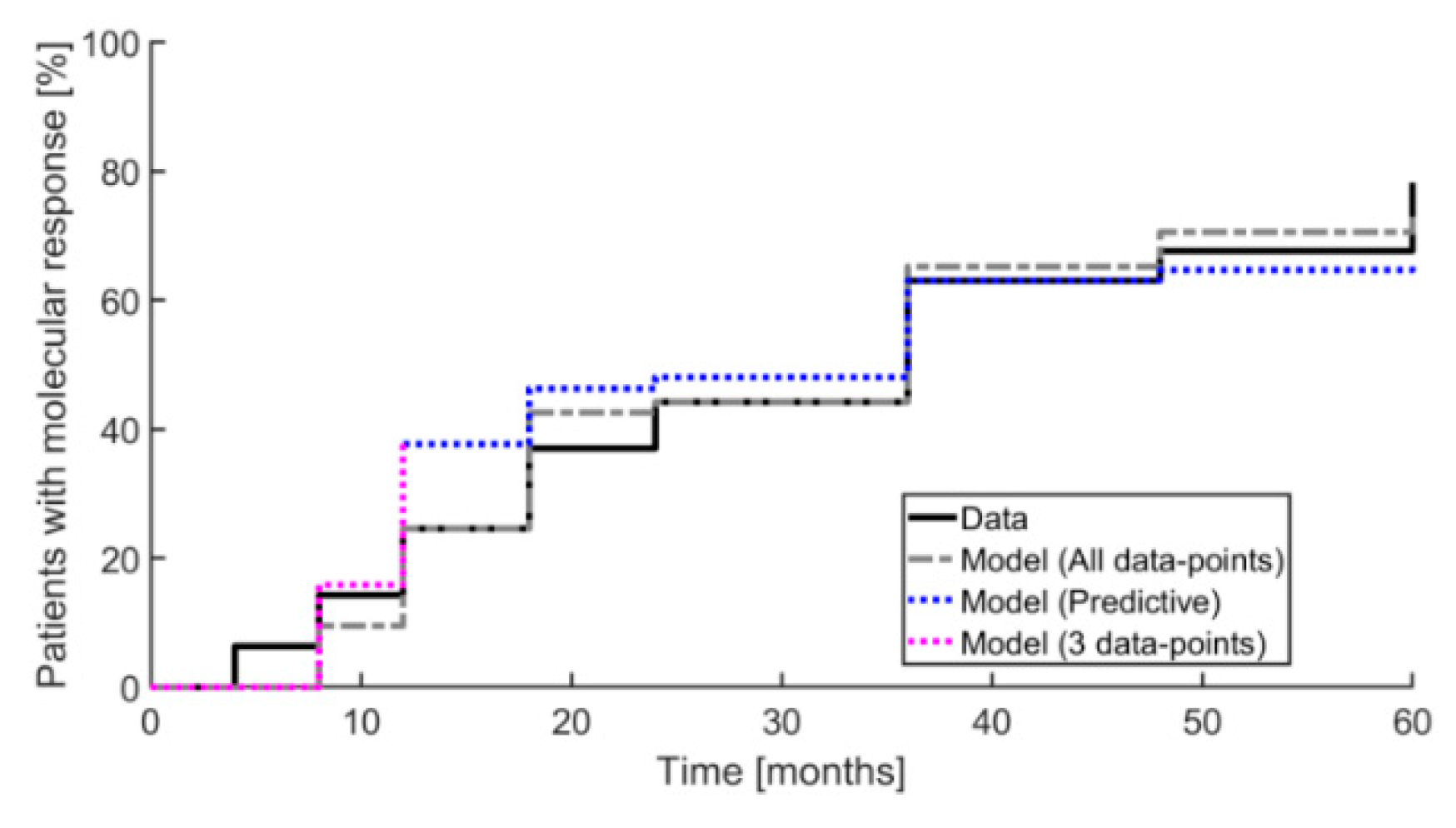
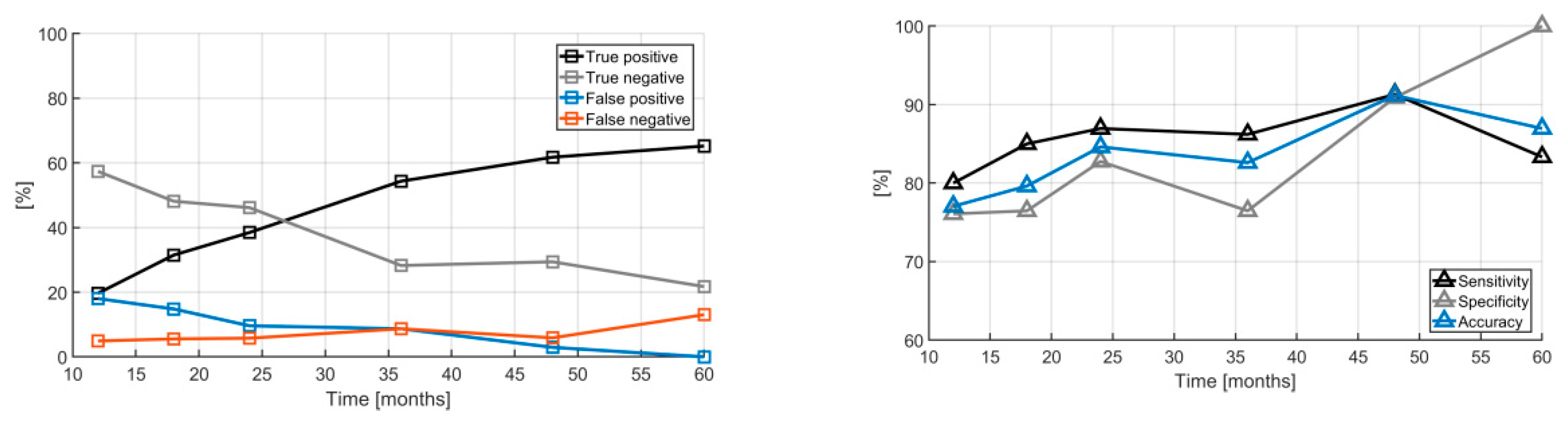
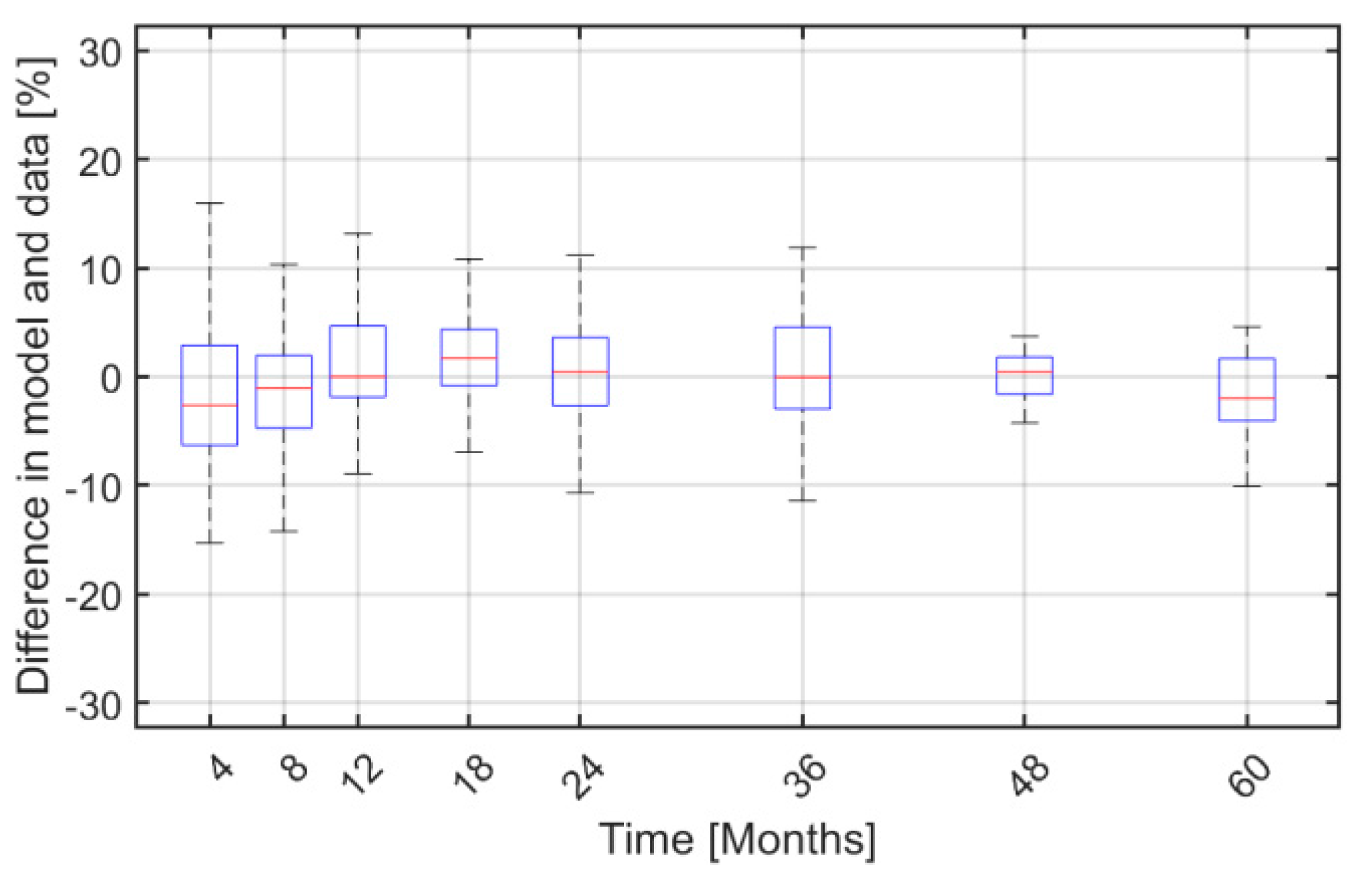
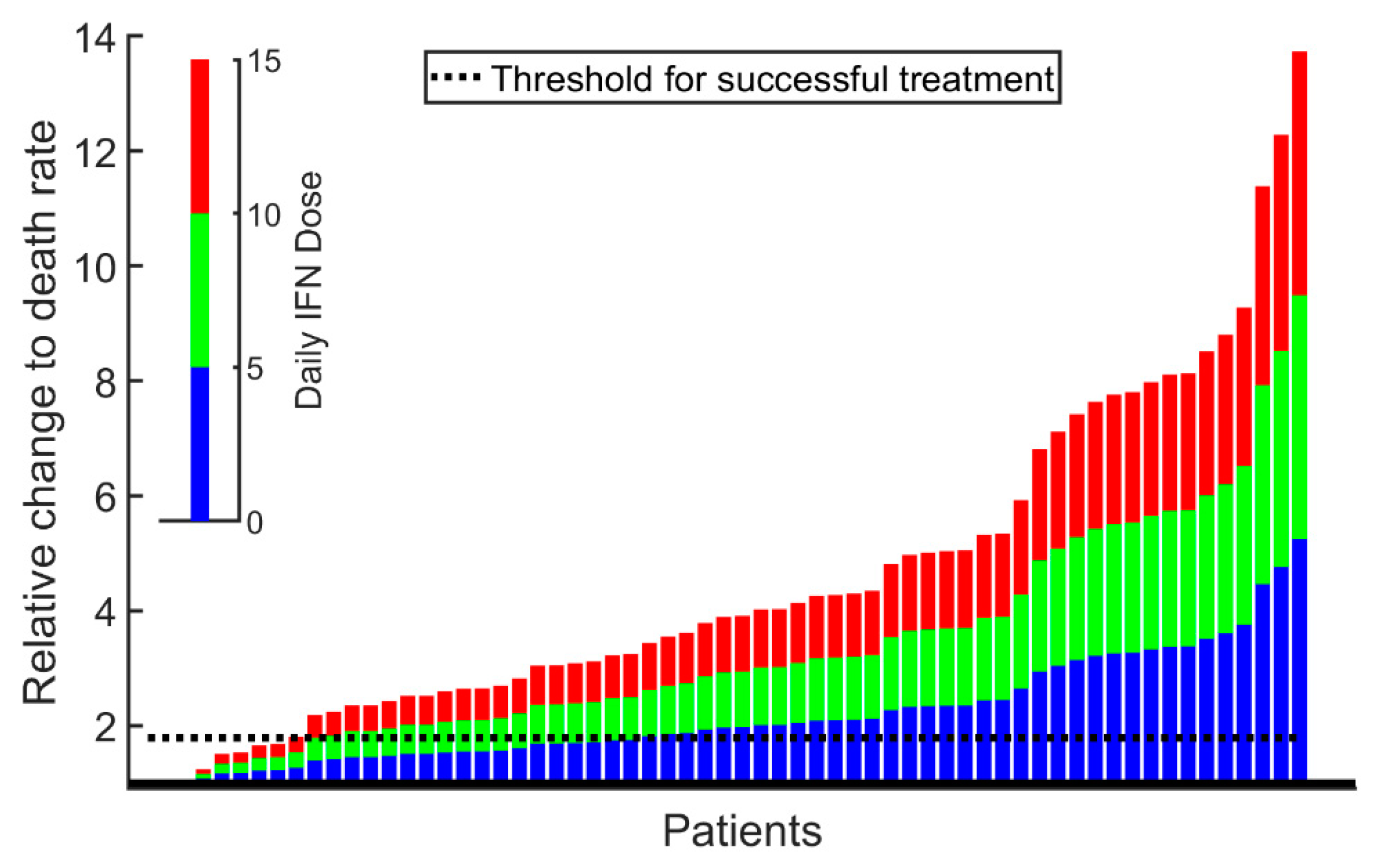
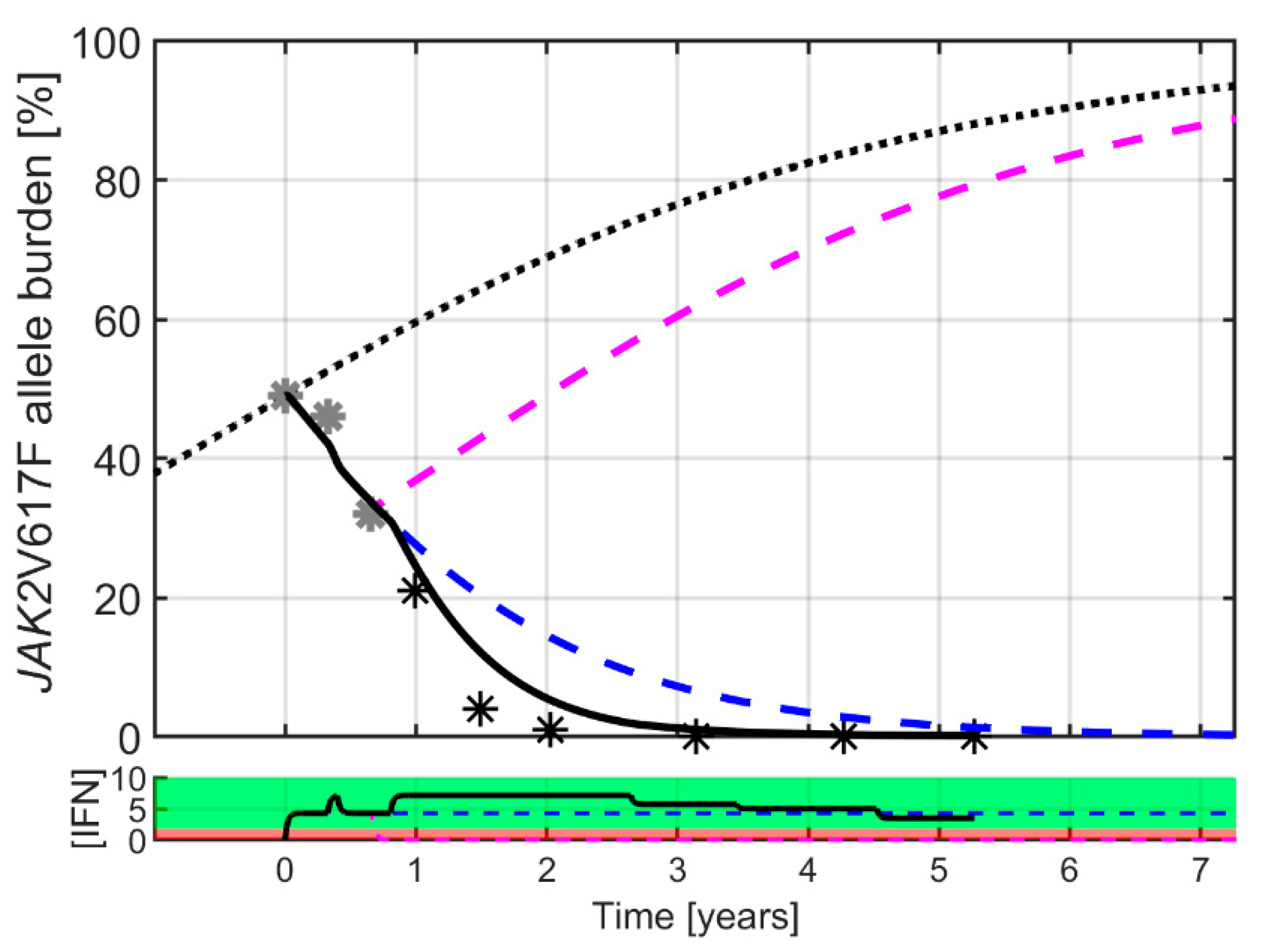
2. Results
3. Discussion
4. Materials and Methods
4.1. Clinical Study Design
4.2. Mathematical Study Design
4.2.1. The Cancitis Model
4.2.2. Mathematical Analysis Design
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Orkin, S.H.; Zon, L.I. Hematopoiesis: An Evolving Paradigm for Stem Cell Biology. Cell 2008, 132, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Vaziri, H.; Dragowska, W.; Allsopp, R.C.; Thomas, T.E.; Harley, C.B.; Lansdorp, P.M. Evidence for a mitotic clock in human hematopoietic stem cells: Loss of telomeric DNA with age. Proc. Natl. Acad. Sci. USA 1994, 91, 9857–9860. [Google Scholar] [CrossRef] [PubMed]
- Anderson, L.A.; McMullin, M.F. Epidemiology of MPN: What Do We Know? Curr. Hematol. Malign- Rep. 2014, 9, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Campbell, P.J.; Green, A.R. The Myeloproliferative Disorders. N. Engl. J. Med. 2006, 355, 2452–2466. [Google Scholar] [CrossRef]
- Marchioli, R.; Finazzi, G.; Landolfi, R.; Kutti, J.; Gisslinger, H.; Patrono, C.; Marilus, R.; Villegas, A.; Tognoni, G.; Barbui, T. Vascular and Neoplastic Risk in a Large Cohort of Patients With Polycythemia Vera. J. Clin. Oncol. 2005, 23, 2224–2232. [Google Scholar] [CrossRef]
- Björkholm, M.; Derolf, Å.R.; Hultcrantz, M.L.; Kristinsson, S.Y.; Ekstrand, C.; Goldin, L.R.; Andreasson, B.; Birgegård, G.; Linder, O.; Malm, C.; et al. Treatment-Related Risk Factors for Transformation to Acute Myeloid Leukemia and Myelodysplastic Syndromes in Myeloproliferative Neoplasms. J. Clin. Oncol. 2011, 29, 2410–2415. [Google Scholar] [CrossRef]
- Roaldsnes, C.; Holst, R.; Frederiksen, H.; Ghanima, W. Myeloproliferative neoplasms: Trends in incidence, prevalence and survival in Norway. Eur. J. Haematol. 2016, 98, 85–93. [Google Scholar] [CrossRef]
- Titmarsh, G.J.; Duncombe, A.S.; McMullin, M.F.; O’Rorke, M.; Mesa, R.; De Vocht, F.; Horan, S.; Fritschi, L.; Clarke, M.; Anderson, L.A. How common are myeloproliferative neoplasms? A systematic review and meta-analysis. Am. J. Hematol. 2014, 89, 581–587. [Google Scholar] [CrossRef]
- Niida, A.; Hasegawa, T.; Innan, H.; Shibata, T.; Mimori, K.; Miyano, S. A unified simulation model for understanding the diversity of cancer evolution. PeerJ 2020, 8, e8842. [Google Scholar] [CrossRef]
- Gisslinger, H.; Klade, C.; Georgiev, P.; Krochmalczyk, D.; Gercheva-Kyuchukova, L.; Egyed, M.; Rossiev, V.; Dulicek, P.; Illes, A.; Pylypenko, H.; et al. Ropeginterferon alfa-2b versus standard therapy for polycythaemia vera (PROUD-PV and CONTINUATION-PV): A randomised, non-inferiority, phase 3 trial and its extension study. Lancet Haematol. 2020, 7, e196–e208. [Google Scholar] [CrossRef]
- Dunn, G.P.; Old, L.J.; Schreiber, R.D. The Three Es of Cancer Immunoediting. Annu. Rev. Immunol. 2004, 22, 329–360. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, R.K.; Andersen, M.; Knudsen, T.A.; Sajid, Z.; Gudmand-Hoeyer, J.; Dam, M.J.B.; Skov, V.; Kjær, L.; Ellervik, C.; Larsen, T.S.; et al. Data-driven analysis of JAK2V617F kinetics during interferon-alpha2 treatment of patients with polycythemia vera and related neoplasms. Cancer Med. 2020, 9, 2039–2051. [Google Scholar] [CrossRef] [PubMed]
- Altrock, P.M.; Liu, L.L.; Michor, F. The mathematics of cancer: Integrating quantitative models. Nat. Rev. Cancer 2015, 15, 730–745. [Google Scholar] [CrossRef] [PubMed]
- Dingli, D.; Michor, F. Successful Therapy Must Eradicate Cancer Stem Cells. Stem Cells 2006, 24, 2603–2610. [Google Scholar] [CrossRef] [PubMed]
- Byrne, H.M. Dissecting cancer through mathematics: From the cell to the animal model. Nat. Rev. Cancer 2010, 10, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Stiehl, T.; Baran, N.; Ho, A.D.; Marciniak-Czochra, A. Cell Division Patterns in Acute Myeloid Leukemia Stem-like Cells Determine Clinical Course: A Model to Predict Patient Survival. Cancer Res. 2015, 75, 940–949. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Fleischman, A.G.; Wodarz, M.; Komarova, N.L. Determining the role of inflammation in the selection of JAK2 mutant cells in myeloproliferative neoplasms. J. Theor. Boil. 2017, 425, 43–52. [Google Scholar] [CrossRef]
- Haeno, H.; Levine, R.L.; Gilliland, D.G.; Michor, F. A progenitor cell origin of myeloid malignancies. Proc. Natl. Acad. Sci. USA 2009, 106, 16616–16621. [Google Scholar] [CrossRef]
- Andersen, M.; Sajid, Z.; Pedersen, R.K.; Gudmand-Hoeyer, J.; Ellervik, C.; Skov, V.; Kjær, L.; Pallisgaard, N.; Kruse, T.A.; Thomassen, M.; et al. Mathematical modelling as a proof of concept for MPNs as a human inflammation model for cancer development. PLoS ONE 2017, 12, e0183620. [Google Scholar] [CrossRef]
- Ottesen, J.T.; Pedersen, R.K.; Sajid, Z.; Gudmand-Hoeyer, J.; Bangsgaard, K.O.; Skov, V.; Kjær, L.; Knudsen, T.A.; Pallisgaard, N.; Hasselbalch, H.C.; et al. Bridging blood cancers and inflammation: The reduced Cancitis model. J. Theor. Boil. 2019, 465, 90–108. [Google Scholar] [CrossRef]
- Sajid, Z.; Andersen, M.; Ottesen, J.T. Mathematical analysis of the Cancitis model and the role of inflammation in blood cancer progression. Math. Biosci. Eng. 2019, 16, 8268–8289. [Google Scholar] [CrossRef] [PubMed]
- Lane, S.W.; Mullally, A. Jak2V617F myeloproliferative neoplasm stem cells and interferon-alpha. Oncotarget 2013, 4, 500–501. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mullally, A.; Bruedigam, C.; Poveromo, L.; Heidel, F.H.; Purdon, A.; Vu, T.; Austin, R.; Heckl, D.; Breyfogle, L.J.; Kuhn, C.P.; et al. Depletion of Jak2V617F myeloproliferative neoplasm-propagating stem cells by interferon-α in a murine model of polycythemia vera. Blood 2013, 121, 3692–3702. [Google Scholar] [CrossRef] [PubMed]
- King, K.Y.; Matatall, K.A.; Shen, C.-C.; Goodell, M.A.; Swierczek, S.I.; Prchal, J.T. Comparative long-term effects of interferon α and hydroxyurea on human hematopoietic progenitor cells. Exp. Hematol. 2015, 43, 912–918.e2. [Google Scholar] [CrossRef] [PubMed]
- Pietras, E.M.; Lakshminarasimhan, R.; Techner, J.-M.; Fong, S.L.; Flach, J.; Binnewies, M.; Passegué, E. Re-entry into quiescence protects hematopoietic stem cells from the killing effect of chronic exposure to type I interferons. J. Exp. Med. 2014, 211, 245–262. [Google Scholar] [CrossRef]
- Kiladjian, J.-J.; Cassinat, B.; Turlure, P.; Cambier, N.; Roussel, M.; Bellucci, S.; Menot, M.-L.; Massonnet, G.; Dutel, J.-L.; Ghomari, K.; et al. High molecular response rate of polycythemia vera patients treated with pegylated interferon -2a. Blood 2006, 108, 2037–2040. [Google Scholar] [CrossRef]
- Masarova, L.; Patel, K.P.; Newberry, K.J.; Cortes, J.; Borthakur, G.; Konopleva, M.; Estrov, Z.; Kantarjian, H.; Verstovsek, S. Pegylated interferon alfa-2a in patients with essential thrombocythaemia or polycythaemia vera: A post-hoc, median 83 month follow-up of an open-label, phase 2 trial. Lancet Haematol. 2017, 4, e165–e175. [Google Scholar] [CrossRef]
- Kiladjian, J.-J.; Giraudier, S.; Cassinat, B. Interferon-alpha for the therapy of myeloproliferative neoplasms: Targeting the malignant clone. Leukemia 2015, 30, 776–781. [Google Scholar] [CrossRef]
- Quintás-Cardama, A.; Abdel-Wahab, O.; Manshouri, T.; Kilpivaara, O.; Cortes, J.; Roupie, A.L.; Zhang, S.-J.; Harris, D.; Estrov, Z.; Kantarjian, H.; et al. Molecular analysis of patients with polycythemia vera or essential thrombocythemia receiving pegylated interferon α-2a. Blood 2013, 122, 893–901. [Google Scholar] [CrossRef]
- Andersen, M.; Hasselbalch, H.C.; Kjaer, L.; Skov, V.; Ottesen, J.T. Global dynamics of healthy and cancer cells competing in the hematopoietic system. Math. Biosci. 2020, 326, 108372. [Google Scholar] [CrossRef]
- Michor, F.; Hughes, T.P.; Iwasa, Y.; Branford, S.; Shah, N.P.; Sawyers, C.L.; Nowak, M.A. Dynamics of chronic myeloid leukaemia. Nature 2005, 435, 1267–1270. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Gonen, M.; Quintas-Cardama, A.; Cortes, J.; Kantarjian, H.; Field, C.; Hughes, T.; Branford, S.; Michor, F. Dynamics of chronic myeloid leukemia response to long-term targeted therapy reveal treatment effects on leukemic stem cells. Blood 2011, 118, 1622–1631. [Google Scholar] [CrossRef] [PubMed]
- Schürch, C.M.; Riether, C.; Ochsenbein, A.F. Interferons in hematopoiesis and leukemia. OncoImmunology 2013, 2, e24572. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhao, E.; Xu, H.; Wang, L.; Kryczek, I.; Wu, K.; Hu, Y.; Wang, G.; Zou, W. Bone marrow and the control of immunity. Cell. Mol. Immunol. 2011, 9, 11–19. [Google Scholar] [CrossRef]
- Mead, A.J.; Mullally, A. Myeloproliferative neoplasm stem cells. Blood 2017, 129, 1607–1616. [Google Scholar] [CrossRef]
- Foster, G.R. Pegylated Interferons for the Treatment of Chronic Hepatitis C Pharmacological and Clinical Differences between Peginterferon-a-2a and Peginterferon-a-2b. Drugs 2010, 70, 147–165. [Google Scholar] [CrossRef]
- Bruno, R.; Sacchi, P.; Scagnolari, C.; Torriani, F.; Maiocchi, L.; Patruno, S.; Bellomi, F.; Filice, G.; Antonelli, G. Pharmacodynamics of peginterferon alfa-2a and peginterferon alfa-2b in interferon-naïve patients with chronic hepatitis C: A randomized, controlled study. Aliment. Pharmacol. Ther. 2007, 26, 369–376. [Google Scholar] [CrossRef]
- Michallet, M.; Maloisel, F.; DeLain, M.; Hellmann, A.; Rosas, A.; Silver, R.T.; Tendler, C.; for the PEG-Intron CML Study Group. Pegylated recombinant interferon alpha-2b vs recombinant interferon alpha-2b for the initial treatment of chronic-phase chronic myelogenous leukemia: A phase III study. Leukemia 2003, 18, 309–315. [Google Scholar] [CrossRef][Green Version]
- Saito, T.; Iida, S.; Kawanishi, T. Population pharmacokinetic-pharmacodynamic modeling and simulation of platelet decrease induced by peg-interferon-alpha 2a. Drug Metab. Pharmacokinet. 2012, 27, 614–620. [Google Scholar] [CrossRef]
- Tefferi, A.; Cervantes, F.; Mesa, R.; Passamonti, F.; Verstovsek, S.; Vannucchi, A.M.; Gotlib, J.; Dupriez, B.; Pardanani, A.; Harrison, C.; et al. Revised response criteria for myelofibrosis: International Working Group-Myeloproliferative Neoplasms Research and Treatment (IWG-MRT) and European LeukemiaNet (ELN) consensus report. Blood 2013, 122, 1395–1398. [Google Scholar] [CrossRef]
- Barosi, G.; Mesa, R.; Finazzi, G.; Harrison, C.; Kiladjian, J.-J.; Lengfelder, E.; McMullin, M.F.; Passamonti, F.; Vannucchi, A.M.; Besses, C.; et al. Revised response criteria for polycythemia vera and essential thrombocythemia: An ELN and IWG-MRT consensus project. Blood 2013, 121, 4778–4781. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ottesen, J.T.; Pedersen, R.K.; Dam, M.J.B.; Knudsen, T.A.; Skov, V.; Kjær, L.; Andersen, M. Mathematical Modeling of MPNs Offers Understanding and Decision Support for Personalized Treatment. Cancers 2020, 12, 2119. https://doi.org/10.3390/cancers12082119
Ottesen JT, Pedersen RK, Dam MJB, Knudsen TA, Skov V, Kjær L, Andersen M. Mathematical Modeling of MPNs Offers Understanding and Decision Support for Personalized Treatment. Cancers. 2020; 12(8):2119. https://doi.org/10.3390/cancers12082119
Chicago/Turabian StyleOttesen, Johnny T., Rasmus K. Pedersen, Marc J. B. Dam, Trine A. Knudsen, Vibe Skov, Lasse Kjær, and Morten Andersen. 2020. "Mathematical Modeling of MPNs Offers Understanding and Decision Support for Personalized Treatment" Cancers 12, no. 8: 2119. https://doi.org/10.3390/cancers12082119
APA StyleOttesen, J. T., Pedersen, R. K., Dam, M. J. B., Knudsen, T. A., Skov, V., Kjær, L., & Andersen, M. (2020). Mathematical Modeling of MPNs Offers Understanding and Decision Support for Personalized Treatment. Cancers, 12(8), 2119. https://doi.org/10.3390/cancers12082119




