MicroRNA-21-Enriched Exosomes as Epigenetic Regulators in Melanomagenesis and Melanoma Progression: The Impact of Western Lifestyle Factors
Abstract
1. Introduction
2. Methodological Approach
3. Mechanism of Action of MiR-21 in Melanoma and Melanoma Cells
3.1. MiR-21 in Melanoma Pathogenesis and Progression
3.2. MiR-21 Targets in Melanoma Cells
3.3. Endogenous Upregulation of MiR-21 in Melanoma
3.3.1. Signal Transducer and Activator of Transcription 3
3.3.2. Hippo Pathway
3.3.3. Long Non-Coding RNAs
4. Exosome-Mediated MiR-21 Transport
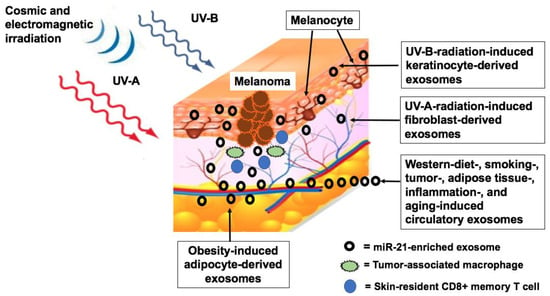
4.1. Exosome-Mediated Transfer of MiR-21 to Melanocytic Lesions
4.2. Melanoma-Derived MiR-21-Enriched Exosomes and Melanoma Progression
4.3. Immunological Surveillance
4.4. MiR-21 Overexpression in Melanoma-Related Tumors
5. Environmental Factors Upregulating MiR-21
5.1. Radiation
5.1.1. Ultraviolet Irradiation
5.1.2. Cosmic Ionizing Irradiation
5.1.3. Electromagnetic Radiation
5.2. Metabolic Deviations Upregulating MiR-21
5.2.1. Metabolic Syndrome and Melanoma Risk
5.2.2. Birth Weight and Height in Childhood
5.2.3. Overweight and Obesity
5.2.4. Diabetes Mellitus
5.2.5. Arterial Hypertension
5.2.6. Western Diet
5.2.7. Smoking and Pollution
5.3. Hormonal Factors
5.3.1. Androgens
5.3.2. Growth Hormone
5.3.3. Vitamin D
5.4. Aging and Chronic Inflammation
5.4.1. Aging
5.4.2. Chronic Inflammation
6. Therapeutic Suppression of MiR-21
6.1. Vemurafenib
6.2. Metformin
6.3. Beta-Blocker
6.4. Anti-MiR-21
6.5. Interferons
6.6. High-Intensity Focused Ultrasound
6.7. Iontophoretic Co-Delivery of STAT3 siRNA and Imatinib
6.8. Curcumin
6.9. Sulforaphane
6.10. Epigallocatechin-3-Gallate
6.11. Vitamin D
6.12. Exercise
7. Conclusions and Perspectives
Funding
Conflicts of Interest
Abbreviations
References
- Rastrelli, M.; Tropea, S.; Rossi, C.R.; Alaibac, M. Melanoma: Epidemiology, risk factors, pathogenesis, diagnosis and classification. In Vivo 2014, 28, 1005–1011. [Google Scholar]
- Hallberg, O.; Johansson, O. Malignant melanoma of the skin—Not a sunshine story! Med. Sci. Monit. 2004, 10, CR336–CR340. [Google Scholar]
- Whiteman, D.C.; Green, A.C.; Olsen, C.M. The growing burden of invasive melanoma: Projections of incidence rates and numbers of new cases in six susceptible populations through 2031. J. Investig. Dermatol. 2016, 136, 1161–1171. [Google Scholar] [CrossRef]
- Garbe, C.; Keim, U.; Eigentler, T.K.; Amaral, T.; Katalinic, A.; Holleczek, B.; Martus, P.; Leiter, U. Time trends in incidence and mortality of cutaneous melanoma in Germany. J. Eur. Acad. Dermatol. Venereol. 2019, 33, 1272–1280. [Google Scholar] [CrossRef] [PubMed]
- Garraway, L.A.; Widlund, H.R.; Rubin, M.A.; Getz, G.; Berger, A.J.; Ramaswamy, S.; Beroukhim, R.; Milner, D.A.; Granter, S.R.; Du, J.; et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 2005, 436, 117–122. [Google Scholar] [CrossRef]
- Berger, M.F.; Garraway, L.A. Applications of genomics in melanoma oncogene discovery. Hematol. Oncol. Clin. N. Am. 2009, 23, 397–414. [Google Scholar] [CrossRef] [PubMed]
- Bastian, B.C. The molecular pathology of melanoma: An integrated taxonomy of melanocytic neoplasia. Annu. Rev. Pathol. 2014, 9, 239–271. [Google Scholar] [CrossRef] [PubMed]
- Kunz, M. Oncogenes in melanoma: An update. Eur. J. Cell Biol. 2014, 93, 1–10. [Google Scholar] [CrossRef]
- Leonardi, G.C.; Falzone, L.; Salemi, R.; Zanghì, A.; Spandidos, D.A.; Mccubrey, J.A.; Candido, S.; Libra, M. Cutaneous melanoma: From pathogenesis to therapy (Review). Int. J. Oncol. 2018, 52, 1071–1080. [Google Scholar] [CrossRef]
- Lorentzen, H.F. Targeted therapy for malignant melanoma. Curr. Opin. Pharmacol. 2019, 46, 116–121. [Google Scholar] [CrossRef]
- Reu, F.J.; Bae, S.I.; Cherkassky, L.; Leaman, D.W.; Lindner, D.; Beaulieu, N.; MacLeod, A.R.; Borden, E.C. Overcoming resistance to interferon-induced apoptosis of renal carcinoma and melanoma cells by DNA demethylation. J. Clin. Oncol. 2006, 24, 3771–3779. [Google Scholar] [CrossRef] [PubMed]
- De Vries, I.J.; Castelli, C.; Huygens, C.; Jacobs, J.F.; Stockis, J.; Schuler-Thurner, B.; Adema, G.J.; Punt, C.J.; Rivoltini, L.; Schuler, G.; et al. Frequency of circulating Tregs with demethylated FOXP3 intron 1 in melanoma patients receiving tumor vaccines and potentially Treg-depleting agents. Clin. Cancer Res. 2011, 17, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Micevic, G.; Theodosakis, N.; Bosenberg, M. Aberrant DNA methylation in melanoma: Biomarker and therapeutic opportunities. Clin. Epigenet. 2017, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- De Unamuno Bustos, B.; Murria Estal, R.; Pérez Simó, G.; Simarro Farinos, J.; Pujol Marco, C.; Navarro Mira, M.; Alegre de Miquel, V.; Ballester Sánchez, R.; Sabater Marco, V.; Llavador Ros, M.; et al. Aberrant DNA methylation is associated with aggressive clinicopathological features and poor survival in cutaneous melanoma. Br. J. Dermatol. 2018, 179, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Bonazzi, V.F.; Stark, M.S.; Hayward, N.K. MicroRNA regulation of melanoma progression. Melanoma Res. 2012, 22, 101–113. [Google Scholar] [CrossRef]
- Völler, D.; Ott, C.; Bosserhoff, A. MicroRNAs in malignant melanoma. Clin. Biochem. 2013, 46, 909–917. [Google Scholar] [CrossRef]
- Luo, C.; Weber, C.E.; Osen, W.; Bosserhoff, A.K.; Eichmüller, S.B. The role of microRNAs in melanoma. Eur. J. Cell Biol. 2014, 93, 11–22. [Google Scholar] [CrossRef]
- Sun, V.; Zhou, W.B.; Majid, S.; Kashani-Sabet, M.; Dar, A.A. MicroRNA-mediated regulation of melanoma. Br. J. Dermatol 2014, 171, 234–241. [Google Scholar] [CrossRef]
- Sarkar, D.; Leung, E.Y.; Baguley, B.C.; Finlay, G.J.; Askarian-Amiri, M.E. Epigenetic regulation in human melanoma: Past and future. Epigenetics 2016, 10, 103–121. [Google Scholar] [CrossRef]
- Mirzaei, H.; Gholamin, S.; Shahidsales, S.; Sahebkar, A.; Jaafari, M.R.; Mirzaei, H.R.; Hassanian, S.M.; Avan, A. MicroRNAs as potential diagnostic and prognostic biomarkers in melanoma. Eur. J. Cancer 2016, 53, 25–32. [Google Scholar] [CrossRef]
- Fattore, L.; Costantini, S.; Malpicci, D.; Ruggiero, C.F.; Ascierto, P.A.; Croce, C.M.; Mancini, R.; Ciliberto, G. MicroRNAs in melanoma development and resistance to target therapy. Oncotarget 2017, 8, 22262–22278. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Sendra, B.; Martinez-Ciarpaglini, C.; González-Muñoz, J.F.; Murgui, A.; Terrádez, L.; Monteagudo, C. Downregulation of intratumoral expression of miR-205, miR-200c and miR-125b in primary human cutaneous melanomas predicts shorter survival. Sci. Rep. 2018, 8, 17076. [Google Scholar] [CrossRef] [PubMed]
- Varrone, F.; Caputo, E. The miRNAs role in melanoma and in its resistance to therapy. Int. J. Mol. Sci. 2020, 2, E878. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.H.; Tsao, C.J. Emerging role of microRNA-21 in cancer. Biomed. Rep. 2016, 5, 395–402. [Google Scholar] [CrossRef]
- Pan, X.; Wang, Z.X.; Wang, R. MicroRNA-21: A novel therapeutic target in human cancer. Cancer Biol. Ther. 2010, 10, 1224–1232. [Google Scholar] [CrossRef] [PubMed]
- Latchana, N.; Del Campo, S.E.; Grignol, V.P.; Clark, J.R.; Albert, S.P.; Zhang, J.; Wei, L.; Aldrink, J.H.; Nicol, K.K.; Ranalli, M.A.; et al. Classification of indeterminate melanocytic lesions by microRNA profiling. Ann. Surg. Oncol. 2017, 24, 347–354. [Google Scholar] [CrossRef]
- Wandler, A.; Riber-Hansen, R.; Hager, H.; Hamilton-Dutoit, S.J.; Schmidt, H.; Nielsen, B.S.; Stougaard, M.; Steiniche, T. Quantification of microRNA-21 and microRNA-125b in melanoma tissue. Melanoma Res. 2017, 27, 417–428. [Google Scholar] [CrossRef]
- Ferracin, M.; Lupini, L.; Salamon, I.; Saccenti, E.; Zanzi, M.V.; Rocchi, A.; Da Ros, L.; Zagatti, B.; Musa, G.; Bassi, C. Absolute quantification of cell-free microRNAs in cancer patients. Oncotarget 2015, 6, 14545–14555. [Google Scholar] [CrossRef]
- Carpi, S.; Polini, B.; Fogli, S.; Podestà, A.; Ylösmäki, E.; Cerullo, V.; Romanini, A.; Nieri, P. Circulating microRNAs as biomarkers for early diagnosis of cutaneous melanoma. Expert Rev. Mol. Diagn. 2020, 20, 19–30. [Google Scholar] [CrossRef]
- Neagu, M.; Constantin, C.; Cretoiu, S.M.; Zurac, S. miRNAs in the diagnosis and prognosis of skin cancer. Front. Cell Dev. Biol. 2020, 8, 71. [Google Scholar] [CrossRef]
- Satzger, I.; Mattern, A.; Kuettler, U.; Weinspach, D.; Niebuhr, M.; Kapp, A.; Gutzmer, R. microRNA-21 is upregulated in malignant melanoma and influences apoptosis of melanocytic cells. Exp. Dermatol. 2012, 21, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Liao, B.; Xiang, X.; Ke, S. MiR-21-5p promotes cell proliferation and G1/S transition in melanoma by targeting CDKN2C. FEBS Open Bio 2020, 10, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Lv, X.; Li, J.; Li, J.; Li, X.; Li, W.; Li, Y. The status of microRNA-21 expression and its clinical significance in human cutaneous malignant melanoma. Acta Histochem. 2012, 114, 582–588. [Google Scholar] [CrossRef]
- Grignol, V.; Fairchild, E.T.; Zimmerer, J.M.; Lesinski, G.B.; Walker, M.J.; Magro, C.M.; Kacher, J.E.; Karpa, V.I.; Clark, J.; Nuovo, G.; et al. miR-21 and miR-155 are associated with mitotic activity and lesion depth of borderline melanocytic lesions. Br. J. Cancer 2011, 105, 1023–1029. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wie, W. The miRNA expression profile of the uveal melanoma. Sci. China Life Sci. 2011, 54, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Torres, R.; Lang, U.E.; Hejna, M.; Shelton, S.J.; Joseph, N.M.; Shain, A.H.; Yeh, I.; Wei, M.L.; Oldham, M.C.; Batian, B.C.; et al. MicroRNA ratios distinguish melanomas from nevi. J. Investig. Dermatol. 2020, 140, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Mo, H.; Guan, J.; Yuan, Z.C.; Lin, X.; Wu, Z.J.; Liu, B.; He, J.L. Expression and predictive value of miR-489 and miR-21 in melanoma metastasis. World J. Clin. Cases 2019, 7, 2930–2941. [Google Scholar] [CrossRef] [PubMed]
- Xi, J.; Huang, Q.; Wang, L.; Ma, X.; Deng, Q.; Kumar, M.; Zhou, Z.; Li, L.; Zeng, Z.; Young, K.H.; et al. miR-21 depletion in macrophages promotes tumoricidal polarization and enhances PD-1 immunotherapy. Oncogene 2018, 37, 3151–3165. [Google Scholar] [CrossRef]
- Saldanha, G.; Potter, L.; Lee, Y.S.; Watson, S.; Shendge, P.; Pringle, J.H. MicroRNA-21 expression and its pathogenetic significance in cutaneous melanoma. Melanoma Res. 2016, 26, 21–28. [Google Scholar] [CrossRef]
- Mao, X.H.; Chen, M.; Wang, Y.; Cui, P.G.; Liu, S.B.; Xu, Z.Y. MicroRNA-21 regulates the ERK/NF-κB signaling pathway to affect the proliferation, migration, and apoptosis of human melanoma A375 cells by targeting SPRY1, PDCD4, and PTEN. Mol. Carcinog. 2017, 56, 886–894. [Google Scholar] [CrossRef]
- Wang, J.H.; Zheng, W.W.; Cheng, S.T.; Liu, B.X.; Liu, F.R.; Song, J.Q. Correlation between microRNA-21 and sprouty homolog 2 gene expression in multiple myeloma. Mol. Med. Rep. 2015, 11, 4220–4224. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, X.; Cao, W.; Hua, Z.C. Tumor-specifically hypoxia-induced therapy of SPRY1/2 displayed differential therapeutic efficacy for melanoma. Am. J. Cancer Res. 2015, 5, 792–801. [Google Scholar] [PubMed]
- Ruan, Q.; Wang, P.; Wang, T.; Qi, J.; Wei, M.; Wang, S.; Fan, T.; Johnson, D.; Wan, X.; Shi, W.; et al. MicroRNA-21 regulates T-cell apoptosis by directly targeting the tumor suppressor gene Tipe2. Cell Death Dis. 2014, 5, e1095. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jia, W.; Niu, J.; Zhang, L. Understanding the roles of negative immune regulator TIPE2 in different diseases and tumourigenesis. Histol. Histopathol. 2018, 33, 919–928. [Google Scholar] [PubMed]
- Gus-Brautbar, Y.; Johnson, D.; Zhang, L.; Sun, H.; Wang, P.; Zhang, S.; Zhang, L.; Chen, Y.H. The anti-inflammatory TIPE2 is an inhibitor of the oncogenic Ras. Mol. Cell. 2012, 45, 610–618. [Google Scholar] [CrossRef]
- Lito, P.; Pratilas, C.A.; Joseph, E.W.; Tadi, M.; Halilovic, E.; Zubrowski, M.; Huang, A.; Wong, W.L.; Callahan, M.K.; Merghoub, T.; et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell. 2012, 22, 668–682. [Google Scholar] [CrossRef]
- Hatley, M.E.; Patrick, D.M.; Garcia, M.R.; Richardson, J.A.; Bassel-Duby, R.; van Rooij, E.; Olson, E.N. Modulation of K-Ras-dependent lung tumorigenesis by microRNA-21. Cancer Cell 2010, 18, 282–293. [Google Scholar] [CrossRef]
- Boiko, A.D.; Porteous, S.; Razorenova, O.V.; Krivokrysenko, V.I.; Williams, B.R.; Gudkov, A.V. A systematic search for downstream mediators of tumor suppressor function of p53 reveals a major role of BTG2 in suppression of Ras-induced transformation. Genes Dev. 2006, 20, 236–252. [Google Scholar] [CrossRef]
- Buscaglia, L.E.; Li, Y. Apoptosis and the target genes of microRNA-21. Chin. J. Cancer 2011, 30, 371–380. [Google Scholar] [CrossRef]
- Yang, C.H.; Pfeffer, S.R.; Sims, M.; Yue, J.; Wang, Y.; Linga, V.G.; Paulus, E.; Davidoff, A.M.; Pfeffer, L.M. The oncogenic microRNA-21 inhibits the tumor suppressive activity of FBXO11 to promote tumorigenesis. J. Biol. Chem. 2015, 290, 6037–6046. [Google Scholar] [CrossRef]
- Wang, P.; Zhao, Y.; Fan, R.; Chen, T.; Dong, C. MicroRNA-21a-5p functions on the regulation of melanogenesis by targeting Sox5 in mouse skin melanocytes. Int. J. Mol. Sci. 2016, 17, E959. [Google Scholar] [CrossRef]
- Kordaß, T.; Weber, C.E.; Oswald, M.; Ast, V.; Bernhardt, M.; Novak, D.; Utikal, J.; Eichmüller, S.B.; König, R. SOX5 is involved in balanced MITF regulation in human melanoma cells. BMC Med. Genomics. 2016, 9, 10. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, A.; Gordon, W.; Dizon, D.; Hopkin, A.S.; Gordon, E.; Yu, Z.; Andersen, B. The Grainyhead transcription factor Grhl3/Get1 suppresses miR-21 expression and tumorigenesis in skin: Modulation of the miR-21 target MSH2 by RNA-binding protein DND1. Oncogene 2013, 32, 1497–1507. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.T.; Wheelwright, M.; Teles, R.; Komisopoulou, E.; Edfeldt, K.; Ferguson, B.; Mehta, M.D.; Vazirnia, A.; Rea, T.H.; Sarno, E.N.; et al. MicroRNA-21 targets the vitamin D-dependent antimicrobial pathway in leprosy. Nat. Med. 2012, 18, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Reis, S.T.; Pontes-Junior, J.; Antunes, A.A.; Dall′Oglio, M.F.; Dip, N.; Passerotti, C.C.; Rossini, G.A.; Morais, D.R.; Nesrallah, A.J.; Piantino, C.; et al. miR-21 may acts as an oncomir by targeting RECK, a matrix metalloproteinase regulator, in prostate cancer. BMC Urol. 2012, 12, 14. [Google Scholar] [CrossRef]
- Martin del Campo, S.E.; Latchana, N.; Levine, K.M.; Grignol, V.P.; Fairchild, E.T.; Jaime-Ramirez, A.C.; Dao, T.V.; Karpa, V.I.; Carson, M.; Ganju, A.; et al. MiR-21 enhances melanoma invasiveness via inhibition of tissue inhibitor of metalloproteinases 3 expression: In vivo effects of miR-21 inhibitor. PLoS ONE 2015, 10, e0115919. [Google Scholar] [CrossRef]
- Gutsaeva, D.R.; Thounaojam, M.; Rajpurohit, S.; Powell, F.L.; Martin, P.M.; Goei, S.; Duncan, M.; Bartoli, M. STAT3-mediated activation of miR-21 is involved in down-regulation of TIMP3 and neovascularization in the ischemic retina. Oncotarget 2017, 8, 103568–103580. [Google Scholar] [CrossRef]
- Wu, L.; Zhu, L.; Li, Y.; Zheng, Z.; Lin, X.; Yang, C. LncRNA MEG3 promotes melanoma growth, metastasis and formation through modulating miR-21/E-cadherin axis. Cancer Cell Int. 2020, 20, 12. [Google Scholar] [CrossRef]
- Lu, T.X.; Munitz, A.; Rothenberg, M.E. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J. Immunol. 2009, 182, 4994–5002. [Google Scholar] [CrossRef]
- Dey, N.; Das, F.; Mariappan, M.M.; Mandal, C.C.; Ghosh-Choudhury, N.; Kasinath, B.S.; Choudhury, G.G. MicroRNA-21 orchestrates high glucose-induced signals to TOR complex 1, resulting in renal cell pathology in diabetes. J. Biol. Chem. 2011, 286, 25586–25603. [Google Scholar] [CrossRef]
- Iliopoulos, D.; Jaeger, S.A.; Hirsch, H.A.; Bulyk, M.L.; Struhl, K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol. Cell. 2010, 39, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Logotheti, S.; Pützer, B.M. STAT3 and STAT5 targeting for simultaneous management of melanoma and autoimmune diseases. Cancers 2019, 11, E1448. [Google Scholar] [CrossRef] [PubMed]
- Huynh, J.; Etemadi, N.; Hollande, F.; Ernst, M.; Buchert, M. The JAK/STAT3 axis: A comprehensive drug target for solid malignancies. Semin. Cancer Biol. 2017, 45, 13–22. [Google Scholar] [CrossRef]
- Sun, X.; Cao, N.; Mu, L.; Cao, W. Stress induced phosphoprotein 1 promotes tumor growth and metastasis of melanoma via modulating JAK2/STAT3 pathway. Biomed. Pharmacother. 2019, 116, 108962. [Google Scholar] [CrossRef]
- Kulesza, D.W.; Przanowski, P.; Kaminska, B. Knockdown of STAT3 targets a subpopulation of invasive melanoma stem-like cells. Cell. Biol. Int. 2019, 43, 613–622. [Google Scholar] [CrossRef]
- Wang, X.; Qu, H.; Dong, Y.; Wang, G.; Zhen, Y.; Zhang, L. Targeting signal-transducer-and-activator-of-transcription 3 sensitizes human cutaneous melanoma cells to BRAF inhibitor. Cancer Biomark. 2018, 23, 67–77. [Google Scholar] [CrossRef]
- Pan, J.; Ruan, W.; Qin, M.; Long, Y.; Wan, T.; Yu, K.; Zhai, Y.; Wu, C.; Xu, Y. Intradermal delivery of STAT3 siRNA to treat melanoma via dissolving microneedles. Sci. Rep. 2018, 8, 1117. [Google Scholar] [CrossRef]
- Delyon, J.; Chevret, S.; Jouary, T.; Dalac, S.; Dalle, S.; Guillot, B.; Arnault, J.P.; Avril, M.F.; Bedane, C.; Bens, G.; et al. STAT3 mediates nilotinib response in KIT-altered melanoma: A phase II multicenter trial of the French Skin Cancer Network. J. Investig. Dermatol. 2018, 138, 58–67. [Google Scholar] [CrossRef]
- Yang, C.H.; Yue, J.; Fan, M.; Pfeffer, L.M. IFN induces miR-21 through a signal transducer and activator of transcription 3-dependent pathway as a suppressive negative feedback on IFN-induced apoptosis. Cancer Res. 2010, 70, 8108–8116. [Google Scholar] [CrossRef]
- Tscherner, A.; Brown, A.C.; Stalker, L.; Kao, J.; Dufort, I.; Sirard, M.A.; LaMarre, J. STAT3 signaling stimulates miR-21 expression in bovine cumulus cells during in vitro oocyte maturation. Sci. Rep. 2018, 8, 11527. [Google Scholar] [CrossRef]
- Yang, C.H.; Yue, J.; Pfeffer, S.R.; Handorf, C.R.; Pfeffer, L.M. MicroRNA miR-21 regulates the metastatic behavior of B16 melanoma cells. J. Biol. Chem. 2011, 286, 39172–39178. [Google Scholar] [CrossRef]
- Koo, J.H.; Plouffe, S.W.; Meng, Z.; Lee, D.H.; Yang, D.; Lim, D.S.; Wang, C.Y.; Guan, K. Induction of AP-1 by YAP/TAZ contributes to cell proliferation and organ growth. Genes Dev. 2020, 34, 72–86. [Google Scholar] [CrossRef]
- Nallet-Staub, F.; Marsaud, V.; Li, L.; Gilbert, C.; Dodier, S.; Bataille, V.; Sudol, M.; Herlyn, M.; Mauviel, A. Pro-invasive activity of the Hippo pathway effectors YAP and TAZ in cutaneous melanoma. J. Investig. Dermatol. 2014, 134, 123–132. [Google Scholar] [CrossRef]
- Zhang, X.; Tang, J.Z.; Vergara, I.A.; Zhang, Y.; Szeto, P.; Yang, L.; Mintoff, C.; Colebatch, A.; McIntosh, L.; Mitchell, K.A.; et al. Somatic hypermutation of the YAP oncogene in a human cutaneous melanoma. Mol. Cancer Res. 2019, 17, 1435–1449. [Google Scholar] [CrossRef]
- Li, H.; Li, Q.; Dang, K.; Ma, S.; Cotton, J.L.; Yang, S.; Zhu, L.J.; Deng, A.C.; Ip, Y.T.; Johnson, R.L.; et al. YAP/TAZ activation drives uveal melanoma initiation and progression. Cell Rep. 2019, 29, 3200–3211. [Google Scholar] [CrossRef]
- Menzel, M.; Meckbach, D.; Weide, B.; Toussaint, N.C.; Schilbach, K.; Noor, S.; Eigentler, T.; Ikenberg, K.; Busch, C.; Quintanilla-Martinez, L.; et al. In melanoma, Hippo signaling is affected by copy number alterations and YAP1 overexpression impairs patient survival. Pigment Cell Melanoma Res. 2014, 27, 671–673. [Google Scholar] [CrossRef]
- Yu, X.; Zheng, H.; Tse, G.; Chan, M.T.; Wu, W.K. Long non-coding RNAs in melanoma. Cell Prolif. 2018, 51, e12457. [Google Scholar] [CrossRef]
- Yang, Z.; Jiang, X.; Jiang, X.; Zhao, H. X-inactive-specific transcript: A long noncoding RNA with complex roles in human cancers. Gene 2018, 679, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xia, T. Long non-coding RNA XIST regulates PDCD4 expression by interacting with miR-21-5p and inhibits osteosarcoma cell growth and metastasis. Int. J. Oncol. 2017, 51, 1460–1470. [Google Scholar] [CrossRef]
- Pan, B.M.; Lin, X.; Zhang, L.; Hong, W.; Zhang, Y. Long noncoding RNA X-inactive specific transcript promotes malignant melanoma progression and oxaliplatin resistance. Melanoma Res. 2019, 29, 254–262. [Google Scholar] [CrossRef]
- Yao, Y.; Ma, J.; Xue, Y.; Wang, P.; Li, Z.; Liu, J.; Chen, L.; Xi, Z.; Teng, H.; Wang, Z.; et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015, 359, 75–86. [Google Scholar] [CrossRef]
- Cheng, Z.; Luo, C.; Guo, Z. LncRNA-XIST/microRNA-126 sponge mediates cell proliferation and glucose metabolism through the IRS1/PI3K/Akt pathway in glioma. J. Cell Biochem. 2020, 121, 2170–2183. [Google Scholar] [CrossRef]
- Li, J.; Bian, E.B.; He, X.J.; Ma, C.C.; Zong, G.; Wang, H.L.; Zhao, B. Epigenetic repression of long non-coding RNA MEG3 mediated by DNMT1 represses the p53 pathway in gliomas. Int. J. Oncol. 2016, 48, 723–733. [Google Scholar] [CrossRef]
- Chen, L.; Yang, H.; Xiao, Y.; Tang, X.; Li, Y.; Han, Q.; Fu, J.; Yang, Y.; Zhu, Y. LncRNA GAS5 is a critical regulator of metastasis phenotype of melanoma cells and inhibits tumor growth in vivo. Onco Targets Ther. 2016, 9, 4075–4087. [Google Scholar] [CrossRef]
- Bian, D.; Shi, W.; Shao, Y.; Li, P.; Song, G. Long non-coding RNA GAS5 inhibits tumorigenesis via miR-137 in melanoma. Am. J. Transl. Res. 2017, 9, 1509–1520. [Google Scholar]
- Chen, L.; Yang, H.; Xiao, Y.; Tang, X.; Li, Y.; Han, Q.; Fu, J.; Yang, Y.; Zhu, Y. Lentiviral-mediated overexpression of long non-coding RNA GAS5 reduces invasion by mediating MMP2 expression and activity in human melanoma cells. Int. J. Oncol. 2016, 48, 1509–1518. [Google Scholar] [CrossRef]
- Tao, H.; Zhang, J.G.; Qin, R.H.; Dai, C.; Shi, P.; Yang, J.J.; Deng, Z.Y.; Shi, K.H. LncRNA GAS5 controls cardiac fibroblast activation and fibrosis by targeting miR-21 via PTEN/MMP-2 signaling pathway. Toxicology 2017, 386, 11–18. [Google Scholar] [CrossRef]
- Hu, L.; Ye, H.; Huang, G.; Luo, F.; Liu, Y.; Liu, Y.; Yang, X.; Shen, J.; Liu, Q.; Zhang, J. Long noncoding RNA GAS5 suppresses the migration and invasion of hepatocellular carcinoma cells via miR-21. Tumour Biol. 2016, 37, 2691–2702. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhu, Z.; Watabe, K.; Zhang, X.; Bai, C.; Xu, M.; Wu, F.; Mo, Y.Y. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013, 20, 1558–1568. [Google Scholar] [CrossRef]
- Fan, Q.; Yang, L.; Zhang, X.; Peng, X.; Wei, S.; Su, D.; Zhai, Z.; Hua, X.; Li, H. The emerging role of exosome-derived non-coding RNAs in cancer biology. Cancer Lett. 2018, 414, 107–115. [Google Scholar] [CrossRef]
- Wang, M.; Zhou, L.; Yu, F.; Zhang, Y.; Li, P.; Wang, K. The functional roles of exosomal long non-coding RNAs in cancer. Cell Mol. Life Sci. 2019, 76, 2059–2076. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Narumi, T.; Watanabe, T.; Otaki, Y.; Takahashi, T.; Aono, T.; Goto, J.; Toshima, T.; Sugai, T.; Wanezaki, M.; et al. The association between microRNA-21 and hypertension-induced cardiac remodeling. PLoS ONE 2020, 15, e0226053. [Google Scholar] [CrossRef] [PubMed]
- Ribas, J.; Ni, X.; Haffner, M.; Wentzel, E.A.; Salmasi, A.H.; Chowdhury, W.H.; Kudrolli, T.A.; Yegnasubramanian, S.; Luo, J.; Rodriguez, R.; et al. miR-21: An androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 2009, 69, 7165–7169. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, X.; Jiang, G.; Wang, J.; Qian, Y. Vitamin D down-regulates microRNA-21 expression to promote human placental trophoblast cell migration and invasion in vitro. Nan Fang Yi Ke Da Xue Xue Bao 2019, 39, 437–442. [Google Scholar]
- Sheane, B.J.; Smyth, P.; Scott, K.; Aziz, R.; Buckley, M.; Lodge, E.; Kiely, N.; Kingston, M.; McGovern, E.; Healy, M.; et al. An association between microRNA-21 expression and vitamin D deficiency in coronary artery disease. Microrna 2015, 4, 57–63. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, H.; Wang, J. Exosomes as a novel pathway for regulating development and diseases of the skin. Biomed. Rep. 2018, 8, 207–214. [Google Scholar] [CrossRef]
- Wang, W.M.; Wu, C.; Jin, H.Z. Exosomes in chronic inflammatory skin diseases and skin tumors. Exp. Dermatol. 2019, 28, 213–218. [Google Scholar] [CrossRef]
- Xiao, D.; Ohlendorf, J.; Chen, Y.; Taylor, D.D.; Rai, S.N.; Waigel, S.; Zacharias, W.; Hao, H.; McMasters, K.M. Identifying mRNA, microRNA and protein profiles of melanoma exosomes. PLoS ONE 2012, 7, e46874. [Google Scholar] [CrossRef]
- Gowda, R.; Robertson, B.M.; Iyer, S.; Barry, J.; Dinavahi, S.S.; Robertson, G.P. The role of exosomes in metastasis and progression of melanoma. Cancer Treat. Rev. 2020, 85, 101975. [Google Scholar] [CrossRef]
- Tian, T.; Zhu, Y.L.; Zhou, Y.Y.; Liang, G.F.; Wang, Y.Y.; Hu, F.H.; Xiao, Z.D. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J. Biol. Chem. 2014, 289, 22258–22267. [Google Scholar] [CrossRef]
- Mannavola, F.; D′Oronzo, S.; Cives, M.; Stucci, L.S.; Ranieri, G.; Silvestris, F.; Tucci, M. Extracellular vesicles and epigenetic modifications are hallmarks of melanoma progression. Int. J. Mol. Sci. 2019, 21, 52. [Google Scholar] [CrossRef] [PubMed]
- Hood, J.L. Natural melanoma-derived extracellular vesicles. Semin. Cancer Biol. 2019, 59, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Harmati, M.; Gyukity-Sebestyen, E.; Dobra, G.; Janovak, L.; Dekany, I.; Saydam, O.; Hunyadi-Gulyas, E.; Nagy, I.; Farkas, A.; Pankotai, T.; et al. Small extracellular vesicles convey the stress-induced adaptive responses of melanoma cells. Sci. Rep. 2019, 9, 15329. [Google Scholar] [CrossRef]
- Hu, T.; Hu, J. Melanoma-derived exosomes induce reprogramming fibroblasts into cancer-associated fibroblasts via Gm26809 delivery. Cell Cycle 2019, 18, 3085–3094. [Google Scholar] [CrossRef]
- Boussadia, Z.; Lamberti, J.; Mattei, F.; Pizzi, E.; Puglisi, R.; Zanetti, C.; Pasquini, L.; Fratini, F.; Fantozzi, L.; Felicetti, F.; et al. Acidic microenvironment plays a key role in human melanoma progression through a sustained exosome mediated transfer of clinically relevant metastatic molecules. J. Exp. Clin. Cancer Res. 2018, 37, 245. [Google Scholar] [CrossRef]
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef]
- Zhou, X.; Yan, T.; Huang, C.; Xu, Z.; Wang, L.; Jiang, E.; Wang, H.; Chen, Y.; Liu, K.; Shao, Z.; et al. Melanoma cell-secreted exosomal miR-155-5p induce proangiogenic switch of cancer-associated fibroblasts via SOCS1/JAK2/STAT3 signaling pathway. J. Exp. Clin. Cancer Res. 2018, 37, 242. [Google Scholar] [CrossRef] [PubMed]
- Abels, E.R.; Maas, S.L.N.; Nieland, L.; Wie, Z.; Cheah, P.S.; Tai, E.; Kolsteeg, C.J.; Dusoswa, S.A.; Ting, D.T.; Hickman, S.; et al. Glioblastoma-associated microglia reprogramming is mediated by functional transfer of extracellular miR-21. Cell Rep. 2019, 28, 3105–3119. [Google Scholar] [CrossRef]
- Gajos-Michniewicz, A.; Czyz, M. Role of miRNAs in melanoma metastasis. Cancers 2019, 11, 326. [Google Scholar] [CrossRef]
- Xiao, D.; Barry, S.; Kmetz, D.; Egger, M.; Pan, J.; Rai, S.N.; Qu, J.; McMasters, K.M.; Hao, H. Melanoma cell-derived exosomes promote epithelial-mesenchymal transition in primary melanocytes through paracrine/autocrine signaling in the tumor microenvironment. Cancer Lett. 2016, 376, 318–327. [Google Scholar] [CrossRef]
- Pfeffer, S.R.; Grossmann, K.F.; Cassidy, P.B.; Yang, C.H.; Fan, M.; Kopelovich, L.; Leachman, S.A.; Pfeffer, L.M. Detection of exosomal miRNAs in the plasma of melanoma patients. J. Clin. Med. 2015, 4, 2012–2027. [Google Scholar] [CrossRef]
- Saldanha, G.; Potter, L.; Shendge, P.; Osborne, J.; Nicholson, S.; Yii, N.; Varma, S.; Aslam, M.I.; Elshaw, S.; Papadogeorgakis, E.; et al. Plasma microRNA-21 is associated with tumor burden in cutaneous melanoma. J. Investig. Dermatol. 2013, 133, 1381–1384. [Google Scholar] [CrossRef]
- Ragusa, M.; Barbagallo, C.; Statello, L.; Caltabiano, R.; Russo, A.; Puzzo, L.; Avitabile, T.; Longo, A.; Toro, M.D.; Barbagallo, D.; et al. miRNA profiling in vitreous humor, vitreal exosomes and serum from uveal melanoma patients: Pathological and diagnostic implications. Cancer Biol. Ther. 2015, 16, 1387–1396. [Google Scholar] [CrossRef]
- Korabiowska, M.; Dengler, H.; Kellner, S.; Stachura, J.; Schauer, A. Decreased expression of MLH1, MSH2, PMS1 and PMS2 in pigmented lesions indicates accumulation of failed DNA repair along with malignant transformation and tumour progression. Oncol. Rep. 1997, 4, 653–655. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Z.; Feng, L.; Jiang, P.; Li, X.; Wang, X. Ionizing radiation-inducible microRNA-21 induces angiogenesis by directly targeting PTEN. Asian Pac. J. Cancer Prev. 2019, 20, 1587–1593. [Google Scholar] [CrossRef]
- Gabriely, G.; Wurdinger, T.; Kesari, S.; Esau, C.C.; Burchard, J.; Linsley, P.S.; Krichevsky, A.M. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol. Cell Biol. 2008, 28, 5369–5380. [Google Scholar] [CrossRef]
- Liu, L.Z.; Li, C.; Chen, Q.; Jing, Y.; Carpenter, R.; Jiang, Y.; Kung, H.F.; Lai, L.; Jiang, B.H. MiR-21 induced angiogenesis through AKT and ERK activation and HIF-1α expression. PLoS ONE 2011, 6, e19139. [Google Scholar] [CrossRef]
- Hermansen, S.K.; Nielsen, B.S.; Aaberg-Jessen, C.; Kristensen, B.W. miR-21 is linked to glioma angiogenesis: A co-localization study. J. Histochem. Cytochem. 2016, 64, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Hong, L.; Sun, L.; Sang, H.; Qian, A.; Li, W.; Zhuang, H.; Liang, H.; Song, D.; Li, C.; et al. miR-21 induces endothelial progenitor cells proliferation and angiogenesis via targeting FASLG and is a potential prognostic marker in deep venous thrombosis. J. Transl. Med. 2019, 17, 270. [Google Scholar] [CrossRef]
- Chen, L.Y.; Wang, X.; Qu, X.L.; Pan, L.N.; Wang, Z.Y.; Lu, Y.H.; Hu, H.Y. Activation of the STAT3/microRNA-21 pathway participates in angiotensin II-induced angiogenesis. J. Cell Physiol. 2019, 234, 19640–19654. [Google Scholar] [CrossRef]
- O′reilly, A.; Larkin, J. Checkpoint inhibitors in advanced melanoma: Effect on the field of immunotherapy. Expert Rev. Anticancer Ther. 2017, 17, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Furue, M.; Ito, T.; Wada, N.; Wada, M.; Kadono, T.; Uchi, H. Melanoma and immune checkpoint inhibitors. Curr. Oncol. Rep. 2018, 20, 29. [Google Scholar] [CrossRef]
- Tokunaga, A.; Sugiyama, D.; Maeda, Y.; Warner, A.B.; Panageas, K.S.; Ito, S.; Togashi, Y.; Sakai, C.; Wolchok, J.D.; Nishikawa, H. Selective inhibition of low-affinity memory CD8+ T cells by corticosteroids. J. Exp. Med. 2019, 216, 2701–2713. [Google Scholar] [CrossRef] [PubMed]
- Molodtsov, A.; Turk, M.J. Tissue resident CD8 memory T cell responses in cancer and autoimmunity. Front. Immunol. 2018, 9, 2810. [Google Scholar] [CrossRef]
- Malik, B.T.; Byrne, K.T.; Vella, J.L.; Zhang, P.; Shabaneh, T.B.; Steinberg, S.M.; Molodtsov, A.K.; Bowers, J.S.; Angeles, C.V.; Paulos, C.M.; et al. Resident memory T cells in the skin mediate durable immunity to melanoma. Sci. Immunol. 2017, 2, eaam6346. [Google Scholar] [CrossRef]
- Willemsen, M.; Linkutė, R.; Luiten, R.M.; Matos, T.R. Skin-resident memory T cells as a potential new therapeutic target in vitiligo and melanoma. Pigment Cell Melanoma Res. 2019, 32, 612–622. [Google Scholar] [CrossRef]
- Massi, D.; Rulli, E.; Cossa, M.; Valeri, B.; Rodolfo, M.; Merelli, B.; De Logu, F.; Nassini, R.; Del Vecchio, M.; Di Guardo, L.; et al. The density and spatial tissue distribution of CD8+ and CD163+ immune cells predict response and outcome in melanoma patients receiving MAPK inhibitors. J. Immunother. Cancer 2019, 7, 308. [Google Scholar] [CrossRef]
- Kim, C.; Hu, B.; Jadhav, R.R.; Jin, J.; Zhang, H.; Cavanagh, M.M.; Akondy, R.S.; Ahmed, R.; Weyand, C.M.; Goronzy, J.J. Activation of miR-21-regulated pathways in immune aging selects against signatures characteristic of memory T Cells. Cell Rep. 2018, 25, 2148–2162. [Google Scholar] [CrossRef]
- Sahraei, M.; Chaube, B.; Liu, Y.; Sun, J.; Kaplan, A.; Price, N.L.; Ding, W.; Oyaghire, S.; García-Milian, R.; Mehta, S.; et al. Suppressing miR-21 activity in tumor-associated macrophages promotes an antitumor immune response. J. Clin. Investig. 2019, 129, 5518–5536. [Google Scholar] [CrossRef]
- Mathsyaraja, H.; Thies, K.; Taffany, D.A.; Deighan, C.; Liu, T.; Yu, L.; Fernandez, S.A.; Shapiro, C.; Otero, J.; Timmers, C.; et al. CSF1-ETS2-induced microRNA in myeloid cells promote metastatic tumor growth. Oncogene 2015, 34, 3651–3661. [Google Scholar] [CrossRef]
- Wolf, S.F.; Sieburth, D.; Sypek, J. Interleukin 12: A key modulator of immune function. Stem Cells 1994, 12, 154–168. [Google Scholar] [CrossRef] [PubMed]
- Lu, X. Impact of IL-12 in cancer. Curr. Cancer Drug Targets 2017, 17, 682–697. [Google Scholar] [CrossRef] [PubMed]
- Cocco, C.; Pistoia, V.; Airoldi, I. New perspectives for melanoma immunotherapy: Role of IL-12. Curr. Mol. Med. 2009, 9, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Nagai, H.; Oniki, S.; Fujiwara, S.; Yoshimoto, T.; Nishigori, C. Antimelanoma immunotherapy: Clinical and preclinical applications of IL-12 family members. Immunotherapy 2010, 2, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Tucci, M.; Mannavola, F.; Passarelli, A.; Stucci, L.S.; Cives, M.; Silvestris, F. Exosomes in melanoma: A role in tumor progression, metastasis and impaired immune system activity. Oncotarget 2018, 9, 20826–20837. [Google Scholar] [CrossRef]
- Sharma, P.; Diergaarde, B.; Ferrone, S.; Kirkwood, J.M.; Whiteside, T.L. Melanoma cell-derived exosomes in plasma of melanoma patients suppress functions of immune effector cells. Sci. Rep. 2020, 10, 92. [Google Scholar] [CrossRef]
- Cordonnier, M.; Nardin, C.; Chanteloup, G.; Derangere, V.; Algros, M.P.; Arnould, L.; Garrido, C.; Aubin, F.; Gobbo, J. Tracking the evolution of circulating exosomal-PD-L1 to monitor melanoma patients. J. Extracell. Vesicles 2020, 9, 1710899. [Google Scholar] [CrossRef]
- Vignard, V.; Labbé, M.; Marec, N.; André-Grégoire, G.; Jouand, N.; Fonteneau, J.F.; Labarrière, N.; Fradin, D. MicroRNAs in tumor exosomes drive immune escape in melanoma. Cancer Immunol. Res. 2020, 8, 255–267. [Google Scholar] [CrossRef]
- Yang, C.H.; Yue, J.; Pfeffer, S.R.; Fan, M.; Paulus, E.; Hosni-Ahmed, A.; Sims, M.; Qayyum, S.; Davidoff, A.M.; Handorf, C.R.; et al. MicroRNA-21 promotes glioblastoma tumorigenesis by down-regulating insulin-like growth factor-binding protein-3 (IGFBP3). J. Biol. Chem. 2014, 289, 25079–25087. [Google Scholar] [CrossRef]
- Masoudi, M.S.; Mehrabian, E.; Mirzaei, H. MiR-21: A key player in glioblastoma pathogenesis. J. Cell. Biochem. 2018, 119, 1285–1290. [Google Scholar] [CrossRef]
- Ribas, J.; Lupold, S.E. The transcriptional regulation of miR-21, its multiple transcripts, and their implication in prostate cancer. Cell Cycle 2010, 9, 923–929. [Google Scholar] [CrossRef] [PubMed]
- Guan, C.; Zhang, L.; Wang, S.; Long, L.; Zhou, H.; Qian, S.; Ma, M.; Bai, F.; Meng, Q.H.; Lyu, J. Upregulation of MicroRNA-21 promotes tumorigenesis of prostate cancer cells by targeting KLF5. Cancer Biol. Ther. 2019, 20, 1149–1161. [Google Scholar] [CrossRef]
- Scarbrough, P.M.; Akushevich, I.; Wrensch, M.; Il′yasova, D. Exploring the association between melanoma and glioma risks. Ann. Epidemiol. 2014, 24, 469–474. [Google Scholar] [CrossRef]
- Patasius, A.; Urbonas, V.; Smailyte, G. Skin melanoma and subsequent risk of prostate cancer: A Lithuanian Cancer Registry Study. Int. J. Environ. Res. Public Health 2019, 16, E3915. [Google Scholar] [CrossRef]
- Cole-Clark, D.; Nair-Shalliker, V.; Bang, A.; Rasiah, K.; Chalasani, V.; Smith, D.P. An initial melanoma diagnosis may increase the subsequent risk of prostate cancer: Results from the New South Wales Cancer Registry. Sci. Rep. 2018, 8, 7167. [Google Scholar] [CrossRef]
- Sutcliffe, S.; Giovannucci, E.; Isaacs, W.B.; Willett, W.C.; Platz, E.A. Acne and risk of prostate cancer. Int. J. Cancer 2007, 121, 2688–2692. [Google Scholar] [CrossRef]
- Ugge, H.; Udumyan, R.; Carlsson, J.; Andrén, O.; Montgomery, S.; Davidsson, S.; Fall, K. Acne in late adolescence and risk of prostate cancer. Int. J. Cancer 2018, 142, 1580–1585. [Google Scholar] [CrossRef]
- Zhang, M.; Qureshi, A.A.; Fortner, R.T.; Hankinson, S.E.; Wei, Q.; Wang, L.E.; Eliassen, A.H.; Willett, W.C.; Hunter, D.J.; Han, J. Teenage acne and cancer risk in US women: A prospective cohort study. Cancer 2015, 121, 1681–1687. [Google Scholar] [CrossRef]
- Mota Garcia, T.; Hiyoshi, A.; Udumyan, R.; Sjöqvist, H.; Fall, K.; Montgomery, S. Acne in late adolescence is not associated with a raised risk of subsequent malignant melanoma among men. Cancer Epidemiol. 2017, 51, 44–48. [Google Scholar] [CrossRef]
- Dréno, B.; Bettoli, V.; Araviiskaia, E.; Sanchez Viera, M.; Bouloc, A. The influence of exposome on acne. J. Eur. Acad. Dermatol. Venereol. 2018, 32, 812–819. [Google Scholar] [CrossRef]
- Melnik, B.C. MiR-21: An environmental driver of malignant melanoma? J. Transl. Med. 2015, 13, 202. [Google Scholar] [CrossRef]
- Puckett, Y.; Wilson, A.M.; Thevenin, C. Cancer, Melanoma Pathology; SourceStatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Sample, A.; He, Y.Y. Mechanisms and prevention of UV-induced melanoma. Photodermatol. Photoimmunol. Photomed. 2018, 34, 13–24. [Google Scholar] [CrossRef]
- Ellerhorst, J.A.; Greene, V.R.; Ekmekcioglu, S.; Warneke, C.L.; Johnson, M.M.; Cooke, C.P.; Wang, L.E.; Prieto, V.G.; Gershenwald, J.E.; Wei, Q.; et al. Clinical correlates of NRAS and BRAF mutations in primary human melanoma. Clin. Cancer Res. 2011, 17, 229–235. [Google Scholar] [CrossRef]
- Curtin, J.A.; Fridlyand, J.; Kageshita, T.; Patel, H.N.; Busam, K.J.; Kutzner, H.; Cho, K.H.; Aiba, S.; Bröcker, E.B.; LeBoit, P.E.; et al. Distinct sets of genetic alterations in melanoma. N. Engl. J. Med. 2005, 353, 2135–2147. [Google Scholar] [CrossRef]
- Maldonado, J.L.; Fridlyand, J.; Patel, H.; Jain, A.N.; Busam, K.; Kageshita, T.; Ono, T.; Albertson, D.G.; Pinkel, D.; Bastian, B.C. Determinants of BRAF mutations in primary melanomas. J. Natl. Cancer Inst. 2003, 95, 1878–1890. [Google Scholar] [CrossRef]
- Edwards, R.H.; Ward, M.R.; Wu, H.; Medina, C.A.; Brose, M.S.; Volpe, P.; Nussen-Lee, S.; Haupt, H.M.; Martin, A.M.; Herlyn, M.; et al. Absence of BRAF mutations in UV-protected mucosal melanomas. J. Med. Genet. 2004, 41, 270–272. [Google Scholar] [CrossRef]
- Bauer, J.; Büttner, P.; Murali, R.; Okamoto, I.; Kolaitis, N.A.; Landi, M.T.; Scolder, R.A.; Bastian, B.C. BRAF mutations in cutaneous melanoma are independently associated with age, anatomic site of the primary tumor, and the degree of solar elastosis at the primary tumor site. Pigment Cell Melanoma Res. 2011, 24, 345–351. [Google Scholar] [CrossRef]
- Whiteman, D.C.; Stickle, M.; Watt, P.; Hughes, M.C.; Davis, M.B.; Green, A.C. Anatomic site, sun exposure, and risk of cutaneous melanoma. J. Clin. Oncol. 2006, 24, 3172–3177. [Google Scholar] [CrossRef]
- Lo Cicero, A.; Delevoye, C.; Gilles-Marsens, F.; Loew, D.; Dingli, F.; Guéré, C.; André, N.; Vié, K.; van Niel, G.; Raposo, G. Exosomes released by keratinocytes modulate melanocyte pigmentation. Nat. Commun. 2015, 6, 7506. [Google Scholar] [CrossRef]
- Liu, Y.; Xue, L.; Gao, H.; Chang, L.; Yu, X.; Zhu, Z.; He, X.; Geng, J.; Dong, Y.; Li, H.; et al. Exosomal miRNA derived from keratinocytes regulates pigmentation in melanocytes. J. Dermatol. Sci. 2019, 93, 159–167. [Google Scholar] [CrossRef]
- Wäster, P.; Eriksson, I.; Vainikka, L.; Öllinger, K. Extracellular vesicles released by melanocytes after UVA irradiation promote intercellular signaling via miR21. Pigment Cell Melanoma Res. 2020, 33, 542–555. [Google Scholar] [CrossRef] [PubMed]
- Syed, D.N.; Khan, M.I.; Shabbir, M.; Mukhtar, H. MicroRNAs in skin response to UV radiation. Curr. Drug Targets 2013, 14, 1128–1134. [Google Scholar] [CrossRef]
- Hou, L.; Bowman, L.; Meighan, T.G.; Pratheeshkumar, P.; Shi, X.; Ding, M. Induction of miR-21-PDCD4 signaling by UVB in JB6 cells involves ROS-mediated MAPK pathways. Exp. Toxicol. Pathol. 2013, 65, 1145–1148. [Google Scholar] [CrossRef]
- Guo, L.; Huang, Z.X.; Chen, X.W.; Deng, Q.K.; Yan, W.; Zhou, M.J.; Ou, C.S.; Ding, Z.H. Differential expression profiles of microRNAs in NIH3T3 cells in response to UVB irradiation. Photochem. Photobiol. 2009, 85, 765–773. [Google Scholar] [CrossRef]
- Lin, K.Y.; Chen, C.M.; Lu, C.Y.; Cheng, C.Y.; Wu, Y.H. Regulation of miR-21 expression in human melanoma via UV-ray-induced melanin pigmentation. Environ. Toxicol. 2017, 32, 2064–2069. [Google Scholar] [CrossRef]
- Degueurce, G.; D′Errico, I.; Pich, C.; Ibberson, M.; Schütz, F.; Montagner, A.; Sgandurra, M.; Mury, L.; Jafari, P.; Boda, A.; et al. Identification of a novel PPARβ/δ/miR-21-3p axis in UV-induced skin inflammation. EMBO Mol. Med. 2016, 8, 919–936. [Google Scholar] [CrossRef]
- Hartman, M.L.; Czyz, M. MITF in melanoma: Mechanisms behind its expression and activity. Cell Mol. Life Sci. 2015, 72, 1249–1260. [Google Scholar] [CrossRef]
- Fishel, R.; Ewel, A.; Lee, S.; Lescoe, M.K.; Griffith, J. Binding of mismatched microsatellite DNA sequences by the human MSH2 protein. Science 1994, 266, 1403–1405. [Google Scholar] [CrossRef]
- Kubeček, O.; Kopecký, J. Microsatellite instability in melanoma: A comprehensive review. Melanoma Res. 2016, 26, 545–550. [Google Scholar]
- Korabiowska, M.; König, F.; Verheggen, R.; Schlott, T.; Cordon-Cardo, C.; Romeike, B.; Brinck, U. Altered expression and new mutations in DNA mismatch repair genes MLH1 and MSH2 in melanoma brain metastases. Anticancer Res. 2004, 24, 981–986. [Google Scholar]
- Korabiowska, M.; Brinck, U.; Stachura, J.; Jawien, J.; Hasse, F.M.; Cordon-Cardos, C.; Fischer, G. Exonic deletions of mismatch repair genes MLH1 and MSH2 correlate with prognosis and protein expression levels in malignant melanomas. Anticancer Res. 2006, 26, 1231–1235. [Google Scholar] [PubMed]
- Hussein, M.R.; Wood, G.S. hMLH1 and hMSH2 gene mutations are present in radial growth-phase cutaneous malignant melanoma cell lines and can be induced further by ultraviolet-B irradiation. Exp. Dermatol. 2003, 12, 872–875. [Google Scholar] [CrossRef] [PubMed]
- Sanlorenzo, M.; Wehner, M.R.; Linos, E.; Kornak, J.; Kainz, W.; Posch, C.; Vujic, I.; Johnston, K.; Gho, D.; Monico, G.; et al. The risk of melanoma in airline pilots and cabin crew: A meta-analysis. JAMA Dermatol. 2015, 151, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Olsen, C.M.; Rea, S.; Marsden, J.; Green, A.C. Do airline pilots and cabin crew have raised risks of melanoma and other skin cancers? Systematic review and meta-analysis. Br. J. Dermatol. 2019, 181, 55–64. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, X.; Wang, P.; Wang, X.; Farris, A.B., III; Wang, Y. Lessons learned using different mouse models during space radiation-induced lung tumorigenesis experiments. Life Sci. Space Res. 2016, 9, 48–55. [Google Scholar] [CrossRef]
- Shi, Y.; Zhang, X.; Tang, X.; Wang, P.; Wang, H.; Wang, Y. MiR-21 is continually elevated long-term in the brain after exposure to ionizing radiation. Radiat. Res. 2012, 177, 124–128. [Google Scholar] [CrossRef]
- Zhu, Y.; Yu, X.; Fu, H.; Wang, H.; Wang, P.; Zheng, X.; Wang, Y. MicroRNA-21 is involved in ionizing radiation-promoted liver carcinogenesis. Int. J. Clin. Exp. Med. 2010, 3, 211–222. [Google Scholar]
- Liu, Z.; Liang, X.; Li, X.; Liu, X.; Zhu, M.; Gu, Y.; Zhou, P. MiRNA-21 functions in ionizing radiation-induced epithelium-to-mesenchymal transition (EMT) by downregulating PTEN. Toxicol. Res. 2019, 8, 328–340. [Google Scholar] [CrossRef]
- Xu, S.; Ding, N.; Pei, H.; Hu, W.; Wie, W.; Zhang, X.; Zhou, G.; Wang, J. MiR-21 is involved in radiation-induced bystander effects. RNA Biol. 2014, 11, 1161–1170. [Google Scholar] [CrossRef]
- Xu, S.; Wang, J.; Ding, N.; Hu, W.; Zhang, X.; Wang, B.; Hua, J.; Wie, W.; Zhu, Q. Exosome-mediated microRNA transfer plays a role in radiation-induced bystander effect. RNA Biol. 2015, 12, 1355–1363. [Google Scholar] [CrossRef]
- Nicholas, J.S.; Lackland, D.T.; Butler, G.C.; Mohr, L.C., Jr.; Dunbar, J.B.; Kaune, W.T.; Grosche, B.; Hoel, D.G. Cosmic radiation and magnetic field exposure to airline flight crews. Am. J. Ind. Med. 1998, 34, 574–580. [Google Scholar] [CrossRef]
- Selvamurugan, N.; He, Z.; Rifkin, D.; Dabovic, B.; Partridge, N.C. Pulsed electromagnetic field regulates microRNA 21 expression to activate TGF-β signaling in human bone marrow stromal cells to enhance osteoblast differentiation. Stem Cells Int. 2017, 2017, 2450327. [Google Scholar] [CrossRef] [PubMed]
- Sanlorenzo, M.; Vujic, I.; Posch, C.; Cleaver, J.E.; Quaglino, P.; Ortiz-Urda, S. The risk of melanoma in pilots and cabin crew: UV measurements in flying airplanes. JAMA Dermatol. 2015, 151, 450–452. [Google Scholar] [CrossRef][Green Version]
- Hallberg, Ö. Cancer incidence vs. FM radio transmitter density. Electromagn. Biol. Med. 2016, 35, 343–347. [Google Scholar] [CrossRef] [PubMed]
- Irvine, D.; Davies, D.M. The mortality of British Airways pilots, 1966–1989: A proportional mortality study. Aviat. Space Environ. Med. 1992, 63, 276–279. [Google Scholar] [PubMed]
- Yong, L.C.; Pinkerton, L.E.; Yiin, J.H.; Anderson, J.L.; Deddens, J.A. Mortality among a cohort of U.S. commercial airline cockpit crew. Am. J. Ind. Med. 2014, 57, 906–914. [Google Scholar] [CrossRef]
- Dupin, E.; Real, C.; Glavieux-Pardanaud, C.; Vaigot, P.; Le Douarin, N.M. Reversal of developmental restrictions in neural crest lineages: Transition from Schwann cells to glial-melanocytic precursors in vitro. Proc. Natl. Acad. Sci. USA 2003, 100, 5229–5233. [Google Scholar] [CrossRef]
- Yang, M.; Guo, W.; Yang, C.; Tang, J.; Huang, Q.; Feng, S.; Jiang, A.; Xu, X.; Jiang, G. Mobile phone use and glioma risk: A systematic review and meta-analysis. PLoS ONE 2017, 12, e0175136. [Google Scholar] [CrossRef]
- Morgan, L.L.; Miller, A.B.; Sasco, A.; Davis, D.L. Mobile phone radiation causes brain tumors and should be classified as a probable human carcinogen (2A) (review). Int. J. Oncol. 2015, 46, 1865–1871. [Google Scholar] [CrossRef]
- Poulsen, A.H.; Friis, S.; Johansen, C.; Jensen, A.; Frei, P.; Kjaear, S.K.; Dalton, S.O.; Schüz, J. Mobile phone use and the risk of skin cancer: A nationwide cohort study in Denmark. Am. J. Epidemiol. 2013, 178, 190–197. [Google Scholar] [CrossRef]
- Zeller, J.; Strack, C.; Fenk, S.; Mohr, M.; Loew, T.; Schmitz, G.; Maier, L.; Fischer, M.; Baessler, A. Relation between obesity, metabolic syndrome, successful long-term weight reduction, and right ventricular function. Int. Heart J. 2016, 57, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Sherling, D.H.; Perumareddi, P.; Hennekens, C.H. Metabolic syndrome. J. Cardiovasc. Pharmacol. Ther. 2017, 22, 365–367. [Google Scholar] [CrossRef]
- Hoch, D.; Gauster, M.; Hauguel-de Mouzon, S.; Desoye, G. Diabesity-associated oxidative and inflammatory stress signalling in the early human placenta. Mol. Aspects Med. 2019, 66, 21–30. [Google Scholar] [CrossRef]
- Alarcón, S.; Niechi, I.; Toledo, F.; Sobrevia, L.; Quezada, C. Glioma progression in diabesity. Mol. Aspects Med. 2019, 66, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.B.; Han, S.P.; Zhu, G.Z.; Zhu, C.; Wang, X.J.; Cao, X.G.; Guo, X.R. Birth weight and subsequent risk of obesity: A systematic review and meta-analysis. Obes. Rev. 2011, 12, 525–542. [Google Scholar] [CrossRef] [PubMed]
- Werneck, A.O.; Silva, D.R.P.; Collings, P.J.; Fernandes, R.A.; Ronque, E.R.V.; Coelho-E-Silva, M.J.; Sardinha, L.B.; Cyrino, E.S. Birth weight, biological maturation and obesity in adolescents: A mediation analysis. J. Dev. Orig. Health Dis. 2017, 8, 502–507. [Google Scholar] [CrossRef] [PubMed]
- Knop, M.R.; Geng, T.T.; Gorny, A.W.; Ding, R.; Li, C.; Ley, S.H.; Huang, T. Birth weight and risk of type 2 diabetes mellitus, cardiovascular disease, and hypertension in adults: A meta-analysis of 7 646 267 participants from 135 studies. J. Am. Heart Assoc. 2018, 7, e008870. [Google Scholar] [CrossRef]
- Ward, Z.J.; Long, M.W.; Resch, S.C.; Giles, C.M.; Cradock, A.L.; Gortmaker, S.L. Simulation of growth trajectories of childhood obesity into adulthood. N. Engl. J. Med. 2017, 377, 2145–2153. [Google Scholar] [CrossRef]
- Salihu, H.M.; Dongarwar, D.; King, L.M.; Yusuf, K.K.; Ibrahimi, S.; Salinas-Miranda, A.A. Trends in the incidence of fetal macrosomia and its phenotypes in the United States, 1971-2017. Arch. Gynecol. Obstet. 2020, 301, 415–426. [Google Scholar] [CrossRef]
- Hermann, G.M.; Dallas, L.M.; Haskell, S.E.; Roghair, R.D. Neonatal macrosomia is an independent risk factor for adult metabolic syndrome. Neonatology 2010, 98, 238–244. [Google Scholar] [CrossRef]
- Wojcik, K.Y.; Escobedo, L.A.; Wysong, A.; Heck, J.E.; Ritz, B.; Hamilton, A.S.; Milam, J.; Cockburn, M.G. High birth weight, early UV exposure, and melanoma risk in children, adolescents, and young adults. Epidemiology 2019, 30, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Wu, W.; Zhang, M.; Li, J.; Peng, Y.; Miao, T.T.; Zhu, H.; Xu, G. Aberrant upregulation of miR-21 in placental tissues of macrosomia. J. Perinatol. 2014, 34, 658–663. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.T.; Cai, Q.Y.; Ji, S.S.; Zhang, H.X.; Wang, Y.H.; Yan, H.T.; Yang, X.J. Decreased miR-143 and increased miR-21 placental expression levels are associated with macrosomia. Mol. Med. Rep. 2016, 13, 3273–3280. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M.D.; Peiris, H.N.; Kobayashi, M.; Koh, Y.Q.; Duncombe, G.; Illanes, S.E.; Rice, G.E.; Salomon, C. Placental exosomes in normal and complicated pregnancy. Am. J. Obstet. Gynecol. 2015, 213, S173–S181. [Google Scholar] [CrossRef] [PubMed]
- Pillay, P.; Moodley, K.; Moodley, J.; Mackraj, I. Placenta-derived exosomes: Potential biomarkers of preeclampsia. Int. J. Nanomedicine 2017, 12, 8009–8023. [Google Scholar] [CrossRef] [PubMed]
- Meyle, K.D.; Gamborg, M.; Sørensen, T.I.A.; Baker, J.L. Childhood body size and the risk of malignant melanoma in adulthood. Am. J. Epidemiol. 2017, 185, 673–680. [Google Scholar] [CrossRef] [PubMed]
- Dusingize, J.C.; Olsen, C.M.; An, J.; Pandeya, N.; Law, M.H.; Thompson, B.S.; Goldstein, A.M.; Iles, M.M.; Webb, P.M.; Neale, R.E.; et al. Body mass index and height and risk of cutaneous melanoma: Mendelian randomization analyses. Int. J. Epidemiol. 2020, dyaa009. [Google Scholar] [CrossRef]
- Hoppe, C.; Mølgaard, C.; Michaelsen, K.F. Cow′s milk and linear growth in industrialized and developing countries. Annu. Rev. Nutr. 2006, 26, 131–173. [Google Scholar] [CrossRef]
- Chen, X.; Gao, C.; Li, H.; Huang, L.; Sun, Q.; Dong, Y.; Tian, C.; Gao, S.; Dong, H.; Guan, D.; et al. Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res. 2010, 20, 1128–1137. [Google Scholar] [CrossRef]
- Melnik, B.C. Milk—A nutrient system of mammalian evolution promoting mTORC1-dependent translation. Int. J. Mol. Sci. 2015, 16, 17048–17087. [Google Scholar] [CrossRef]
- Yu, S.; Zhao, Z.; Sun, L.; Li, P. Fermentation results in quantitative changes in milk-derived exosomes and different effects on cell growth and survival. J. Agric. Food Chem. 2017, 65, 1220–1228. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C.; Schmitz, G. Exosomes of pasteurized milk: Potential pathogens of Western diseases. J. Transl. Med. 2019, 17, 3. [Google Scholar] [CrossRef]
- Melnik, B.C. Western diet-induced imbalances of FoxO1 and mTORC1 signalling promote the sebofollicular inflammasomopathy acne vulgaris. Exp. Dermatol. 2016, 25, 103–104. [Google Scholar] [CrossRef] [PubMed]
- Agamia, N.F.; Abdallah, D.M.; Sorour, O.; Mourad, B.; Younan, D.N. Skin expression of mammalian target of rapamycin and forkhead box transcription factor O1, and serum insulin-like growth factor-1 in patients with acne vulgaris and their relationship with diet. Br. J. Dermatol. 2016, 174, 1299–1307. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Liang, R.; Yang, Y.; Hou, X.; Wang, Z.; Zhu, S.; Wang, C.; Tang, Z.; Li, K. MicroRNA-21 regulates PI3K/Akt/mTOR signaling by targeting TGFβI during skeletal muscle development in pigs. PLoS ONE 2015, 10, e0119396. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Liu, X.; Zhao, K.; Zhu, Y.; Hu, B.; Zhou, Y.; Wang, M.; Wu, Y.; Zhang, C.; Xu, J.; et al. MiRNA-21 promotes osteogenesis via the PTEN/PI3K/Akt/HIF-1α pathway and enhances bone regeneration in critical size defects. Stem Cell Res. Ther. 2019, 10, 65. [Google Scholar] [CrossRef]
- Damsky, W.; Micevic, G.; Meeth, K.; Muthusamy, V.; Curley, D.P.; Santhanakrishnan, M.; Erdelyi, I.; Platt, J.T.; Huang, L.; Theodosakis, N.; et al. mTORC1 activation blocks BrafV600E-induced growth arrest but is insufficient for melanoma formation. Cancer Cell 2015, 27, 41–56. [Google Scholar] [CrossRef]
- Dey, N.; Das, F.; Ghosh-Choudhury, N.; Mandal, C.C.; Parekh, D.J.; Block, K.; Kasinath, B.S.; Abboud, H.E.; Choudhury, G.G. microRNA-21 governs TORC1 activation in renal cancer cell proliferation and invasion. PLoS ONE 2012, 7, e37366. [Google Scholar] [CrossRef]
- Brandon, E.L.; Gu, J.W.; Cantwell, L.; He, Z.; Wallace, G.; Hall, J.E. Obesity promotes melanoma tumor growth: Role of leptin. Cancer Biol. Ther. 2009, 8, 1871–1879. [Google Scholar] [CrossRef]
- Pandey, V.; Vijayakumar, M.V.; Ajay, A.K.; Malvi, P.; Bhat, M.K. Diet-induced obesity increases melanoma progression: Involvement of Cav-1 and FASN. Int. J. Cancer 2012, 130, 497–508. [Google Scholar] [CrossRef]
- Chen, J.; Chi, M.; Chen, C.; Zhang, X.D. Obesity and melanoma: Exploring molecular links. J. Cell. Biochem. 2013, 114, 1955–1961. [Google Scholar] [CrossRef] [PubMed]
- Malvi, P.; Chaube, B.; Pandey, V.; Vijayakumar, M.V.; Boreddy, P.R.; Mohammad, N.; Singh, S.V.; Bhat, M.K. Obesity induced rapid melanoma progression is reversed by orlistat treatment and dietary intervention: Role of adipokines. Mol. Oncol. 2015, 9, 689–703. [Google Scholar] [CrossRef]
- Karimi, K.; Lindgren, T.H.; Koch, C.A.; Brodell, R.T. Obesity as a risk factor for malignant melanoma and non-melanoma skin cancer. Rev. Endocr. Metab. Disord. 2016, 17, 389–403. [Google Scholar] [CrossRef] [PubMed]
- De Giorgi, V.; Gori, A.; Savarese, I.; D′Errico, A.; Scarfì, F.; Papi, F.; Maio, V.; Covarelli, P.; Massi, D.; Gandini, S. Role of BMI and hormone therapy in melanoma risk: A case-control study. J. Cancer Res. Clin. Oncol. 2017, 143, 1191–1197. [Google Scholar] [CrossRef] [PubMed]
- Coelho, P.; Almeida, J.; Prudêncio, C.; Fernandes, R.; Soares, R. Effect of adipocyte secretome in melanoma progression and vasculogenic mimicry. J. Cell. Biochem. 2016, 117, 1697–1706. [Google Scholar] [CrossRef]
- Ko, J.H.; Um, J.Y.; Lee, S.G.; Yang, W.M.; Sethi, G.; Ahn, K.S. Conditioned media from adipocytes promote proliferation, migration, and invasion in melanoma and colorectal cancer cells. J. Cell. Physiol. 2019, 234, 18249–18261. [Google Scholar] [CrossRef]
- Lazar, I.; Clement, E.; Dauvillier, S.; Milhas, D.; Ducoux-Petit, M.; LeGonidec, S.; Moro, C.; Soldan, V.; Dalle, S.; Balor, S.; et al. Adipocyte exosomes promote melanoma aggressiveness through fatty acid oxidation: A novel mechanism linking obesity and cancer. Cancer Res. 2016, 76, 4051–4057. [Google Scholar] [CrossRef] [PubMed]
- Clement, E.; Lazar, I.; Muller, C.; Nieto, L. Obesity and melanoma: Could fat be fueling malignancy? Pigment Cell Melanoma Res. 2017, 30, 294–306. [Google Scholar] [CrossRef]
- Kim, Y.J.; Hwang, S.H.; Cho, H.H.; Shin, K.K.; Bae, Y.C.; Jung, J.S. MicroRNA 21 regulates the proliferation of human adipose tissue-derived mesenchymal stem cells and high-fat diet-induced obesity alters microRNA 21 expression in white adipose tissues. J. Cell. Physiol. 2012, 227, 183–193. [Google Scholar] [CrossRef]
- Chartoumpekis, D.V.; Zaravinos, A.; Ziros, P.G.; Iskrenova, R.P.; Psyrogiannis, A.I.; Kyriazopoulou, V.E.; Habeos, I.G. Differential expression of microRNAs in adipose tissue after long-term high-fat diet-induced obesity in mice. PLoS ONE 2012, 7, e34872. [Google Scholar] [CrossRef]
- An, Y.; Zhao, J.; Nie, F.; Qin, Z.; Xue, H.; Wang, G.; Li, D. Exosomes from adipose-derived stem cells (ADSCs) overexpressing miR-21 promote vascularization of endothelial cells. Sci. Rep. 2019, 9, 12861. [Google Scholar] [CrossRef]
- Yang, C.; Luo, L.; Bai, X.; Shen, K.; Liu, K.; Wang, J.; Hu, D. Highly-expressed micoRNA-21 in adipose derived stem cell exosomes can enhance the migration and proliferation of the HaCaT cells by increasing the MMP-9 expression through the PI3K/AKT pathway. Arch. Biochem. Biophys. 2020, 681, 108259. [Google Scholar] [CrossRef] [PubMed]
- Amjadi, F.; Javanmard, S.H.; Zarkesh-Esfahani, H.; Khazaei, M.; Narimani, M. Leptin promotes melanoma tumor growth in mice related to increasing circulating endothelial progenitor cells numbers and plasma NO production. J. Exp. Clin. Cancer 2011, 30, 21. [Google Scholar] [CrossRef]
- Park, H.K.; Ahima, R.S. Leptin signaling. F1000Prime Rep. 2014, 6, 73. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhang, N.; Song, L.; Xie, H.; Zhao, C.; Li, S.; Zhao, W.; Zhao, Y.; Gao, C.; Xu, G. Adipokines and free fatty acids regulate insulin sensitivity by increasing microRNA-21 expression in human mature adipocytes. Mol. Med. Rep. 2017, 16, 2254–2258. [Google Scholar] [CrossRef][Green Version]
- Nieman, K.M.; Romero, I.L.; Van Houten, B.; Lengyel, E. Adipose tissue and adipocytes support tumorigenesis and metastasis. Biochim. Biophys. Acta 2013, 1831, 1533–1541. [Google Scholar] [CrossRef] [PubMed]
- Au Yeung, C.L.; Co, N.N.; Tsuruga, T.; Yeung, T.L.; Kwan, S.Y.; Leung, C.S.; Li, Y.; Lu, E.S.; Kwan, K.; Wong, K.K.; et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat. Commun. 2016, 7, 11150. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Qi, X.; Xiong, H.; Liu, Q.; Li, J.; Zhang, Y.; Ma, X.; Wu, N.; Liu, Q.; Feng, L. Type 2 diabetes mellitus and risk of malignant melanoma: A systematic review and meta-analysis of cohort studies. Iran J. Public Health 2014, 43, 857–866. [Google Scholar]
- Sekar, D.; Venugopal, B.; Sekar, P.; Ramalingam, K. Role of microRNA 21 in diabetes and associated/related diseases. Gene 2016, 582, 14–18. [Google Scholar] [CrossRef]
- Nunez Lopez, Y.O.; Garufi, G.; Seyhan, A.A. Altered levels of circulating cytokines and microRNAs in lean and obese individuals with prediabetes and type 2 diabetes. Mol. Biosyst. 2016, 13, 106–121. [Google Scholar] [CrossRef]
- Seyhan, A.A.; Nunez Lopez, Y.O.; Xie, H.; Yi, F.; Mathews, C.; Pasarica, M.; Pratley, R.E. Pancreas-enriched miRNAs are altered in the circulation of subjects with diabetes: A pilot cross-sectional study. Sci. Rep. 2016, 6, 31479. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Kong, L.; Tan, Y.; Epstein, P.N.; Zeng, J.; Gu, J.; Liang, G.; Kong, M.; Chen, X.; Miao, L.; et al. C66 ameliorates diabetic nephropathy in mice by both upregulating NRF2 function via increase in miR-200a and inhibiting miR-21. Diabetologia 2016, 59, 1558–1568. [Google Scholar] [CrossRef] [PubMed]
- Lakhter, A.J.; Pratt, R.E.; Moore, R.E.; Doucette, K.K.; Maier, B.F.; DiMeglio, L.A.; Sims, E.K. Beta cell extracellular vesicle miR-21-5p cargo is increased in response to inflammatory cytokines and serves as a biomarker of type 1 diabetes. Diabetologia 2018, 61, 1124–1134. [Google Scholar] [CrossRef] [PubMed]
- Stocks, T.; Van Hemelrijck, M.; Manjer, J.; Bjørge, T.; Ulmer, H.; Hallmans, G.; Lindkvist, B.; Selmer, R.; Nagel, G.; Tretli, S.; et al. Blood pressure and risk of cancer incidence and mortality in the Metabolic Syndrome and Cancer Project. Hypertension 2012, 59, 802–810. [Google Scholar] [CrossRef] [PubMed]
- Nagel, G.; Bjørge, T.; Stocks, T.; Manjer, J.; Hallmans, G.; Edlinger, M.; Häggström, C.; Engeland, A.; Johansen, D.; Kleiner, A.; et al. Metabolic risk factors and skin cancer in the Metabolic Syndrome and Cancer Project (Me-Can). Br. J. Dermatol. 2012, 167, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Radišauskas, R.; Kuzmickienė, I.; Milinavičienė, E.; Everatt, R. Hypertension, serum lipids and cancer risk: A review of epidemiological evidence. Medicina 2016, 52, 89–98. [Google Scholar] [CrossRef]
- Warner, A.B.; McQuade, J.L. Modifiable host factors in melanoma: Emerging evidence for obesity, diet, exercise, and the microbiome. Curr. Oncol. Rep. 2019, 21, 72. [Google Scholar] [CrossRef]
- Hall, K.D. Did the food environment cause the obesity epidemic? Obesity 2018, 26, 11–13. [Google Scholar] [CrossRef]
- Chen, G.L.; Luo, Y.; Eriksson, D.; Meng, X.; Qian, C.; Bäuerle, T.; Chen, X.X.; Schett, G.; Bozec, A. High fat diet increases melanoma cell growth in the bone marrow by inducing osteopontin and interleukin 6. Oncotarget 2016, 7, 26653–26669. [Google Scholar] [CrossRef] [PubMed]
- Malagoli, C.; Malavolti, M.; Farnetani, F.; Longo, C.; Filippini, T.; Pellacani, G.; Vinceti, M. Food and beverage consumption and melanoma risk: A population-based case-control study in northern Italy. Nutrients 2019, 11, 2206. [Google Scholar] [CrossRef]
- Malavolti, M.; Malagoli, C.; Crespi, C.M.; Brighenti, F.; Agnoli, C.; Sieri, S.; Krogh, V.; Fiorentini, C.; Farnetani, F.; Longo, C.; et al. Glycaemic index, glycaemic load and risk of cutaneous melanoma in a population-based, case-control study. Br. J. Nutr. 2017, 117, 432–438. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Burris, J.; Shikany, J.M.; Rietkerk, W.; Woolf, K. A low glycemic index and glycemic load diet decreases insulin-like growth factor-1 among adults with moderate and severe acne: A short-duration, 2-week randomized controlled trial. J. Acad. Nutr. Diet 2018, 118, 1874–1885. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B. Dietary intervention in acne: Attenuation of increased mTORC1 signaling promoted by Western diet. Dermatoendocrinology 2012, 4, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Xiong, Y.; Li, G.; Liu, M.; He, T.; Tang, Y.; Chen, Y.; Cai, L.; Jiang, R.; Tao, J. MiR-21 is overexpressed in response to high glucose and protects endothelial cells from apoptosis. Exp. Clin. Endocrinol. Diabetes 2013, 121, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Hanousková, B.; Neprašová, B.; Skálová, L.; Maletínská, L.; Zemanová, K.; Ambrož, M.; Matoušková, P. High-fructose drinks affect microRNAs expression differently in lean and obese mice. J. Nutr. Biochem. 2019, 68, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Castellana, M.; Conte, E.; Cignarelli, A.; Perrini, S.; Giustina, A.; Giovanella, L.; Giorgino, F.; Trimboli, P. Efficacy and safety of very low calorie ketogenic diet (VLCKD) in patients with overweight and obesity: A systematic review and meta-analysis. Rev. Endocr. Metab. Disord. 2020, 21, 5–16. [Google Scholar] [CrossRef]
- McDonald, T.J.W.; Cervenka, M.C. Lessons learned from recent clinical trials of ketogenic diet therapies in adults. Curr. Opin. Clin. Nutr. Metab. Care 2019, 22, 418–424. [Google Scholar] [CrossRef]
- Xia, S.; Lin, R.; Jin, L.; Zhao, L.; Kang, H.B.; Pan, Y.; Liu, S.; Qian, G.; Qian, Z.; Konstantakou, E.; et al. Prevention of dietary-fat-fueled ketogenesis attenuates BRAF V600E tumor growth. Cell Metab. 2017, 25, 358–373. [Google Scholar] [CrossRef]
- Kang, H.B.; Fan, J.; Lin, R.; Elf, S.; Ji, Q.; Zhao, L.; Jin, L.; Seo, J.H.; Shan, C.; Arbiser, J.L.; et al. Metabolic rewiring by oncogenic BRAF V600E links ketogenesis pathway to BRAF-MEK1 signaling. Mol. Cell. 2015, 59, 345–358. [Google Scholar] [CrossRef]
- Zhao, L.; Fan, J.; Xia, S.; Pan, Y.; Liu, S.; Qian, G.; Qian, Z.; Kang, H.B.; Arbiser, J.L.; Pollack, B.P.; et al. HMG-CoA synthase 1 is a synthetic lethal partner of BRAFV600E in human cancers. J. Biol. Chem. 2017, 292, 10142–10152. [Google Scholar] [CrossRef]
- Pratilas, C.A.; Taylor, B.S.; Ye, Q.; Viale, A.; Sander, C.; Solit, D.B.; Rosen, N. (V600E) BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc. Natl. Acad. Sci. USA 2009, 106, 4519–4524. [Google Scholar] [CrossRef]
- Banikazemi, Z.; Haji, H.A.; Mohammadi, M.; Taheripak, G.; Iranifar, E.; Poursadeghiyan, M.; Moridikia, A.; Rashidi, B.; Taghizadeh, M.; Mirzaei, H. Diet and cancer prevention: Dietary compounds, dietary microRNAs, and dietary exosomes. J. Cell. Biochem. 2018, 119, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Munir, J.; Lee, M.; Ryu, S. Exosomes in food: Health benefits and clinical relevance in diseases. Adv. Nutr. 2020, 11, 687–696. [Google Scholar] [CrossRef]
- Manca, S.; Upadhyaya, B.; Mutai, E.; Desaulniers, A.T.; Cederberg, R.A.; White, B.R.; Zempleni, J. Milk exosomes are bioavailable and distinct microRNA cargos have unique tissue distribution patterns. Sci. Rep. 2018, 8, 11321. [Google Scholar] [CrossRef] [PubMed]
- Zempleni, J.; Sukreet, S.; Zhou, F.; Wu, D.; Mutai, E. Milk-derived exosomes and metabolic regulation. Annu. Rev. Anim. Biosci. 2019, 7, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Golan-Gerstl, R.; Elbaum Shiff, Y.; Moshayoff, V.; Schecter, D.; Leshkowitz, D.; Reif, S. Characterization and biological function of milk-derived miRNAs. Mol. Nutr. Food Res. 2017, 61, 10. [Google Scholar] [CrossRef] [PubMed]
- Tucker, L.A. Milk fat intake and telomere length in U.S. women and men: The role of the milk fat fraction. Oxid. Med. Cell. Longev. 2019, 2019, 1574021. [Google Scholar] [CrossRef]
- Zhu, H.Y.; Li, C.; Bai, W.D.; Su, L.L.; Liu, J.Q.; Li, Y.; Shi, J.H.; Cai, W.X.; Bai, X.Z.; Jia, Y.H.; et al. MicroRNA-21 regulates hTERT via PTEN in hypertrophic scar fibroblasts. PLoS ONE 2014, 9, e97114. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, J.J.; Tao, H.; Jin, W.S. MicroRNA-21 controls hTERT via PTEN in human colorectal cancer cell proliferation. J. Physiol. Biochem. 2015, 71, 59–68. [Google Scholar] [CrossRef]
- De Unamuno Bustos, B.; Sahuquillo Torralba, A.; Moles Poveda, P.; Pérez Simó, G.; Simarro Farinos, J.; Llavador Ros, M.; Palanca Suela, S.; Botella Estrada, R. Telomerase expression in a series of melanocytic neoplasms. Actas Dermosifiliogr. 2019, 110, 212–219. [Google Scholar] [CrossRef]
- Shaughnessy, M.; Njauw, C.N.; Artomov, M.; Tsao, H. Classifying melanoma by TERT promoter mutational status. J. Investig. Dermatol. 2020, 140, 390–394. [Google Scholar] [CrossRef] [PubMed]
- Grant, B.F.; Chou, S.P.; Saha, T.D.; Pickering, R.P.; Kerridge, B.T.; Ruan, W.J.; Huang, B.; Jung, J.; Zhang, H.; Fan, A.; et al. Prevalence of 12-month alcohol use, high-risk drinking, and DSM-IV alcohol use disorder in the United States, 2001–2002 to 2012–2013: Results from the National Epidemiologic Survey on Alcohol and Related Conditions. JAMA Psychiatry 2017, 74, 911–923. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Fung, T.T.; Nan, H. An epidemiological review of diet and cutaneous malignant melanoma. Cancer Epidemiol. Biomarkers Prev. 2018, 27, 1115–1122. [Google Scholar] [CrossRef] [PubMed]
- Lefèvre, A.; Adler, H.; Lieber, C.S. Effect of ethanol on ketone metabolism. J. Clin. Investig. 1970, 49, 1775–1782. [Google Scholar] [CrossRef] [PubMed]
- Beech, R.D.; Leffert, J.J.; Lin, A.; Hong, K.A.; Hansen, J.; Umlauf, S.; Mane, S.; Zhao, H.; Sinha, R. Stress-related alcohol consumption in heavy drinkers correlates with expression of miR-10a, miR-21, and components of the TAR-RNA-binding protein-associated complex. Alcohol Clin. Exp. Res. 2014, 38, 2743–2753. [Google Scholar] [CrossRef]
- Bian, J.T.; Piano, M.R.; Kotlo, K.U.; Mahmoud, A.M.; Phillips, S.A. MicroRNA-21 contributes to reduced microvascular function in binge drinking young adults. Alcohol Clin. Exp. Res. 2018, 42, 278–285. [Google Scholar] [CrossRef]
- Donat-Vargas, C.; Berglund, M.; Glynn, A.; Wolk, A.; Åkesson, A. Dietary polychlorinated biphenyls, long-chain n-3 polyunsaturated fatty acids and incidence of malignant melanoma. Eur. J. Cancer 2017, 72, 137–143. [Google Scholar] [CrossRef]
- Ju, L.; Zhou, Z.; Jiang, B.; Lou, Y.; Zhang, Z. miR-21 is involved in skeletal deficiencies of zebrafish embryos exposed to polychlorinated biphenyls. Environ. Sci. Pollut. Res. Int. 2017, 24, 886–891. [Google Scholar] [CrossRef]
- Wahlang, B.; Petriello, M.C.; Perkins, J.T.; Shen, S.; Hennig, B. Polychlorinated biphenyl exposure alters the expression profile of microRNAs associated with vascular diseases. Toxicol. In Vitro 2016, 35, 180–187. [Google Scholar] [CrossRef][Green Version]
- Furman, D.; Campisi, J.; Verdin, E.; Carrera-Bastos, P.; Targ, S.; Franceschi, C.; Ferrucci, L.; Gilroy, D.W.; Fasano, A.; Miller, G.W.; et al. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 2019, 25, 1822–1832. [Google Scholar] [CrossRef]
- Newton-Bishop, J.A.; Davies, J.R.; Latheef, F.; Randerson-Moor, J.; Chan, M.; Gascoyne, J.; Waseem, S.; Haynes, S.; O′Donovan, C.; Bishop, D.T. 25-Hydroxyvitamin D2/D3 levels and factors associated with systemic inflammation and melanoma survival in the Leeds Melanoma Cohort. Int. J. Cancer 2015, 136, 2890–2899. [Google Scholar] [CrossRef] [PubMed]
- Hardie, C.M.; Elliott, F.; Chan, M.; Rogers, Z.; Bishop, D.T.; Newton-Bishop, J.A. Environmental exposures such as smoking and low vitamin D are predictive of poor outcome in cutaneous melanoma rather than other deprivation measures. J. Investig. Dermatol. 2020, 140, 327–337. [Google Scholar] [CrossRef]
- Dusingize, J.C.; Olsen, C.M.; Pandeya, N.; Thompson, B.S.; Webb, P.M.; Green, A.C.; Neale, R.E.; Whiteman, D.C. QSkin Study. Smoking and cutaneous melanoma: Findings from the QSkin Sun and Health Cohort Study. Cancer Epidemiol. Biomarkers Prev. 2018, 27, 874–881. [Google Scholar] [CrossRef] [PubMed]
- Gibson, J.A.G.; Dobbs, T.D.; Griffiths, R.; Song, J.; Akbari, A.; Whitaker, S.; Watkins, A.; Langan, S.M.; Hutchings, H.A.; Lyons, R.A.; et al. The association of smoking and socioeconomic status on cutaneous melanoma: A population based, data linkage, case-control study. Br. J. Dermatol. 2020, 182, 1136–1147. [Google Scholar] [CrossRef] [PubMed]
- Sondermeijer, L.; Lamboo, L.G.E.; de Waal, A.C.; Galesloot, T.E.; Kiemeney, L.A.L.M.; van Rossum, M.; Aben, K.H. Cigarette smoking and the risk of cutaneous melanoma: A case-control study. Dermatology 2020, 236, 228–236. [Google Scholar] [CrossRef]
- Zhang, Y.; Pan, T.; Zhong, X.; Cheng, C. Nicotine upregulates microRNA-21 and promotes TGF-β-dependent epithelial-mesenchymal transition of esophageal cancer cells. Tumour Biol. 2014, 35, 7063–7072. [Google Scholar] [CrossRef]
- Xu, H.; Ling, M.; Xue, J.; Dai, X.; Sun, Q.; Chen, C.; Liu, Y.; Zhou, L.; Liu, J.; Luo, F.; et al. Exosomal microRNA-21 derived from bronchial epithelial cells is involved in aberrant epithelium-fibroblast cross-talk in COPD induced by cigarette smoking. Theranostics 2018, 8, 5419–5433. [Google Scholar] [CrossRef]
- Zhu, J.; Liu, B.; Wang, Z.; Wang, D.; Ni, H.; Zhang, L.; Wang, Y. Exosomes from nicotine-stimulated macrophages accelerate atherosclerosis through miR-21-3p/PTEN-mediated VSMC migration and proliferation. Theranostics 2019, 9, 6901–6919. [Google Scholar] [CrossRef]
- Zhou, F.; Li, S.; Jia, W.; Lv, G.; Song, C.; Kang, C.; Zhang, Q. Effects of diesel exhaust particles on microRNA-21 in human bronchial epithelial cells and potential carcinogenic mechanisms. Mol. Med. Rep. 2015, 12, 2329–2335. [Google Scholar] [CrossRef]
- Goldenberg, A.; Jiang, S.I.; Cohen, P.R. A possible association between melanoma and prostate cancer. Results from a case-control-study. Cancers 2015, 7, 670–678. [Google Scholar] [CrossRef]
- Wang, Y.; Ou, Z.; Sun, Y.; Yeh, S.; Wang, X.; Long, J.; Chang, C. Androgen receptor promotes melanoma metastasis via altering the miRNA-539-3p/USP13/MITF/AXL signals. Oncogene 2017, 36, 1644–1654. [Google Scholar] [CrossRef] [PubMed]
- Rutkowski, K.; Sowa, P.; Rutkowska-Talipska, J.; Kuryliszyn-Moskal, A.; Rutkowski, R. Dehydroepiandrosterone (DHEA): Hypes and hopes. Drugs 2014, 74, 1195–1207. [Google Scholar] [CrossRef]
- Teng, Y.; Litchfield, L.M.; Ivanova, M.M.; Prough, R.A.; Clark, B.J.; Klinge, C.M. Dehydroepiandrosterone-induces miR-21 transcription in HepG2 cells through estrogen receptor β and androgen receptor. Mol. Cell. Endocrinol. 2014, 392, 23–36. [Google Scholar] [CrossRef]
- Sustarsic, E.G.; Junnila, R.K.; Kopchick, J.J. Human metastatic melanoma cell lines express high levels of growth hormone receptor and respond to GH treatment. Biochem. Biophys. Res. Commun. 2013, 441, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Basu, R.; Wu, S.; Kopchick, J.J. Targeting growth hormone receptor in human melanoma cells attenuates tumor progression and epithelial mesenchymal transition via suppression of multiple oncogenic pathways. Oncotarget 2017, 8, 21579–21598. [Google Scholar] [CrossRef] [PubMed]
- Basu, R.; Kulkarni, P.; Qian, Y.; Walsh, C.; Arora, P.; Davis, E.; Duran-Ortiz, S.; Funk, K.; Ibarra, D.; Kruse, C.; et al. Growth hormone upregulates melanocyte-inducing transcription factor expression and activity via JAK2-STAT5 and SRC signaling in GH receptor-positive human melanoma. Cancers 2019, 11, 1352. [Google Scholar] [CrossRef]
- Palabiyik, O.; Tastekin, E.; Doganlar, Z.B.; Tayfur, P.; Dogan, A.; Vardar, S.A. Alteration in cardiac PI3K/Akt/mTOR and ERK signaling pathways with the use of growth hormone and swimming, and the roles of miR21 and miR133. Biomed. Rep. 2019, 10, 97–106. [Google Scholar] [CrossRef]
- Caldarola, G.; Battista, C.; Pellicano, R. Melanoma onset after estrogen, thyroid, and growth hormone replacement therapy. Clin. Ther. 2010, 32, 57–59. [Google Scholar] [CrossRef]
- Handler, M.Z.; Ross, A.L.; Shiman, M.I.; Elgart, G.W.; Grichnik, J.M. Potential role of human growth hormone in melanoma growth promotion. Arch. Dermatol. 2012, 148, 1179–1182. [Google Scholar] [CrossRef][Green Version]
- Melnik, B.C.; John, S.M.; Schmitz, G. Milk is not just food but most likely a genetic transfection system activating mTORC1 signaling for postnatal growth. Nutr. J. 2013, 12, 103. [Google Scholar] [CrossRef]
- Rich-Edwards, J.W.; Ganmaa, D.; Pollak, M.N.; Nakamoto, E.K.; Kleinman, K.; Tserendolgor, U.; Willett, W.C.; Frazier, A.L. Milk consumption and the prepubertal somatotropic axis. Nutr. J. 2007, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Van Vught, A.J.; Nieuwenhuizen, A.G.; Veldhorst, M.A.; Brummer, R.J.; Westerterp-Plantenga, M.S. The effects of dietary protein on the somatotropic axis: A comparison of soy, gelatin, alpha-lactalbumin and milk. Eur. J. Clin. Nutr. 2010, 64, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C. Androgen abuse in the community. Curr. Opin. Endocrinol. Diabetes Obes. 2009, 16, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Fearfield, L.; Nobbs, J.; Petruckevitch, A.; Harland, C. Severe vitamin D deficiency associated with BRAF-mutated melanoma. Br. J. Dermatol. 2019, 181, 1343. [Google Scholar] [CrossRef] [PubMed]
- Newton-Bishop, J.A.; Beswick, S.; Randerson-Moor, J.; Chang, Y.M.; Affleck, P.; Elliott, F.; Chan, M.; Leake, S.; Karpavicius, B.; Haynes, S.; et al. Serum 25-hydroxyvitamin D3 levels are associated with Breslow thickness at presentation and survival from melanoma. J. Clin. Oncol. 2009, 27, 5439–5444. [Google Scholar] [CrossRef]
- Bade, B.; Zdebik, A.; Wagenpfeil, S.; Gräber, S.; Geisel, J.; Vogt, T.; Reichrath, J. Low serum 25-hydroxyvitamin D concentrations are associated with increased risk for melanoma and unfavourable prognosis. PLoS ONE 2014, 9, e112863. [Google Scholar] [CrossRef]
- Slominski, A.T.; Brożyna, A.A.; Zmijewski, M.A.; Jóźwicki, W.; Jetten, A.M.; Mason, R.S.; Tuckey, R.C.; Elmets, C.A. Vitamin D signaling and melanoma: Role of vitamin D and its receptors in melanoma progression and management. Lab. Investig. 2017, 97, 706–724. [Google Scholar] [CrossRef]
- Slominski, A.T.; Brożyna, A.A.; Skobowiat, C.; Zmijewski, M.A.; Kim, T.K.; Janjetovic, Z.; Oak, A.S.; Jozwicki, W.; Jetten, A.M.; Mason, R.S.; et al. On the role of classical and novel forms of vitamin D in melanoma progression and management. J. Steroid Biochem. Mol. Biol. 2018, 177, 159–170. [Google Scholar] [CrossRef]
- Stucci, L.S.; D′Oronzo, S.; Tucci, M.; Macerollo, A.; Ribero, S.; Spagnolo, F.; Marra, E.; Picasso, V.; Orgiano, L.; Marconcini, R.; et al. Vitamin D in melanoma: Controversies and potential role in combination with immune check-point inhibitors. Cancer Treat. Rev. 2018, 69, 21–28. [Google Scholar] [CrossRef]
- Dambal, S.; Giangreco, A.A.; Acosta, A.M.; Fairchild, A.; Richards, Z.; Deaton, R.; Wagner, D.; Vieth, R.; Gann, P.H.; Kajdacsy-Balla, A.; et al. microRNAs and DICER1 are regulated by 1,25-dihydroxyvitamin D in prostate stroma. J. Steroid Biochem. Mol. Biol. 2017, 167, 192–202. [Google Scholar] [CrossRef]
- Xu, Y.; Qian, J.; Yu, Z. Budesonide up-regulates vitamin D receptor expression in human bronchial fibroblasts and enhances the inhibitory effect of calcitriol on airway remodeling. Allergol. Immunopathol. (Madr) 2019, 47, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; BinHumaid, F.S.; Alzahrani, A.S.; Baitei, E.Y.; Al-Mohanna, F.A.; Meyer, B.F.; Shi, Y. Increased CYP24A1 expression is associated with BRAF(V600E) mutation and advanced stages in papillary thyroid carcinoma. Clin. Endocrinol. (Oxf.) 2014, 81, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, Y.; Noshiro, M.; Okuda, K. Cloning and expression of cDNA encoding 25-hydroxyvitamin D3 24-hydroxylase. FEBS Lett. 1991, 278, 195–198. [Google Scholar] [CrossRef]
- Brożyna, A.A.; Jochymski, C.; Janjetovic, Z.; Jóźwicki, W.; Tuckey, R.C.; Slominski, A.T. CYP24A1 expression inversely correlates with melanoma progression: Clinic-pathological studies. Int. J. Mol. Sci. 2014, 15, 19000–19017. [Google Scholar] [CrossRef]
- Wacker, M.; Holick, M.F. Sunlight and vitamin D: A global perspective for health. Dermatoendocrinol. 2013, 5, 51–108. [Google Scholar] [CrossRef]
- Tuckey, R.C.; Li, W.; Ma, D.; Cheng, C.Y.S.; Wang, K.M.; Kim, T.K.; Jeayeng, S.; Slominski, A.T. CYP27A1 acts on the pre-vitamin D3 photoproduct, lumisterol, producing biologically active hydroxy-metabolites. J. Steroid. Biochem. Mol. Biol. 2018, 181, 1–10. [Google Scholar] [CrossRef]
- Slominski, A.T.; Kim, T.K.; Hobrath, J.V.; Janjetovic, Z.; Oak, A.S.W.; Postlethwaite, A.; Lin, Z.; Li, W.; Takeda, Y.; Jetten, A.M.; et al. Characterization of a new pathway that activates lumisterol in vivo to biologically active hydroxylumisterols. Sci. Rep. 2017, 7, 11434. [Google Scholar] [CrossRef]
- Slominski, A.; Kim, T.K.; Zmijewski, M.A.; Janjetovic, Z.; Li, W.; Chen, J.; Kusniatsova, E.I.; Semak, I.; Postlethwaite, A.; Miller, D.D.; et al. Novel vitamin D photoproducts and their precursors in the skin. Dermatoendocrinology 2013, 5, 7–19. [Google Scholar] [CrossRef]
- Merrill, S.J.; Ashrafi, S.; Subramanian, M.; Godar, D.E. Exponentially increasing incidences of cutaneous malignant melanoma in Europe correlate with low personal annual UV doses and suggests 2 major risk factors. Dermatoendocrinology 2015, 7, e1004018. [Google Scholar] [CrossRef]
- Montero, I.; Requena, C.; Traves, V.; García-Casado, Z.; Kumar, R.; Nagore, E. Age-related characteristics of cutaneous melanoma in a Spanish Mediterranean population. Int. J. Dermatol. 2015, 54, 778–784. [Google Scholar] [CrossRef]
- Cavanaugh-Hussey, M.W.; Mu, E.W.; Kang, S.; Balch, C.M.; Wang, T. Older age is associated with a higher incidence of melanoma death but a lower incidence of sentinel lymph node metastasis in the SEER databases (2003–2011). Ann. Surg. Oncol. 2015, 22, 2120–2126. [Google Scholar] [CrossRef]
- Ribero, S.; Stucci, L.S.; Marra, E.; Marconcini, R.; Spagnolo, F.; Orgiano, L.; Picasso, V.; Queirolo, P.; Palmieri, G.; Quaglino, P.; et al. Effect of age on melanoma risk, prognosis and treatment response. Acta Derm. Venereol. 2018, 98, 624–629. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, F.; Spazzafumo, L.; Santini, G.; Lazzarini, R.; Albertini, M.C.; Rippo, M.R.; Galeazzi, R.; Abbatecola, A.M.; Marcheselli, F.; Monti, D.; et al. Age-related differences in the expression of circulating microRNAs: miR-21 as a new circulating marker of inflammaging. Mech. Ageing Dev. 2012, 133, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Halper, B.; Hofmann, M.; Oesen, S.; Franzke, B.; Stuparits, P.; Vidotto, C.; Tschan, H.; Bachl, N.; Strasser, E.M.; Quittan, M.; et al. Influence of age and physical fitness on miRNA-21, TGF-β and its receptors in leukocytes of healthy women. Exerc. Immunol. Rev. 2015, 21, 154–163. [Google Scholar] [PubMed]
- Zitvogel, L.; Pietrocola, F.; Kroemer, G. Nutrition, inflammation and cancer. Nat. Immunol. 2017, 18, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef]
- Fan, Y.; Mao, R.; Yang, J. NF-κB and STAT3 signaling pathways collaboratively link inflammation to cancer. Protein Cell. 2013, 4, 176–185. [Google Scholar] [CrossRef]
- Xue, Z.; Xi, Q.; Liu, H.; Guo, X.; Zhang, J.; Zhang, Z.; Li, Y.; Yang, G.; Zhou, D.; Yang, H.; et al. miR-21 promotes NLRP3 inflammasome activation to mediate pyroptosis and endotoxic shock. Cell Death Dis 2019, 10, 461. [Google Scholar] [CrossRef]
- Ning, Z.W.; Luo, X.Y.; Wang, G.Z.; Li, Y.; Pan, M.X.; Yang, R.Q.; Ling, X.G.; Huang, S.; Ma, X.X.; Jin, S.Y.; et al. MicroRNA-21 mediates angiotensin II-induced liver fibrosis by activating NLRP3 inflammasome/IL-1β axis via targeting Smad7 and Spry1. Antioxid. Redox Signal. 2017, 27, 1–20. [Google Scholar] [CrossRef]
- Loboda, A.; Sobczak, M.; Jozkowicz, A.; Dulak, J. TGF-β1/Smads and miR-21 in renal fibrosis and inflammation. Mediators Inflamm. 2016, 2016, 8319283. [Google Scholar] [CrossRef]
- Specjalski, K.; Jassem, E. MicroRNAs: Potential biomarkers and targets of therapy in allergic diseases? Arch. Immunol. Ther. Exp. (Warsz.) 2019, 67, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Van den Berge, M.; Tasena, H. Role of microRNAs and exosomes in asthma. Curr. Opin. Pulm. Med. 2019, 25, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Isanejad, A.; Alizadeh, A.M.; Amani Shalamzari, S.; Khodayari, H.; Khodayari, S.; Khori, V.; Khojastehnjad, N. MicroRNA-206, let-7a and microRNA-21 pathways involved in the anti-angiogenesis effects of the interval exercise training and hormone therapy in breast cancer. Life Sci. 2016, 151, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.; Åkerström, T.; Rinnov, A.; Yfanti, C.; Scheele, C.; Pedersen, B.K.; Laye, M.J. The miRNA plasma signature in response to acute aerobic exercise and endurance training. PLoS ONE 2014, 9, e87308. [Google Scholar] [CrossRef] [PubMed]
- Pinto, R.; Strippoli, S.; De Summa, S.; Albano, A.; Azzariti, A.; Guida, G.; Popescu, O.; Lorusso, V.; Guida, M.; Tommasi, S. MicroRNA expression in BRAF-mutated and wild-type metastatic melanoma and its correlation with response duration to BRAF inhibitors. Expert Opin. Ther. Targets 2015, 19, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Lunavat, T.R.; Cheng, L.; Einarsdottir, B.O.; Olofsson Bagge, R.; Veppil Muralidharan, S.; Sharples, R.A.; Lässer, C.; Gho, Y.S.; Hill, A.F.; Nilsson, J.A.; et al. BRAFV600 inhibition alters the microRNA cargo in the vesicular secretome of malignant melanoma cells. Proc. Natl. Acad. Sci. USA 2017, 114, E5930–E5939. [Google Scholar] [CrossRef]
- Jaune, E.; Rocchi, S. Metformin: Focus on melanoma. Front. Endocrinol. 2018, 9, 472. [Google Scholar] [CrossRef]
- De Souza Neto, F.P.; Bernardes, S.S.; Marinello, P.C.; Melo, G.P.; Luiz, R.C.; Cecchini, R.; Cecchini, A. Metformin: Oxidative and proliferative parameters in-vitro and in-vivo models of murine melanoma. Melanoma Res. 2017, 27, 536–544. [Google Scholar] [CrossRef]
- Li, K.; Zhang, T.T.; Wang, F.; Cui, B.; Zhao, C.X.; Yu, J.J.; Lv, X.X.; Zhang, X.W.; Yang, Z.N.; Huang, B.; et al. Metformin suppresses melanoma progression by inhibiting KAT5-mediated SMAD3 acetylation, transcriptional activity and TRIB3 expression. Oncogene 2018, 37, 2967–2981. [Google Scholar] [CrossRef]
- Ryabaya, O.; Prokofieva, A.; Akasov, R.; Khochenkov, D.; Emelyanova, M.; Burov, S.; Markvicheva, E.; Inshakov, A.; Stepanova, E. Metformin increases antitumor activity of MEK inhibitor binimetinib in 2D and 3D models of human metastatic melanoma cells. Biomed. Pharmacother. 2019, 109, 2548–2560. [Google Scholar] [CrossRef]
- Montaudié, H.; Cerezo, M.; Bahadoran, P.; Roger, C.; Passeron, T.; Machet, L.; Arnault, J.P.; Verneuil, L.; Maubec, E.; Aubin, F.; et al. Metformin monotherapy in melanoma: A pilot, open-label, prospective, and multicentric study indicates no benefit. Pigment Cell Melanoma Res. 2017, 30, 378–380. [Google Scholar] [CrossRef] [PubMed]
- Vujic, I.; Sanlorenzo, M.; Posch, C.; Esteve-Puig, R.; Yen, A.J.; Kwong, A.; Tsumura, A.; Murphy, R.; Rappersberger, K.; Ortiz-Urda, S. Metformin and trametinib have synergistic effects on cell viability and tumor growth in NRAS mutant cancer. Oncotarget 2015, 6, 969–978. [Google Scholar] [CrossRef] [PubMed]
- Afzal, M.Z.; Mercado, R.R.; Shirai, K. Efficacy of metformin in combination with immune checkpoint inhibitors (anti-PD-1/anti-CTLA-4) in metastatic malignant melanoma. J. Immunother. Cancer 2018, 6, 64. [Google Scholar] [CrossRef] [PubMed]
- Tseng, H.W.; Li, S.C.; Tsai, K.W. Metformin treatment suppresses melanoma cell growth and motility through modulation of microRNA expression. Cancers 2019, 11, 209. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, H.A.; Iliopoulos, D.; Struhl, K. Metformin inhibits the inflammatory response associated with cellular transformation and cancer stem cell growth. Proc. Natl. Acad. Sci. USA 2013, 110, 972–977. [Google Scholar] [CrossRef] [PubMed]
- Leidgens, V.; Proske, J.; Rauer, L.; Moeckel, S.; Renner, K.; Bogdahn, U.; Riemenschneider, M.J.; Proescholdt, M.; Vollmann-Zwerenz, A.; Hau, P.; et al. Stattic and metformin inhibit brain tumor initiating cells by reducing STAT3-phosphorylation. Oncotarget 2017, 8, 8250–8263. [Google Scholar] [CrossRef]
- Esparza-López, J.; Alvarado-Muñoz, J.F.; Escobar-Arriaga, E.; Ulloa-Aguirre, A.; de Jesús Ibarra-Sánchez, M. Metformin reverses mesenchymal phenotype of primary breast cancer cells through STAT3/NF-κB pathways. BMC Cancer 2019, 19, 728. [Google Scholar] [CrossRef]
- Wang, J.; Gao, Y.; Duan, L.; Wei, S.; Liu, J.; Tian, L.; Quan, J.; Zhang, Q.; Liu, J.; Yang, J. Metformin ameliorates skeletal muscle insulin resistance by inhibiting miR-21 expression in a high-fat dietary rat model. Oncotarget 2017, 8, 98029–98039. [Google Scholar] [CrossRef]
- Luo, M.; Tan, X.; Mu, L.; Luo, Y.; Li, R.; Deng, X.; Chen, N.; Ren, M.; Li, Y.; Wang, L.; et al. MiRNA-21 mediates the antiangiogenic activity of metformin through targeting PTEN and SMAD7 expression and PI3K/AKT pathway. Sci. Rep. 2017, 7, 43427. [Google Scholar] [CrossRef]
- Deng, Y.; Ma, W. Metformin inhibits HaCaT cell viability via the miR-21/PTEN/Akt signaling pathway. Mol. Med. Rep. 2018, 17, 4062–4066. [Google Scholar] [CrossRef]
- Demirsoy, İ.H.; Ertural, D.Y.; Balci, Ş.; Çınkır, Ü.; Sezer, K.; Tamer, L.; Aras, N. Profiles of circulating miRNAs following metformin treatment in patients with type 2 diabetes. J. Med. Biochem. 2018, 37, 499–506. [Google Scholar] [PubMed]
- Bao, B.; Azmi, A.S.; Ali, S.; Zaiem, F.; Sarkar, F.H. Metformin may function as anti-cancer agent via targeting cancer stem cells: The potential biological significance of tumor-associated miRNAs in breast and pancreatic cancers. Ann. Transl. Med. 2014, 2, 59. [Google Scholar] [PubMed]
- Kokolus, K.M.; Zhang, Y.; Sivik, J.M.; Schmeck, C.; Zhu, J.; Repasky, E.A.; Drabick, J.J.; Schell, T.D. Beta blocker use correlates with better overall survival in metastatic melanoma patients and improves the efficacy of immunotherapies in mice. Oncoimmunology 2017, 7, e1405205. [Google Scholar] [CrossRef] [PubMed]
- De Giorgi, V.; Grazzini, M.; Benemei, S.; Marchionni, N.; Botteri, E.; Pennacchioli, E.; Geppetti, P.; Gandini, S. Propranolol for off-label treatment of patients with melanoma: Results from a cohort study. JAMA Oncol. 2018, 4, e172908. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, P.; Miyamoto, D.; Goyeneche, A.; de Alba Graue, P.G.; Jin, E.; Tsering, T.; Dias, A.B.; Burnier, M.N.; Burnier, J.V. Beta-blockers exert potent anti-tumor effects in cutaneous and uveal melanoma. Cancer Med. 2019, 8, 7265–7277. [Google Scholar] [CrossRef]
- Sayed, D.; Rane, S.; Lypowy, J.; He, M.; Chen, I.Y.; Vashistha, H.; Yan, L.; Malhotra, A.; Vatner, D.; Abdellatif, M. MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol. Biol. Cell 2008, 19, 3272–3282. [Google Scholar] [CrossRef]
- Hou, Y.; Sun, Y.; Shan, H.; Li, X.; Zhang, M.; Zhou, X.; Xing, S.; Sun, H.; Chu, W.; Qiao, G.; et al. β-adrenoceptor regulates miRNA expression in rat heart. Med. Sci. Monit. 2012, 18, BR309–BR314. [Google Scholar] [CrossRef]
- Liu, D.; Yang, Z.; Wang, T.; Yang, Z.; Chen, H.; Hu, Y.; Hu, C.; Guo, L.; Deng, Q.; Liu, Y.; et al. β2-AR signaling controls trastuzumab resistance-dependent pathway. Oncogene 2016, 35, 47–58. [Google Scholar]
- Gryshkova, V.; Fleming, A.; McGhan, P.; De Ron, P.; Fleurance, R.; Valentin, J.P.; Nogueira da Costa, A. miR-21-5p as a potential biomarker of inflammatory infiltration in the heart upon acute drug-induced cardiac injury in rats. Toxicol. Lett. 2018, 286, 31–38. [Google Scholar] [CrossRef]
- Carrillo, E.D.; Escobar, Y.; González, G.; Hernández, A.; Galindo, J.M.; García, M.C.; Sánchez, J.A. Posttranscriptional regulation of the β2-subunit of cardiac L-type Ca2+ channels by MicroRNAs during long-term exposure to isoproterenol in rats. J. Cardiovasc. Pharmacol. 2011, 58, 470–478. [Google Scholar] [CrossRef]
- Zhang, W.; Qu, X.; Chen, B.; Snyder, M.; Wang, M.; Li, B.; Tang, Y.; Chen, H.; Zhu, W.; Zhan, L.; et al. Critical roles of STAT3 in β-adrenergic functions in the heart. Circulation 2016, 133, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Balligand, J.L. β-Adrenergic receptors cooperate with transcription factors: The “STAT” of their union. Circulation 2016, 133, 4–7. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chimenti, I.; Pagano, F.; Cavarretta, E.; Angelini, F.; Peruzzi, M.; Barretta, A.; Greco, E.; De Falco, E.; Marullo, A.G.; Sciarretta, S.; et al. Β-blockers treatment of cardiac surgery patients enhances isolation and improves phenotype of cardiosphere-derived cells. Sci. Rep. 2016, 6, 36774. [Google Scholar] [CrossRef]
- Ji, Y.; Chen, S.; Xu, C.; Li, L.; Xiang, B. The use of propranolol in the treatment of infantile haemangiomas: An update on potential mechanisms of action. Br. J. Dermatol. 2015, 172, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Miroshnichenko, S.K.; Patutina, O.A.; Burakova, E.A.; Chelobanov, B.P.; Fokina, A.A.; Vlassov, V.V.; Altman, S.; Zenkova, M.A.; Stetsenko, D.A. Mesyl phosphoramidate antisense oligonucleotides as an alternative to phosphorothioates with improved biochemical and biological properties. Proc. Natl. Acad. Sci. USA 2019, 116, 1229–1234. [Google Scholar] [CrossRef]
- Patutina, O.A.; Miroshnichenko, S.K.; Mironova, N.L.; Sen′kova, A.V.; Bichenkova, E.V.; Clarke, D.J.; Vlassov, V.V.; Zenkova, M.A. Catalytic knockdown of miR-21 by artificial ribonuclease: Biological performance in tumor model. Front. Pharmacol. 2019, 10, 879. [Google Scholar] [CrossRef]
- Bhere, D.; Arghiani, N.; Lechtich, E.R.; Yao, Y.; Alsaab, S.; Bei, F.; Matin, M.M.; Shah, K. Simultaneous downregulation of miR-21 and upregulation of miR-7 has anti-tumor efficacy. Sci. Rep. 2020, 10, 1779. [Google Scholar] [CrossRef]
- Wei, X.; You, X.; Zhang, J.; Zhou, C. miR-21 inhibitor facilitates the anticancer activity of doxorubicin loaded nanometer in melanoma. Oncol. Rep. 2019, 42, 414–424. [Google Scholar] [CrossRef]
- Zhang, H.L.; Si, L.B.; Zeng, A.; Long, F.; Qi, Z.; Zhao, R.; Bai, M. MicroRNA-21 antisense oligonucleotide improves the sensitivity of A375 human melanoma cell to Cisplatin: An in vitro study. J. Cell. Biochem. 2018, 119, 3129–3141. [Google Scholar] [CrossRef]
- Rui, M.; Qu, Y.; Gao, T.; Ge, Y.; Feng, C.; Xu, X. Simultaneous delivery of anti-miR21 with doxorubicin prodrug by mimetic lipoprotein nanoparticles for synergistic effect against drug resistance in cancer cells. Int. J. Nanomed. 2016, 12, 217–237. [Google Scholar] [CrossRef]
- Liang, G.; Zhu, Y.; Ali, D.J.; Tian, T.; Xu, H.; Si, K.; Sun, B.; Chen, B.; Xiao, Z. Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J. Nanobiotechnol. 2020, 18, 10. [Google Scholar] [CrossRef] [PubMed]
- Agha, A.; Tarhini, A.A. Adjuvant therapy for melanoma. Curr. Oncol. Rep. 2017, 19, 36. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bazhin, A.V.; von Ahn, K.; Fritz, J.; Werner, J.; Karakhanova, S. Interferon-α up-regulates the expression of PD-L1 molecules on immune cells through STAT3 and p38 signaling. Front. Immunol. 2018, 9, 2129. [Google Scholar] [CrossRef] [PubMed]
- Reinsbach, S.; Nazarov, P.V.; Philippidou, D.; Schmitt, M.; Wienecke-Baldacchino, A.; Muller, A.; Vallar, L.; Behrmann, I.; Kreis, S. Dynamic regulation of microRNA expression following interferon-γ-induced gene transcription. RNA Biol. 2012, 9, 978–989. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yuan, S.M.; Yang, M.; Zha, H.; Li, X.R.; Sun, H.; Duan, L.; Gu, Y.; Li, A.F.; Weng, Y.G.; et al. High intensity focused ultrasound inhibits melanoma cell migration and metastasis through attenuating microRNA-21-mediated PTEN suppression. Oncotarget 2016, 7, 50450–50460. [Google Scholar] [CrossRef]
- Labala, S.; Jose, A.; Chawla, S.R.; Khan, M.S.; Bhatnagar, S.; Kulkarni, O.P.; Venuganti, V.V.K. Effective melanoma cancer suppression by iontophoretic co-delivery of STAT3 siRNA and imatinib using gold nanoparticles. Int. J. Pharm. 2017, 525, 407–417. [Google Scholar] [CrossRef]
- Zhou, X.; Yuan, P.; Liu, Q.; Liu, Z. LncRNA MEG3 regulates imatinib resistance in chronic myeloid leukemia via suppressing microRNA-21. Biomol. Ther. (Seoul) 2017, 25, 490–496. [Google Scholar] [CrossRef]
- Nabavi, S.M.; Russo, G.L.; Tedesco, I.; Daglia, M.; Orhan, I.E.; Nabavi, S.F.; Bishayee, A.; Nagulapalli Venkata, K.C.; Abdollahi, M.; Hajheydari, Z. Curcumin and melanoma: From chemistry to medicine. Nutr. Cancer 2018, 70, 164–175. [Google Scholar] [CrossRef]
- Yang, C.H.; Yue, J.; Sims, M.; Pfeffer, L.M. The curcumin analog EF24 targets NF-κB and miRNA-21, and has potent anticancer activity in vitro and in vivo. PLoS ONE 2013, 8, e71130. [Google Scholar] [CrossRef]
- Lelli, D.; Pedone, C.; Sahebkar, A. Curcumin and treatment of melanoma: The potential role of microRNAs. Biomed. Pharmacother. 2017, 88, 832–834. [Google Scholar] [CrossRef]
- Mudduluru, G.; George-William, J.N.; Muppala, S.; Asangani, I.A.; Kumarswamy, R.; Nelson, L.D.; Allgayer, H. Curcumin regulates miR-21 expression and inhibits invasion and metastasis in colorectal cancer. Biosci. Rep. 2011, 31, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xu, T.; Chen, C. The critical roles of miR-21 in anti-cancer effects of curcumin. Ann. Transl. Med. 2015, 3, 330. [Google Scholar] [PubMed]
- Tahata, S.; Singh, S.V.; Lin, Y.; Hahm, E.R.; Beumer, J.H.; Christner, S.M.; Rao, U.N.; Sander, C.; Tarhini, A.A.; Tawbi, H.; et al. Evaluation of biodistribution of sulforaphane after administration of oral broccoli sprout extract in melanoma patients with multiple atypical nevi. Cancer Prev. Res. 2018, 11, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Arcidiacono, P.; Ragonese, F.; Stabile, A.; Pistilli, A.; Kuligina, E.; Rende, M.; Bottoni, U.; Calvieri, S.; Crisanti, A.; Spaccapelo, R. Antitumor activity and expression profiles of genes induced by sulforaphane in human melanoma cells. Eur. J. Nutr. 2018, 57, 2547–2569. [Google Scholar] [CrossRef]
- Rudolf, K.; Cervinka, M.; Rudolf, E. Sulforaphane-induced apoptosis involves p53 and p38 in melanoma cells. Apoptosis 2014, 19, 734–747. [Google Scholar] [CrossRef]
- Mitsiogianni, M.; Koutsidis, G.; Mavroudis, N.; Trafalis, D.T.; Botaitis, S.; Franco, R.; Zoumpourlis, V.; Amery, T.; Galanis, A.; Pappa, A.; et al. The role of isothiocyanates as cancer chemo-preventive, chemo-therapeutic and anti-melanoma agents. Antioxidants 2019, 8, 106. [Google Scholar] [CrossRef]
- Martin, S.L.; Kala, R.; Tollefsbol, T.O. Mechanisms for the inhibition of colon cancer cells by sulforaphane through epigenetic modulation of microRNA-21 and human telomerase reverse transcriptase (hTERT) down-regulation. Curr. Cancer Drug Targets 2018, 18, 97–106. [Google Scholar] [CrossRef]
- Lan, F.; Pan, Q.; Yu, H.; Yue, X. Sulforaphane enhances temozolomide-induced apoptosis because of down-regulation of miR-21 via Wnt/β-catenin signaling in glioblastoma. J. Neurochem. 2015, 134, 811–818. [Google Scholar] [CrossRef]
- Shen, Q.; Tian, F.; Jiang, P.; Li, Y.; Zhang, L.; Lu, J.; Li, J. EGCG enhances TRAIL-mediated apoptosis in human melanoma A375 cell line. J. Huazhong Univ. Sci. Technolog. Med. Sci. 2009, 29, 771–775. [Google Scholar] [CrossRef]
- Ellis, L.Z.; Liu, W.; Luo, Y.; Okamoto, M.; Qu, D.; Dunn, J.H.; Fujita, M. Green tea polyphenol epigallocatechin-3-gallate suppresses melanoma growth by inhibiting inflammasome and IL-1β secretion. Biochem. Biophys. Res. Commun. 2011, 414, 551–556. [Google Scholar] [CrossRef]
- Zhang, J.; Lei, Z.; Huang, Z.; Zhang, X.; Zhou, Y.; Luo, Z.; Zeng, W.; Su, J.; Peng, C.; Chen, X. Epigallocatechin-3-gallate (EGCG) suppresses melanoma cell growth and metastasis by targeting TRAF6 activity. Oncotarget 2016, 7, 79557–79571. [Google Scholar] [CrossRef] [PubMed]
- Fujiki, H.; Watanabe, T.; Sueoka, E.; Rawangkan, A.; Suganuma, M. Cancer prevention with green tea and its principal constituent, EGCG: From early investigations to current focus on human cancer stem cells. Mol. Cells 2018, 41, 73–82. [Google Scholar] [PubMed]
- Chen, X.; Chang, L.; Qu, Y.; Liang, J.; Jin, W.; Xia, X. Tea polyphenols inhibit the proliferation, migration, and invasion of melanoma cells through the down-regulation of TLR4. Int. J. Immunopathol. Pharmacol. 2018, 32. [Google Scholar] [CrossRef] [PubMed]
- Khoi, P.N.; Park, J.S.; Kim, J.H.; Xia, Y.; Kim, N.H.; Kim, K.K.; Jung, Y.D. (-)-Epigallocatechin-3-gallate blocks nicotine-induced matrix metalloproteinase-9 expression and invasiveness via suppression of NF-κB and AP-1 in endothelial cells. Int. J. Oncol. 2013, 43, 868–876. [Google Scholar] [CrossRef]
- Kim, J.E.; Shin, M.H.; Chung, J.H. Epigallocatechin-3-gallate prevents heat shock-induced MMP-1 expression by inhibiting AP-1 activity in human dermal fibroblasts. Arch. Dermatol. Res. 2013, 305, 595–602. [Google Scholar] [CrossRef]
- Siddiqui, I.A.; Asim, M.; Hafeez, B.B.; Adhami, V.M.; Tarapore, R.S.; Mukhtar, H. Green tea polyphenol EGCG blunts androgen receptor function in prostate cancer. FASEB J. 2011, 25, 1198–1207. [Google Scholar] [CrossRef]
- Fix, L.N.; Shah, M.; Efferth, T.; Farwell, M.A.; Zhang, B. MicroRNA expression profile of MCF-7 human breast cancer cells and the effect of green tea polyphenon-60. Cancer Genom. Proteom. 2010, 7, 261–277. [Google Scholar]
- Schlumpf, M.; Reichrath, J.; Lehmann, B.; Sigmundsdottir, H.; Feldmeyer, L.; Hofbauer, G.F.; Lichtensteiger, W. Fundamental questions to sun protection: A continuous education symposium on vitamin D, immune system and sun protection at the University of Zürich. Dermatoendocrinology 2010, 2, 19–25. [Google Scholar] [CrossRef]
- Reichrath, J.; Reichrath, S. Sunlight, vitamin D and malignant melanoma: An update. Adv. Exp. Med. Biol. 2014, 810, 390–405. [Google Scholar]
- Huerter, C.J.; Vaudreuil, A.; Agrawal, D.K.; Nguyen, A.H. Has vitamin D had its “time in the sun” for melanoma? J. Clin. Aesthet. Dermatol. 2016, 9, 11–12. [Google Scholar]
- De Smedt, J.; Van Kelst, S.; Boecxstaens, V.; Stas, M.; Bogaerts, K.; Vanderschueren, D.; Aura, C.; Vandenberghe, K.; Lambrechts, D.; Wolter, P.; et al. Vitamin D supplementation in cutaneous malignant melanoma outcome (ViDMe): A randomized controlled trial. BMC Cancer 2017, 17, 562. [Google Scholar] [CrossRef] [PubMed]
- Brożyna, A.A.; Hoffman, R.M.; Slominski, A.T. Relevance of vitamin D in melanoma development, progression and therapy. Anticancer Res. 2020, 40, 473–489. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.C.; Lee, I.M.; Weiderpass, E.; Campbell, P.T.; Sampson, J.N.; Kitahara, C.M.; Keadle, S.K.; Arem, H.; Berrington de Gonzalez, A.; Hartge, P.; et al. Association of leisure-time physical activity with risk of 26 types of cancer in 1.44 million adults. JAMA Intern. Med. 2016, 176, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Shors, A.R.; Solomon, C.; McTiernan, A.; White, E. Melanoma risk in relation to height, weight, and exercise (United States). Cancer Causes Control 2001, 12, 599–606. [Google Scholar] [CrossRef]
- Gogas, H.; Trakatelli, M.; Dessypris, N.; Terzidis, A.; Katsambas, A.; Chrousos, G.P.; Petridou, E.T. Melanoma risk in association with serum leptin levels and lifestyle parameters: A case-control study. Ann. Oncol. 2008, 19, 384–389. [Google Scholar] [CrossRef]
- Ruiz-Casado, A.; Martín-Ruiz, A.; Pérez, L.M.; Provencio, M.; Fiuza-Luces, C.; Lucia, A. Exercise and the hallmarks of cancer. Trends Cancer 2017, 3, 423–441. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell 2015, 161, 1681–1696. [Google Scholar] [CrossRef]
- Zhang, T.; Dutton-Regester, K.; Brown, K.M.; Haywar, N.K. The genomic landscape of cutaneous melanoma. Pigment Cell Melanoma Res. 2016, 29, 266–283. [Google Scholar] [CrossRef]
- Hayward, N.K.; Wilmott, J.S.; Waddell, N.; Johansson, P.A.; Field, M.A.; Nones, K.; Patch, A.M.; Kakavand, H.; Alexandrov, L.B.; Burke, H.; et al. Whole-genome landscapes of major melanoma subtypes. Nature 2017, 545, 175–180. [Google Scholar] [CrossRef]
- Shain, A.H.; Joseph, N.M.; Yu, R.; Benhamida, J.; Liu, S.; Prow, T.; Ruben, B.; North, J.; Pincus, L.; Yeh, I.; et al. Genomic and transcriptomic analysis reveals incremental disruption of key signaling pathways during melanoma evolution. Cancer Cell 2018, 34, 45–55. [Google Scholar] [CrossRef]
- Moon, H.; Donahue, L.R.; Choi, E.; Scumpia, P.O.; Lowry, W.E.; Grenier, J.K.; Zhu, J.; White, A.C. Melanocyte stem cell activation and translocation initiate cutaneous melanoma in response to UV exposure. Cell Stem Cell 2017, 21, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Lorusso, C.; De Summa, S.; Pinto, R.; Danza, K.; Tommas, S. miRNAs as key players in the management of cutaneous melanoma. Cells 2020, 9, 415. [Google Scholar] [CrossRef] [PubMed]
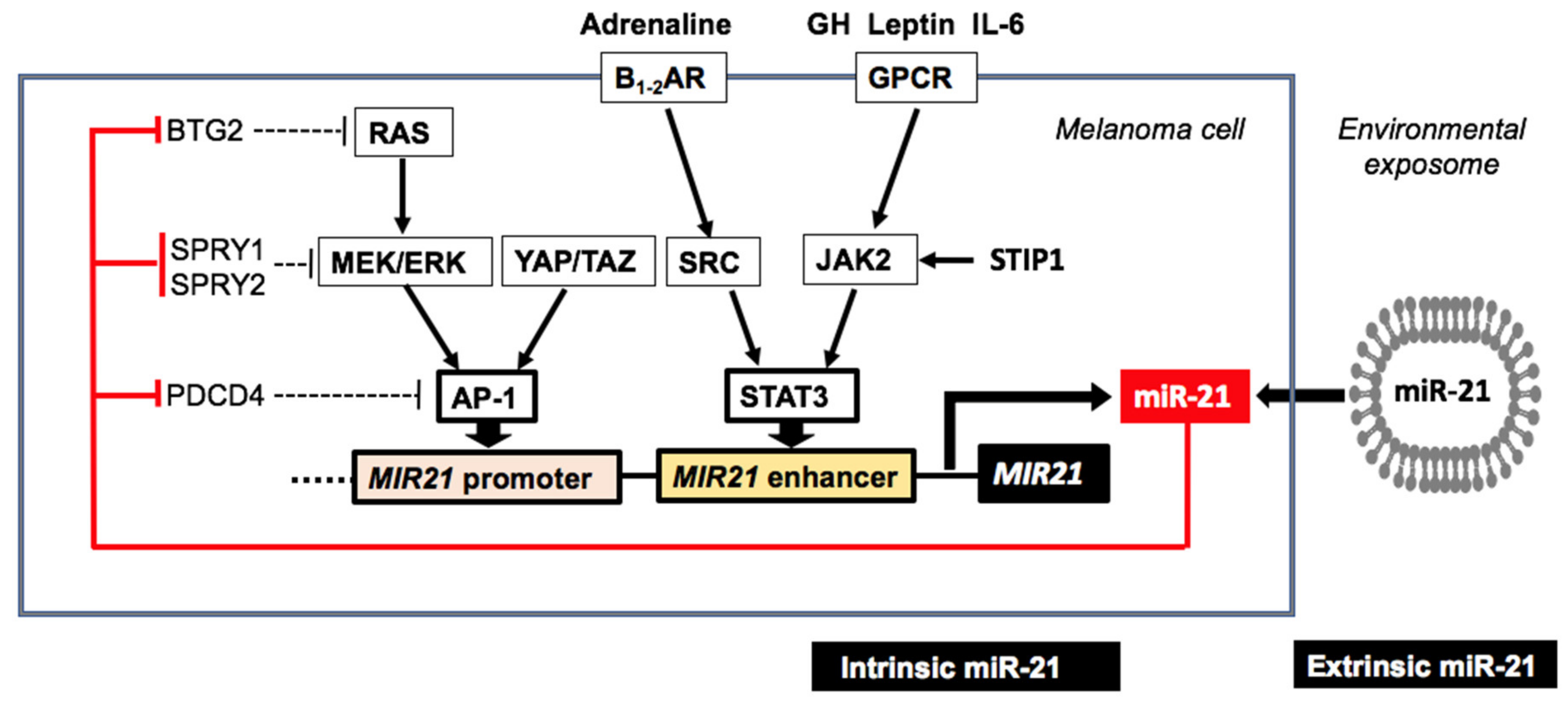
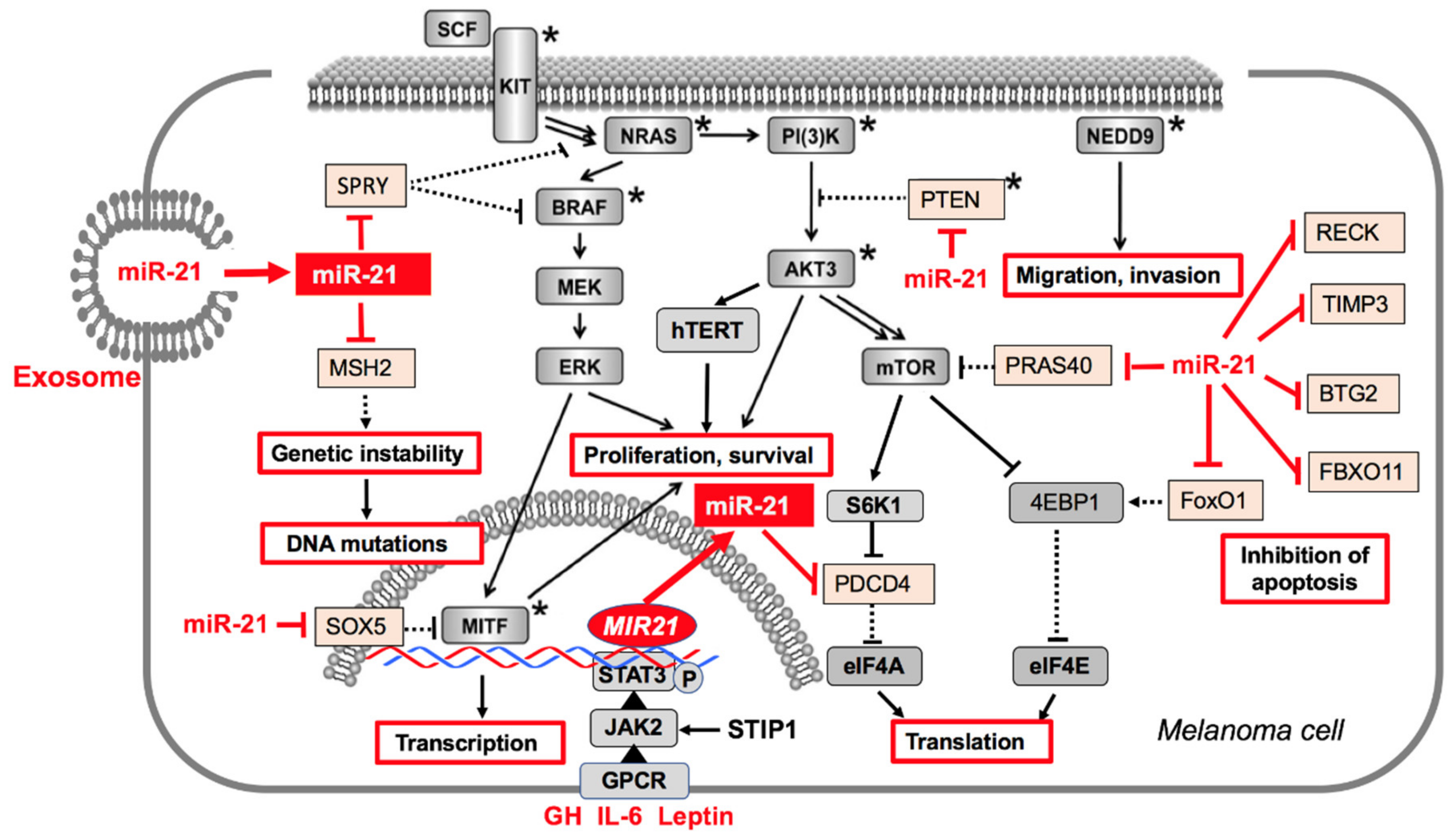
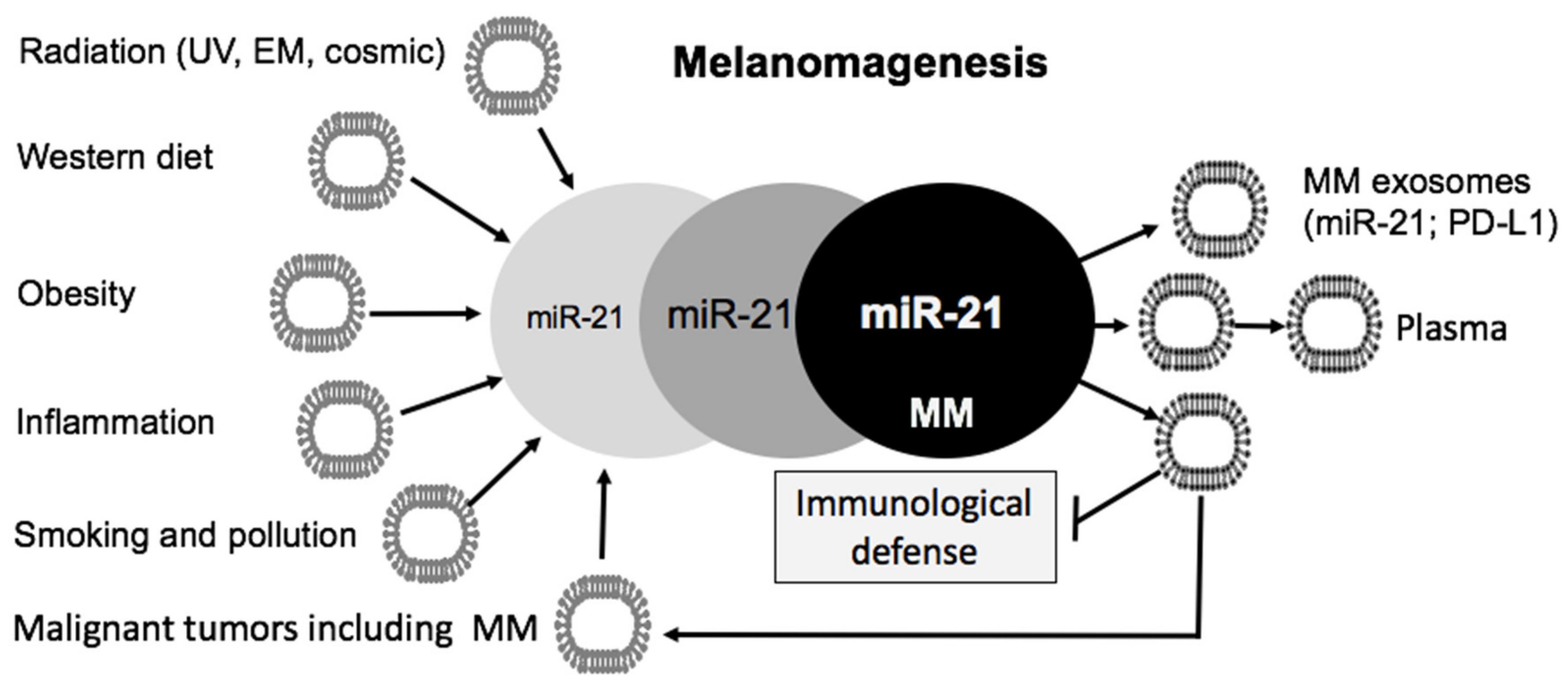
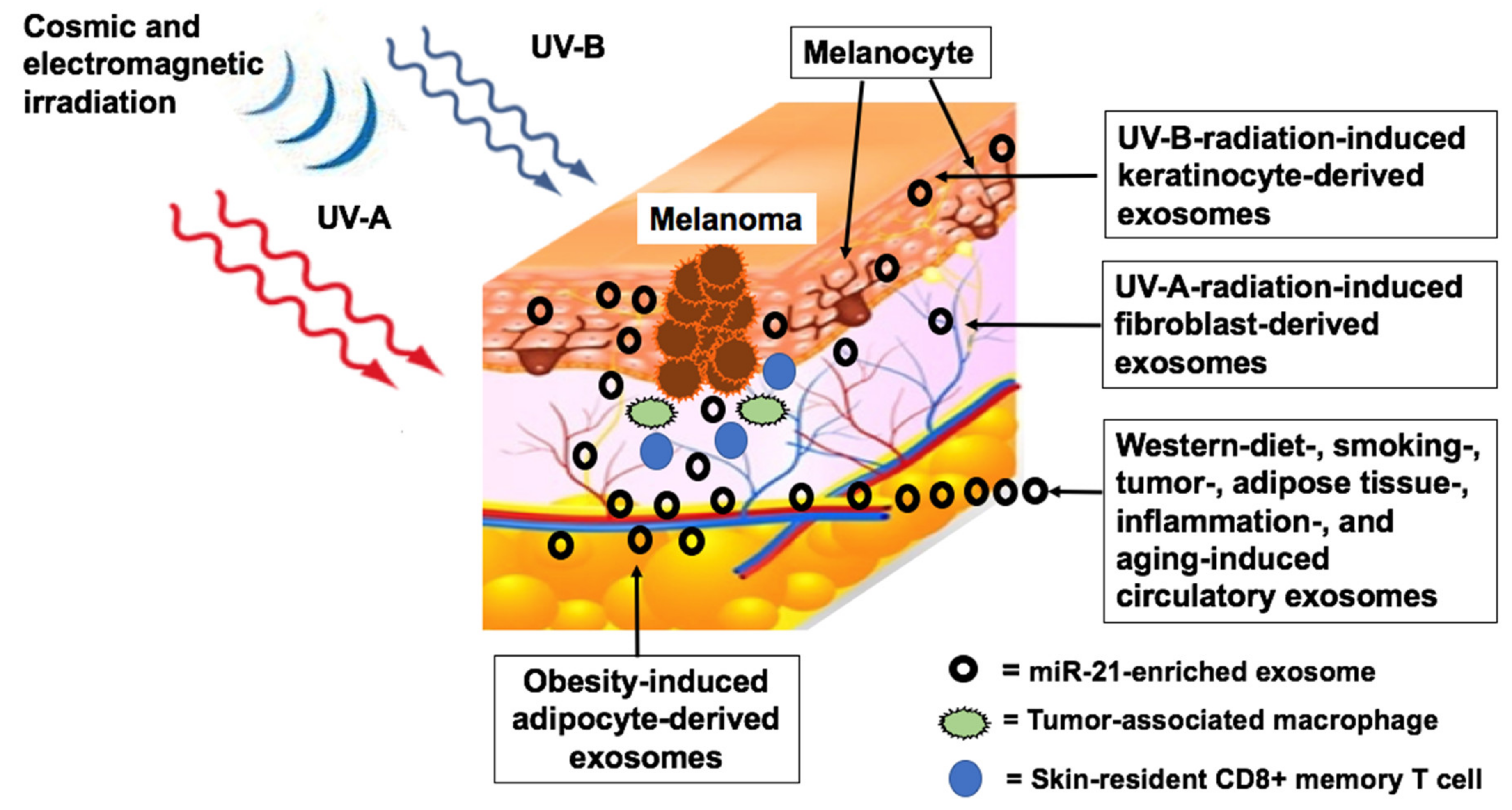
| Associations of miR-21 with Melanoma Pathology | References |
|---|---|
| MiR-21 expression increases from benign nevi to MM and metastatic MM | [31,33] |
| MiR-21 expression correlates with mitotic activity in MM | [34] |
| MiR-21 levels correlate with MM cellularity | [36] |
| MiR-21 promotes proliferation, migration, and inhibits apoptosis of MM cells | [31,32] |
| MiR-21 expression correlates with Breslow thickness and advanced clinical stage | [31,33] |
| MiR-21 expression correlates with positive sentinel lymph node biopsy | [34] |
| MiR-21 promotes MM invasion and metastasis | [37] |
| MiR-21 expression correlates with shorter 5-year disease-free or overall survival | [33] |
| MiR-21 inhibits PD-L1 expression of MM-associated macrophages | [38] |
| Antisense-mediated miR-21 inhibition suppresses growth, increases apoptosis and enhances chemo- or radiosensitivity of human MM cells | [33] |
| Target Genes | Proteins | Functions | References |
|---|---|---|---|
| TIPE2 | Tumor necrosis factor-α-induced protein 8 (TNFAIP8)-like 2 | Inhibition of RAS | [43,44,45] |
| SPRY1 | Sprouty RTK signaling antagonist 1 | Inhibition of RAS and RAF | [40,42] |
| SPRY2 | Sprouty RTK signaling antagonist 2 | Inhibition of RAS and RAF | [42,49] |
| PTEN | Phosphatase and tensin homolog | Inhibition of PI3K and downstream AKT-mTORC1 signaling | [40,49] |
| PDCD4 | Programmed cell death 4 | Inhibition of translation initiation | [40,49] |
| FBXO11 | F-box only protein 11 | Tumor suppression promoting apoptosis, exhibiting decreased expression in higher Clark level MM | [50] |
| SOX5 | SRY-box 5 | Suppression of MITF | [51,52] |
| CDKN2C | Cyclin-dependent kinase inhibitor 2C | Inhibition of G1/S transition, proliferation | [32,49] |
| MSH2 | DNA mismatch repair protein 2 | DNA repair, prevention of microsatellite instability | [49,53] |
| CYP27B1 | 25-hydroxyvitamin D3-1-α-hydroxylase | Conversion of 25(OH) vitamin D to active 1,25(OH)2 vitamin D3 | [54] |
| RECK | Reversion-inducing cysteine-rich protein with Kazal motifs | Extracellular matrix integrity and regulation of angiogenesis | [49,55] |
| TIMP1 | Tissue inhibitor of metalloproteinase 1 | Inhibition of MMP1-mediated matrix degradation | [49,56] |
| TIMP3 | Tissue inhibitor of metalloproteinase 3 | Inhibition of MMP3-mediated matrix degradation | [49,57] |
| CDH1 | E-cadherin | Cell-cell adhesion | [58] |
| IL12A | p35 subunit of interleukin 12 | Anti-tumor activities via NK- and cytotoxic T cell activation | [59] |
| AKT1S1 | AKT1 substrate 1, proline-rich | Negative regulator of mTORC1 | [60] |
| Regulatory Agent | Transcriptional Regulator | MiR-21 Expression | References |
|---|---|---|---|
| AP1 | Activator protein 1 (Fos, Jun) | Upregulation | [25,92] |
| STAT3 | Signal transducer and activator of transcription 3 | Upregulation | [25,57,69,70,71] |
| p65 | Nuclear factor kappa-B, subunit 3 | Upregulation | [25] |
| AR | Androgen receptor | Upregulation | [25,93] |
| PU.1 | ETS-domain transcription factor PU.1 | Upregulation | [25] |
| C/EBPα | CCATT/enhancer binding protein α | Upregulation | [25] |
| TGFβ1 | Transforming growth factor β1 | Upregulation | [25] |
| NFIB | Nuclear factor IB | Downregulation | [25] |
| VDR | Vitamin D receptor | Downregulation | [94,95] |
| XIST | X inactivation-specific transcript (lncRNA) | Downregulation | [79] |
| MEG3 | Maternally expressed gene 3 (lncRNA) | Downregulation | [58] |
| GAS5 | Growth arrest specific transcript 5 (lncRNA) | Downregulation | [87,88,89] |
| Lifestyle Factor | Biological Responses | References |
|---|---|---|
| Ultraviolet radiation | Keratinocyte-derived release of miR-21-enriched exosomes; increased miR-21 expression of melanocytes | [163,164,165,166,167] |
| Cosmic irradiation | Increase of exosomal miR-21 | [177,178,179,180,181] |
| Electromagnetic radiation | Increased expression of miR-21 | [183] |
| Obese adipose tissue | Adipocyte secretome with increased release of miR-21-enriched exosomes | [232,233,234,235,236] |
| High-fat diet-induced obesity | Increase of circulatory and adipocyte miR-21 | [230,231] |
| High glucose intake | Increase of circulatory miR-21 | [60] |
| High fructose intake | Increase of circulatory miR-21 | [256] |
| Alcohol consumption | Increase of circulatory miR-21 | [276,277] |
| Milk consumption | Increase of circulatory exosomal miR-21 | [210,211,212,213] |
| Vitamin D deficiency | Increased expression of miR-21 | [95] |
| Smoking | Increased expression of exosomal miR-21 in airway epithelial cells | [287,288,289] |
| Air pollution (Diesel) | Increased expression of exosomal miR-21 in airway epithelial cells | [290] |
| Sedentary lifestyle | Increase of circulatory miR-21 | [334,335] |
| Aging | Increase of circulatory miR-21 | [324,325] |
| Chronic inflammation | Increase of circulatory miR-21 | [331,332,333] |
| Therapeutic Factors | Potential Benefits for Melanoma Prevention and Therapy | References |
|---|---|---|
| Anti-miR-21 | Direct suppression of miR-21 signaling in melanocytes, activation of skin-resident CD8+ memory T-cells; reduction of miR-21 in tumor-associated macrophages associated with improved cytotoxic T-cell responses | [33,366,367,368,369,370,371] |
| BRAF inhibition | Attenuation of miR-21 expression | [336] |
| Sunscreen | Reduction of keratinocyte-derived exosomal miR-21 | [163,164,165,166,167] |
| Restriction of electro-magnetic radiation | Limitation of smart phone radiation on miR-21 expression | [189,190] |
| Control of birth weight and body weight | Balanced expression of miR-21 during fetal and postnatal life | [203,204] |
| Reduction of glycemic load and fat intake | Reduction of circulating and adipocyte-derived miR-21 | [60,230,231,256] |
| Cessation of smoking | Reduction of airway epithelial cell-derived exosomal miR-21 | [287,288,289] |
| Restriction of alcohol intake | Reduction of miR-21 expression | [276,277] |
| Metformin | Reduction of STAT3 activation and miR-21 expression | [349,350,352,353] |
| Beta-blocker | Suppression of STAT3 activation and miR-21 expression | [364] |
| Curcumin | Suppression of STAT3 activation and inactivation of AP-1 resulting in reduced expression of miR-21 | [380,381,382,383] |
| EGCG | Reduction of miR-21 expression | [397,398] |
| Sulforaphane | Reduction of miR-21 expression | [388,389] |
| Vitamin D | Reduction of miR-21 expression | [54,95,307] |
| Exercise | Reduction of miR-21 expression | [334,335] |
| HIFU | Reduction of miR-21 in metastatic melanoma tissue | [376] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melnik, B.C.; John, S.M.; Carrera-Bastos, P.; Schmitz, G. MicroRNA-21-Enriched Exosomes as Epigenetic Regulators in Melanomagenesis and Melanoma Progression: The Impact of Western Lifestyle Factors. Cancers 2020, 12, 2111. https://doi.org/10.3390/cancers12082111
Melnik BC, John SM, Carrera-Bastos P, Schmitz G. MicroRNA-21-Enriched Exosomes as Epigenetic Regulators in Melanomagenesis and Melanoma Progression: The Impact of Western Lifestyle Factors. Cancers. 2020; 12(8):2111. https://doi.org/10.3390/cancers12082111
Chicago/Turabian StyleMelnik, Bodo C., Swen Malte John, Pedro Carrera-Bastos, and Gerd Schmitz. 2020. "MicroRNA-21-Enriched Exosomes as Epigenetic Regulators in Melanomagenesis and Melanoma Progression: The Impact of Western Lifestyle Factors" Cancers 12, no. 8: 2111. https://doi.org/10.3390/cancers12082111
APA StyleMelnik, B. C., John, S. M., Carrera-Bastos, P., & Schmitz, G. (2020). MicroRNA-21-Enriched Exosomes as Epigenetic Regulators in Melanomagenesis and Melanoma Progression: The Impact of Western Lifestyle Factors. Cancers, 12(8), 2111. https://doi.org/10.3390/cancers12082111