The Tumor Suppressor CYLD Inhibits Mammary Epithelial to Mesenchymal Transition by the Coordinated Inhibition of YAP/TAZ and TGFβ Signaling
Abstract
1. Introduction
2. Results
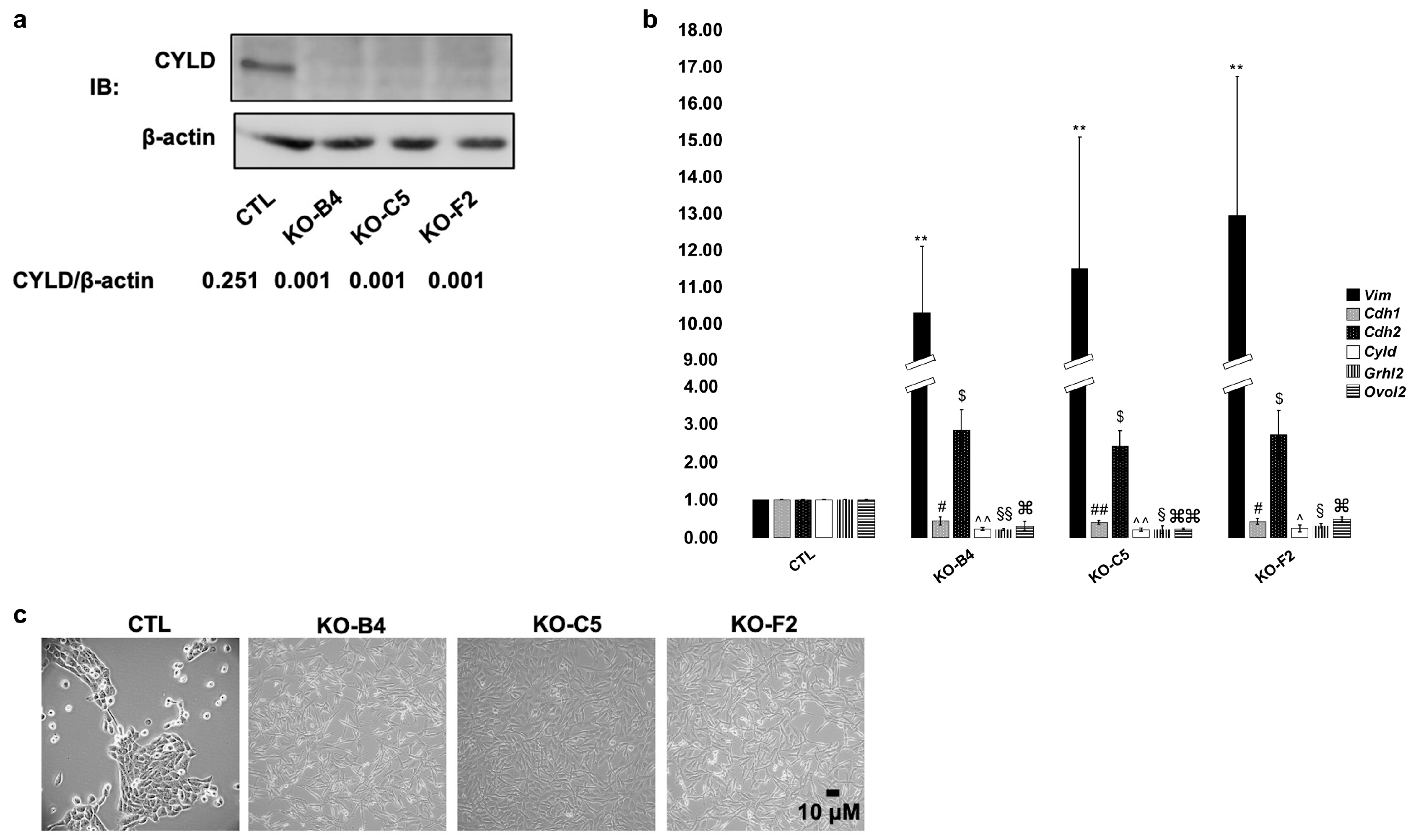
2.1. Downregulation of CYLD in Mammary Epithelial Cells Promotes Mesenchymal Phenotypic Characteristics
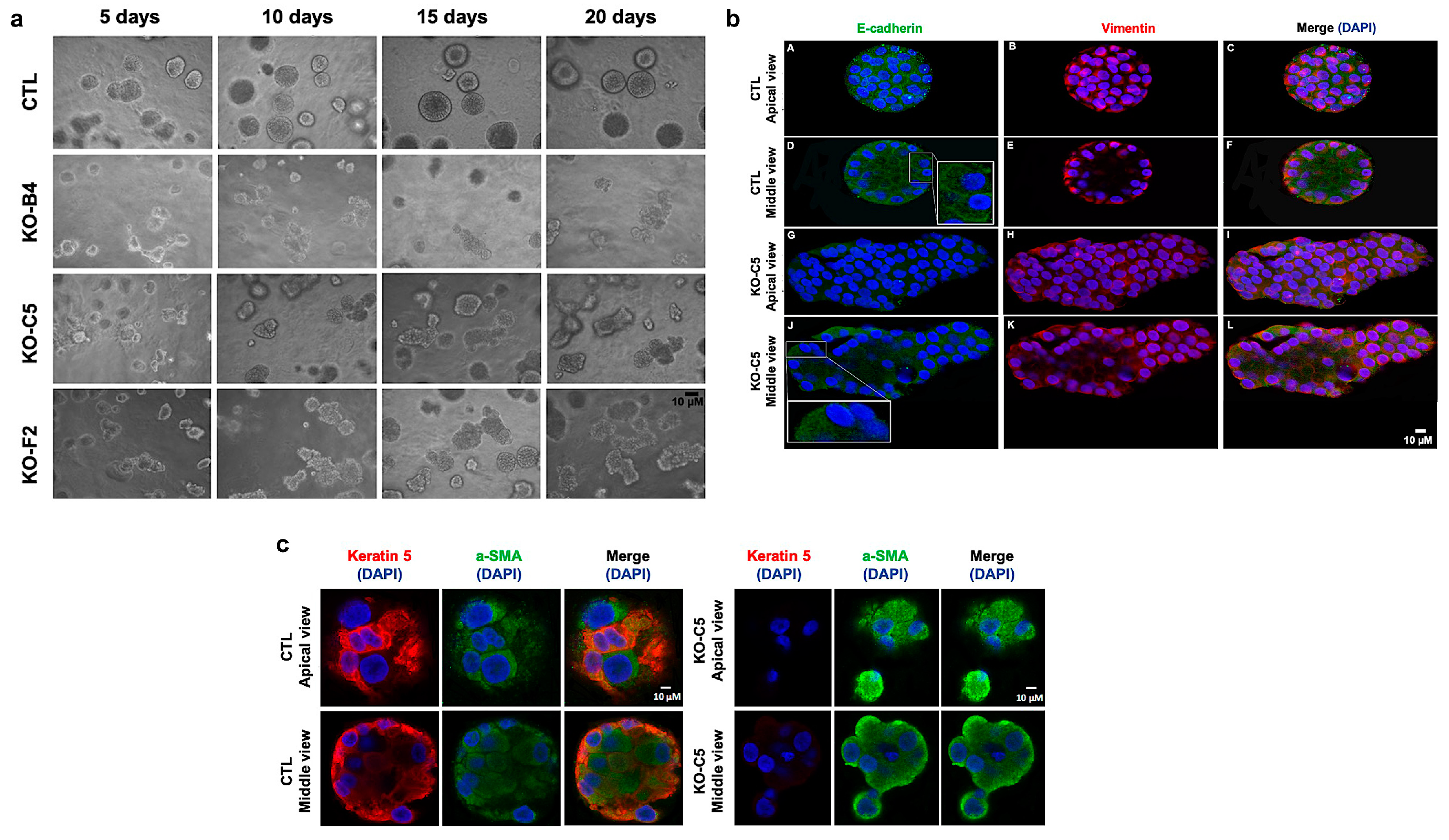
2.2. CYLD Deficiency Impairs the Proper Development and Organization of Mammary Spheroids
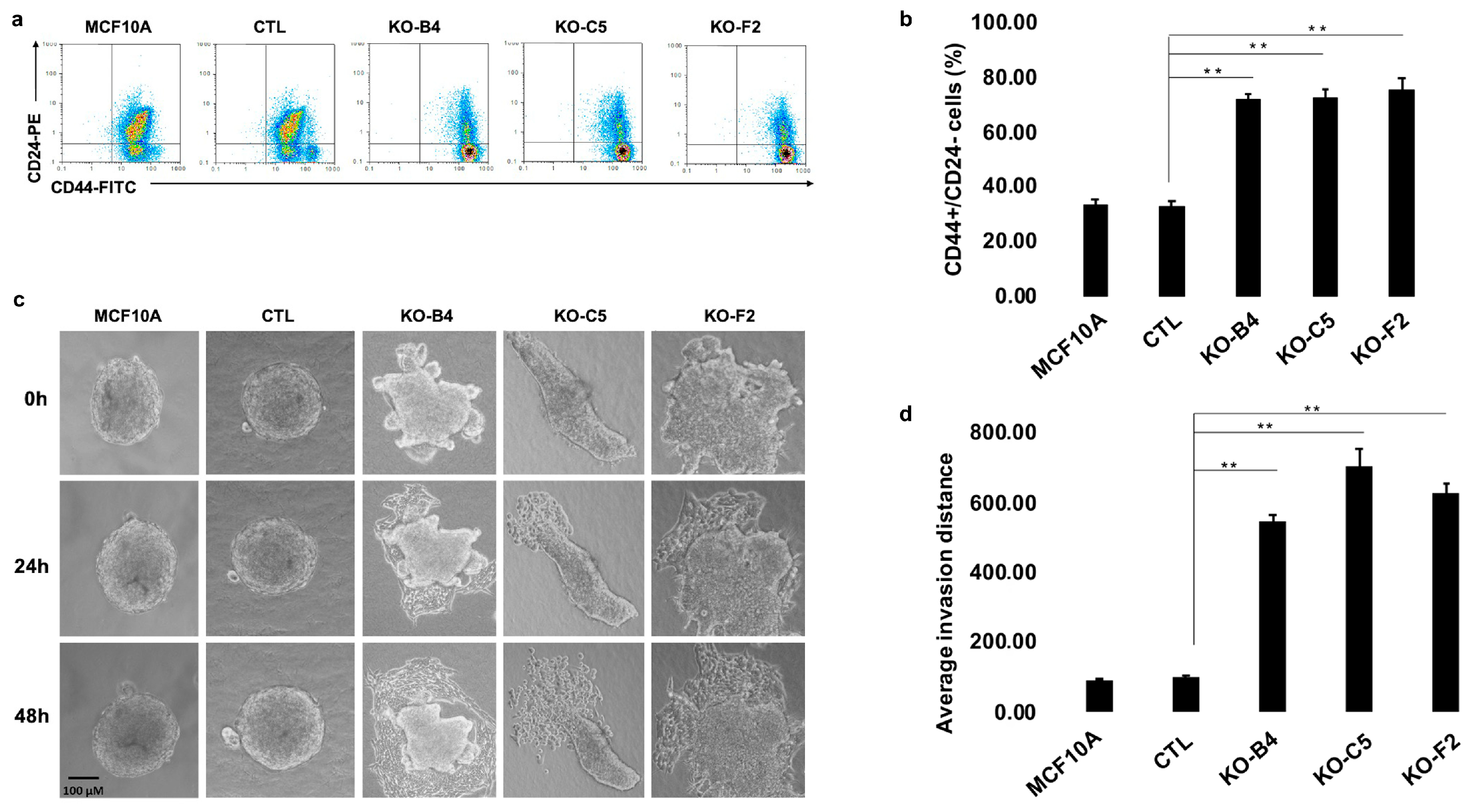
2.3. Inactivation of CYLD in Mammary Epithelial Cells Promotes the Development of Stem Cell and Tumorigenic Characteristics
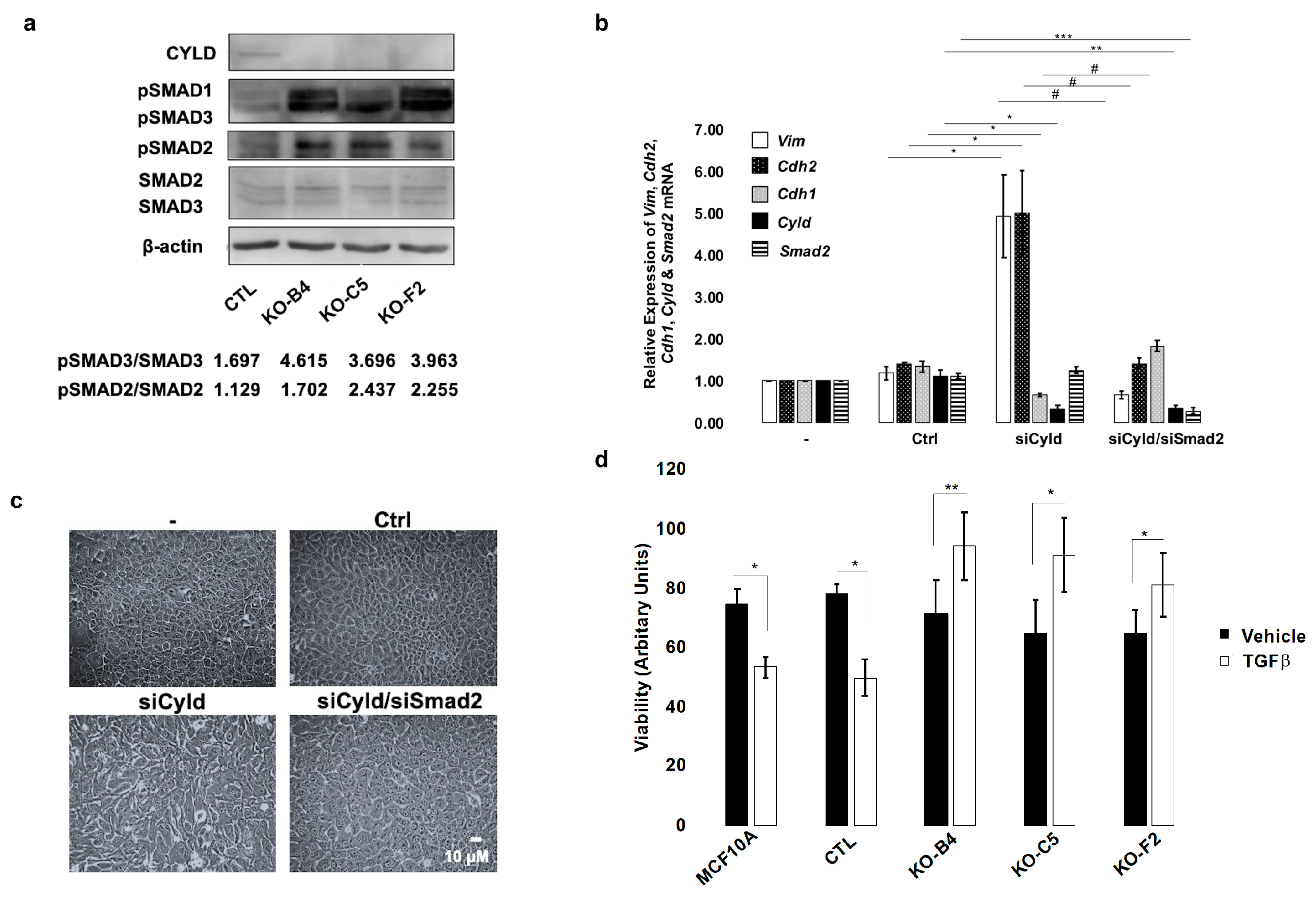
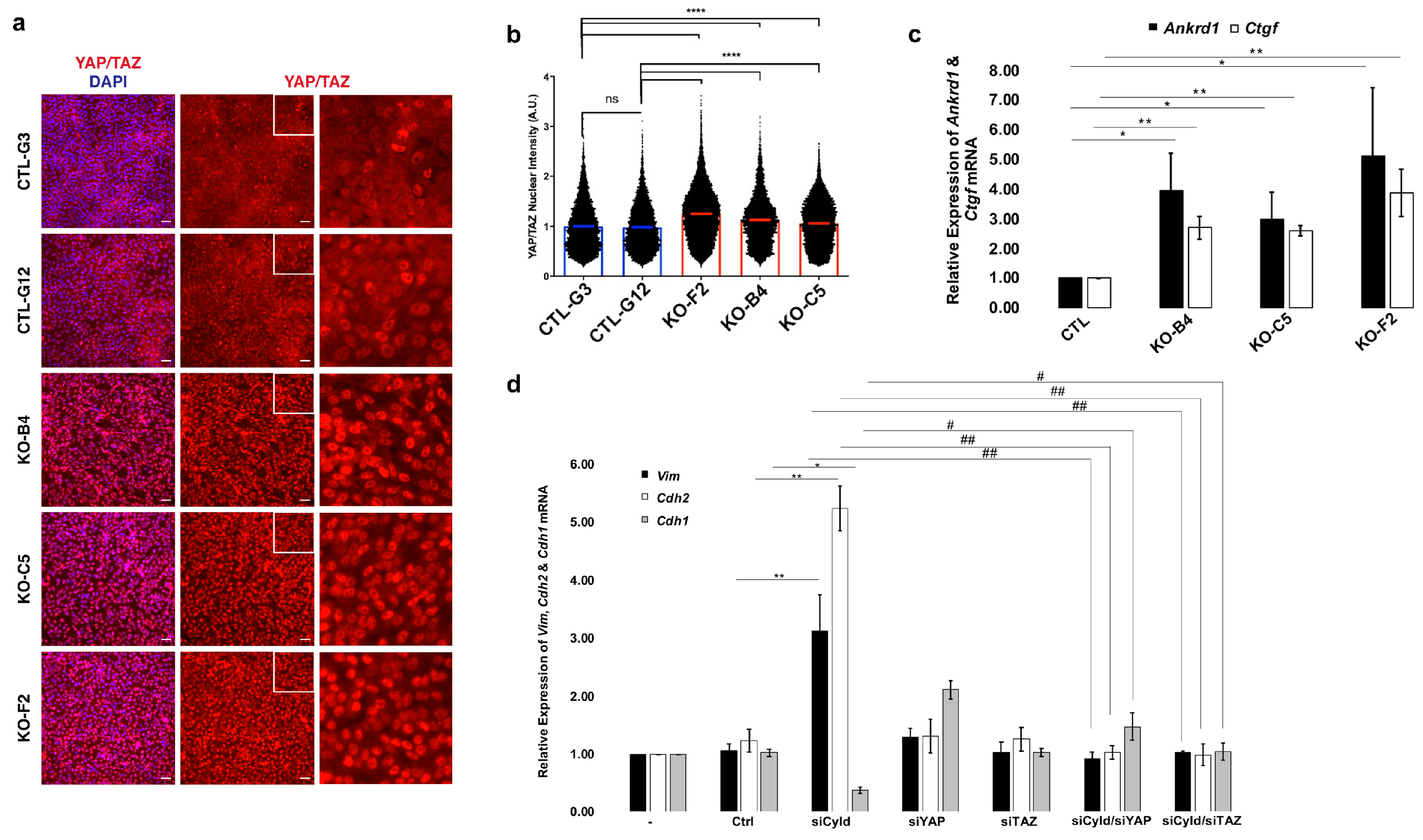
2.4. Inactivation of CYLD in Mammary Epithelial Cells Coordinately Activates the TGFβ and YAP/TAZ Pathways
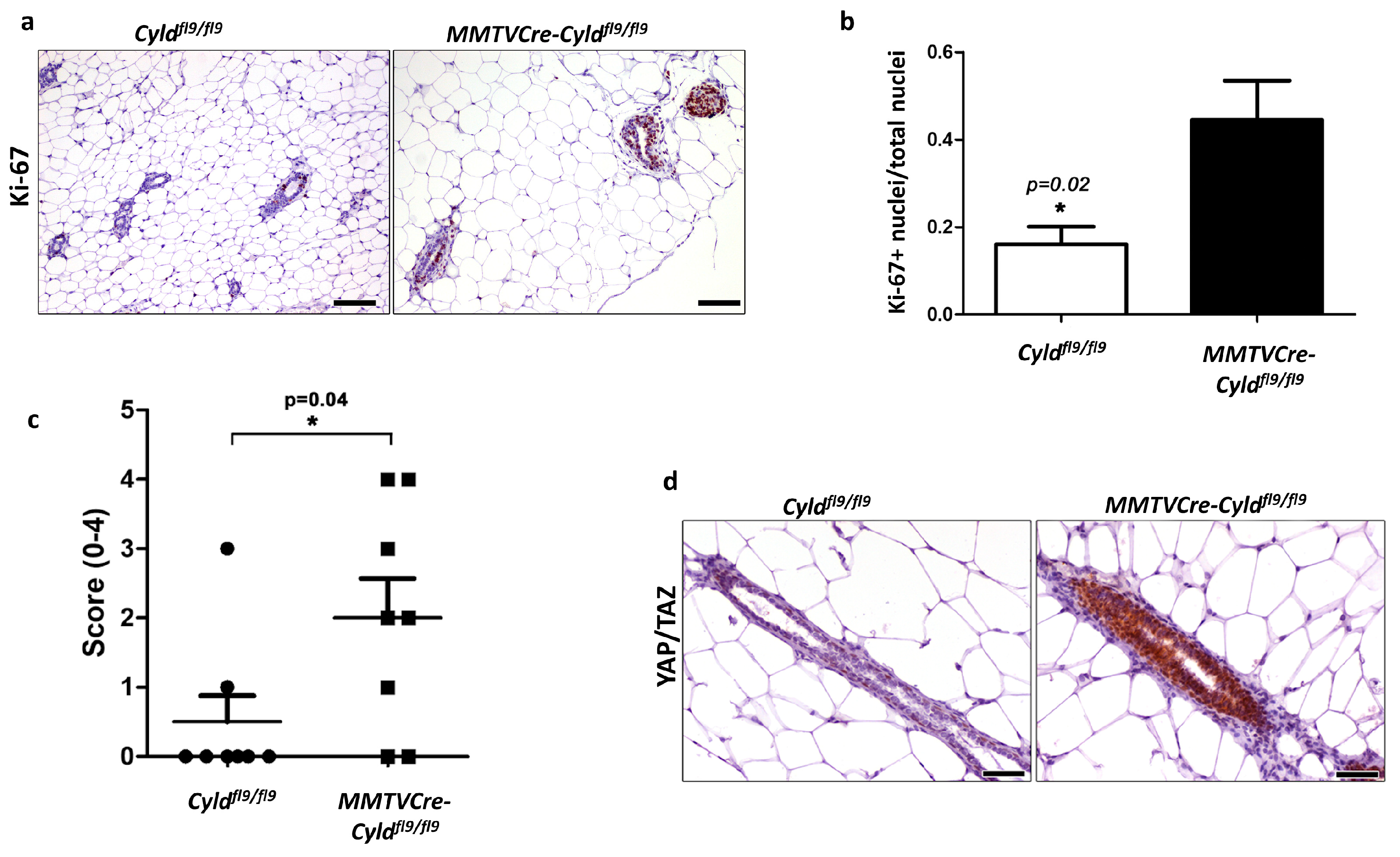
2.5. Inactivation of CYLD in the Mouse Mammary Epithelium Results in Hyperplastic Alterations
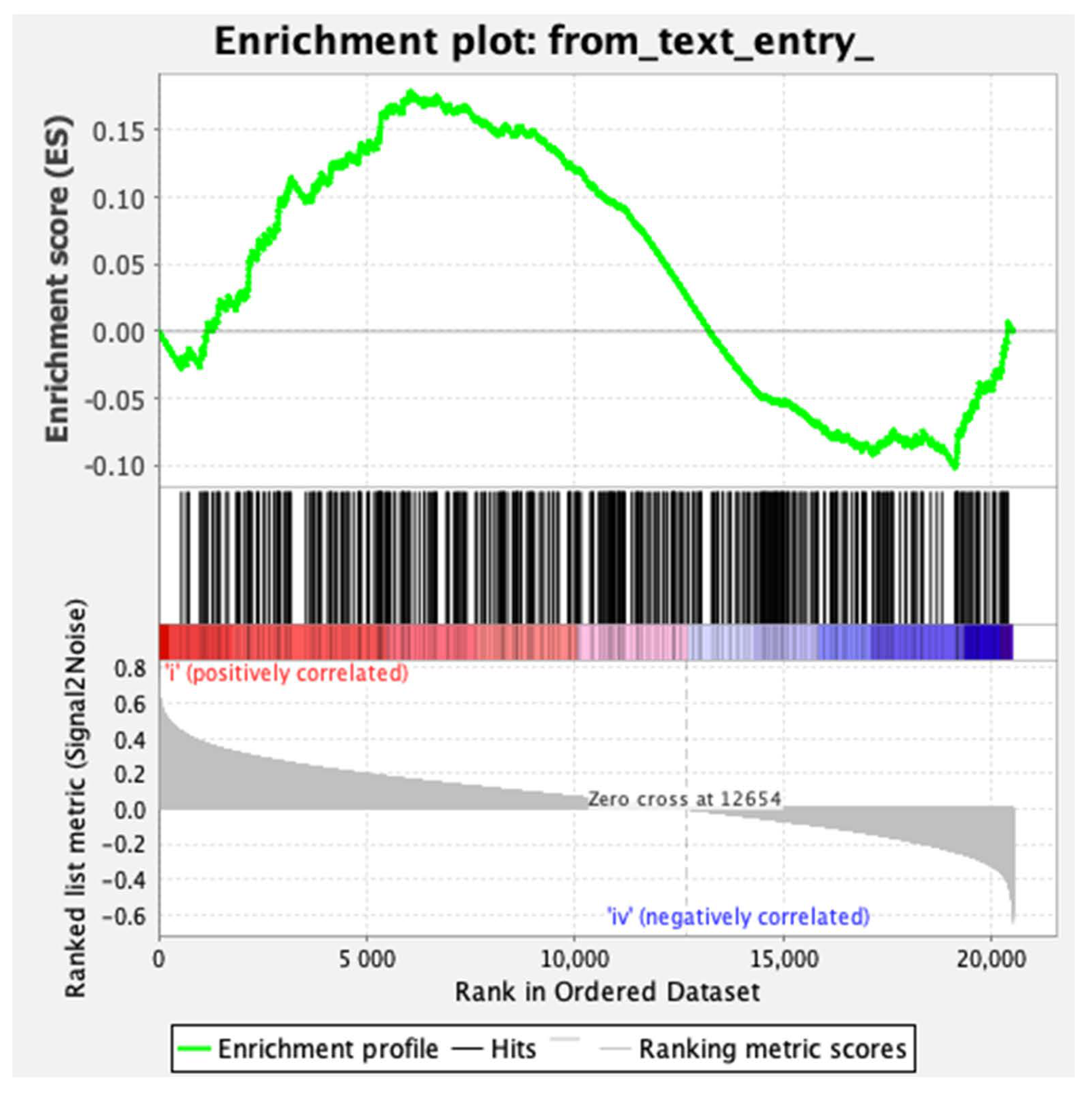
2.6. CYLD Downregulation in Human Breast Cancer Patient Samples Correlates with the Activation of the YAP/TAZ Pathway.
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Mouse Models
4.3. Generation of CYLD-Mutated MCF10A Cell Lines
4.4. TGFβ Treatment and Cell Viability Assay
4.5. Small Interfering RNA (siRNA) Knockdown
4.6. Immunoblotting
4.7. RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR (qPCR)
4.8. Flow Cytometric Analysis for CD44/CD24 Markers
4.9. Three-Dimensional Mammosphere Formation
4.10. Three-Dimensional Invasion Assay
4.11. Immunofluorescence
4.12. Immunohistochemistry (IHC) and Morphometry
4.13. Statistics
4.14. Gene Set Enrichment Analysis (GSEA)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Shibue, T.; Weinberg, R.A. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nature reviews. Clin. Oncol. 2017, 14, 611–629. [Google Scholar] [CrossRef]
- Bignell, G.R.; Warren, W.; Seal, S.; Takahashi, M.; Rapley, E.; Barfoot, R.; Green, H.; Brown, C.; Biggs, P.J.; Lakhani, S.R.; et al. Identification of the familial cylindromatosis tumour-suppressor gene. Nat. Gen. 2000, 25, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Hellerbrand, C.; Bumes, E.; Bataille, F.; Dietmaier, W.; Massoumi, R.; Bosserhoff, A.K. Reduced expression of CYLD in human colon and hepatocellular carcinomas. Carcinogenesis 2007, 28, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Annunziata, C.M.; Davis, R.E.; Demchenko, Y.; Bellamy, W.; Gabrea, A.; Zhan, F.; Lenz, G.; Hanamura, I.; Wright, G.; Xiao, W.; et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 2007, 12, 115–130. [Google Scholar] [CrossRef] [PubMed]
- Keats, J.J.; Fonseca, R.; Chesi, M.; Schop, R.; Baker, A.; Chng, W.J.; Van Wier, S.; Tiedemann, R.; Shi, C.X.; Sebag, M.; et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell 2007, 12, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Jenner, M.W.; Leone, P.E.; Walker, B.A.; Ross, F.M.; Johnson, D.C.; Gonzalez, D.; Chiecchio, L.; Dachs Cabanas, E.; Dagrada, G.P.; Nightingale, M.; et al. Gene mapping and expression analysis of 16q loss of heterozygosity identifies WWOX and CYLD as being important in determining clinical outcome in multiple myeloma. Blood 2007, 110, 3291–3300. [Google Scholar] [CrossRef] [PubMed]
- Massoumi, R.; Kuphal, S.; Hellerbrand, C.; Haas, B.; Wild, P.; Spruss, T.; Pfeifer, A.; Fassler, R.; Bosserhoff, A.K. Down-regulation of CYLD expression by Snail promotes tumor progression in malignant melanoma. J. Exp. Med. 2009, 206, 221–232. [Google Scholar] [CrossRef]
- Hajek, M.; Sewell, A.; Kaech, S.; Burtness, B.; Yarbrough, W.G.; Issaeva, N. TRAF3/CYLD mutations identify a distinct subset of human papillomavirus-associated head and neck squamous cell carcinoma. Cancer 2017, 123, 1778–1790. [Google Scholar] [CrossRef]
- Li, Y.Y.; Chung, G.T.; Lui, V.W.; To, K.F.; Ma, B.B.; Chow, C.; Woo, J.K.; Yip, K.Y.; Seo, J.; Hui, E.P.; et al. Exome and genome sequencing of nasopharynx cancer identifies NF-kappaB pathway activating mutations. Nat. Commun. 2017, 8, 14121. [Google Scholar] [CrossRef]
- Jiang, K.; Xie, L.F.; Xiao, T.Z.; Qiu, M.Y.; Wang, W.L. MiR-181d inhibits cell proliferation and metastasis through PI3K/AKT pathway in gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8861–8869. [Google Scholar] [CrossRef]
- Ling, T.; Yu, F.; Cao, H. MiR-182 controls cell growth in gastrointestinal stromal tumors by negatively regulating CYLD expression. Oncol. Rep. 2018, 40, 3705–3713. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.D.; Zhou, P.J.; Wang, Y.; Zhang, L.; Fu, W.Y.; Ruan, B.B.; Xu, H.P.; Hu, C.Z.; Tian, L.; Qin, J.H.; et al. Reciprocal activation between STAT3 and miR-181b regulates the proliferation of esophageal cancer stem-like cells via the CYLD pathway. Mol. Cancer 2016, 15, 40. [Google Scholar] [CrossRef] [PubMed]
- Massoumi, R.; Chmielarska, K.; Hennecke, K.; Pfeifer, A.; Fassler, R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell 2006, 125, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Stirling, B.; Temmerman, S.T.; Ma, C.A.; Fuss, I.J.; Derry, J.M.; Jain, A. Impaired regulation of NF-kappaB and increased susceptibility to colitis-associated tumorigenesis in CYLD-deficient mice. J. Clin. Invest. 2006, 116, 3042–3049. [Google Scholar] [CrossRef]
- Nikolaou, K.; Tsagaratou, A.; Eftychi, C.; Kollias, G.; Mosialos, G.; Talianidis, I. Inactivation of the deubiquitinase CYLD in hepatocytes causes apoptosis, inflammation, fibrosis, and cancer. Cancer Cell 2012, 21, 738–750. [Google Scholar] [CrossRef]
- Pannem, R.R.; Dorn, C.; Ahlqvist, K.; Bosserhoff, A.K.; Hellerbrand, C.; Massoumi, R. CYLD controls c-MYC expression through the JNK-dependent signaling pathway in hepatocellular carcinoma. Carcinogenesis 2014, 35, 461–468. [Google Scholar] [CrossRef]
- Karatzas, D.N.; Xanthopoulos, K.; Kotantaki, P.; Pseftogas, A.; Teliousis, K.; Hatzivassiliou, E.G.; Kontoyiannis, D.L.; Poutahidis, T.; Mosialos, G. Inactivation of CYLD in intestinal epithelial cells exacerbates colitis-associated colorectal carcinogenesis—A short report. Cell. Oncol. 2016, 39, 287–293. [Google Scholar] [CrossRef]
- Fernandez-Majada, V.; Welz, P.S.; Ermolaeva, M.A.; Schell, M.; Adam, A.; Dietlein, F.; Komander, D.; Buttner, R.; Thomas, R.K.; Schumacher, B.; et al. The tumour suppressor CYLD regulates the p53 DNA damage response. Nat. Commun. 2016, 7, 12508. [Google Scholar] [CrossRef]
- Komander, D.; Reyes-Turcu, F.; Licchesi, J.D.; Odenwaelder, P.; Wilkinson, K.D.; Barford, D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009, 10, 466–473. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, J. CYLD—A deubiquitylase that acts to fine-tune microtubule properties and functions. J. Cell Sci. 2016, 129, 2289–2295. [Google Scholar] [CrossRef]
- Wickstrom, S.A.; Masoumi, K.C.; Khochbin, S.; Fassler, R.; Massoumi, R. CYLD negatively regulates cell-cycle progression by inactivating HDAC6 and increasing the levels of acetylated tubulin. EMBO J. 2010, 29, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, M.; Li, D.; Ran, J.; Gao, J.; Suo, S.; Sun, S.C.; Zhou, J. CYLD regulates spindle orientation by stabilizing astral microtubules and promoting dishevelled-NuMA-dynein/dynactin complex formation. Proc. Natl. Acad. Sci. USA 2014, 111, 2158–2163. [Google Scholar] [CrossRef] [PubMed]
- Stegmeier, F.; Sowa, M.E.; Nalepa, G.; Gygi, S.P.; Harper, J.W.; Elledge, S.J. The tumor suppressor CYLD regulates entry into mitosis. Proc. Natl. Acad. Sci. USA 2007, 104, 8869–8874. [Google Scholar] [CrossRef] [PubMed]
- Trompouki, E.; Hatzivassiliou, E.; Tsichritzis, T.; Farmer, H.; Ashworth, A.; Mosialos, G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature 2003, 424, 793–796. [Google Scholar] [CrossRef]
- Brummelkamp, T.R.; Nijman, S.M.; Dirac, A.M.; Bernards, R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature 2003, 424, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Kovalenko, A.; Chable-Bessia, C.; Cantarella, G.; Israel, A.; Wallach, D.; Courtois, G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature 2003, 424, 801–805. [Google Scholar] [CrossRef]
- Moquin, D.M.; McQuade, T.; Chan, F.K. CYLD deubiquitinates RIP1 in the TNFalpha-induced necrosome to facilitate kinase activation and programmed necrosis. PLoS ONE 2013, 8, e76841. [Google Scholar] [CrossRef]
- Song, H.; Li, D.; Wu, T.; Xie, D.; Hua, K.; Hu, J.; Deng, X.; Ji, C.; Deng, Y.; Fang, L. MicroRNA-301b promotes cell proliferation and apoptosis resistance in triple-negative breast cancer by targeting CYLD. BMB Rep. 2018, 51, 602–607. [Google Scholar] [CrossRef]
- Pseftogas, A.; Gonidas, C.; Mosialos, G. Activation of peroxisome proliferator-activated receptor gamma in mammary epithelial cells upregulates the expression of tumor suppressor Cyld to mediate growth inhibition and anti-inflammatory effects. Int. J. Biochem. Cell Biol. 2017, 82, 49–56. [Google Scholar] [CrossRef]
- Wo, L.; Lu, D.; Gu, X. Knockdown of miR-182 promotes apoptosis via regulating RIP1 deubiquitination in TNF-alpha-treated triple-negative breast cancer cells. Tumour Biol. 2016, 37, 13733–13742. [Google Scholar] [CrossRef]
- Ni, F.; Gui, Z.; Guo, Q.; Hu, Z.; Wang, X.; Chen, D.; Wang, S. Downregulation of miR-362-5p inhibits proliferation, migration and invasion of human breast cancer MCF7 cells. Oncol. Lett. 2016, 11, 1155–1160. [Google Scholar] [CrossRef] [PubMed]
- Hutti, J.E.; Shen, R.R.; Abbott, D.W.; Zhou, A.Y.; Sprott, K.M.; Asara, J.M.; Hahn, W.C.; Cantley, L.C. Phosphorylation of the tumor suppressor CYLD by the breast cancer oncogene IKKepsilon promotes cell transformation. Mol. Cell 2009, 34, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Baiocchi, R.A.; Pal, S.; Mosialos, G.; Caligiuri, M.; Sif, S. The BRG1- and hBRM-associated factor BAF57 induces apoptosis by stimulating expression of the cylindromatosis tumor suppressor gene. Mol. Cell. Biol. 2005, 25, 7953–7965. [Google Scholar] [CrossRef] [PubMed]
- Orfanidou, T.; Xanthopoulos, K.; Dafou, D.; Pseftogas, A.; Hadweh, P.; Psyllaki, C.; Hatzivassiliou, E.; Mosialos, G. Down-regulation of the tumor suppressor CYLD enhances the transformed phenotype of human breast cancer cells. Anticancer Res. 2017, 37, 3493–3503. [Google Scholar] [CrossRef]
- Hayashi, M.; Jono, H.; Shinriki, S.; Nakamura, T.; Guo, J.; Sueta, A.; Tomiguchi, M.; Fujiwara, S.; Yamamoto-Ibusuki, M.; Murakami, K.; et al. Clinical significance of CYLD downregulation in breast cancer. Breast Cancer Res. Treat. 2014, 143, 447–457. [Google Scholar] [CrossRef]
- Kurosaki, T.; Maquat, L.E. Nonsense-mediated mRNA decay in humans at a glance. J. Cell Sci. 2016, 129, 461–467. [Google Scholar] [CrossRef]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef]
- Morel, A.P.; Lievre, M.; Thomas, C.; Hinkal, G.; Ansieau, S.; Puisieux, A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS ONE 2008, 3, e2888. [Google Scholar] [CrossRef]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef]
- Brabletz, T.; Kalluri, R.; Nieto, M.A.; Weinberg, R.A. EMT in cancer. Nat. Rev. Cancer 2018, 18, 128–134. [Google Scholar] [CrossRef]
- Hiemer, S.E.; Szymaniak, A.D.; Varelas, X. The transcriptional regulators TAZ and YAP direct transforming growth factor beta-induced tumorigenic phenotypes in breast cancer cells. J. Biol. Chem. 2014, 289, 13461–13474. [Google Scholar] [CrossRef]
- Misra, J.R.; Irvine, K.D. The Hippo signaling network and its biological functions. Annu. Rev. Gen. 2018. [Google Scholar] [CrossRef] [PubMed]
- Varelas, X.; Samavarchi-Tehrani, P.; Narimatsu, M.; Weiss, A.; Cockburn, K.; Larsen, B.G.; Rossant, J.; Wrana, J.L. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-beta-SMAD pathway. Dev. Cell 2010, 19, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Trompouki, E.; Tsagaratou, A.; Kosmidis, S.K.; Dolle, P.; Qian, J.; Kontoyiannis, D.L.; Cardoso, W.V.; Mosialos, G. Truncation of the catalytic domain of the cylindromatosis tumor suppressor impairs lung maturation. Neoplasia 2009, 11, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Andrechek, E.R.; Hardy, W.R.; Siegel, P.M.; Rudnicki, M.A.; Cardiff, R.D.; Muller, W.J. Amplification of the neu/erbB-2 oncogene in a mouse model of mammary tumorigenesis. Proc. Natl. Acad. Sci. USA 2000, 97, 3444–3449. [Google Scholar] [CrossRef] [PubMed]
- Gregorieff, A.; Liu, Y.; Inanlou, M.R.; Khomchuk, Y.; Wrana, J.L. Yap-dependent reprogramming of Lgr5+ stem cells drives intestinal regeneration and cancer. Nature 2015, 526, 715. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.H.; Jono, H.; Komatsu, K.; Woo, C.H.; Lee, J.; Miyata, M.; Matsuno, T.; Xu, X.; Huang, Y.; Zhang, W.; et al. CYLD negatively regulates transforming growth factor-beta-signalling via deubiquitinating Akt. Nat. Commun. 2012, 3, 771. [Google Scholar] [CrossRef]
- Ge, W.L.; Xu, J.F.; Hu, J. Regulation of oral squamous cell carcinoma proliferation through crosstalk between SMAD7 and CYLD. Cell. Physiol. Biochem. 2016, 38, 1209–1217. [Google Scholar] [CrossRef]
- Shinriki, S.; Jono, H.; Maeshiro, M.; Nakamura, T.; Guo, J.; Li, J.D.; Ueda, M.; Yoshida, R.; Shinohara, M.; Nakayama, H.; et al. Loss of CYLD promotes cell invasion via ALK5 stabilization in oral squamous cell carcinoma. J. Pathol. 2017. [Google Scholar] [CrossRef]
- Zhang, Y.; Alexander, P.B.; Wang, X.F. TGF-beta family signaling in the control of cell proliferation and survival. Cold Spring Harb. Perspec. Biol. 2017, 9. [Google Scholar] [CrossRef]
- Wu, L.; Yang, X. Targeting the Hippo pathway for breast cancer therapy. Cancers 2018, 10, 422. [Google Scholar] [CrossRef] [PubMed]
- Yao, F.; Zhou, Z.; Kim, J.; Hang, Q.; Xiao, Z.; Ton, B.N.; Chang, L.; Liu, N.; Zeng, L.; Wang, W.; et al. SKP2- and OTUD1-regulated non-proteolytic ubiquitination of YAP promotes YAP nuclear localization and activity. Nat. Commun. 2018, 9, 2269. [Google Scholar] [CrossRef]
- Jolly, M.K.; Somarelli, J.A.; Sheth, M.; Biddle, A.; Tripathi, S.C.; Armstrong, A.J.; Hanash, S.M.; Bapat, S.A.; Rangarajan, A.; Levine, H. Hybrid epithelial/mesenchymal phenotypes promote metastasis and therapy resistance across carcinomas. Pharmacol. Ther. 2019, 194, 161–184. [Google Scholar] [CrossRef] [PubMed]
- Pastushenko, I.; Blanpain, C. EMT Transition states during tumor progression and metastasis. Trends Cell Biol. 2019, 29, 212–226. [Google Scholar] [CrossRef] [PubMed]
- De Carcer, G.; Venkateswaran, S.V.; Salgueiro, L.; El Bakkali, A.; Somogyi, K.; Rowald, K.; Montanes, P.; Sanclemente, M.; Escobar, B.; de Martino, A.; et al. Plk1 overexpression induces chromosomal instability and suppresses tumor development. Nat. Commun. 2018, 9, 3012. [Google Scholar] [CrossRef]
- Lobry, C.; Oh, P.; Aifantis, I. Oncogenic and tumor suppressor functions of Notch in cancer: It’s NOTCH what you think. J. Exp. Med. 2011, 208, 1931–1935. [Google Scholar] [CrossRef]
- Popeda, M.; Stokowy, T.; Bednarz-Knoll, N.; Jurek, A.; Niemira, M.; Bielska, A.; Kretowski, A.; Kalinowski, L.; Szade, J.; Markiewicz, A.; et al. NF-kappa B signaling-related signatures are connected with the mesenchymal phenotype of circulating tumor cells in non-metastatic breast cancer. Cancers 2019, 11, 1961. [Google Scholar] [CrossRef]
- Sanjana, N.E.; Shalem, O.; Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 2014, 11, 783–784. [Google Scholar] [CrossRef]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelson, T.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef]
- Zufferey, R.; Nagy, D.; Mandel, R.J.; Naldini, L.; Trono, D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 1997, 15, 871–875. [Google Scholar] [CrossRef]
- Gu, J.; Kagawa, S.; Takakura, M.; Kyo, S.; Inoue, M.; Roth, J.A.; Fang, B. Tumor-specific transgene expression from the human telomerase reverse transcriptase promoter enables targeting of the therapeutic effects of the Bax gene to cancers. Cancer Res. 2000, 60, 5359–5364. [Google Scholar] [PubMed]
- Dull, T.; Zufferey, R.; Kelly, M.; Mandel, R.J.; Nguyen, M.; Trono, D.; Naldini, L. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 1998, 72, 8463–8471. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; He, X.; Zheng, Z.; Zhang, Z.; Wei, C.; Guan, K.; Hou, L.; Zhang, B.; Zhu, L.; Cao, Y.; et al. Downregulation of microRNA miR-526a by enterovirus inhibits RIG-I-dependent innate immune response. J. Virol. 2014, 88, 11356–11368. [Google Scholar] [CrossRef] [PubMed]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Berens, E.B.; Holy, J.M.; Riegel, A.T.; Wellstein, A. A Cancer cell spheroid assay to assess invasion in a 3D setting. J. Vis. Exp. 2015. [Google Scholar] [CrossRef]
- Carpenter, A.E.; Jones, T.R.; Lamprecht, M.R.; Clarke, C.; Kang, I.H.; Friman, O.; Guertin, D.A.; Chang, J.H.; Lindquist, R.A.; Moffat, J.; et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7, R100. [Google Scholar] [CrossRef]
- Kingston, N.M.; Tilston-Lunel, A.M.; Hicks-Berthet, J.; Varelas, X. Immunofluorescence microscopy to study endogenous TAZ in mammalian cells. Methods Mol. Biol. 2019, 1893, 107–113. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstrale, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pseftogas, A.; Xanthopoulos, K.; Poutahidis, T.; Ainali, C.; Dafou, D.; Panteris, E.; Kern, J.G.; Varelas, X.; Hardas, A.; Gonidas, C.; et al. The Tumor Suppressor CYLD Inhibits Mammary Epithelial to Mesenchymal Transition by the Coordinated Inhibition of YAP/TAZ and TGFβ Signaling. Cancers 2020, 12, 2047. https://doi.org/10.3390/cancers12082047
Pseftogas A, Xanthopoulos K, Poutahidis T, Ainali C, Dafou D, Panteris E, Kern JG, Varelas X, Hardas A, Gonidas C, et al. The Tumor Suppressor CYLD Inhibits Mammary Epithelial to Mesenchymal Transition by the Coordinated Inhibition of YAP/TAZ and TGFβ Signaling. Cancers. 2020; 12(8):2047. https://doi.org/10.3390/cancers12082047
Chicago/Turabian StylePseftogas, Athanasios, Konstantinos Xanthopoulos, Theofilos Poutahidis, Chrysanthi Ainali, Dimitra Dafou, Emmanuel Panteris, Joseph G. Kern, Xaralabos Varelas, Alexander Hardas, Christos Gonidas, and et al. 2020. "The Tumor Suppressor CYLD Inhibits Mammary Epithelial to Mesenchymal Transition by the Coordinated Inhibition of YAP/TAZ and TGFβ Signaling" Cancers 12, no. 8: 2047. https://doi.org/10.3390/cancers12082047
APA StylePseftogas, A., Xanthopoulos, K., Poutahidis, T., Ainali, C., Dafou, D., Panteris, E., Kern, J. G., Varelas, X., Hardas, A., Gonidas, C., Tsingotjidou, A., Hatzivassiliou, E., & Mosialos, G. (2020). The Tumor Suppressor CYLD Inhibits Mammary Epithelial to Mesenchymal Transition by the Coordinated Inhibition of YAP/TAZ and TGFβ Signaling. Cancers, 12(8), 2047. https://doi.org/10.3390/cancers12082047






