Inhibition of TLR4 Signaling Affects Mitochondrial Fitness and Overcomes Bortezomib Resistance in Myeloma Plasma Cells
Abstract
1. Introduction
2. Results
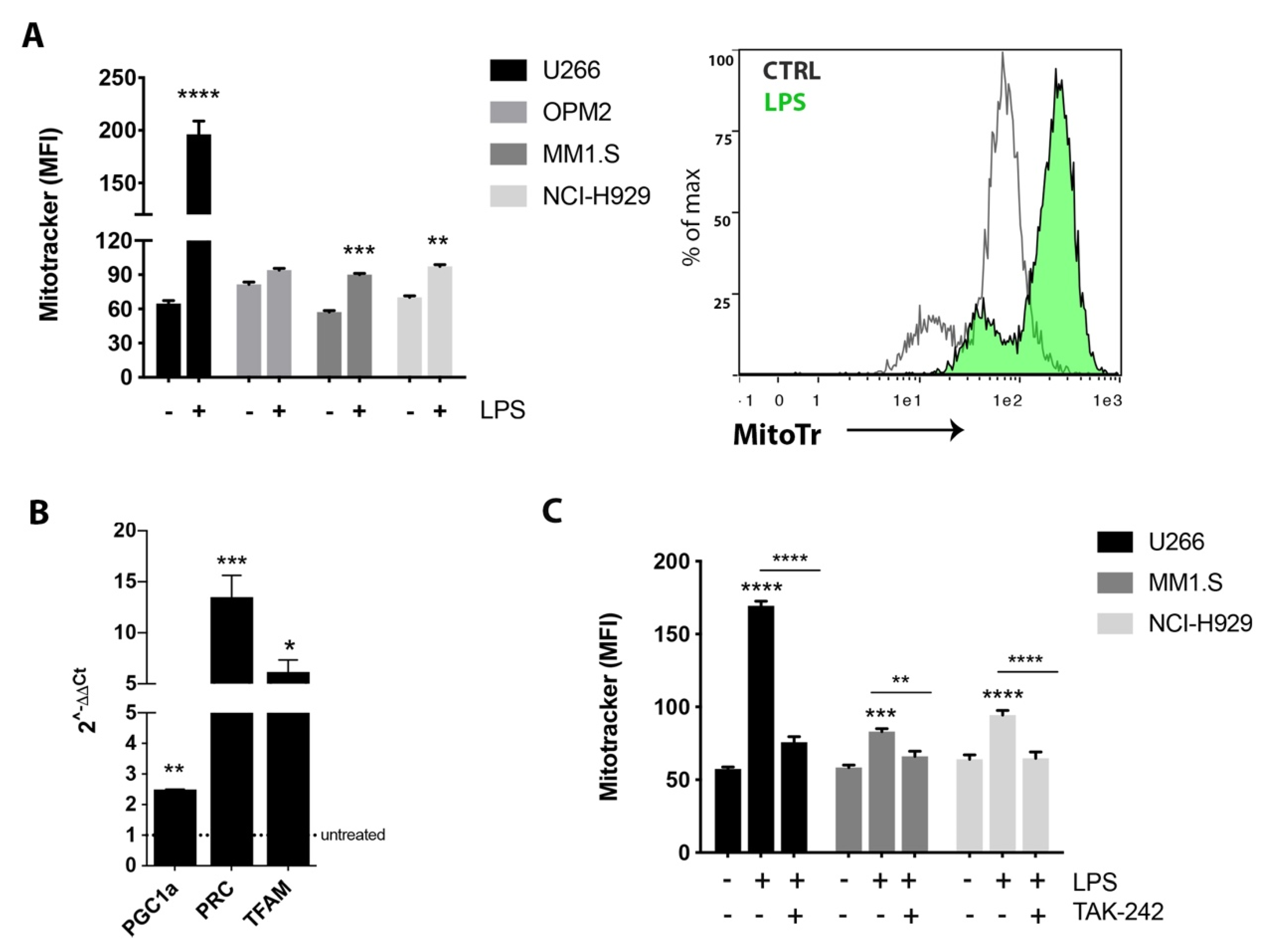
2.1. TLR4 Activation Induced Mitochondrial Biogenesis in MM Cell Lines
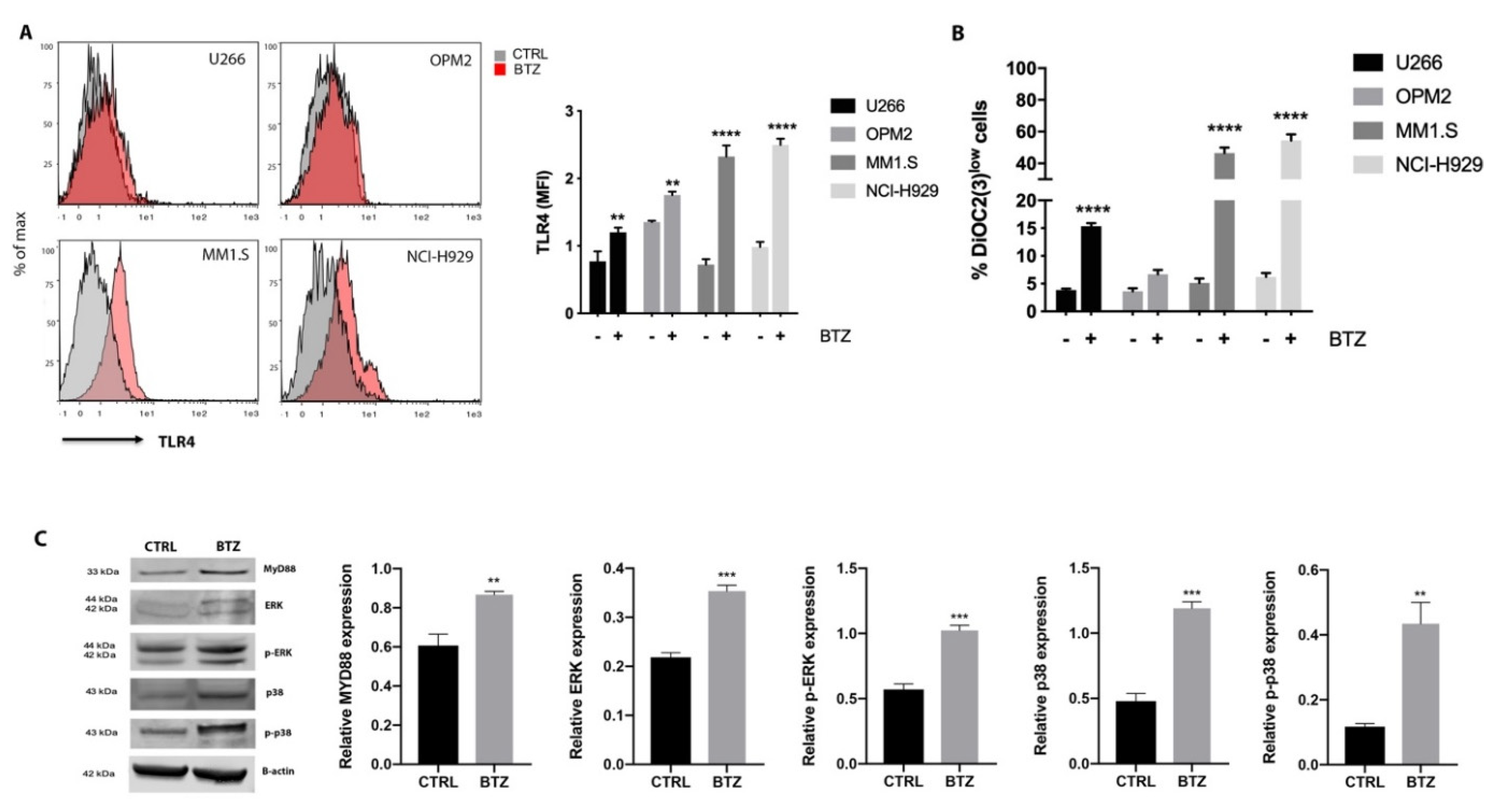
2.2. BTZ Treatment Led to Activation of TLR4 Signaling
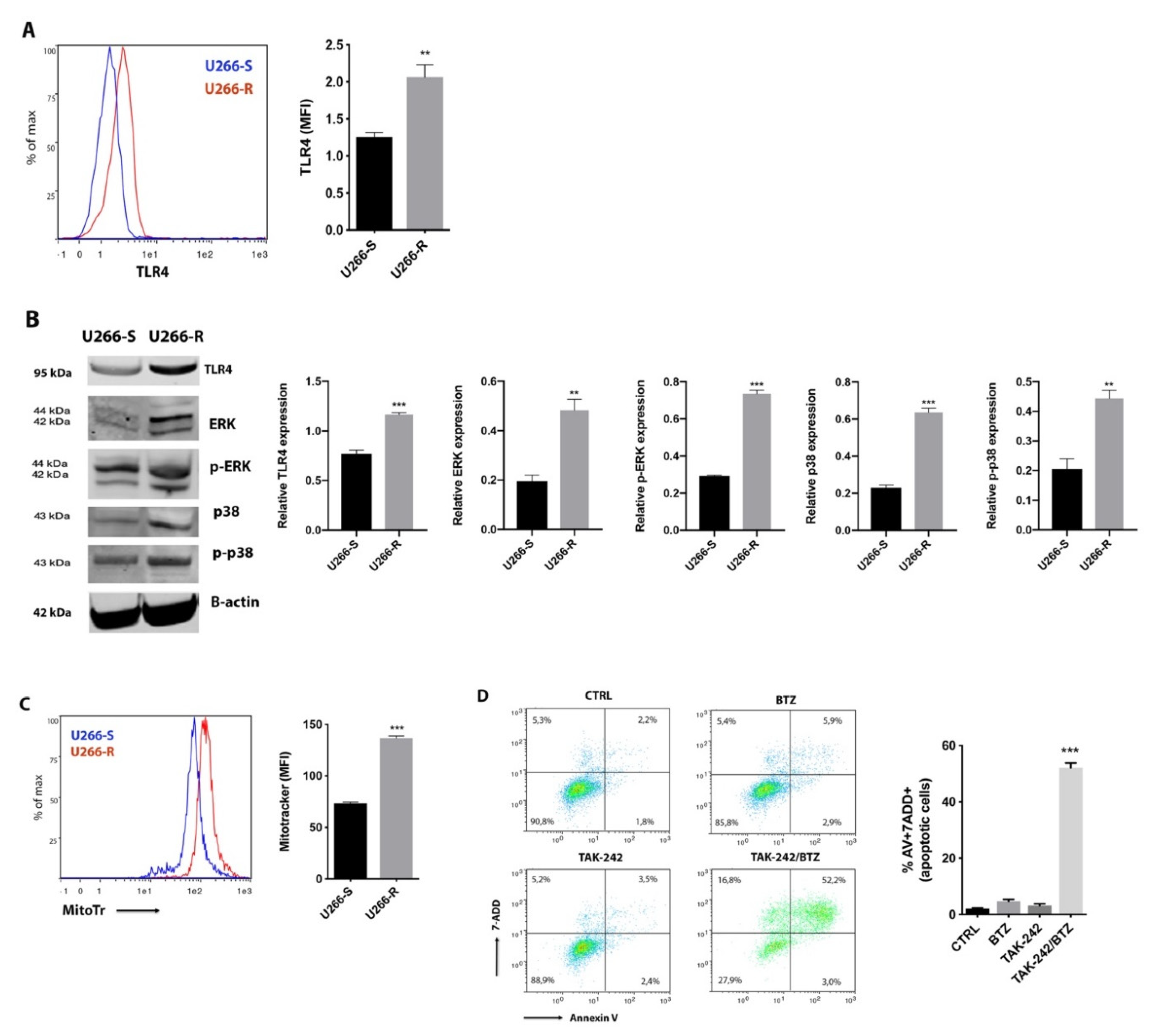
2.3. TLR4 Inhibition Restores Pharmacological Response of U266-R to BTZ-Induced Apoptosis
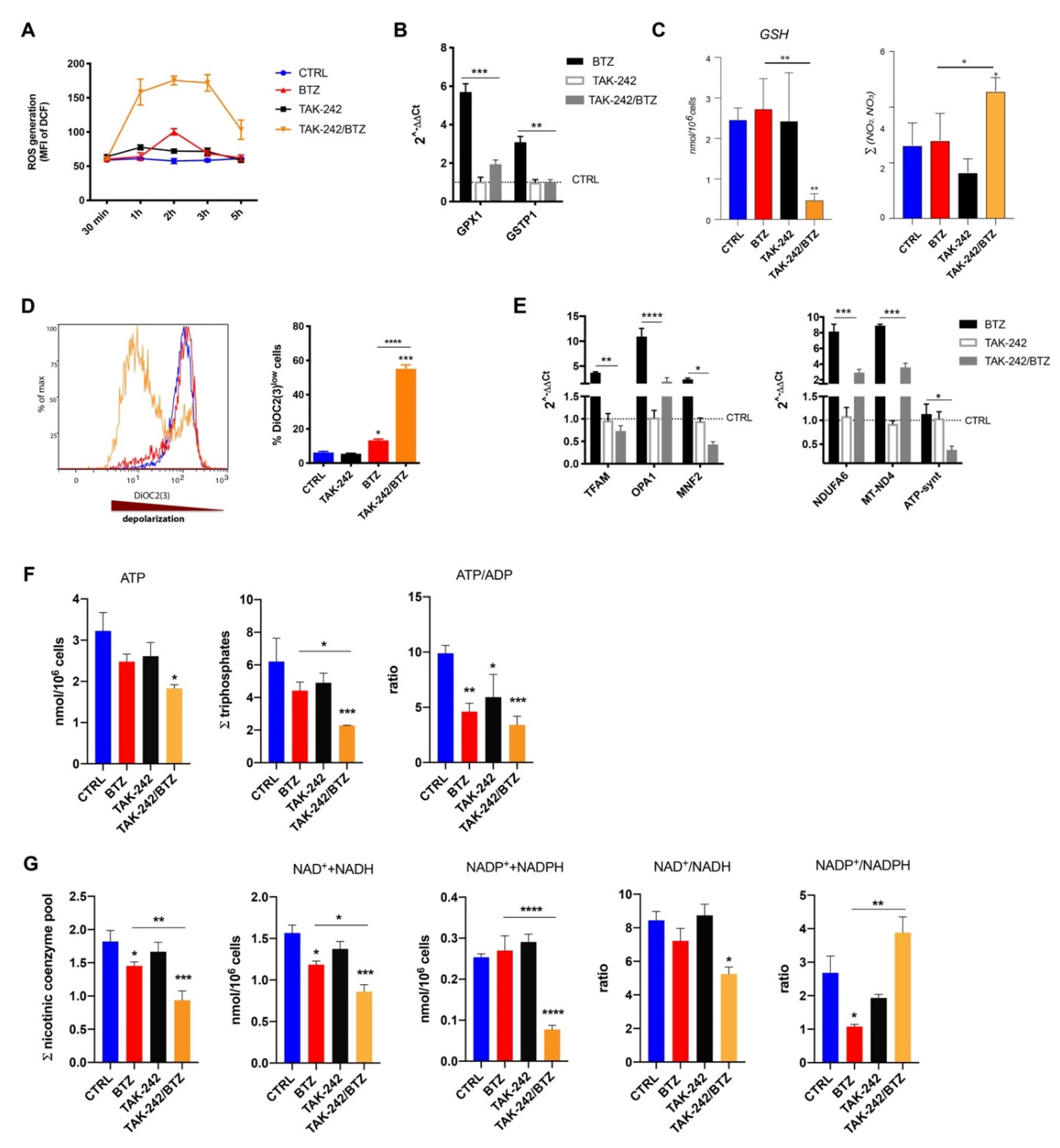
2.4. Combined Treatment with TLR4 Inhibitor and BTZ Induced Oxidative Stress and Mitochondrial Depolarization Damaging Mitochondrial Dynamics and Energy Metabolism
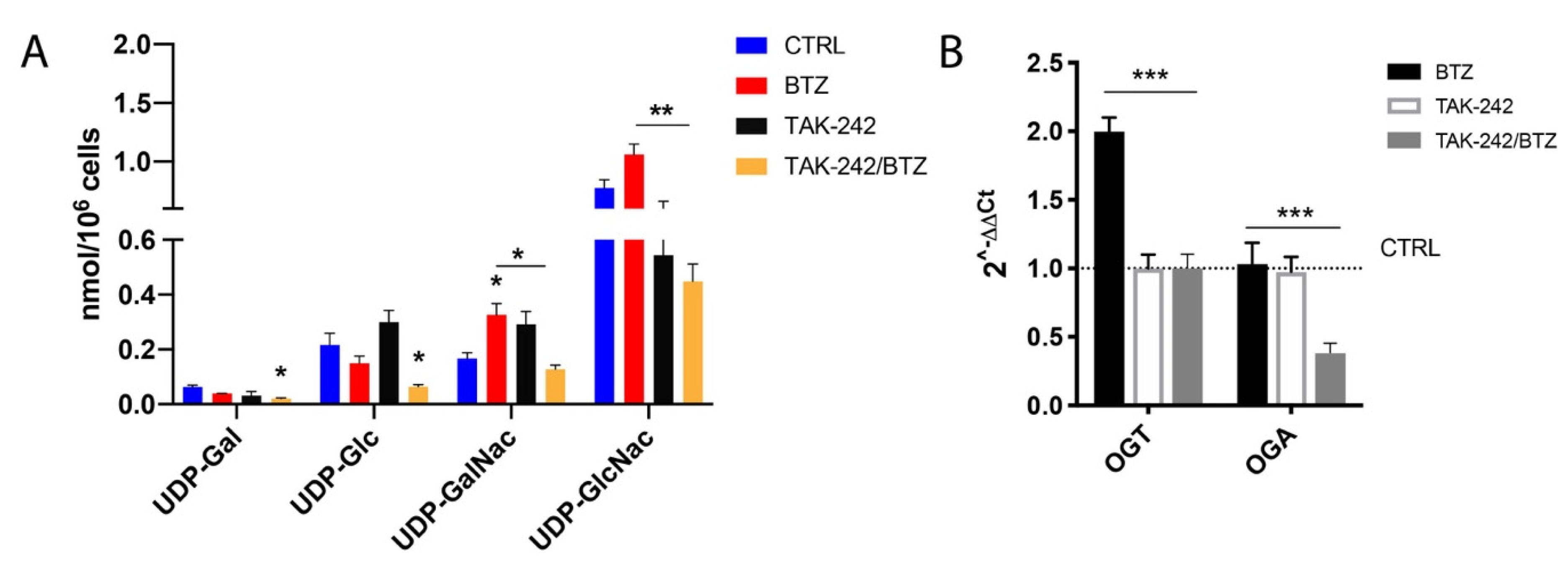
2.5. Combination of TAK-242/BTZ Decreased the Hexosamine Biosynthetic Pathway Products
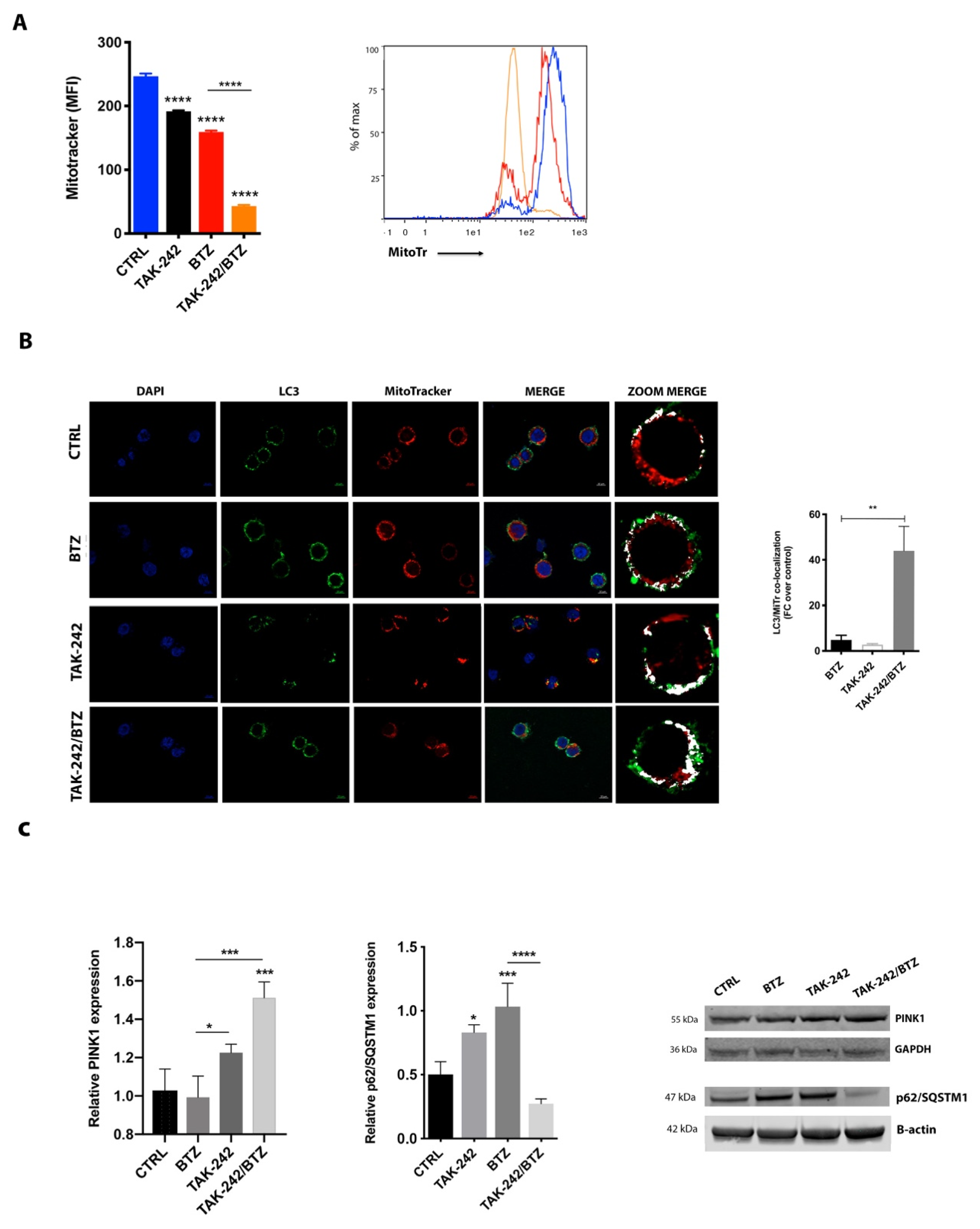
2.6. Mitochondrial Impairment Induced by TAK-242/BTZ Cotreatment Increased Mitophagy
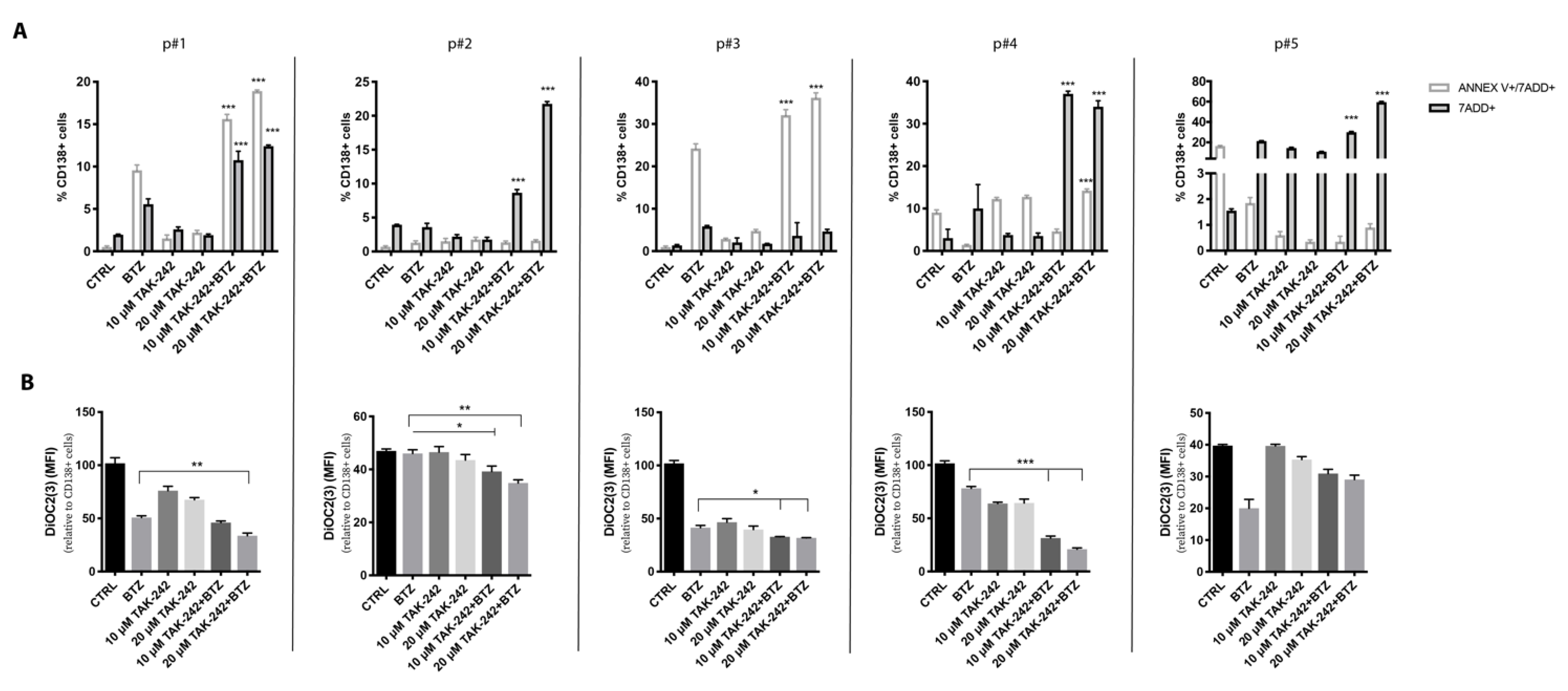
2.7. Targeting TLR4 and Proteasome Activities Resensitized CD138+ Cells from Refractory MM Patients
3. Discussion
4. Material and Methods
4.1. MM Cell Lines and CD138+ Plasma Cells
4.2. Flow Cytometry
4.3. RT-qPCR
4.4. Western Blot Analysis
4.5. HPLC Materials and Method
4.6. Immunofluorescence
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sonneveld, P. Management of multiple myeloma in the relapsed/refractory patient. Hematology Am. Soc. Hematol. Educ. Program. 2017, 2017, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Abdi, J.; Mutis, T.; Garssen, J.; Redegeld, F. Characterization of the Toll-like receptor expression profile in human multiple myeloma cells. PLoS ONE 2013, 8, e60671. [Google Scholar] [CrossRef] [PubMed]
- Kutikhin, A.G. Association of polymorphisms in TLR genes and in genes of the Toll-like receptor signaling pathway with cancer risk. Hum. Immunol. 2011, 72, 1095–1116. [Google Scholar] [CrossRef] [PubMed]
- Oblak, A.; Jerala, R. Toll-like receptor 4 activation in cancer progression and therapy. Clin. Dev. Immunol. 2011, 2011, 609579. [Google Scholar] [CrossRef]
- Lu, Y.C.; Yeh, W.C.; Ohashi, P.S. LPS/TLR4 signal transduction pathway. Cytokine 2008, 42, 145–151. [Google Scholar] [CrossRef]
- Akira, S.; Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef]
- Yang, H.; Wang, B.; Wang, T.; Xu, L.; He, C.; Wen, H.; Yan, J.; Su, H.; Zhu, X. Toll-like receptor 4 prompts human breast cancer cells invasiveness via lipopolysaccharide stimulation and is overexpressed in patients with lymph node metastasis. PLoS ONE 2014, 9, e109980. [Google Scholar] [CrossRef]
- Domenis, R.; Cifu, A.; Marino, D.; Fabris, M.; Niazi, K.R.; Soon-Shiong, P.; Curcio, F. Toll-like Receptor-4 Activation Boosts the Immunosuppressive Properties of Tumor Cells-derived Exosomes. Sci. Rep. 2019, 9, 8457. [Google Scholar] [CrossRef]
- Wang, W.; Wang, J. Toll-Like Receptor 4 (TLR4)/Cyclooxygenase-2 (COX-2) Regulates Prostate Cancer Cell Proliferation, Migration, and Invasion by NF-kappaB Activation. Med. Sci. Monit. 2018, 24, 5588–5597. [Google Scholar] [CrossRef]
- Ran, S. The Role of TLR4 in Chemotherapy-Driven Metastasis. Cancer Res. 2015, 75, 2405–2410. [Google Scholar] [CrossRef]
- Tang, X.; Zhu, Y. TLR4 signaling promotes immune escape of human colon cancer cells by inducing immunosuppressive cytokines and apoptosis resistance. Oncol. Res. 2012, 20, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Zhan, F.; Huang, Y.; Colla, S.; Stewart, J.P.; Hanamura, I.; Gupta, S.; Epstein, J.; Yaccoby, S.; Sawyer, J.; Burington, B.; et al. The molecular classification of multiple myeloma. Blood 2006, 108, 2020–2028. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Wang, J. Andrographolide inhibits multiple myeloma cells by inhibiting the TLR4/NF-kappaB signaling pathway. Mol. Med. Rep. 2016, 13, 1827–1832. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhao, Y.; Huang, H.; Chen, G.; Wu, X.; Wang, Y.; Chang, W.; Zhu, Z.; Feng, Y.; Wu, D. Expression and function of toll-like receptors in multiple myeloma patients: Toll-like receptor ligands promote multiple myeloma cell growth and survival via activation of nuclear factor-kappaB. Br. J. Haematol. 2010, 150, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hamrouni, A.; Wolowiec, D.; Coiteux, V.; Kuliczkowski, K.; Hetuin, D.; Saudemont, A.; Quesnel, B. Plasma cells from multiple myeloma patients express B7-H1 (PD-L1) and increase expression after stimulation with IFN-{gamma} and TLR ligands via a MyD88-, TRAF6-, and MEK-dependent pathway. Blood 2007, 110, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.; Lu, P.; Li, Y.; Wang, L.; Li, H.; He, D.; Yang, Y.; Zhao, Y.; Yang, L.; Wang, M.; et al. Triggering of toll-like receptor-4 in human multiple myeloma cells promotes proliferation and alters cell responses to immune and chemotherapy drug attack. Cancer Biol. Ther. 2011, 11, 58–67. [Google Scholar] [CrossRef]
- Giallongo, C.; Tibullo, D.; Camiolo, G.; Parrinello, N.L.; Romano, A.; Puglisi, F.; Barbato, A.; Conticello, C.; Lupo, G.; Anfuso, C.D.; et al. Correction: TLR4 signaling drives mesenchymal stromal cells commitment to promote tumor microenvironment transformation in multiple myeloma. Cell Death Dis. 2019, 10, 820. [Google Scholar] [CrossRef]
- Gu, H.; Chen, X.; Gao, G.; Dong, H. Caspase-2 functions upstream of mitochondria in endoplasmic reticulum stress-induced apoptosis by bortezomib in human myeloma cells. Mol. Cancer Ther. 2008, 7, 2298–2307. [Google Scholar] [CrossRef]
- Tibullo, D.; Barbagallo, I.; Giallongo, C.; Vanella, L.; Conticello, C.; Romano, A.; Saccone, S.; Godos, J.; Di Raimondo, F.; Li Volti, G. Heme oxygenase-1 nuclear translocation regulates bortezomibinduced cytotoxicity and mediates genomic instability in myeloma cells. Oncotarget 2016, 7, 28868–28880. [Google Scholar] [CrossRef]
- Canestraro, M.; Galimberti, S.; Savli, H.; Palumbo, G.A.; Tibullo, D.; Nagy, B.; Guerrini, F.; Piaggi, S.; Cine, N.; Metelli, M.R.; et al. Synergistic antiproliferative effect of arsenic trioxide combined with bortezomib in HL60 cell line and primary blasts from patients affected by myeloproliferative disorders. Cancer Genet. Cytogenet. 2010, 199, 110–120. [Google Scholar] [CrossRef]
- Obeng, E.A.; Carlson, L.M.; Gutman, D.M.; Harrington, W.J., Jr.; Lee, K.P.; Boise, L.H. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood 2006, 107, 4907–4916. [Google Scholar] [CrossRef] [PubMed]
- Bagratuni, T.; Sklirou, A.D.; Kastritis, E.; Liacos, C.I.; Spilioti, C.; Eleutherakis-Papaiakovou, E.; Kanellias, N.; Gavriatopoulou, M.; Terpos, E.; Trougakos, I.P.; et al. Toll-Like Receptor 4 Activation Promotes Multiple Myeloma Cell Growth and Survival Via Suppression of The Endoplasmic Reticulum Stress Factor Chop. Sci. Rep. 2019, 9, 3245. [Google Scholar] [CrossRef] [PubMed]
- Bosc, C.; Selak, M.A.; Sarry, J.E. Resistance Is Futile: Targeting Mitochondrial Energetics and Metabolism to Overcome Drug Resistance in Cancer Treatment. Cell Metab. 2017, 26, 705–707. [Google Scholar] [CrossRef] [PubMed]
- Farge, T.; Saland, E.; de Toni, F.; Aroua, N.; Hosseini, M.; Perry, R.; Bosc, C.; Sugita, M.; Stuani, L.; Fraisse, M.; et al. Chemotherapy-Resistant Human Acute Myeloid Leukemia Cells Are Not Enriched for Leukemic Stem Cells but Require Oxidative Metabolism. Cancer Discov. 2017, 7, 716–735. [Google Scholar] [CrossRef] [PubMed]
- Soriano, G.P.; Besse, L.; Li, N.; Kraus, M.; Besse, A.; Meeuwenoord, N.; Bader, J.; Everts, B.; den Dulk, H.; Overkleeft, H.S.; et al. Proteasome inhibitor-adapted myeloma cells are largely independent from proteasome activity and show complex proteomic changes, in particular in redox and energy metabolism. Leukemia 2016, 30, 2198–2207. [Google Scholar] [CrossRef]
- Thompson, R.M.; Dytfeld, D.; Reyes, L.; Robinson, R.M.; Smith, B.; Manevich, Y.; Jakubowiak, A.; Komarnicki, M.; Przybylowicz-Chalecka, A.; Szczepaniak, T.; et al. Glutaminase inhibitor CB-839 synergizes with carfilzomib in resistant multiple myeloma cells. Oncotarget 2017, 8, 35863–35876. [Google Scholar] [CrossRef]
- Zhan, X.; Yu, W.; Franqui-Machin, R.; Bates, M.L.; Nadiminti, K.; Cao, H.; Amendt, B.A.; Jethava, Y.; Frech, I.; Zhan, F.; et al. Alteration of mitochondrial biogenesis promotes disease progression in multiple myeloma. Oncotarget 2017, 8, 111213–111224. [Google Scholar] [CrossRef]
- Tibullo, D.; Giallongo, C.; Romano, A.; Vicario, N.; Barbato, A.; Puglisi, F.; Parenti, R.; Amorini, A.M.; Wissam Saab, M.; Tavazzi, B.; et al. Mitochondrial Functions, Energy Metabolism and Protein Glycosylation are Interconnected Processes Mediating Resistance to Bortezomib in Multiple Myeloma Cells. Biomolecules 2020, 10, 696. [Google Scholar] [CrossRef]
- Ling, Y.H.; Liebes, L.; Zou, Y.; Perez-Soler, R. Reactive oxygen species generation and mitochondrial dysfunction in the apoptotic response to Bortezomib, a novel proteasome inhibitor, in human H460 non-small cell lung cancer cells. J. Biol. Chem. 2003, 278, 33714–33723. [Google Scholar] [CrossRef]
- Li, B.; Kohler, J.J. Glycosylation of the nuclear pore. Traffic 2014, 15, 347–361. [Google Scholar] [CrossRef]
- Widdrington, J.D.; Gomez-Duran, A.; Pyle, A.; Ruchaud-Sparagano, M.H.; Scott, J.; Baudouin, S.V.; Rostron, A.J.; Lovat, P.E.; Chinnery, P.F.; Simpson, A.J. Exposure of Monocytic Cells to Lipopolysaccharide Induces Coordinated Endotoxin Tolerance, Mitochondrial Biogenesis, Mitophagy, and Antioxidant Defenses. Front. Immunol. 2018, 9, 2217. [Google Scholar] [CrossRef] [PubMed]
- Okatsu, K.; Saisho, K.; Shimanuki, M.; Nakada, K.; Shitara, H.; Sou, Y.S.; Kimura, M.; Sato, S.; Hattori, N.; Komatsu, M.; et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells 2010, 15, 887–900. [Google Scholar] [CrossRef] [PubMed]
- Xiao, B.; Deng, X.; Lim, G.G.Y.; Zhou, W.; Saw, W.T.; Zhou, Z.D.; Lim, K.L.; Tan, E.K. p62-Mediated mitochondrial clustering attenuates apoptosis induced by mitochondrial depolarization. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 1308–1317. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhou, J.; Luo, P.; Gao, H.; Ma, Y.; Chen, Y.S.; Li, L.; Zou, D.; Zhang, Y.; Jing, Z. Prosaposin promotes the proliferation and tumorigenesis of glioma through toll-like receptor 4 (TLR4)-mediated NF-kappaB signaling pathway. EBioMedicine 2018, 37, 78–90. [Google Scholar] [CrossRef]
- Wang, N.; Meng, X.; Liu, Y.; Chen, Y.; Liang, Q. LPS promote Osteosarcoma invasion and migration through TLR4/HOTAIR. Gene 2019, 680, 1–8. [Google Scholar] [CrossRef]
- Molteni, M.; Gemma, S.; Rossetti, C. The Role of Toll-Like Receptor 4 in Infectious and Noninfectious Inflammation. Mediators Inflamm. 2016, 2016, 6978936. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Wang, W.; Tamaki, Z.; Shi, B.; Yeldandi, A.; Tsukimi, Y.; Yamasaki, M.; Varga, J. Pharmacological Inhibition of Toll-Like Receptor-4 Signaling by TAK242 Prevents and Induces Regression of Experimental Organ Fibrosis. Front. Immunol. 2018, 9, 2434. [Google Scholar] [CrossRef]
- Lackner, L.L. The Expanding and Unexpected Functions of Mitochondria Contact Sites. Trends Cell Biol. 2019, 29, 580–590. [Google Scholar] [CrossRef]
- Gupta, S.; Cuffe, L.; Szegezdi, E.; Logue, S.E.; Neary, C.; Healy, S.; Samali, A. Mechanisms of ER Stress-Mediated Mitochondrial Membrane Permeabilization. Int. J. Cell Biol. 2010, 2010, 170215. [Google Scholar] [CrossRef]
- Chauhan, D.; Li, G.; Podar, K.; Hideshima, T.; Mitsiades, C.; Schlossman, R.; Munshi, N.; Richardson, P.; Cotter, F.E.; Anderson, K.C. Targeting mitochondria to overcome conventional and bortezomib/proteasome inhibitor PS-341 resistance in multiple myeloma (MM) cells. Blood 2004, 104, 2458–2466. [Google Scholar] [CrossRef]
- Barresi, V.; Romano, A.; Musso, N.; Capizzi, C.; Consoli, C.; Martelli, M.P.; Palumbo, G.; Di Raimondo, F.; Condorelli, D.F. Broad copy neutral-loss of heterozygosity regions and rare recurring copy number abnormalities in normal karyotype-acute myeloid leukemia genomes. Genes Chromosomes Cancer 2010, 49, 1014–1023. [Google Scholar] [CrossRef] [PubMed]
- Wolf, D.A. Is reliance on mitochondrial respiration a “chink in the armor” of therapy-resistant cancer? Cancer Cell 2014, 26, 788–795. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.Y.; Zhang, T.; Kraus, W.L. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005, 19, 1951–1967. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Fang, Y.Z.; Yang, S.; Lupton, J.R.; Turner, N.D. Glutathione metabolism and its implications for health. J. Nutr. 2004, 134, 489–492. [Google Scholar] [CrossRef] [PubMed]
- Flannery, P.J.; Trushina, E. Mitochondrial dynamics and transport in Alzheimer’s disease. Mol. Cell Neurosci. 2019, 98, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Twig, G.; Shirihai, O.S. The interplay between mitochondrial dynamics and mitophagy. Antioxid Redox. Signal. 2011, 14, 1939–1951. [Google Scholar] [CrossRef]
- Liu, J.; Liu, W.; Li, R.; Yang, H. Mitophagy in Parkinson’s Disease: From Pathogenesis to Treatment. Cells 2019, 8, 712. [Google Scholar] [CrossRef]
- Valera-Alberni, M.; Canto, C. Mitochondrial stress management: A dynamic journey. Cell Stress 2018, 2, 253–274. [Google Scholar] [CrossRef]
- Tibullo, D.; Barbagallo, I.; Giallongo, C.; La Cava, P.; Branca, A.; Conticello, C.; Stagno, F.; Chiarenza, A.; Palumbo, G.A.; Di Raimondo, F. Effects of second-generation tyrosine kinase inhibitors towards osteogenic differentiation of human mesenchymal cells of healthy donors. Hematol Oncol. 2012, 30, 27–33. [Google Scholar] [CrossRef]
- Lazzarino, G.; Amorini, A.M.; Fazzina, G.; Vagnozzi, R.; Signoretti, S.; Donzelli, S.; Di Stasio, E.; Giardina, B.; Tavazzi, B. Single-sample preparation for simultaneous cellular redox and energy state determination. Anal. Biochem. 2003, 322, 51–59. [Google Scholar] [CrossRef]
- Romitelli, F.; Santini, S.A.; Chierici, E.; Pitocco, D.; Tavazzi, B.; Amorini, A.M.; Lazzarino, G.; Di Stasio, E. Comparison of nitrite/nitrate concentration in human plasma and serum samples measured by the enzymatic batch Griess assay, ion-pairing HPLC and ion-trap GC-MS: The importance of a correct removal of proteins in the Griess assay. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 2007, 851, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Gulino, R.; Vicario, N.; Giunta, M.A.S.; Spoto, G.; Calabrese, G.; Vecchio, M.; Gulisano, M.; Leanza, G.; Parenti, R. Neuromuscular Plasticity in a Mouse Neurotoxic Model of Spinal Motoneuronal Loss. Int. J. Mol. Sci. 2019, 20, 1500. [Google Scholar] [CrossRef] [PubMed]
- Vicario, N.; Pasquinucci, L.; Spitale, F.M.; Chiechio, S.; Turnaturi, R.; Caraci, F.; Tibullo, D.; Avola, R.; Gulino, R.; Parenti, R.; et al. Simultaneous Activation of Mu and Delta Opioid Receptors Reduces Allodynia and Astrocytic Connexin 43 in an Animal Model of Neuropathic Pain. Mol. Neurobiol. 2019, 56, 7338–7354. [Google Scholar] [CrossRef]
- Giusto, E.; Codrich, M.; de Leo, G.; Francardo, V.; Coradazzi, M.; Parenti, R.; Gulisano, M.; Vicario, N.; Gulino, R.; Leanza, G. Compensatory changes in degenerating spinal motoneurons sustain functional sparing in the SOD1-G93A mouse model of amyotrophic lateral sclerosis. J. Comp. Neurol. 2020, 528, 231–243. [Google Scholar] [CrossRef] [PubMed]
| pts | Sex | Age | Status Disease | Ig Type | R-ISS Stage | LDH (U/L) | Hb (g/dL) | ANC/mmc | ALC/mmc | AMC/mmc | k-lambda sFLC Ratio | FISH |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p#1 | M | 49 | Ref | IgAlambda | 1 | 213 | 11.6 | 900 | 540 | 800 | 0.01 | t (4;14) |
| p#2 | M | 72 | Ref | IgGk | 3 | 315 | 9.8 | 1670 | 560 | 870 | 230 | t (11;14) |
| p#3 | M | 58 | Ref | IgGk | 3 | 280 | 9.7 | 1890 | 700 | 910 | 120 | t (4;14) |
| p#4 | M | 73 | Ref | IgGk | 3 | 654 | 6.5 | 310 | 560 | 650 | 210 | del17p13 |
| p#5 | F | 68 | Ref | IgGlambda | 3 | 310 | 8.9 | 1100 | 780 | 760 | 0.01 | del17p13 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giallongo, C.; Tibullo, D.; Puglisi, F.; Barbato, A.; Vicario, N.; Cambria, D.; Parrinello, N.L.; Romano, A.; Conticello, C.; Forte, S.; et al. Inhibition of TLR4 Signaling Affects Mitochondrial Fitness and Overcomes Bortezomib Resistance in Myeloma Plasma Cells. Cancers 2020, 12, 1999. https://doi.org/10.3390/cancers12081999
Giallongo C, Tibullo D, Puglisi F, Barbato A, Vicario N, Cambria D, Parrinello NL, Romano A, Conticello C, Forte S, et al. Inhibition of TLR4 Signaling Affects Mitochondrial Fitness and Overcomes Bortezomib Resistance in Myeloma Plasma Cells. Cancers. 2020; 12(8):1999. https://doi.org/10.3390/cancers12081999
Chicago/Turabian StyleGiallongo, Cesarina, Daniele Tibullo, Fabrizio Puglisi, Alessandro Barbato, Nunzio Vicario, Daniela Cambria, Nunziatina Laura Parrinello, Alessandra Romano, Concetta Conticello, Stefano Forte, and et al. 2020. "Inhibition of TLR4 Signaling Affects Mitochondrial Fitness and Overcomes Bortezomib Resistance in Myeloma Plasma Cells" Cancers 12, no. 8: 1999. https://doi.org/10.3390/cancers12081999
APA StyleGiallongo, C., Tibullo, D., Puglisi, F., Barbato, A., Vicario, N., Cambria, D., Parrinello, N. L., Romano, A., Conticello, C., Forte, S., Parenti, R., Amorini, A. M., Lazzarino, G., Li Volti, G., Palumbo, G. A., & Di Raimondo, F. (2020). Inhibition of TLR4 Signaling Affects Mitochondrial Fitness and Overcomes Bortezomib Resistance in Myeloma Plasma Cells. Cancers, 12(8), 1999. https://doi.org/10.3390/cancers12081999














