DEK Expression in Breast Cancer Cells Leads to the Alternative Activation of Tumor Associated Macrophages
Abstract
1. Introduction
2. Results
2.1. Dek Expression Dictates the Differential Expression of Immune Genes
2.2. Dek Expression in Breast Cancer Cells Promotes M2-Like Macrophage Polarization In Vitro and In Vivo
2.3. Dek Expression in Breast Cancers Promotes Macrophage Polarization to an Iron-Recycling M2 Subtype
2.4. DEK Expression in Human Primary Breast Cancers Is Associated with Macrophage Regulation and Poor Survival
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Cell Culture
4.3. RNA-Sequencing
4.4. Quantitative RT-PCR
4.5. Western Blotting
4.6. Iron Assay
4.7. Luminex Assay
4.8. Flow Cytometry
4.9. Immunohistochemistry and Immunofluorescence
4.10. Perl’s Prussian Blue Stain
4.11. Human Breast Cancer Databases
4.12. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lanca, T.; Silva-Santos, B. The split nature of tumor-infiltrating leukocytes: Implications for cancer surveillance and immunotherapy. Oncoimmunology 2012, 1, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Shankaran, V.; Ikeda, H.; Bruce, A.T.; White, J.M.; Swanson, P.E.; Old, L.J.; Schreiber, R.D. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature 2001, 410, 1107–1111. [Google Scholar] [CrossRef] [PubMed]
- Chittezhath, M.; Dhillon, M.K.; Lim, J.Y.; Laoui, D.; Shalova, I.N.; Teo, Y.L.; Chen, J.; Kamaraj, R.; Raman, L.; Lum, J.; et al. Molecular profiling reveals a tumor-promoting phenotype of monocytes and macrophages in human cancer progression. Immunity 2014, 41, 815–829. [Google Scholar] [CrossRef]
- Condamine, T.; Ramachandran, I.; Youn, J.I.; Gabrilovich, D.I. Regulation of tumor metastasis by myeloid-derived suppressor cells. Annu. Rev. Med. 2015, 66, 97–110. [Google Scholar] [CrossRef]
- Zhang, Q.W.; Liu, L.; Gong, C.Y.; Shi, H.S.; Zeng, Y.H.; Wang, X.Z.; Zhao, Y.W.; Wei, Y.Q. Prognostic significance of tumor-associated macrophages in solid tumor: A meta-analysis of the literature. PLoS ONE 2012, 7, e50946. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Qu, J.; Sun, Y.; Wang, J.; Liu, X.; Wang, F.; Zhang, H.; Wang, W.; Ma, X.; Gao, X.; et al. Prognostic significance of tumor-associated macrophages in breast cancer: A meta-analysis of the literature. Oncotarget 2017, 8, 30576–30586. [Google Scholar] [CrossRef] [PubMed]
- Gwak, J.M.; Jang, M.H.; Kim, D.I.; Seo, A.N.; Park, S.Y. Prognostic value of tumor-associated macrophages according to histologic locations and hormone receptor status in breast cancer. PLoS ONE 2015, 10, e0125728. [Google Scholar] [CrossRef] [PubMed]
- Xuan, Q.J.; Wang, J.X.; Nanding, A.; Wang, Z.P.; Liu, H.; Lian, X.; Zhang, Q.Y. Tumor-associated macrophages are correlated with tamoxifen resistance in the postmenopausal breast cancer patients. Pathol. Oncol. Res. 2014, 20, 619–624. [Google Scholar] [CrossRef]
- Martinez, F.O.; Sica, A.; Mantovani, A.; Locati, M. Macrophage activation and polarization. Front. Biosci. 2008, 13, 453–461. [Google Scholar] [CrossRef]
- Aras, S.; Zaidi, M.R. TAMeless traitors: Macrophages in cancer progression and metastasis. Br. J. Cancer 2017, 117, 1583–1591. [Google Scholar] [CrossRef] [PubMed]
- Roszer, T. Understanding the Mysterious M2 Macrophage through Activation Markers and Effector Mechanisms. Mediat. Inflamm. 2015, 2015, 816460. [Google Scholar] [CrossRef]
- Chanmee, T.; Ontong, P.; Konno, K.; Itano, N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers 2014, 6, 1670–1690. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.Y.; Pollard, J.W. Tumor-associated macrophages press the angiogenic switch in breast cancer. Cancer Res. 2007, 67, 5064–5066. [Google Scholar] [CrossRef] [PubMed]
- Wyckoff, J.B.; Wang, Y.; Lin, E.Y.; Li, J.F.; Goswami, S.; Stanley, E.R.; Segall, J.E.; Pollard, J.W.; Condeelis, J. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007, 67, 2649–2656. [Google Scholar] [CrossRef]
- Riabov, V.; Gudima, A.; Wang, N.; Mickley, A.; Orekhov, A.; Kzhyshkowska, J. Role of tumor associated macrophages in tumor angiogenesis and lymphangiogenesis. Front. Physiol. 2014, 5, 75. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.Y.; Li, J.F.; Gnatovskiy, L.; Deng, Y.; Zhu, L.; Grzesik, D.A.; Qian, H.; Xue, X.N.; Pollard, J.W. Macrophages regulate the angiogenic switch in a mouse model of breast cancer. Cancer Res. 2006, 66, 11238–11246. [Google Scholar] [CrossRef] [PubMed]
- Ying, G.; Wu, Y. DEK: A novel early screening and prognostic marker for breast cancer. Mol. Med. Rep. 2015, 12, 7491–7495. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, X.; Sun, F.; Kong, J.; Li, Z.; Lin, Z. DEK overexpression is correlated with the clinical features of breast cancer. Pathol. Int. 2012, 62, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.C.; Liu, Y.L.; You, P.; Pan, J.S.; Zhou, J.Y.; Liu, Z.J.; Zhang, Z.Y. Overexpression of DEK gene is correlated with poor prognosis in hepatocellular carcinoma. Mol. Med. Rep. 2015, 11, 1318–1323. [Google Scholar] [CrossRef]
- Lee, S.Y.; Jung, W.; Lee, J.; Kim, A.; Kim, H.K.; Kim, B.H. High expression of DEK is associated with poor prognosis in hepatocellular carcinoma. Histol. Histopathol. 2019, 34, 1279–1288. [Google Scholar] [CrossRef]
- Datta, A.; Adelson, M.E.; Mogilevkin, Y.; Mordechai, E.; Sidi, A.A.; Trama, J.P. Oncoprotein DEK as a tissue and urinary biomarker for bladder cancer. BMC Cancer 2011, 11, 234. [Google Scholar] [CrossRef] [PubMed]
- Privette Vinnedge, L.M.; McClaine, R.; Wagh, P.K.; Wikenheiser-Brokamp, K.A.; Waltz, S.E.; Wells, S.I. The human DEK oncogene stimulates beta-catenin signaling, invasion and mammosphere formation in breast cancer. Oncogene 2011, 30, 2741–2752. [Google Scholar] [CrossRef]
- Adams, A.K.; Hallenbeck, G.E.; Casper, K.A.; Patil, Y.J.; Wilson, K.M.; Kimple, R.J.; Lambert, P.F.; Witte, D.P.; Xiao, W.; Gillison, M.L.; et al. DEK promotes HPV-positive and -negative head and neck cancer cell proliferation. Oncogene 2014, 34, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Dong, X.; Wang, K.; Wyatt, A.W.; Crea, F.; Xue, H.; Wang, Y.; Wu, R.; Bell, R.H.; Haegert, A.; et al. Identification of DEK as a potential therapeutic target for neuroendocrine prostate cancer. Oncotarget 2015, 6, 1806–1820. [Google Scholar] [CrossRef]
- Lin, L.; Piao, J.; Gao, W.; Piao, Y.; Jin, G.; Ma, Y.; Li, J.; Lin, Z. DEK over expression as an independent biomarker for poor prognosis in colorectal cancer. BMC Cancer 2013, 13, 366. [Google Scholar] [CrossRef] [PubMed]
- Carro, M.S.; Spiga, F.M.; Quarto, M.; Di Ninni, V.; Volorio, S.; Alcalay, M.; Muller, H. DEK Expression is controlled by E2F and deregulated in diverse tumor types. Cell Cycle 2006, 5, 1202–1207. [Google Scholar] [CrossRef]
- Privette Vinnedge, L.M.; Ho, S.M.; Wikenheiser-Brokamp, K.A.; Wells, S.I. The DEK Oncogene Is a Target of Steroid Hormone Receptor Signaling in Breast Cancer. PLoS ONE 2012, 7, e46985. [Google Scholar] [CrossRef]
- Sitwala, K.V.; Adams, K.; Markovitz, D.M. YY1 and NF-Y binding sites regulate the transcriptional activity of the dek and dek-can promoter. Oncogene 2002, 21, 8862–8870. [Google Scholar] [CrossRef]
- Khodadoust, M.S.; Verhaegen, M.; Kappes, F.; Riveiro-Falkenbach, E.; Cigudosa, J.C.; Kim, D.S.; Chinnaiyan, A.M.; Markovitz, D.M.; Soengas, M.S. Melanoma proliferation and chemoresistance controlled by the DEK oncogene. Cancer Res. 2009, 69, 6405–6413. [Google Scholar] [CrossRef]
- Liu, K.; Feng, T.; Liu, J.; Zhong, M.; Zhang, S. Silencing of the DEK gene induces apoptosis and senescence in CaSki cervical carcinoma cells via the up-regulation of NF-kappaB p65. Biosci. Rep. 2012, 32, 323–332. [Google Scholar] [CrossRef]
- Sammons, M.; Wan, S.S.; Vogel, N.L.; Mientjes, E.J.; Grosveld, G.; Ashburner, B.P. Negative regulation of the RelA/p65 transactivation function by the product of the DEK proto-oncogene. J. Biol. Chem. 2006, 281, 26802–26812. [Google Scholar] [CrossRef] [PubMed]
- Pease, N.A.; Wise-Draper, T.; Privette Vinnedge, L. Dissecting the Potential Interplay of DEK Functions in Inflammation and Cancer. J. Oncol. 2015, 2015, 106517. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Cigdem, S.; Okuwaki, M.; Nagata, K. Leukemia-Associated Nup214 Fusion Proteins Disturb the XPO1-Mediated Nuclear-Cytoplasmic Transport Pathway and Thereby the NF-kappaB Signaling Pathway. Mol. Cell Biol. 2016, 36, 1820–1835. [Google Scholar] [CrossRef] [PubMed]
- Adams, A.K.; Bolanos, L.C.; Dexheimer, P.J.; Karns, R.A.; Aronow, B.J.; Komurov, K.; Jegga, A.G.; Casper, K.A.; Patil, Y.J.; Wilson, K.M.; et al. IRAK1 is a novel DEK transcriptional target and is essential for head and neck cancer cell survival. Oncotarget 2015, 6, 43395–43407. [Google Scholar] [CrossRef]
- Privette Vinnedge, L.M.; Kappes, F.; Nassar, N.; Wells, S.I. Stacking the DEK: From chromatin topology to cancer stem cells. Cell Cycle 2013, 12, 51–66. [Google Scholar] [CrossRef]
- Deutzmann, A.; Ganz, M.; Schonenberger, F.; Vervoorts, J.; Kappes, F.; Ferrando-May, E. The human oncoprotein and chromatin architectural factor DEK counteracts DNA replication stress. Oncogene 2015, 34, 4270–4277. [Google Scholar] [CrossRef] [PubMed]
- Ivanauskiene, K.; Delbarre, E.; McGhie, J.D.; Kuntziger, T.; Wong, L.H.; Collas, P. The PML-associated protein DEK regulates the balance of H3.3 loading on chromatin and is important for telomere integrity. Genome Res. 2014, 24, 1584–1594. [Google Scholar] [CrossRef]
- Sanden, C.; Jarvstrat, L.; Lennartsson, A.; Brattas, P.L.; Nilsson, B.; Gullberg, U. The DEK oncoprotein binds to highly and ubiquitously expressed genes with a dual role in their transcriptional regulation. Mol. Cancer 2014, 13, 215. [Google Scholar] [CrossRef]
- Shibata, T.; Kokubu, A.; Miyamoto, M.; Hosoda, F.; Gotoh, M.; Tsuta, K.; Asamura, H.; Matsuno, Y.; Kondo, T.; Imoto, I.; et al. DEK oncoprotein regulates transcriptional modifiers and sustains tumor initiation activity in high-grade neuroendocrine carcinoma of the lung. Oncogene 2010, 29, 4671–4681. [Google Scholar] [CrossRef]
- Hollenbach, A.D.; McPherson, C.J.; Mientjes, E.J.; Iyengar, R.; Grosveld, G. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J. Cell Sci. 2002, 115, 3319–3330. [Google Scholar]
- Cleary, J.; Sitwala, K.V.; Khodadoust, M.S.; Kwok, R.P.; Mor-Vaknin, N.; Cebrat, M.; Cole, P.A.; Markovitz, D.M. p300/CBP-associated factor drives DEK into interchromatin granule clusters. J. Biol. Chem. 2005, 280, 31760–31767. [Google Scholar] [CrossRef] [PubMed]
- Koleva, R.I.; Ficarro, S.B.; Radomska, H.S.; Carrasco-Alfonso, M.J.; Alberta, J.A.; Webber, J.T.; Luckey, C.J.; Marcucci, G.; Tenen, D.G.; Marto, J.A. C/EBPalpha and DEK coordinately regulate myeloid differentiation. Blood 2012, 119, 4878–4888. [Google Scholar] [CrossRef]
- Campillos, M.; Garcia, M.A.; Valdivieso, F.; Vazquez, J. Transcriptional activation by AP-2alpha is modulated by the oncogene DEK. Nucleic Acids Res. 2003, 31, 1571–1575. [Google Scholar] [CrossRef] [PubMed]
- Gamble, M.J.; Fisher, R.P. SET and PARP1 remove DEK from chromatin to permit access by the transcription machinery. Nat. Struct. Mol. Biol. 2007, 14, 548–555. [Google Scholar] [CrossRef] [PubMed]
- Sawatsubashi, S.; Murata, T.; Lim, J.; Fujiki, R.; Ito, S.; Suzuki, E.; Tanabe, M.; Zhao, Y.; Kimura, S.; Fujiyama, S.; et al. A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes Dev. 2010, 24, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Kavanaugh, G.M.; Wise-Draper, T.M.; Morreale, R.J.; Morrison, M.A.; Gole, B.; Schwemberger, S.; Tichy, E.D.; Lu, L.; Babcock, G.F.; Wells, J.M.; et al. The human DEK oncogene regulates DNA damage response signaling and repair. Nucleic Acids Res. 2011, 39, 7465–7476. [Google Scholar] [CrossRef]
- Smith, E.A.; Gole, B.; Willis, N.A.; Soria, R.; Starnes, L.M.; Krumpelbeck, E.F.; Jegga, A.G.; Ali, A.M.; Guo, H.; Meetei, A.R.; et al. DEK is required for homologous recombination repair of DNA breaks. Sci. Rep. 2017, 7, 44662. [Google Scholar] [CrossRef]
- Le Hir, H.; Gatfield, D.; Izaurralde, E.; Moore, M.J. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. Embo J. 2001, 20, 4987–4997. [Google Scholar] [CrossRef]
- Soares, L.M.; Zanier, K.; Mackereth, C.; Sattler, M.; Valcarcel, J. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science 2006, 312, 1961–1965. [Google Scholar] [CrossRef]
- Yue, L.; Wan, R.; Luan, S.; Zeng, W.; Cheung, T.H. Dek Modulates Global Intron Retention during Muscle Stem Cells Quiescence Exit. Dev. Cell 2020, 53, 661–676.e6. [Google Scholar] [CrossRef]
- Vinnedge, L.P.; Benight, N.M.; Wagh, P.K.; Pease, N.A.; Nashu, M.A.; Serrano-Lopez, J.; Adams, A.K.; Cancelas, J.; Waltz, S.E.; Wells, S.I. The DEK oncogene promotes cellular proliferation through paracrine Wnt signaling in Ron receptor positive breast cancers. Oncogene 2014, 34, 2325–2336. [Google Scholar] [CrossRef] [PubMed]
- Zinser, G.M.; Leonis, M.A.; Toney, K.; Pathrose, P.; Thobe, M.; Kader, S.A.; Peace, B.E.; Beauman, S.R.; Collins, M.H.; Waltz, S.E. Mammary-specific Ron receptor overexpression induces highly metastatic mammary tumors associated with beta-catenin activation. Cancer Res. 2006, 66, 11967–11974. [Google Scholar] [CrossRef] [PubMed]
- Wise-Draper, T.M.; Mintz-Cole, R.A.; Morris, T.A.; Simpson, D.S.; Wikenheiser-Brokamp, K.A.; Currier, M.A.; Cripe, T.P.; Grosveld, G.C.; Wells, S.I. Overexpression of the cellular DEK protein promotes epithelial transformation in vitro and in vivo. Cancer Res. 2009, 69, 1792–1799. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, J.; Wang, S.; Luo, X.; Li, Y.; Lv, Z.; Zhu, J.; Lin, J.; Ding, L.; Ye, Q. The DEK oncogene activates VEGF expression and promotes tumor angiogenesis and growth in HIF-1alpha-dependent and -independent manners. Oncotarget 2016, 7, 23740–23756. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Huang, X.; Zhang, W.; Zhao, H.; Wu, G.; Lv, F.; Shi, L.; Teng, Y. Critical role of DEK and its regulation in tumorigenesis and metastasis of hepatocellular carcinoma. Oncotarget 2016, 7, 26844–26855. [Google Scholar] [CrossRef]
- Yang, M.Q.; Bai, L.L.; Lei, L.; Zheng, Y.W.; Wang, Z.; Li, Z.H.; Liu, C.C.; Huang, W.J.; Xu, H.T. DEK promotes the proliferation and invasion of lung cancers and indicates poor prognosis in lung adenocarcinomas. Oncol. Rep. 2020, 43, 1338–1348. [Google Scholar] [CrossRef]
- Han, H.; Headley, M.B.; Xu, W.; Comeau, M.R.; Zhou, B.; Ziegler, S.F. Thymic stromal lymphopoietin amplifies the differentiation of alternatively activated macrophages. J. Immunol. 2013, 190, 904–912. [Google Scholar] [CrossRef]
- An, G.; Wu, F.; Huang, S.; Feng, L.; Bai, J.; Gu, S.; Zhao, X. Effects of CCL5 on the biological behavior of breast cancer and the mechanisms of its interaction with tumorassociated macrophages. Oncol. Rep. 2019, 42, 2499–2511. [Google Scholar] [CrossRef]
- Liu, D.; Guo, M.; Zhou, P.; Xiao, J.; Ji, X. TSLP promote M2 macrophages polarization and cardiac healing after myocardial infarction. Biochem. Biophys. Res. Commun. 2019, 516, 437–444. [Google Scholar] [CrossRef]
- Wang, N.; Liang, H.; Zen, K. Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front. Immunol. 2014, 5, 614. [Google Scholar] [CrossRef]
- Xuan, W.; Qu, Q.; Zheng, B.; Xiong, S.; Fan, G.H. The chemotaxis of M1 and M2 macrophages is regulated by different chemokines. J. Leukoc. Biol. 2015, 97, 61–69. [Google Scholar] [CrossRef]
- Lacey, D.C.; Achuthan, A.; Fleetwood, A.J.; Dinh, H.; Roiniotis, J.; Scholz, G.M.; Chang, M.W.; Beckman, S.K.; Cook, A.D.; Hamilton, J.A. Defining GM-CSF- and macrophage-CSF-dependent macrophage responses by in vitro models. J. Immunol. 2012, 188, 5752–5765. [Google Scholar] [CrossRef] [PubMed]
- Neamatallah, T. Mitogen-Activated Protein Kinase Pathway: A Critical Regulator in Tumor-associated Macrophage Polarization. J. Microsc. Ultrastruct. 2019, 7, 53–56. [Google Scholar] [CrossRef]
- Jin, Y.; Liu, Y.; Nelin, L.D. Extracellular signal-regulated kinase mediates expression of arginase II but not inducible nitric-oxide synthase in lipopolysaccharide-stimulated macrophages. J. Biol. Chem. 2015, 290, 2099–2111. [Google Scholar] [CrossRef] [PubMed]
- Corna, G.; Campana, L.; Pignatti, E.; Castiglioni, A.; Tagliafico, E.; Bosurgi, L.; Campanella, A.; Brunelli, S.; Manfredi, A.A.; Apostoli, P.; et al. Polarization dictates iron handling by inflammatory and alternatively activated macrophages. Haematologica 2010, 95, 1814–1822. [Google Scholar] [CrossRef] [PubMed]
- Recalcati, S.; Locati, M.; Marini, A.; Santambrogio, P.; Zaninotto, F.; De Pizzol, M.; Zammataro, L.; Girelli, D.; Cairo, G. Differential regulation of iron homeostasis during human macrophage polarized activation. Eur. J. Immunol. 2010, 40, 824–835. [Google Scholar] [CrossRef] [PubMed]
- Mor-Vaknin, N.; Saha, A.; Legendre, M.; Carmona-Rivera, C.; Amin, M.A.; Rabquer, B.J.; Gonzales-Hernandez, M.J.; Jorns, J.; Mohan, S.; Yalavarthi, S.; et al. DEK-targeting DNA aptamers as therapeutics for inflammatory arthritis. Nat. Commun. 2017, 8, 14252. [Google Scholar] [CrossRef]
- Mor-Vaknin, N.; Punturieri, A.; Sitwala, K.; Faulkner, N.; Legendre, M.; Khodadoust, M.S.; Kappes, F.; Ruth, J.H.; Koch, A.; Glass, D.; et al. The DEK nuclear autoantigen is a secreted chemotactic factor. Mol. Cell Biol. 2006, 26, 9484–9496. [Google Scholar] [CrossRef]
- Forciniti, S.; Greco, L.; Grizzi, F.; Malesci, A.; Laghi, L. Iron Metabolism in Cancer Progression. Int. J. Mol. Sci. 2020, 21, 2257. [Google Scholar] [CrossRef]
- Medrek, C.; Ponten, F.; Jirstrom, K.; Leandersson, K. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer 2012, 12, 306. [Google Scholar] [CrossRef]
- Sullivan, C.; Brown, N.E.; Vasiliauskas, J.; Pathrose, P.; Starnes, S.L.; Waltz, S.E. Prostate Epithelial RON Signaling Promotes M2 Macrophage Activation to Drive Prostate Tumor Growth and Progression. Mol. Cancer Res. 2020. [Google Scholar] [CrossRef] [PubMed]
- Takegawa, S.; Jin, Z.; Nakayama, T.; Oyama, T.; Hieshima, K.; Nagakubo, D.; Shirakawa, A.K.; Tsuzuki, T.; Nakamura, S.; Yoshie, O. Expression of CCL17 and CCL22 by latent membrane protein 1-positive tumor cells in age-related Epstein-Barr virus-associated B-cell lymphoproliferative disorder. Cancer Sci. 2008, 99, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Anisowicz, A.; Messineo, M.; Lee, S.W.; Sager, R. An NF-kappa B-like transcription factor mediates IL-1/TNF-alpha induction of gro in human fibroblasts. J. Immunol. 1991, 147, 520–527. [Google Scholar] [PubMed]
- Wickremasinghe, M.I.; Thomas, L.H.; O’Kane, C.M.; Uddin, J.; Friedland, J.S. Transcriptional mechanisms regulating alveolar epithelial cell-specific CCL5 secretion in pulmonary tuberculosis. J. Biol. Chem. 2004, 279, 27199–27210. [Google Scholar] [CrossRef]
- Son, Y.H.; Jeong, Y.T.; Lee, K.A.; Choi, K.H.; Kim, S.M.; Rhim, B.Y.; Kim, K. Roles of MAPK and NF-kappaB in interleukin-6 induction by lipopolysaccharide in vascular smooth muscle cells. J. Cardiovasc. Pharm. 2008, 51, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Hein, H.; Schluter, C.; Kulke, R.; Christophers, E.; Schroder, J.M.; Bartels, J. Genomic organization, sequence, and transcriptional regulation of the human eotaxin gene. Biochem. Biophys. Res. Commun. 1997, 237, 537–542. [Google Scholar] [CrossRef]
- Sun, S.C. The non-canonical NF-kappaB pathway in immunity and inflammation. Nat. Rev. Immunol. 2017, 17, 545–558. [Google Scholar] [CrossRef]
- Dejardin, E.; Bonizzi, G.; Bellahcene, A.; Castronovo, V.; Merville, M.P.; Bours, V. Highly-expressed p100/p52 (NFKB2) sequesters other NF-kappa B-related proteins in the cytoplasm of human breast cancer cells. Oncogene 1995, 11, 1835–1841. [Google Scholar]
- Schneider, G. S1P Signaling in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1223, 129–153. [Google Scholar] [CrossRef]
- Weigert, A.; Tzieply, N.; von Knethen, A.; Johann, A.M.; Schmidt, H.; Geisslinger, G.; Brune, B. Tumor cell apoptosis polarizes macrophages role of sphingosine-1-phosphate. Mol. Biol. Cell 2007, 18, 3810–3819. [Google Scholar] [CrossRef]
- Weigert, A.; Schiffmann, S.; Sekar, D.; Ley, S.; Menrad, H.; Werno, C.; Grosch, S.; Geisslinger, G.; Brune, B. Sphingosine kinase 2 deficient tumor xenografts show impaired growth and fail to polarize macrophages towards an anti-inflammatory phenotype. Int. J. Cancer 2009, 125, 2114–2121. [Google Scholar] [CrossRef]
- Weigert, A.; von Knethen, A.; Thomas, D.; Faria, I.; Namgaladze, D.; Zezina, E.; Fuhrmann, D.; Petcherski, A.; Heringdorf, D.M.Z.; Radeke, H.H.; et al. Sphingosine kinase 2 is a negative regulator of inflammatory macrophage activation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 1235–1246. [Google Scholar] [CrossRef]
- Liu, J.; Zhou, Q.; Wu, C.P.; Xu, Y.W.; Liu, W.L.; Zhao, H.F.; Li, W.P. SPHK2 protein expression, Ki-67 index and infiltration of tumor-associated macrophages (TAMs) in human glioma. Histol. Histopathol. 2018, 33, 987–994. [Google Scholar] [CrossRef]
- Chen, P.; Zuo, H.; Xiong, H.; Kolar, M.J.; Chu, Q.; Saghatelian, A.; Siegwart, D.J.; Wan, Y. Gpr132 sensing of lactate mediates tumor-macrophage interplay to promote breast cancer metastasis. Proc. Natl. Acad. Sci. USA 2017, 114, 580–585. [Google Scholar] [CrossRef]
- Mu, X.; Shi, W.; Xu, Y.; Xu, C.; Zhao, T.; Geng, B.; Yang, J.; Pan, J.; Hu, S.; Zhang, C.; et al. Tumor-derived lactate induces M2 macrophage polarization via the activation of the ERK/STAT3 signaling pathway in breast cancer. Cell Cycle 2018, 17, 428–438. [Google Scholar] [CrossRef]
- Matrka, M.C.; Watanabe, M.; Muraleedharan, R.; Lambert, P.F.; Lane, A.N.; Romick-Rosendale, L.E.; Wells, S.I. Overexpression of the human DEK oncogene reprograms cellular metabolism and promotes glycolysis. PLoS ONE 2017, 12, e0177952. [Google Scholar] [CrossRef]
- Pascual-Garcia, M.; Bonfill-Teixidor, E.; Planas-Rigol, E.; Rubio-Perez, C.; Iurlaro, R.; Arias, A.; Cuartas, I.; Sala-Hojman, A.; Escudero, L.; Martinez-Ricarte, F.; et al. LIF regulates CXCL9 in tumor-associated macrophages and prevents CD8(+) T cell tumor-infiltration impairing anti-PD1 therapy. Nat. Commun. 2019, 10, 2416. [Google Scholar] [CrossRef]
- Weischenfeldt, J.; Porse, B. Bone Marrow-Derived Macrophages (BMM): Isolation and Applications. Csh. Protoc. 2008, 2008, pdb.prot5080. [Google Scholar] [CrossRef]
- Solmaz, M.; Lane, A.; Gonen, B.; Akmamedova, O.; Gunes, M.H.; Komurov, K. Graphical data mining of cancer mechanisms with SEMA. Bioinformatics 2019, 35, 4413–4418. [Google Scholar] [CrossRef]

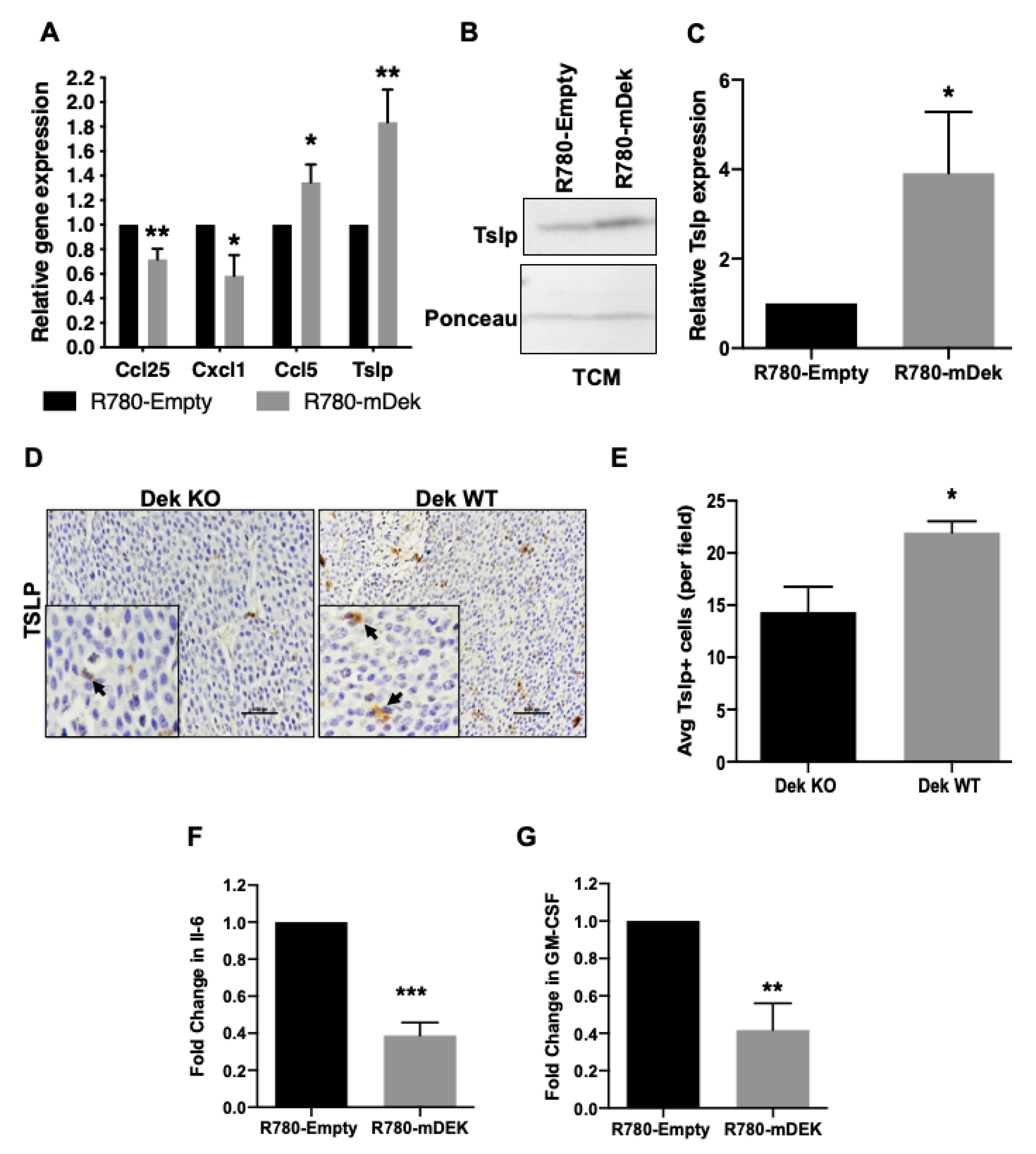
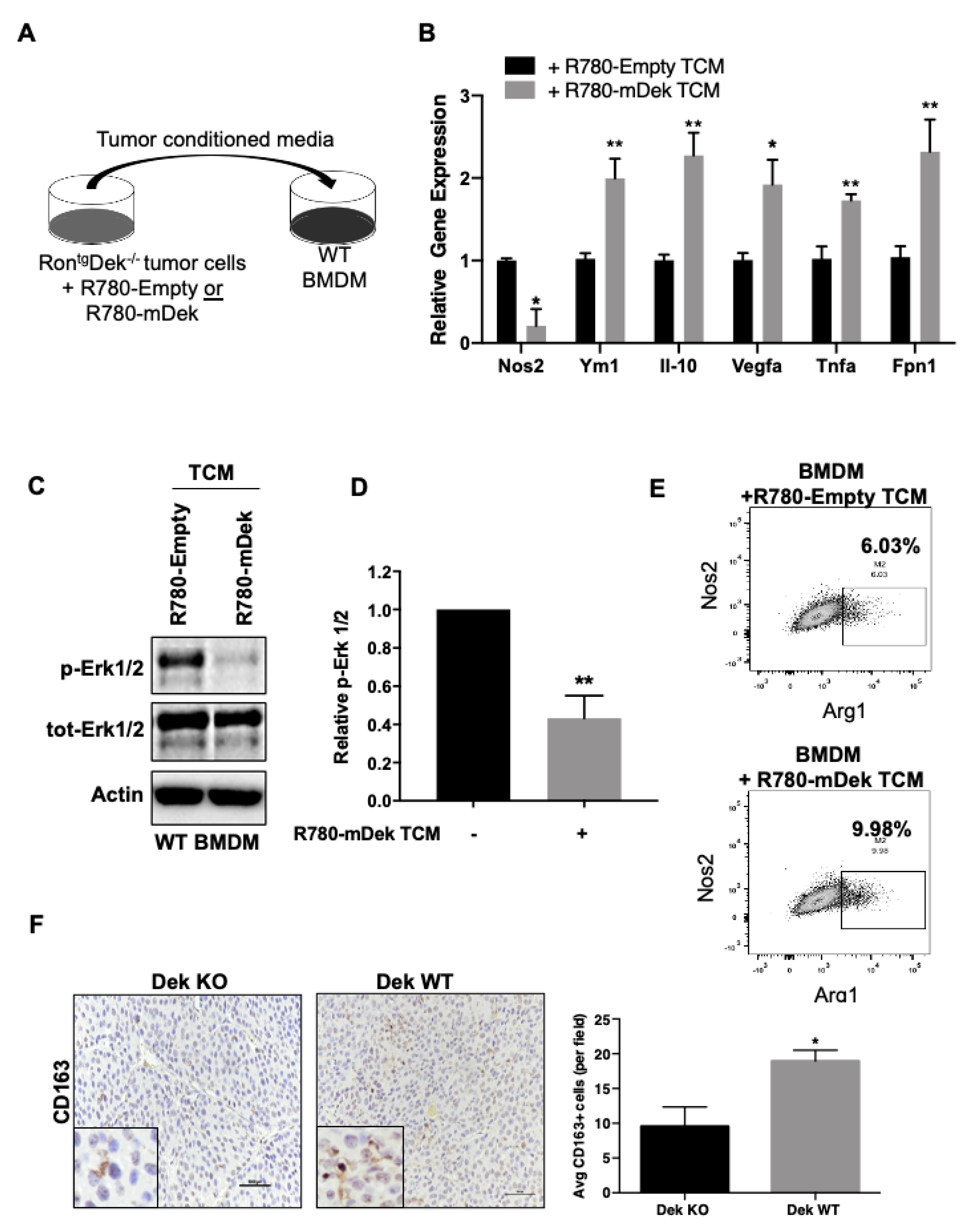

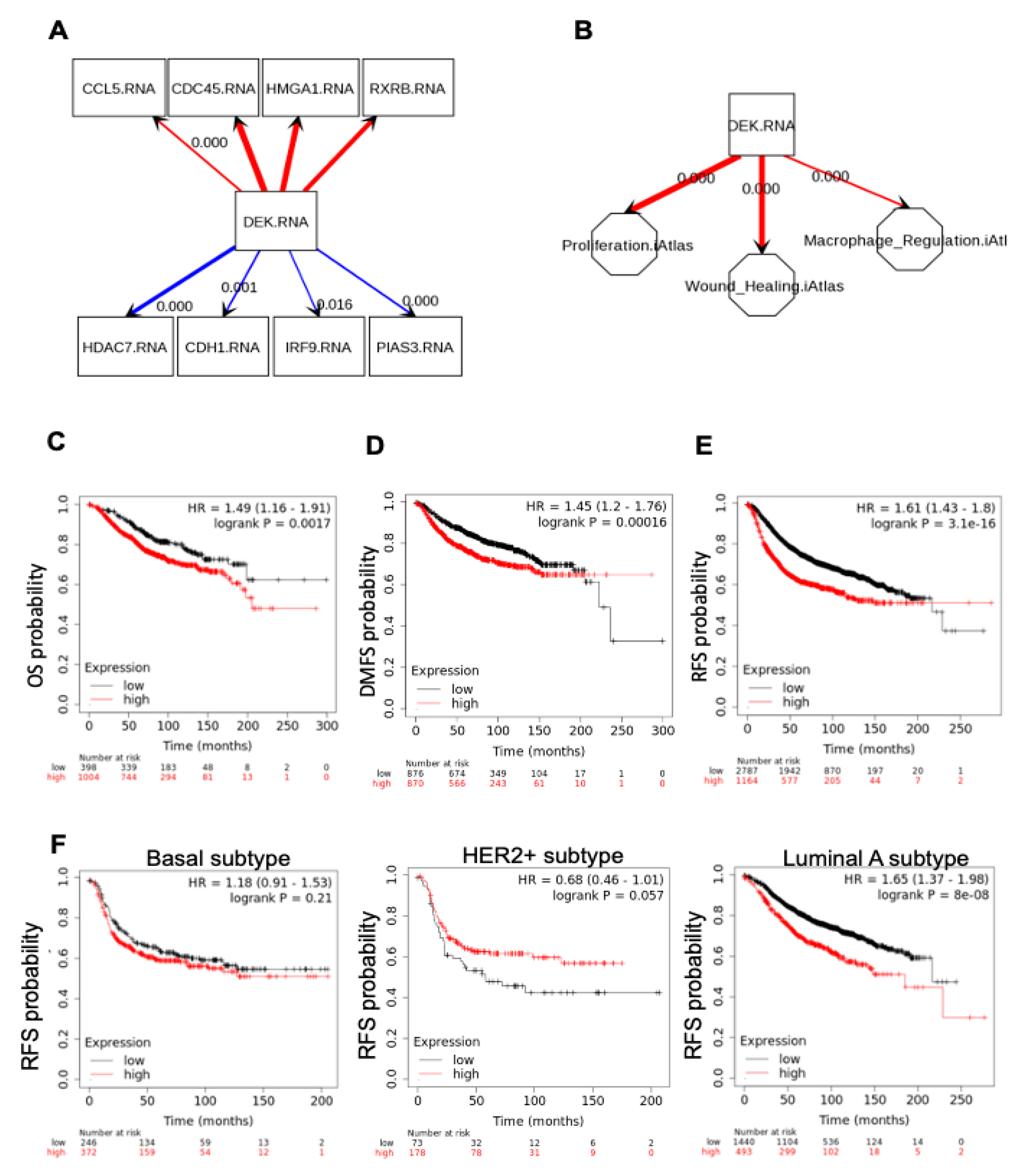
| Gene | Fold Change | Gene | Fold Change |
|---|---|---|---|
| Dek | 21.13 | Hdac7 | 0.24 |
| Cuedc2 | 12.95 | Gas7 | 0.28 |
| Rxrb | 4.83 | Cxcl2 | 0.31 |
| Taz | 3.24 | Cxcl10 | 0.31 |
| Hmga1 | 3.11 | Setdb1 | 0.33 |
| Tufm | 2.91 | Llgl1 | 0.36 |
| Sphk2 | 2.67 | Irf9 | 0.41 |
| Rad54l/ATRX | 2.29 | Ccl17 | 0.41 |
| Vegfa | 2.11 | Socs1 | 0.42 |
| Tslp | 1.84 | Cxcl1 | 0.43 |
| Tcf3 | 1.76 | Ccl25 | 0.46 |
| Cpeb2 | 1.59 | Irf7 | 0.44 |
| Cdc45 | 1.58 | Cav2 | 0.46 |
| Ercc1 | 1.57 | Pias3 | 0.46 |
| Gemin5 | 1.57 | cdh1/E-cadherin | 0.48 |
| Ccl5 | 1.52 | Rap1gap | 0.48 |
| Nfkb2 | 1.49 | Casp8ap2 | 0.50 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pease, N.A.; Shephard, M.S.; Sertorio, M.; Waltz, S.E.; Vinnedge, L.M.P. DEK Expression in Breast Cancer Cells Leads to the Alternative Activation of Tumor Associated Macrophages. Cancers 2020, 12, 1936. https://doi.org/10.3390/cancers12071936
Pease NA, Shephard MS, Sertorio M, Waltz SE, Vinnedge LMP. DEK Expression in Breast Cancer Cells Leads to the Alternative Activation of Tumor Associated Macrophages. Cancers. 2020; 12(7):1936. https://doi.org/10.3390/cancers12071936
Chicago/Turabian StylePease, Nicholas A., Miranda S. Shephard, Mathieu Sertorio, Susan E. Waltz, and Lisa M. Privette Vinnedge. 2020. "DEK Expression in Breast Cancer Cells Leads to the Alternative Activation of Tumor Associated Macrophages" Cancers 12, no. 7: 1936. https://doi.org/10.3390/cancers12071936
APA StylePease, N. A., Shephard, M. S., Sertorio, M., Waltz, S. E., & Vinnedge, L. M. P. (2020). DEK Expression in Breast Cancer Cells Leads to the Alternative Activation of Tumor Associated Macrophages. Cancers, 12(7), 1936. https://doi.org/10.3390/cancers12071936






