New Insights into Diffuse Large B-Cell Lymphoma Pathobiology
Abstract
1. Introduction
2. Bridging the Gaps between Disease Biology and Clinical Translation: New and Old Tricks in DLBCL Classification
2.1. Molecular Pathogenesis: Novel Insights
2.2. Molecular Prognostic Models
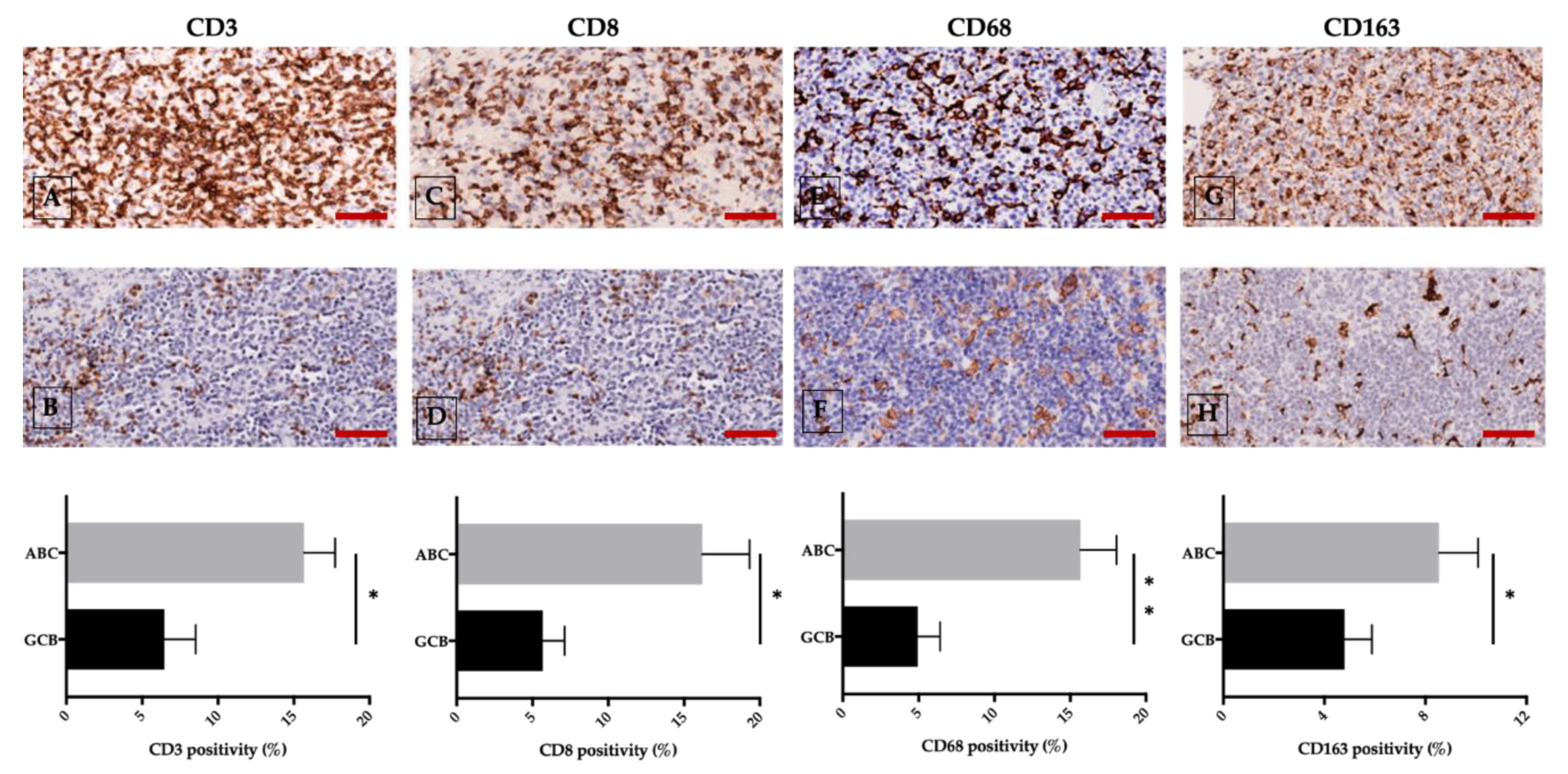
2.3. Tumor Microenvironment and Angiogenesis
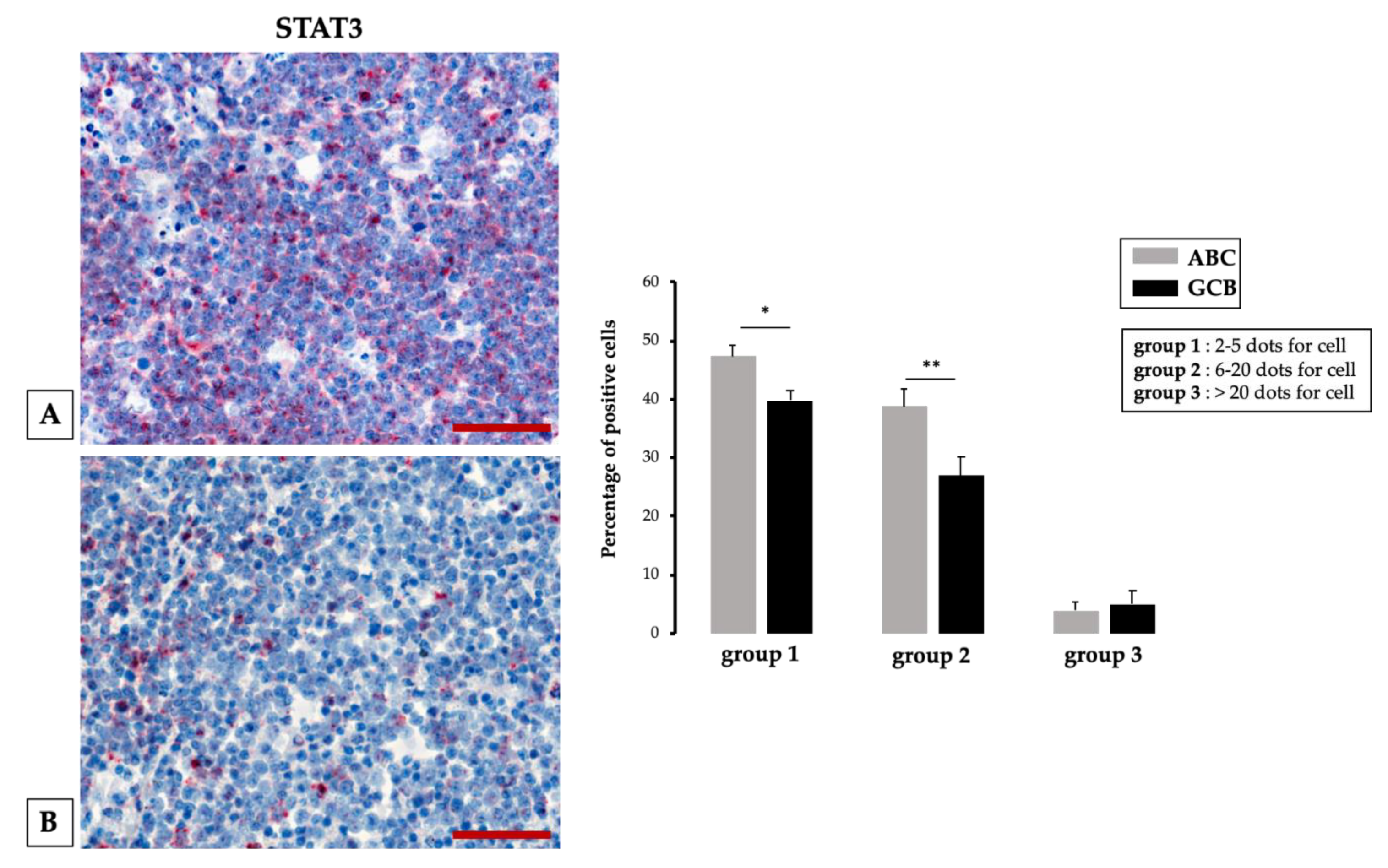
2.4. Increased Vascularization, VEGF Expression and MicroRNA (miRNA)
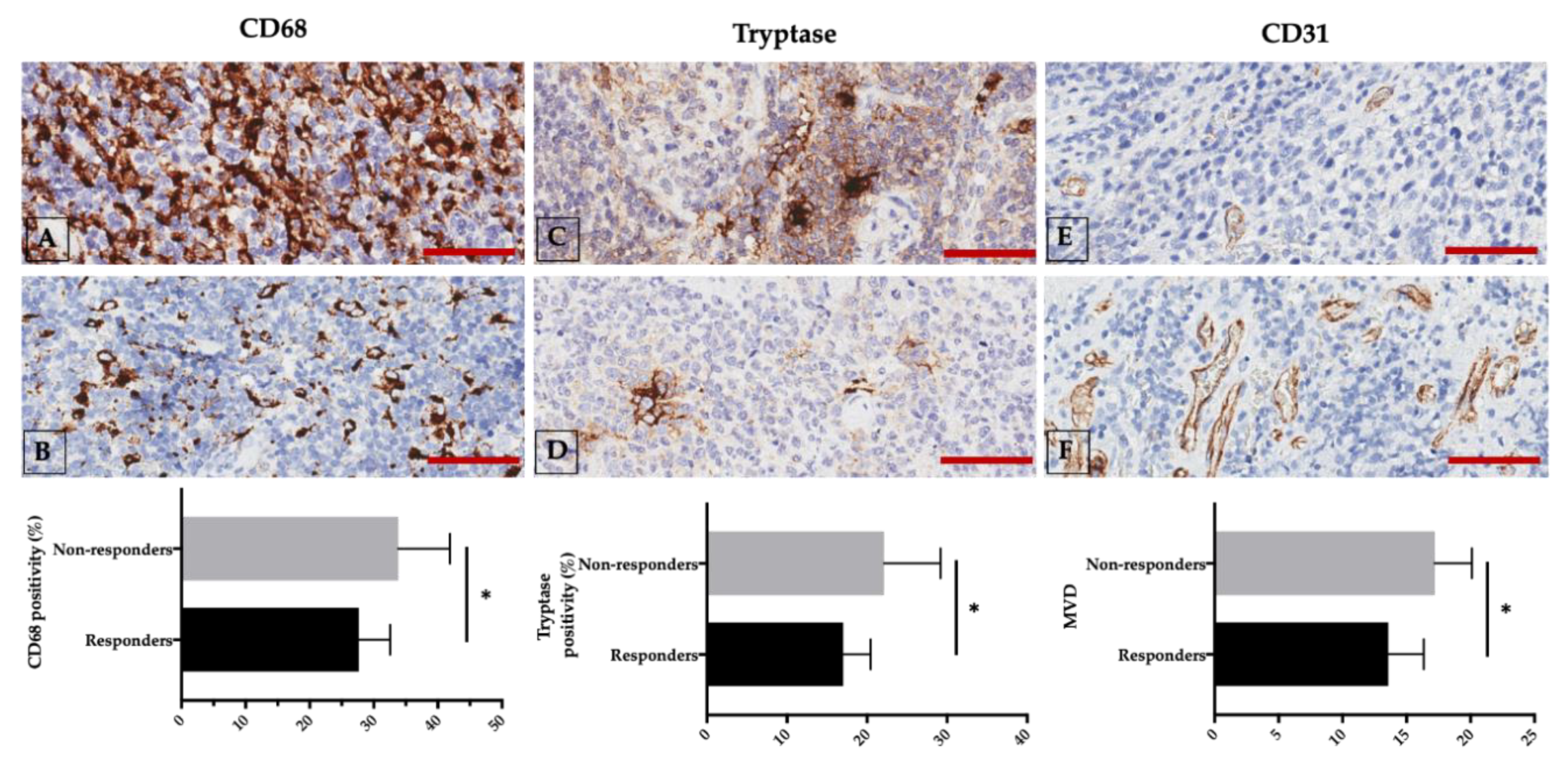
2.5. Correlations among Angiogenesis, VEGF Expression, and Response to Therapy
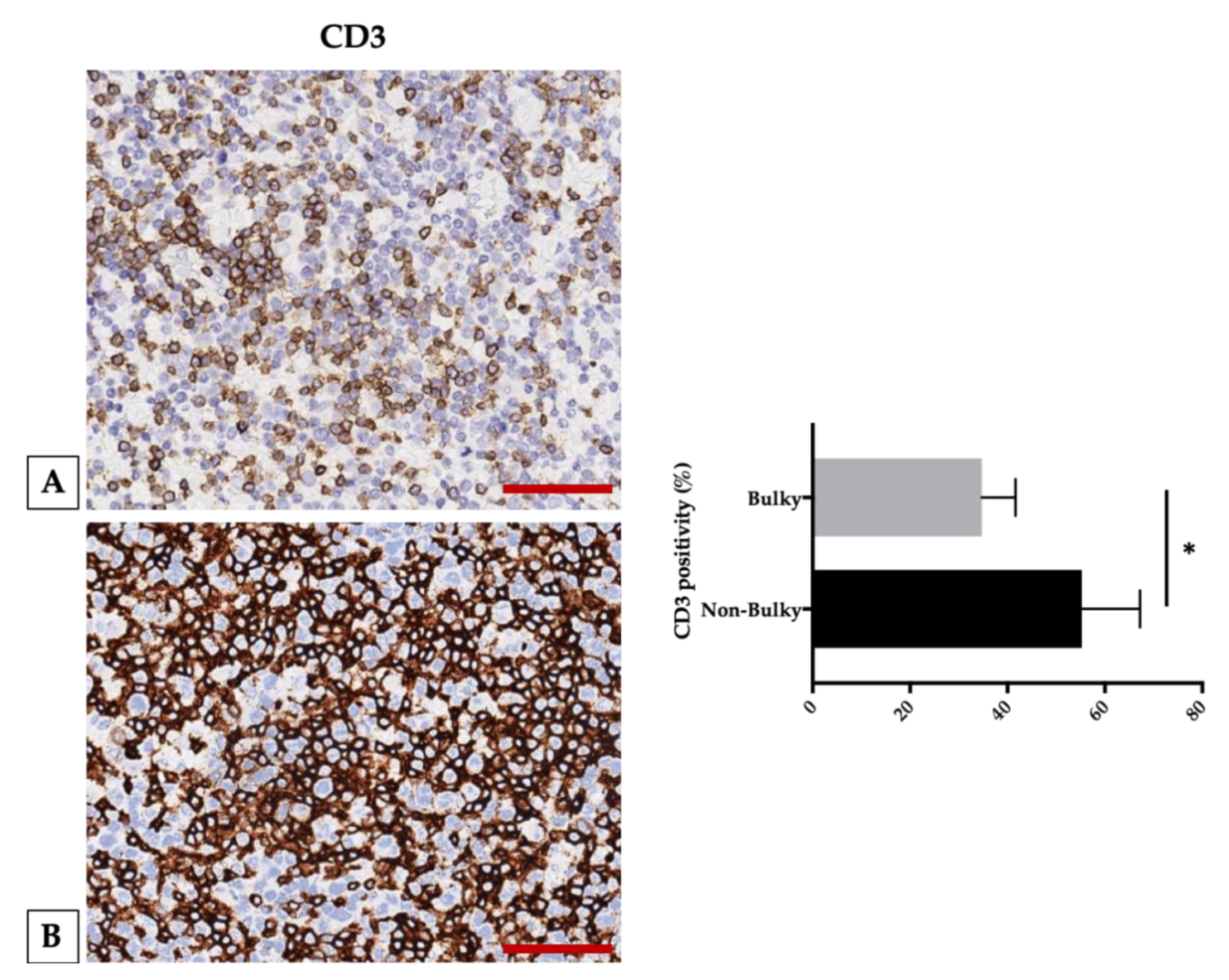
2.6. Targeting Angiogenesis and the Immune System in DLBCL: A Single-Center Experience
3. Discussion
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ADAM17 | Disintegrin and metalloproteinase domain-containing protein 17 |
| BCL2 | B-Cell CLL/Lymphoma 2 |
| BCL6 | B-Cell Lymphoma 6 Protein |
| BCR | B-Cell Receptor |
| BRD | Bromodomain |
| CARD11 | Caspase Recruitment Domain Family Member 11 |
| CD68 | cluster of differentiation 68 |
| CN | Copy number |
| CREB | cAMP-response element-binding protein |
| CREBBP | CREB Binding Protein |
| DDX3X | DEAD-Box Helicase 3 X-Linked |
| DEAD | DEAD-box helicase family |
| EZH2 | Enhancer Of Zeste Homolog 2 |
| FISH | Fluorescence in situ hybridization |
| FVIII | Factor VIII |
| GNA13 | G Protein Subunit Alpha 13 |
| HAT | Histone Acetyltransferase |
| HB-EGF | Heparin-binding EGF-like growth factor |
| Ig | Immunoglobulin |
| IRF4 | interferon regulatory factors 4 |
| ISH | In situ hybridization |
| KMT2D | Lysine Methyltransferase 2D |
| KO | Knock out |
| LMO2 | LIM domain only 2 (rhombotin-like 1) |
| m3D-IPI) | three-risk group model International Prognostic Index |
| MIB1 | Mindbomb E3 Ubiquitin Protein Ligase 1 |
| MUM1 | melanoma associated antigen (mutated) 1 |
| MYC | Myc-Related Translation/Localization Regulatory Factor |
| MYD | Myeloid Differentiation Primary Response Gene |
| MYD88 | Myeloid Differentiation Primary Response Gene (88) |
| NF-κB | Nuclear Factor Kappa B Subunit |
| NGS | next generation sequensing |
| NOS | nitric oxide synthase |
| NOTCH2 | Neurogenic Locus Notch Homolog Protein 2 |
| OS | Overall Survival |
| PFS | Progressiob free Survival |
| PTEN | Phosphatase And Tensin Homolog |
| RAS | Rat Sarcoma Viral Oncogene Homolog |
| SPEN | SPEN family transcriptional repressor ( |
| SPHK1 | Sphingosine Kinase 1 |
| STAT3 | Signal Transducer And Activator Of Transcription 3 |
| SUZ12 | Suppressor Of Zeste 12 Protein Homolog |
| TIM-3 | T-cell immunoglobulin mucin-3 |
| TNFAIP3 | Tumor necrosis factor, alpha-induced protein 3 |
| TP53 | Tumor Protein P53 |
References
- WHO. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, Revised 4th ed.; Swerdlow, S.H., Campo, E., Harris, N.L., Jaffe, E.S., Pileri, S.A., Stein, H., Thiele, J., Eds.; World Health Organization Classification of Tumours; International Agency for Research on Cancer: Lyon, France, 2017; ISBN 978-92-832-4494-3. [Google Scholar]
- Liu, Y.; Barta, S.K. Diffuse large B-cell lymphoma: 2019 update on diagnosis, risk stratification, and treatment. Am. J. Hematol. 2019, 94, 604–616. [Google Scholar] [CrossRef]
- Schneider, C.; Pasqualucci, L.; Dalla-Favera, R. Molecular pathogenesis of diffuse large B-cell lymphoma. Semin. Diagn. Pathol. 2011, 28, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Medeiros, L.J.; Li, Y.; Li, J.; Young, K.H. Genetic alterations and their clinical implications in DLBCL. Nat. Rev. Clin. Oncol. 2019, 16, 634–652. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, A.A.; Eisen, M.B.; Davis, R.E.; Ma, C.; Lossos, I.S.; Rosenwald, A.; Boldrick, J.C.; Sabet, H.; Tran, T.; Yu, X.; et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000, 403, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Dunleavy, K.; Pittaluga, S.; Czuczman, M.S.; Dave, S.S.; Wright, G.; Grant, N.; Shovlin, M.; Jaffe, E.S.; Janik, J.E.; Staudt, L.M.; et al. Differential efficacy of bortezomib plus chemotherapy within molecular subtypes of diffuse large B-cell lymphoma. Blood 2009, 113, 6069–6076. [Google Scholar] [CrossRef]
- Rosenwald, A.; Wright, G.; Chan, W.C.; Connors, J.M.; Campo, E.; Fisher, R.I.; Gascoyne, R.D.; Muller-Hermelink, H.K.; Smeland, E.B.; Giltnane, J.M.; et al. The Use of Molecular Profiling to Predict Survival after Chemotherapy for Diffuse Large-B-Cell Lymphoma. N. Engl. J. Med. 2002, 346, 1937–1947. [Google Scholar] [CrossRef]
- Davis, R.E.; Brown, K.D.; Siebenlist, U.; Staudt, L.M. Constitutive Nuclear Factor κB Activity Is Required for Survival of Activated B Cell–like Diffuse Large B Cell Lymphoma Cells. J. Exp. Med. 2001, 194, 1861–1874. [Google Scholar] [CrossRef]
- Compagno, M.; Lim, W.K.; Grunn, A.; Nandula, S.V.; Brahmachary, M.; Shen, Q.; Bertoni, F.; Ponzoni, M.; Scandurra, M.; Califano, A.; et al. Mutations of multiple genes cause deregulation of NF-κB in diffuse large B-cell lymphoma. Nature 2009, 459, 717–721. [Google Scholar] [CrossRef]
- Zl, W.; Yq, S.; Yf, S.; Zhu, J. High nuclear expression of STAT3 is associated with unfavorable prognosis in diffuse large B-cell lymphoma. J. Hematol. Oncol. J. Hematol. Oncol. 2011, 4, 31. [Google Scholar] [CrossRef] [PubMed]
- Ok, C.Y.; Chen, J.; Xu-Monette, Z.Y.; Tzankov, A.; Manyam, G.C.; Li, L.; Visco, C.; Montes-Moreno, S.; Dybkaer, K.; Chiu, A.; et al. Clinical Implications of Phosphorylated STAT3 Expression in De Novo Diffuse Large B-cell Lymphoma. Clin. Cancer Res. 2014, 20, 5113–5123. [Google Scholar] [CrossRef]
- Ding, B.B.; Yu, J.J.; Yu, R.Y.-L.; Mendez, L.M.; Shaknovich, R.; Zhang, Y.; Cattoretti, G.; Ye, B.H. Constitutively activated STAT3 promotes cell proliferation and survival in the activated B-cell subtype of diffuse large B-cell lymphomas. Blood 2008, 111, 1515–1523. [Google Scholar] [CrossRef]
- Lam, L.T.; Wright, G.; Davis, R.E.; Lenz, G.; Farinha, P.; Dang, L.; Chan, J.W.; Rosenwald, A.; Gascoyne, R.D.; Staudt, L.M. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-κB pathways in subtypes of diffuse large B-cell lymphoma. Blood 2008, 111, 3701–3713. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Kortylewski, M.; Pardoll, D. Crosstalk between cancer and immune cells: Role of STAT3 in the tumour microenvironment. Nat. Rev. Immunol. 2007, 7, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Doucette, T.A.; Kong, L.-Y.; Yang, Y.; Ferguson, S.D.; Yang, J.; Wei, J.; Qiao, W.; Fuller, G.N.; Bhat, K.P.; Aldape, K.; et al. Signal transducer and activator of transcription 3 promotes angiogenesis and drives malignant progression in glioma. Neuro-Oncology 2012, 14, 1136–1145. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Niu, N.; Wei, T.; Tozawa, H.; Chen, X.; Zhang, C.; Zhang, J.; Wada, Y.; Kapron, C.M.; Liu, J. The roles of signal transducer and activator of transcription factor 3 in tumor angiogenesis. Oncotarget 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Lenz, G.; Wright, G.; Dave, S.S.; Xiao, W.; Powell, J.; Zhao, H.; Xu, W.; Tan, B.; Goldschmidt, N.; Iqbal, J.; et al. Stromal Gene Signatures in Large-B-Cell Lymphomas. N. Engl. J. Med. 2008, 359, 2313–2323. [Google Scholar] [CrossRef]
- Sehn, L.H.; Gascoyne, R.D. Diffuse large B-cell lymphoma: Optimizing outcome in the context of clinical and biologic heterogeneity. Blood 2015, 125, 22–32. [Google Scholar] [CrossRef]
- Azzaoui, I.; Uhel, F.; Rossille, D.; Pangault, C.; Dulong, J.; Le Priol, J.; Lamy, T.; Houot, R.; Le Gouill, S.; Cartron, G.; et al. T-cell defect in diffuse large B-cell lymphomas involves expansion of myeloid-derived suppressor cells. Blood 2016, 128, 1081–1092. [Google Scholar] [CrossRef]
- Ji, H.; Niu, X.; Yin, L.; Wang, Y.; Huang, L.; Xuan, Q.; Li, L.; Zhang, H.; Li, J.; Yang, Y.; et al. Ratio of Immune Response to Tumor Burden Predicts Survival Via Regulating Functions of Lymphocytes and Monocytes in Diffuse Large B-Cell Lymphoma. Cell. Physiol. Biochem. 2018, 45, 951–961. [Google Scholar] [CrossRef]
- Rudelius, M.; Rosenfeldt, M.T.; Leich, E.; Rauert-Wunderlich, H.; Solimando, A.G.; Beilhack, A.; Ott, G.; Rosenwald, A. Inhibition of focal adhesion kinase overcomes resistance of mantle cell lymphoma to ibrutinib in the bone marrow microenvironment. Haematologica 2018, 103, 116–125. [Google Scholar] [CrossRef]
- Solimando, A.G.; Da Vià, M.C.; Leone, P.; Borrelli, P.; Croci, G.A.; Tabares, P.; Brandl, A.; Di Lernia, G.; Bianchi, F.P.; Tafuri, S.; et al. Halting the vicious cycle within the multiple myeloma ecosystem: Blocking JAM-A on bone marrow endothelial cells restores the angiogenic homeostasis and suppresses tumor progression. Haematologica 2020. [Google Scholar] [CrossRef] [PubMed]
- Solimando, A.G.; Vacca, A.; Ribatti, D. A Comprehensive Biological and Clinical Perspective Can Drive a Patient-Tailored Approach to Multiple Myeloma: Bridging the Gaps between the Plasma Cell and the Neoplastic Niche. J. Oncol. 2020, 2020, 1–16. [Google Scholar] [CrossRef]
- Plaks, V.; Kong, N.; Werb, Z. The cancer stem cell niche: How essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 2015, 16, 225–238. [Google Scholar] [CrossRef]
- Tilly, H.; Gomes da Silva, M.; Vitolo, U.; Jack, A.; Meignan, M.; Lopez-Guillermo, A.; Walewski, J.; André, M.; Johnson, P.W.; Pfreundschuh, M.; et al. Diffuse large B-cell lymphoma (DLBCL): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015, 26, v116–v125. [Google Scholar] [CrossRef] [PubMed]
- Solimando, A.G.; Ribatti, D.; Vacca, A.; Einsele, H. Targeting B-cell non Hodgkin lymphoma: New and old tricks. Leuk. Res. 2016, 42, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Reagan, P.M.; Davies, A. Current treatment of double hit and double expressor lymphoma. Hematol. Am. Soc. Hematol. Educ. Program 2017, 2017, 295–297. [Google Scholar] [CrossRef] [PubMed]
- Hans, C.P.; Weisenburger, D.D.; Greiner, T.C.; Gascoyne, R.D.; Delabie, J.; Ott, G.; Müller-Hermelink, H.K.; Campo, E.; Braziel, R.M.; Jaffe, E.S.; et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 2004, 103, 275–282. [Google Scholar] [CrossRef]
- Huang, W.; Medeiros, L.J.; Lin, P.; Wang, W.; Tang, G.; Khoury, J.; Konoplev, S.; Yin, C.C.; Xu, J.; Oki, Y.; et al. MYC/BCL2/BCL6 triple hit lymphoma: A study of 40 patients with a comparison to MYC/BCL2 and MYC/BCL6 double hit lymphomas. Mod. Pathol. Off. J. U. S. Can. Acad. Pathol. Inc. 2018, 31, 1470–1478. [Google Scholar] [CrossRef]
- Ennishi, D.; Jiang, A.; Boyle, M.; Collinge, B.; Grande, B.M.; Ben-Neriah, S.; Rushton, C.; Tang, J.; Thomas, N.; Slack, G.W.; et al. Double-Hit Gene Expression Signature Defines a Distinct Subgroup of Germinal Center B-Cell-Like Diffuse Large B-Cell Lymphoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2019, 37, 190–201. [Google Scholar] [CrossRef]
- Reddy, A.; Zhang, J.; Davis, N.S.; Moffitt, A.B.; Love, C.L.; Waldrop, A.; Leppa, S.; Pasanen, A.; Meriranta, L.; Karjalainen-Lindsberg, M.-L.; et al. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell 2017, 171, 481–494.e15. [Google Scholar] [CrossRef]
- Schmitz, R.; Wright, G.W.; Huang, D.W.; Johnson, C.A.; Phelan, J.D.; Wang, J.Q.; Roulland, S.; Kasbekar, M.; Young, R.M.; Shaffer, A.L.; et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2018, 378, 1396–1407. [Google Scholar] [CrossRef] [PubMed]
- Cheah, C.Y.; Fowler, N.H.; Wang, M.L. Breakthrough therapies in B-cell non-Hodgkin lymphoma. Ann. Oncol. 2016, 27, 778–787. [Google Scholar] [CrossRef] [PubMed]
- Ayyappan, S.; Maddocks, K. Novel and emerging therapies for B cell lymphoma. J. Hematol. Oncol. J. Hematol. Oncol. 2019, 12, 82. [Google Scholar] [CrossRef] [PubMed]
- Pasqualucci, L.; Dominguez-Sola, D.; Chiarenza, A.; Fabbri, G.; Grunn, A.; Trifonov, V.; Kasper, L.H.; Lerach, S.; Tang, H.; Ma, J.; et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature 2011, 471, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Pasqualucci, L.; Trifonov, V.; Fabbri, G.; Ma, J.; Rossi, D.; Chiarenza, A.; Wells, V.A.; Grunn, A.; Messina, M.; Elliot, O.; et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat. Genet. 2011, 43, 830–837. [Google Scholar] [CrossRef] [PubMed]
- Morin, R.D.; Mendez-Lago, M.; Mungall, A.J.; Goya, R.; Mungall, K.L.; Corbett, R.D.; Johnson, N.A.; Severson, T.M.; Chiu, R.; Field, M.; et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 2011, 476, 298–303. [Google Scholar] [CrossRef]
- Pasqualucci, L.; Dalla-Favera, R. Genetics of diffuse large B-cell lymphoma. Blood 2018, 131, 2307–2319. [Google Scholar] [CrossRef]
- Meyer, S.N.; Scuoppo, C.; Vlasevska, S.; Bal, E.; Holmes, A.B.; Holloman, M.; Garcia-Ibanez, L.; Nataraj, S.; Duval, R.; Vantrimpont, T.; et al. Unique and Shared Epigenetic Programs of the CREBBP and EP300 Acetyltransferases in Germinal Center B Cells Reveal Targetable Dependencies in Lymphoma. Immunity 2019, 51, 535–547.e9. [Google Scholar] [CrossRef]
- Li, J.; Ding, N.; Wang, X.; Mi, L.; Ping, L.; Jin, X.; Liu, Y.; Ying, Z.; Xie, Y.; Liu, W.; et al. EP300 single nucleotide polymorphism rs20551 correlates with prolonged overall survival in diffuse large B cell lymphoma patients treated with R-CHOP. Cancer Cell Int. 2017, 17, 70. [Google Scholar] [CrossRef]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef]
- Russano, M.; Napolitano, A.; Ribelli, G.; Iuliani, M.; Simonetti, S.; Citarella, F.; Pantano, F.; Dell’Aquila, E.; Anesi, C.; Silvestris, N.; et al. Liquid biopsy and tumor heterogeneity in metastatic solid tumors: The potentiality of blood samples. J. Exp. Clin. Cancer Res. CR 2020, 39, 95. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.; Solimando, A.G.; Kalogirou, C.; Marquardt, A.; Frank, T.; Sokolakis, I.; Hatzichristodoulou, G.; Kneitz, S.; Bargou, R.; Kübler, H.; et al. miR-221-3p Regulates VEGFR2 Expression in High-Risk Prostate Cancer and Represents an Escape Mechanism from Sunitinib In Vitro. J. Clin. Med. 2020, 9, 670. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. Liquid biopsy and minimal residual disease - latest advances and implications for cure. Nat. Rev. Clin. Oncol. 2019, 16, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Desantis, V.; Saltarella, I.; Lamanuzzi, A.; Melaccio, A.; Solimando, A.G.; Mariggiò, M.A.; Racanelli, V.; Paradiso, A.; Vacca, A.; Frassanito, M.A. MicroRNAs-Based Nano-Strategies as New Therapeutic Approach in Multiple Myeloma to Overcome Disease Progression and Drug Resistance. Int. J. Mol. Sci. 2020, 21, 3084. [Google Scholar] [CrossRef]
- Di Lernia, G.; Leone, P.; Solimando, A.G.; Buonavoglia, A.; Saltarella, I.; Ria, R.; Ditonno, P.; Silvestris, N.; Crudele, L.; Vacca, A.; et al. Bortezomib Treatment Modulates Autophagy in Multiple Myeloma. J. Clin. Med. 2020, 9, 552. [Google Scholar] [CrossRef] [PubMed]
- Leone, P.; Buonavoglia, A.; Fasano, R.; Solimando, A.G.; De Re, V.; Cicco, S.; Vacca, A.; Racanelli, V. Insights into the Regulation of Tumor Angiogenesis by Micro-RNAs. J. Clin. Med. 2019, 8, 2030. [Google Scholar] [CrossRef]
- Larrabeiti-Etxebarria, A.; Lopez-Santillan, M.; Santos-Zorrozua, B.; Lopez-Lopez, E.; Garcia-Orad, A. Systematic Review of the Potential of MicroRNAs in Diffuse Large B Cell Lymphoma. Cancers 2019, 11, 144. [Google Scholar] [CrossRef]
- Hutchinson, L. CtDNA—Identifying cancer before it is clinically detectable. Nat. Rev. Clin. Oncol. 2015, 12, 372. [Google Scholar] [CrossRef]
- Snyder, M.W.; Kircher, M.; Hill, A.J.; Daza, R.M.; Shendure, J. Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell 2016, 164, 57–68. [Google Scholar] [CrossRef]
- Rossi, D.; Diop, F.; Spaccarotella, E.; Monti, S.; Zanni, M.; Rasi, S.; Deambrogi, C.; Spina, V.; Bruscaggin, A.; Favini, C.; et al. Diffuse large B-cell lymphoma genotyping on the liquid biopsy. Blood 2017, 129, 1947–1957. [Google Scholar] [CrossRef]
- Kurtz, D.M.; Scherer, F.; Jin, M.C.; Soo, J.; Craig, A.F.M.; Esfahani, M.S.; Chabon, J.J.; Stehr, H.; Liu, C.L.; Tibshirani, R.; et al. Circulating Tumor DNA Measurements As Early Outcome Predictors in Diffuse Large B-Cell Lymphoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2018, 36, 2845–2853. [Google Scholar] [CrossRef] [PubMed]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Pasqualucci, L.; Dalla-Favera, R. The genetic landscape of diffuse large B-cell lymphoma. Semin. Hematol. 2015, 52, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Solimando, A.G.; Da Vià, M.C.; Cicco, S.; Leone, P.; Di Lernia, G.; Giannico, D.; Desantis, V.; Frassanito, M.A.; Morizio, A.; Delgado Tascon, J.; et al. High-Risk Multiple Myeloma: Integrated Clinical and Omics Approach Dissects the Neoplastic Clone and the Tumor Microenvironment. J. Clin. Med. 2019, 8, 997. [Google Scholar] [CrossRef] [PubMed]
- Ventura, R.A.; Martin-Subero, J.I.; Jones, M.; McParland, J.; Gesk, S.; Mason, D.Y.; Siebert, R. FISH analysis for the detection of lymphoma-associated chromosomal abnormalities in routine paraffin-embedded tissue. J. Mol. Diagn. JMD 2006, 8, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Dargent, J.-L.; Lespagnard, L.; Feoli, F.; Debusscher, L.; Greuse, M.; Bron, D. De novo CD5-positive diffuse large B-cell lymphoma of the skin arising in chronic limb lymphedema. Leuk. Lymphoma 2005, 46, 775–780. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.-X.; Miao, Y.; Wu, J.-Z.; Gong, Q.-X.; Liang, J.-H.; Wang, Z.; Wang, L.; Fan, L.; Hua, D.; Chen, Y.-Y.; et al. The distinct clinical features and prognosis of the CD10+MUM1+ and CD10−Bcl6−MUM1− diffuse large B-cell lymphoma. Sci. Rep. 2016, 6, 20465. [Google Scholar] [CrossRef]
- Hu, S.; Xu-Monette, Z.Y.; Tzankov, A.; Green, T.; Wu, L.; Balasubramanyam, A.; Liu, W.; Visco, C.; Li, Y.; Miranda, R.N.; et al. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: A report from The International DLBCL Rituximab-CHOP Consortium Program. Blood 2013, 121, 4021–4031. [Google Scholar] [CrossRef]
- Swerdlow, S.H. Diagnosis of “double hit” diffuse large B-cell lymphoma and B-cell lymphoma, unclassifiable, with features intermediate between DLBCL and Burkitt lymphoma: When and how, FISH versus IHC. Hematol. Am. Soc. Hematol. Educ. Program 2014, 2014, 90–99. [Google Scholar] [CrossRef]
- Hasselblom, S.; Ridell, B.; Sigurdardottir, M.; Hansson, U.; Nilsson-Ehle, H.; Andersson, P.-O. Low rather than high Ki-67 protein expression is an adverse prognostic factor in diffuse large B-cell lymphoma. Leuk. Lymphoma 2008, 49, 1501–1509. [Google Scholar] [CrossRef]
- Song, J.Y.; Perry, A.M.; Herrera, A.F.; Chen, L.; Skrabek, P.; Nasr, M.; Ottesen, R.; Nikowitz, J.; Bedell, V.; Murata-Collins, J.; et al. New Genomic Model Integrating Clinical Factors and Gene Mutations to Predict Overall Survival in Patients with Diffuse Large B-Cell Lymphoma Treated with R-CHOP. Blood 2018, 132, 346. [Google Scholar] [CrossRef]
- Chapuy, B.; Stewart, C.; Dunford, A.J.; Kim, J.; Kamburov, A.; Redd, R.A.; Lawrence, M.S.; Roemer, M.G.M.; Li, A.J.; Ziepert, M.; et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med. 2018, 24, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Bojarczuk, K.; Wienand, K.; Ryan, J.A.; Chen, L.; Villalobos-Ortiz, M.; Mandato, E.; Stachura, J.; Letai, A.; Lawton, L.N.; Chapuy, B.; et al. Targeted inhibition of PI3Kα/δ is synergistic with BCL-2 blockade in genetically defined subtypes of DLBCL. Blood 2019, 133, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Visco, C.; Tzankov, A.; Xu-Monette, Z.Y.; Miranda, R.N.; Tai, Y.C.; Li, Y.; Liu, W.; d’Amore, E.S.G.; Li, Y.; Montes-Moreno, S.; et al. Patients with diffuse large B-cell lymphoma of germinal center origin with BCL2 translocations have poor outcome, irrespective of MYC status: A report from an International DLBCL rituximab-CHOP Consortium Program Study. Haematologica 2013, 98, 255–263. [Google Scholar] [CrossRef]
- Seibold, M.; Stühmer, T.; Kremer, N.; Mottok, A.; Scholz, C.-J.; Schlosser, A.; Leich, E.; Holzgrabe, U.; Brünnert, D.; Barrio, S.; et al. RAL GTPases mediate multiple myeloma cell survival and are activated independently of oncogenic RAS. Haematologica 2019. [Google Scholar] [CrossRef]
- Chapuy, B.; Stewart, C.; Wood, T.; Dunford, A.; Wienand, K.; Getz, G.; Shipp, M.A. Validation of the Genetically-Defined DLBCL Subtypes and Generation of a Parsimonious Probabilistic Classifier. Blood 2019, 134, 920. [Google Scholar] [CrossRef]
- Ruan, J.; Leonard, J.P. Targeting angiogenesis: A novel, rational therapeutic approach for non-Hodgkin lymphoma. Leuk. Lymphoma 2009, 50, 679–681. [Google Scholar] [CrossRef]
- Ciavarella, S.; Vegliante, M.C.; Fabbri, M.; De Summa, S.; Melle, F.; Motta, G.; De Iuliis, V.; Opinto, G.; Enjuanes, A.; Rega, S.; et al. Dissection of DLBCL microenvironment provides a gene expression-based predictor of survival applicable to formalin-fixed paraffin-embedded tissue. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2018, 29, 2363–2370. [Google Scholar] [CrossRef]
- Kinugasa, Y.; Matsui, T.; Takakura, N. CD44 Expressed on Cancer-Associated Fibroblasts Is a Functional Molecule Supporting the Stemness and Drug Resistance of Malignant Cancer Cells in the Tumor Microenvironment: Tumor Stromal CD44. Stem Cells 2014, 32, 145–156. [Google Scholar] [CrossRef]
- Frassanito, M.A.; Desantis, V.; Di Marzo, L.; Craparotta, I.; Beltrame, L.; Marchini, S.; Annese, T.; Visino, F.; Arciuli, M.; Saltarella, I.; et al. Bone marrow fibroblasts overexpress miR-27b and miR-214 in step with multiple myeloma progression, dependent on tumour cell-derived exosomes. J. Pathol. 2019, 247, 241–253. [Google Scholar] [CrossRef]
- Nicholas, N.S.; Apollonio, B.; Ramsay, A.G. Tumor microenvironment (TME)-driven immune suppression in B cell malignancy. Biochim. Biophys. Acta BBA Mol. Cell Res. 2016, 1863, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Hedström, G.; Berglund, M.; Molin, D.; Fischer, M.; Nilsson, G.; Thunberg, U.; Book, M.; Sundström, C.; Rosenquist, R.; Roos, G.; et al. Mast cell infiltration is a favourable prognostic factor in diffuse large B-cell lymphoma. Br. J. Haematol. 2007, 138, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Liao, H.; Lin, S.; Xia, Y.; Wang, X.; Gao, Y.; Lin, Z.; Lu, J.; Huang, H. High expression of tumor-infiltrating macrophages correlates with poor prognosis in patients with diffuse large B-cell lymphoma. Med. Oncol. 2012, 29, 2317–2322. [Google Scholar] [CrossRef] [PubMed]
- Kini, A.R. Angiogenesis in Leukemia and Lymphoma. In Hematopathology in Oncology; Finn, W.G., Peterson, L.C., Eds.; Cancer Treatment and Research; Kluwer Academic Publishers: Boston, MA, USA, 2004; Volume 121, pp. 221–238. ISBN 978-1-4020-7919-1. [Google Scholar]
- Ribatti, D.; Nico, B.; Ranieri, G.; Specchia, G.; Vacca, A. The Role of Angiogenesis in Human Non-Hodgkin Lymphomas. Neoplasia 2013, 15, 231–238. [Google Scholar] [CrossRef]
- Shain, K.H.; Dalton, W.S.; Tao, J. The tumor microenvironment shapes hallmarks of mature B-cell malignancies. Oncogene 2015, 34, 4673–4682. [Google Scholar] [CrossRef] [PubMed]
- Buggy, J.J.; Elias, L. Bruton Tyrosine Kinase (BTK) and Its Role in B-cell Malignancy. Int. Rev. Immunol. 2012, 31, 119–132. [Google Scholar] [CrossRef]
- Fornecker, L.-M.; Muller, L.; Bertrand, F.; Paul, N.; Pichot, A.; Herbrecht, R.; Chenard, M.-P.; Mauvieux, L.; Vallat, L.; Bahram, S.; et al. Multi-omics dataset to decipher the complexity of drug resistance in diffuse large B-cell lymphoma. Sci. Rep. 2019, 9, 895. [Google Scholar] [CrossRef]
- Di Marzo, L.; Desantis, V.; Solimando, A.G.; Ruggieri, S.; Annese, T.; Nico, B.; Fumarulo, R.; Vacca, A.; Frassanito, M.A. Microenvironment drug resistance in multiple myeloma: Emerging new players. Oncotarget 2016, 7, 60698–60711. [Google Scholar] [CrossRef]
- Tomida, A.; Tsuruo, T. Drug resistance mediated by cellular stress response to the microenvironment of solid tumors. Anticancer. Drug Des. 1999, 14, 169–177. [Google Scholar]
- Alizadeh, A.A.; Gentles, A.J.; Alencar, A.J.; Liu, C.L.; Kohrt, H.E.; Houot, R.; Goldstein, M.J.; Zhao, S.; Natkunam, Y.; Advani, R.H.; et al. Prediction of survival in diffuse large B-cell lymphoma based on the expression of 2 genes reflecting tumor and microenvironment. Blood 2011, 118, 1350–1358. [Google Scholar] [CrossRef]
- Argentiero, A.; De Summa, S.; Di Fonte, R.; Iacobazzi, R.M.; Porcelli, L.; Da Vià, M.; Brunetti, O.; Azzariti, A.; Silvestris, N.; Solimando, A.G. Gene Expression Comparison between the Lymph Node-Positive and -Negative Reveals a Peculiar Immune Microenvironment Signature and a Theranostic Role for WNT Targeting in Pancreatic Ductal Adenocarcinoma: A Pilot Study. Cancers 2019, 11, 942. [Google Scholar] [CrossRef] [PubMed]
- Facciabene, A.; Motz, G.T.; Coukos, G. T-regulatory cells: Key players in tumor immune escape and angiogenesis. Cancer Res. 2012, 72, 2162–2171. [Google Scholar] [CrossRef]
- Leone, P.; Di Lernia, G.; Solimando, A.G.; Cicco, S.; Saltarella, I.; Lamanuzzi, A.; Ria, R.; Frassanito, M.A.; Ponzoni, M.; Ditonno, P.; et al. Bone marrow endothelial cells sustain a tumor-specific CD8+ T cell subset with suppressive function in myeloma patients. Oncoimmunology 2019, 8, e1486949. [Google Scholar] [CrossRef] [PubMed]
- Ribatti, D.; Vacca, A.; Dammacco, F.; English, D. Angiogenesis and Anti-Angiogenesis in Hematological Malignancies. J. Hematother. Stem Cell Res. 2003, 12, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Desantis, V.; Frassanito, M.A.; Tamma, R.; Saltarella, I.; Di Marzo, L.; Lamanuzzi, A.; Solimando, A.G.; Ruggieri, S.; Annese, T.; Nico, B.; et al. Rhu-Epo down-regulates pro-tumorigenic activity of cancer-associated fibroblasts in multiple myeloma. Ann. Hematol. 2018, 97, 1251–1258. [Google Scholar] [CrossRef] [PubMed]
- Tonia, T.; Mettler, A.; Robert, N.; Schwarzer, G.; Seidenfeld, J.; Weingart, O.; Hyde, C.; Engert, A.; Bohlius, J. Erythropoietin or darbepoetin for patients with cancer. Cochrane Database Syst. Rev. 2012, 12, CD003407. [Google Scholar] [CrossRef] [PubMed]
- Suhasini, A.N.; Wang, L.; Holder, K.N.; Lin, A.-P.; Bhatnagar, H.; Kim, S.-W.; Moritz, A.W.; Aguiar, R.C.T. A phosphodiesterase 4B-dependent interplay between tumor cells and the microenvironment regulates angiogenesis in B-cell lymphoma. Leukemia 2016, 30, 617–626. [Google Scholar] [CrossRef]
- Serafini, P.; Mgebroff, S.; Noonan, K.; Borrello, I. Myeloid-Derived Suppressor Cells Promote Cross-Tolerance in B-Cell Lymphoma by Expanding Regulatory T Cells. Cancer Res. 2008, 68, 5439–5449. [Google Scholar] [CrossRef]
- Broséus, J.; Mourah, S.; Ramstein, G.; Bernard, S.; Mounier, N.; Cuccuini, W.; Gaulard, P.; Gisselbrecht, C.; Brière, J.; Houlgatte, R.; et al. VEGF121, is predictor for survival in activated B-cell-like diffuse large B-cell lymphoma and is related to an immune response gene signature conserved in cancers. Oncotarget 2017, 8, 90808–90824. [Google Scholar] [CrossRef]
- Burger, J.A.; Ghia, P.; Rosenwald, A.; Caligaris-Cappio, F. The microenvironment in mature B-cell malignancies: A target for new treatment strategies. Blood 2009, 114, 3367–3375. [Google Scholar] [CrossRef]
- Solimando, A.G.; Da Via’, M.C.; Leone, P.; Croci, G.; Borrelli, P.; Tabares Gaviria, P.; Brandl, A.; Di Lernia, G.; Bianchi, F.P.; Tafuri, S.; et al. Adhesion-Mediated Multiple Myeloma (MM) Disease Progression: Junctional Adhesion Molecule a Enhances Angiogenesis and Multiple Myeloma Dissemination and Predicts Poor Survival. Blood 2019, 134, 855. [Google Scholar] [CrossRef]
- Pizzi, M.; Boi, M.; Bertoni, F.; Inghirami, G. Emerging therapies provide new opportunities to reshape the multifaceted interactions between the immune system and lymphoma cells. Leukemia 2016, 30, 1805–1815. [Google Scholar] [CrossRef] [PubMed]
- Scuto, A.; Kujawski, M.; Kowolik, C.; Krymskaya, L.; Wang, L.; Weiss, L.M.; Digiusto, D.; Yu, H.; Forman, S.; Jove, R. STAT3 inhibition is a therapeutic strategy for ABC-like diffuse large B-cell lymphoma. Cancer Res. 2011, 71, 3182–3188. [Google Scholar] [CrossRef] [PubMed]
- Passalidou, E.; Stewart, M.; Trivella, M.; Steers, G.; Pillai, G.; Dogan, A.; Leigh, I.; Hatton, C.; Harris, A.; Gatter, K.; et al. Vascular patterns in reactive lymphoid tissue and in non-Hodgkin’s lymphoma. Br. J. Cancer 2003, 88, 553–559. [Google Scholar] [CrossRef]
- Cardesa-Salzmann, T.M.; Colomo, L.; Gutierrez, G.; Chan, W.C.; Weisenburger, D.; Climent, F.; Gonzalez-Barca, E.; Mercadal, S.; Arenillas, L.; Serrano, S.; et al. High microvessel density determines a poor outcome in patients with diffuse large B-cell lymphoma treated with rituximab plus chemotherapy. Haematologica 2011, 96, 996–1001. [Google Scholar] [CrossRef]
- Woźnialis, N.; Gierej, B.; Popławska, L.; Ziarkiewicz, M.; Wilczek, E.; Kulczycka, E.; Ziarkiewicz-Wróblewska, B. Angiogenesis in CD5-positive Diffuse Large B Cell Lymphoma: A Morphometric Analysis. Adv. Clin. Exp. Med. 2016, 25, 1149–1155. [Google Scholar] [CrossRef]
- Shipp, M.A.; Ross, K.N.; Tamayo, P.; Weng, A.P.; Kutok, J.L.; Aguiar, R.C.T.; Gaasenbeek, M.; Angelo, M.; Reich, M.; Pinkus, G.S.; et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat. Med. 2002, 8, 68–74. [Google Scholar] [CrossRef]
- Gratzinger, D.; Zhao, S.; Tibshirani, R.J.; Hsi, E.D.; Hans, C.P.; Pohlman, B.; Bast, M.; Avigdor, A.; Schiby, G.; Nagler, A.; et al. Prognostic Significance of VEGF, VEGF Receptors, and Microvessel Density in Diffuse Large B Cell Lymphoma Treated with Anthracycline-Based Chemotherapy. Blood 2007, 110, 53. [Google Scholar] [CrossRef]
- Jørgensen, J.M.; Sørensen, F.B.; Bendix, K.; Nielsen, J.L.; Olsen, M.L.; Funder, A.M.D.; D’amore, F. Angiogenesis in non-Hodgkin’s lymphoma: Clinico-pathological correlations and prognostic significance in specific subtypes. Leuk. Lymphoma 2007, 48, 584–595. [Google Scholar] [CrossRef]
- Jiang, L.; Sun, J.; Quan, L.-N.; Tian, Y.-Y.; Jia, C.-M.; Liu, Z.-Q.; Liu, A.-C. Abnormal vascular endothelial growth factor protein expression may be correlated with poor prognosis in diffuse large B-cell lymphoma: A meta-analysis. J. Cancer Res. Ther. 2016, 12, 605. [Google Scholar] [CrossRef]
- Yoon, K.-A.; Kim, M.K.; Eom, H.-S.; Lee, H.; Park, W.S.; Sohn, J.Y.; Kim, M.J.; Kong, S.-Y. Adverse prognostic impact of vascular endothelial growth factor gene polymorphisms in patients with diffuse large B-cell lymphoma. Leuk. Lymphoma 2017, 58, 2677–2682. [Google Scholar] [CrossRef] [PubMed]
- Ganjoo, K.N.; Moore, A.M.; Orazi, A.; Sen, J.A.; Johnson, C.S.; An, C.S. The importance of angiogenesis markers in the outcome of patients with diffuse large B cell lymphoma: A retrospective study of 97 patients. J. Cancer Res. Clin. Oncol. 2008, 134, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; Suh, C.; Chi, H.S.; Cho, H.S.; Bae, Y.K.; Lee, K.H.; Lee, G.-W.; Kim, I.-S.; Eom, H.-S.; Kong, S.-Y.; et al. VEGFA and VEGFR2 genetic polymorphisms and survival in patients with diffuse large B cell lymphoma. Cancer Sci. 2012, 103, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Borges, N.M.; do Elias, M.V.; Fook-Alves, V.L.; Andrade, T.A.; de Conti, M.L.; Macedo, M.P.; Begnami, M.D.; Campos, A.H.J.F.M.; Etto, L.Y.; Bortoluzzo, A.B.; et al. Angiomirs expression profiling in diffuse large B-Cell lymphoma. Oncotarget 2016, 7. [Google Scholar] [CrossRef]
- Lupino, L.; Perry, T.; Margielewska, S.; Hollows, R.; Ibrahim, M.; Care, M.; Allegood, J.; Tooze, R.; Sabbadini, R.; Reynolds, G.; et al. Sphingosine-1-phosphate signalling drives an angiogenic transcriptional programme in diffuse large B cell lymphoma. Leukemia 2019, 33, 2884–2897. [Google Scholar] [CrossRef]
- Pyne, S.; Pyne, N. Sphingosine 1-phosphate signalling via the endothelial differentiation gene family of G-protein-coupled receptors. Pharmacol. Ther. 2000, 88, 115–131. [Google Scholar] [CrossRef]
- Wang, E.S.; Teruya-Feldstein, J.; Wu, Y.; Zhu, Z.; Hicklin, D.J.; Moore, M.A.S. Targeting autocrine and paracrine VEGF receptor pathways inhibits human lymphoma xenografts in vivo. Blood 2004, 104, 2893–2902. [Google Scholar] [CrossRef]
- Aref, S.; Mabed, M.; Zalata, K.; Sakrana, M.; El Askalany, H. The Interplay Between C-Myc Oncogene Expression and Circulating Vascular Endothelial Growth Factor (sVEGF), Its Antagonist Receptor, Soluble Flt-1 in Diffuse Large B Cell Lymphoma (DLBCL): Relationship to Patient Outcome. Leuk. Lymphoma 2004, 45, 499–506. [Google Scholar] [CrossRef]
- Stopeck, A.T.; Unger, J.M.; Rimsza, L.M.; Bellamy, W.T.; Iannone, M.; Persky, D.O.; Leblanc, M.; Fisher, R.I.; Miller, T.P. A phase II trial of single agent bevacizumab in patients with relapsed, aggressive non-Hodgkin lymphoma: Southwest oncology group study S0108. Leuk. Lymphoma 2009, 50, 728–735. [Google Scholar] [CrossRef]
- Marinaccio, C.; Ingravallo, G.; Gaudio, F.; Perrone, T.; Nico, B.; Maoirano, E.; Specchia, G.; Ribatti, D. Microvascular density, CD68 and tryptase expression in human Diffuse Large B-Cell Lymphoma. Leuk. Res. 2014, 38, 1374–1377. [Google Scholar] [CrossRef]
- Marinaccio, C.; Ingravallo, G.; Gaudio, F.; Perrone, T.; Ruggieri, S.; Opinto, G.; Nico, B.; Maiorano, E.; Specchia, G.; Ribatti, D. T cells, mast cells and microvascular density in diffuse large B cell lymphoma. Clin. Exp. Med. 2016, 16, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Song, M.-K.; Chung, J.-S.; Sung-Yong, O.; Lee, G.-W.; Kim, S.-G.; Seol, Y.-M.; Shin, H.-J.; Choi, Y.-J.; Cho, G.-J.; Shin, D.-H.; et al. Clinical impact of bulky mass in the patient with primary extranodal diffuse large B cell lymphoma treated with R-CHOP therapy. Ann. Hematol. 2010, 89, 985–991. [Google Scholar] [CrossRef] [PubMed]
- Tamma, R.; Ingravallo, G.; Albano, F.; Gaudio, F.; Annese, T.; Ruggieri, S.; Lorusso, L.; Errede, M.; Maiorano, E.; Specchia, G.; et al. STAT-3 RNAscope Determination in Human Diffuse Large B-Cell Lymphoma. Transl. Oncol. 2019, 12, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Tamma, R.; Ingravallo, G.; Gaudio, F.; Annese, T.; Albano, F.; Ruggieri, S.; Dicataldo, M.; Maiorano, E.; Specchia, G.; Ribatti, D. STAT3, tumor microenvironment, and microvessel density in diffuse large B cell lymphomas. Leuk. Lymphoma 2020, 61, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Sircar, A.; Chowdhury, S.M.; Hart, A.; Bell, W.C.; Singh, S.; Sehgal, L.; Epperla, N. Impact and Intricacies of Bone Marrow Microenvironment in B-cell Lymphomas: From Biology to Therapy. Int. J. Mol. Sci. 2020, 21, 904. [Google Scholar] [CrossRef]
- De Charette, M.; Houot, R. Hide or defend, the two strategies of lymphoma immune evasion: Potential implications for immunotherapy. Haematologica 2018, 103, 1256–1268. [Google Scholar] [CrossRef] [PubMed]
- Miao, X.; Wu, Y.; Wang, Y.; Zhu, X.; Yin, H.; He, Y.; Li, C.; Liu, Y.; Lu, X.; Chen, Y.; et al. Y-box-binding protein-1 (YB-1) promotes cell proliferation, adhesion and drug resistance in diffuse large B-cell lymphoma. Exp. Cell Res. 2016, 346, 157–166. [Google Scholar] [CrossRef]
- Huang, X.; Bai, X.; Cao, Y.; Wu, J.; Huang, M.; Tang, D.; Tao, S.; Zhu, T.; Liu, Y.; Yang, Y.; et al. Lymphoma endothelium preferentially expresses Tim-3 and facilitates the progression of lymphoma by mediating immune evasion. J. Exp. Med. 2010, 207, 505–520. [Google Scholar] [CrossRef]
- Edwards, J.P.; Zhang, X.; Mosser, D.M. The expression of heparin-binding epidermal growth factor-like growth factor by regulatory macrophages. J. Immunol. Baltim. Md 1950 2009, 182, 1929–1939. [Google Scholar] [CrossRef]
- Rao, L.; Giannico, D.; Leone, P.; Solimando, A.G.; Maiorano, E.; Caporusso, C.; Duda, L.; Tamma, R.; Mallamaci, R.; Susca, N.; et al. HB-EGF-EGFR Signaling in Bone Marrow Endothelial Cells Mediates Angiogenesis Associated with Multiple Myeloma. Cancers 2020, 12, 173. [Google Scholar] [CrossRef]
- Tjin, E.P.M.; Groen, R.W.J.; Vogelzang, I.; Derksen, P.W.B.; Klok, M.D.; Meijer, H.P.; van Eeden, S.; Pals, S.T.; Spaargaren, M. Functional analysis of HGF/MET signaling and aberrant HGF-activator expression in diffuse large B-cell lymphoma. Blood 2006, 107, 760–768. [Google Scholar] [CrossRef]
- Moschetta, M.; Basile, A.; Ferrucci, A.; Frassanito, M.A.; Rao, L.; Ria, R.; Solimando, A.G.; Giuliani, N.; Boccarelli, A.; Fumarola, F.; et al. Novel targeting of phospho-cMET overcomes drug resistance and induces antitumor activity in multiple myeloma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 4371–4382. [Google Scholar] [CrossRef] [PubMed]
- Ferrucci, A.; Moschetta, M.; Frassanito, M.A.; Berardi, S.; Catacchio, I.; Ria, R.; Racanelli, V.; Caivano, A.; Solimando, A.G.; Vergara, D.; et al. A HGF/cMET autocrine loop is operative in multiple myeloma bone marrow endothelial cells and may represent a novel therapeutic target. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 5796–5807. [Google Scholar] [CrossRef] [PubMed]
- Rao, L.; De Veirman, K.; Giannico, D.; Saltarella, I.; Desantis, V.; Frassanito, M.A.; Solimando, A.G.; Ribatti, D.; Prete, M.; Harstrick, A.; et al. Targeting angiogenesis in multiple myeloma by the VEGF and HGF blocking DARPin® protein MP0250: A preclinical study. Oncotarget 2018, 9, 13366–13381. [Google Scholar] [CrossRef] [PubMed]
- Siegler, E.; Li, S.; Kim, Y.J.; Wang, P. Designed Ankyrin Repeat Proteins as Her2 Targeting Domains in Chimeric Antigen Receptor-Engineered T Cells. Hum. Gene Ther. 2017, 28, 726–736. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.-P.; Sun, Y.-F.; Fang, Y.; Song, Q.; Yan, Z.-X.; Chen, Y.; Jiang, X.-F.; Fei, X.-C.; Zhao, Y.; Leboeuf, C.; et al. JAM-A overexpression is related to disease progression in diffuse large B-cell lymphoma and downregulated by lenalidomide. Sci. Rep. 2017, 7, 7433. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Si, M.; Li, L.; He, P.; Fan, Z.; Zhang, Q.; Jiao, X. Biomarkers Reflecting the Destruction of the Blood-Brain Barrier Are Valuable in Predicting the Risk of Lymphomas with Central Nervous System Involvement. Onco Targets Ther. 2019, 12, 9505–9512. [Google Scholar] [CrossRef]
- Karar, J.; Maity, A. PI3K/AKT/mTOR Pathway in Angiogenesis. Front. Mol. Neurosci. 2011, 4, 51. [Google Scholar] [CrossRef]
- Goodman, A.M.; Choi, M.; Wieduwilt, M.; Mulroney, C.; Costello, C.; Frampton, G.; Miller, V.; Kurzrock, R. Next-Generation Sequencing Reveals Potentially Actionable Alterations in the Majority of Patients with Lymphoid Malignancies. JCO Precis. Oncol. 2017, 1–13. [Google Scholar] [CrossRef]
- Todorovic Balint, M.; Jelicic, J.; Mihaljevic, B.; Kostic, J.; Stanic, B.; Balint, B.; Pejanovic, N.; Lucic, B.; Tosic, N.; Marjanovic, I.; et al. Gene Mutation Profiles in Primary Diffuse Large B Cell Lymphoma of Central Nervous System: Next Generation Sequencing Analyses. Int. J. Mol. Sci. 2016, 17, 683. [Google Scholar] [CrossRef]
- Vaqué, J.P.; Martínez, N.; Batlle-López, A.; Pérez, C.; Montes-Moreno, S.; Sánchez-Beato, M.; Piris, M.A. B-cell lymphoma mutations: Improving diagnostics and enabling targeted therapies. Haematologica 2014, 99, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.S.; Vo, T.-T.; Fruman, D.A. Targeting mTOR for the treatment of B cell malignancies. Br. J. Clin. Pharmacol. 2016, 82, 1213–1228. [Google Scholar] [CrossRef] [PubMed]
- Da Vià, M.C.; Solimando, A.G.; Garitano-Trojaola, A.; Barrio, S.; Munawar, U.; Strifler, S.; Haertle, L.; Rhodes, N.; Teufel, E.; Vogt, C.; et al. CIC Mutation as a Molecular Mechanism of Acquired Resistance to Combined BRAF-MEK Inhibition in Extramedullary Multiple Myeloma with Central Nervous System Involvement. Oncologist 2020, 25, 112–118. [Google Scholar] [CrossRef]
- Holderfield, M.; Deuker, M.M.; McCormick, F.; McMahon, M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat. Rev. Cancer 2014, 14, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Lamanuzzi, A.; Saltarella, I.; Desantis, V.; Frassanito, M.A.; Leone, P.; Racanelli, V.; Nico, B.; Ribatti, D.; Ditonno, P.; Prete, M.; et al. Inhibition of mTOR complex 2 restrains tumor angiogenesis in multiple myeloma. Oncotarget 2018, 9, 20563–20577. [Google Scholar] [CrossRef] [PubMed]
- Solimando, A.G.; Brandl, A.; Mattenheimer, K.; Graf, C.; Ritz, M.; Ruckdeschel, A.; Stühmer, T.; Mokhtari, Z.; Rudelius, M.; Dotterweich, J.; et al. JAM-A as a prognostic factor and new therapeutic target in multiple myeloma. Leukemia 2018, 32, 736–743. [Google Scholar] [CrossRef]
- Solimando, A.G.; Da Via’, M.C.; Borrelli, P.; Leone, P.; Di Lernia, G.; Tabares Gaviria, P.; Brandl, A.; Pedone, G.L.; Rauert-Wunderlich, H.; Lapa, C.; et al. Central Function for JAM-a in Multiple Myeloma Patients with Extramedullary Disease. Blood 2018, 132, 4455. [Google Scholar] [CrossRef]
- Mielcarek, M.; Sperling, C.; Shrappe, M.; Meyer, U.; Riehm, H.; Ludwig, W.-D. Expression of intercellular adhesion molecule 1 (ICAM-1) in childhood acute lymphoblastic leukaemia: Correlation with clinical features and outcome. Br. J. Haematol. 1997, 96, 301–307. [Google Scholar] [CrossRef]
- Horstmann, W.G.; Timens, W. Lack of adhesion molecules in testicular diffuse centroblastic and immunoblastic B cell lymphomas as a contributory factor in malignant behaviour. Virchows Arch. 1996, 429, 83–90. [Google Scholar] [CrossRef]
- Da Via’, M.C.; Solimando, A.G.; Garitano-Trojaola, A.; Barrio, S.; Rodhes, N.; Strifler, S.; Teufel, E.; Lapa, C.; Einsele, H.; Beilhack, A.; et al. CIC-Mutation As a Potential Molecular Mechanism of Acquired Resistance to Combined BRAF/MEK Inhibition in CNS Multiple Myeloma. Blood 2018, 132, 3181. [Google Scholar] [CrossRef]
- Pasqualucci, L. The genetic basis of diffuse large B-cell lymphoma. Curr. Opin. Hematol. 2013, 20, 336–344. [Google Scholar] [CrossRef] [PubMed]
- Stopeck, A.T.; Gessner, A.; Miller, T.P.; Hersh, E.M.; Johnson, C.S.; Cui, H.; Frutiger, Y.; Grogan, T.M. Loss of B7.2 (CD86) and intracellular adhesion molecule 1 (CD54) expression is associated with decreased tumor-infiltrating T lymphocytes in diffuse B-cell large-cell lymphoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2000, 6, 3904–3909. [Google Scholar]
- Terol, M.J.; Tormo, M.; Martinez-Climent, J.A.; Marugan, I.; Benet, I.; Ferrandez, A.; Teruel, A.; Ferrer, R.; García-Conde, J. Soluble intercellular adhesion molecule-1 (s-ICAM-1/s-CD54) in diffuse large B-cell lymphoma: Association with clinical characteristics and outcome. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2003, 14, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-H.; Kosek, J.; Wang, M.; Heise, C.; Schafer, P.H.; Chopra, R. Lenalidomide efficacy in activated B-cell-like subtype diffuse large B-cell lymphoma is dependent upon IRF4 and cereblon expression. Br. J. Haematol. 2013, 160, 487–502. [Google Scholar] [CrossRef]
- Gascoyne, D.M.; Banham, A.H. The significance of FOXP1 in diffuse large B-cell lymphoma. Leuk. Lymphoma 2017, 58, 1037–1051. [Google Scholar] [CrossRef]
- Solimando, A.; Brandl, A.; Katharina, M.; Graf, C.; Ritz, M.; Ruckdeschel, A.; Stühmer, T.; Rudelius, M.; Frassanito, M.A.; Andreas, R.; et al. JAM-A as a Prognostic Factor and New Therapeutic Target in Multiple Myeloma. Blood 2016, 128, 307. [Google Scholar] [CrossRef]
- Ebnet, K. Junctional Adhesion Molecules (JAMs): Cell Adhesion Receptors With Pleiotropic Functions in Cell Physiology and Development. Physiol. Rev. 2017, 97, 1529–1554. [Google Scholar] [CrossRef]
- Koenen, R.R.; Pruessmeyer, J.; Soehnlein, O.; Fraemohs, L.; Zernecke, A.; Schwarz, N.; Reiss, K.; Sarabi, A.; Lindbom, L.; Hackeng, T.M.; et al. Regulated release and functional modulation of junctional adhesion molecule A by disintegrin metalloproteinases. Blood 2009, 113, 4799–4809. [Google Scholar] [CrossRef]
- Leech, A.O.; Cruz, R.G.B.; Hill, A.D.K.; Hopkins, A.M. Paradigms lost-an emerging role for over-expression of tight junction adhesion proteins in cancer pathogenesis. Ann. Transl. Med. 2015, 3, 184. [Google Scholar] [CrossRef]
- Zhao, C.; Lu, F.; Chen, H.; Zhao, X.; Sun, J.; Chen, H. Dysregulation of JAM-A plays an important role in human tumor progression. Int. J. Clin. Exp. Pathol. 2014, 7, 7242–7248. [Google Scholar]
- Scheller, J.; Chalaris, A.; Garbers, C.; Rose-John, S. ADAM17: A molecular switch to control inflammation and tissue regeneration. Trends Immunol. 2011, 32, 380–387. [Google Scholar] [CrossRef]
- Katz, E.; Deehan, M.R.; Seatter, S.; Lord, C.; Sturrock, R.D.; Harnett, M.M. B cell receptor-stimulated mitochondrial phospholipase A2 activation and resultant disruption of mitochondrial membrane potential correlate with the induction of apoptosis in WEHI-231 B cells. J. Immunol. Baltim. Md 1950 2001, 166, 137–147. [Google Scholar] [CrossRef]
- Jridi, I.; Catacchio, I.; Majdoub, H.; Shahbazzadeh, D.; El Ayeb, M.; Frassanito, M.A.; Solimando, A.G.; Ribatti, D.; Vacca, A.; Borchani, L. The small subunit of Hemilipin2, a new heterodimeric phospholipase A2 from Hemiscorpius lepturus scorpion venom, mediates the antiangiogenic effect of the whole protein. Toxicon Off. J. Int. Soc. Toxinol. 2017, 126, 38–46. [Google Scholar] [CrossRef]
- Upadhyay, R.; Hammerich, L.; Peng, P.; Brown, B.; Merad, M.; Brody, J.D. Lymphoma: Immune evasion strategies. Cancers 2015, 7, 736–762. [Google Scholar] [CrossRef]
- Tripodo, C.; Sangaletti, S.; Piccaluga, P.P.; Prakash, S.; Franco, G.; Borrello, I.; Orazi, A.; Colombo, M.P.; Pileri, S.A. The bone marrow stroma in hematological neoplasms–a guilty bystander. Nat. Rev. Clin. Oncol. 2011, 8, 456–466. [Google Scholar] [CrossRef]
- Rafii, S.; Lyden, D.; Benezra, R.; Hattori, K.; Heissig, B. Vascular and haematopoietic stem cells: Novel targets for anti-angiogenesis therapy? Nat. Rev. Cancer 2002, 2, 826–835. [Google Scholar] [CrossRef]
- Grunewald, M.; Avraham, I.; Dor, Y.; Bachar-Lustig, E.; Itin, A.; Jung, S.; Yung, S.; Chimenti, S.; Landsman, L.; Abramovitch, R.; et al. VEGF-induced adult neovascularization: Recruitment, retention, and role of accessory cells. Cell 2006, 124, 175–189. [Google Scholar] [CrossRef]
- Seymour, J.F.; Pfreundschuh, M.; Trneny, M.; Sehn, L.H.; Catalano, J.; Csinady, E.; Moore, N.; Coiffier, B. R-CHOP with or without bevacizumab in patients with previously untreated diffuse large B-cell lymphoma: Final MAIN study outcomes. Haematologica 2014, 99, 1343–1349. [Google Scholar] [CrossRef]
- Jiang, L.; Li, N. B-cell non-Hodgkin lymphoma: Importance of angiogenesis and antiangiogenic therapy. Angiogenesis 2020. [Google Scholar] [CrossRef]
- Joyce, J.A.; Fearon, D.T. T cell exclusion, immune privilege, and the tumor microenvironment. Science 2015, 348, 74–80. [Google Scholar] [CrossRef]
- Galon, J.; Bruni, D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019, 18, 197–218. [Google Scholar] [CrossRef]
- Antonio, G.; Oronzo, B.; Vito, L.; Angela, C.; Antonel-la, A.; Roberto, C.; Giovanni, S.A.; Antonella, L. Immune system and bone microenvironment: Rationale for targeted cancer therapies. Oncotarget 2020, 11. [Google Scholar] [CrossRef][Green Version]
- Fukumura, D.; Kloepper, J.; Amoozgar, Z.; Duda, D.G.; Jain, R.K. Enhancing cancer immunotherapy using antiangiogenics: Opportunities and challenges. Nat. Rev. Clin. Oncol. 2018, 15, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Compagno, N.; Cinetto, F.; Semenzato, G.; Agostini, C. Subcutaneous immunoglobulin in lymphoproliferative disorders and rituximab-related secondary hypogammaglobulinemia: A single-center experience in 61 patients. Haematologica 2014, 99, 1101–1106. [Google Scholar] [CrossRef] [PubMed]
- Vacca, A.; Melaccio, A.; Sportelli, A.; Solimando, A.G.; Dammacco, F.; Ria, R. Subcutaneous immunoglobulins in patients with multiple myeloma and secondary hypogammaglobulinemia: A randomized trial. Clin. Immunol. Orlando Fl. 2018, 191, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Li, L.; Zhang, L.; Fu, X.; Li, X.; Wang, X.; Wu, J.; Sun, Z.; Zhang, X.; Feng, X.; et al. Apatinib in Patients with Relapsed or Refractory Diffuse Large B Cell Lymphoma: A Phase II, Open-Label, Single-Arm, Prospective Study. Drug Des. Devel. Ther. 2020, 14, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Deng, M.; Chen, Q.; Li, Y.; Guo, X.; Shi, P.; He, L.; Xie, S.; Yu, L.; Zhang, H.; et al. Apatinib exerts anti-tumor activity to non-Hodgkin lymphoma by inhibition of the Ras pathway. Eur. J. Pharmacol. 2019, 843, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Pfreundschuh, M.; Kloess, M.; Schmits, R.; Zeynalova, S.; Lengfelder, E.; Franke, A.; Steinhauer, H.; Reiser, M.; Clemens, M.; Nickenig, C.; et al. Six, Not Eight Cycles of Bi-Weekly CHOP with Rituximab (R-CHOP-14) Is the Preferred Treatment for Elderly Patients with Diffuse Large B-Cell Lymphoma (DLBCL): Results of the RICOVER-60 Trial of the German High-Grade Non-Hodgkin Lymphoma Study Group (DSHNHL). Blood 2005, 106, 13. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Solimando, A.G.; Annese, T.; Tamma, R.; Ingravallo, G.; Maiorano, E.; Vacca, A.; Specchia, G.; Ribatti, D. New Insights into Diffuse Large B-Cell Lymphoma Pathobiology. Cancers 2020, 12, 1869. https://doi.org/10.3390/cancers12071869
Solimando AG, Annese T, Tamma R, Ingravallo G, Maiorano E, Vacca A, Specchia G, Ribatti D. New Insights into Diffuse Large B-Cell Lymphoma Pathobiology. Cancers. 2020; 12(7):1869. https://doi.org/10.3390/cancers12071869
Chicago/Turabian StyleSolimando, Antonio Giovanni, Tiziana Annese, Roberto Tamma, Giuseppe Ingravallo, Eugenio Maiorano, Angelo Vacca, Giorgina Specchia, and Domenico Ribatti. 2020. "New Insights into Diffuse Large B-Cell Lymphoma Pathobiology" Cancers 12, no. 7: 1869. https://doi.org/10.3390/cancers12071869
APA StyleSolimando, A. G., Annese, T., Tamma, R., Ingravallo, G., Maiorano, E., Vacca, A., Specchia, G., & Ribatti, D. (2020). New Insights into Diffuse Large B-Cell Lymphoma Pathobiology. Cancers, 12(7), 1869. https://doi.org/10.3390/cancers12071869









