The Role of Liquid Biopsies in Detecting Molecular Tumor Biomarkers in Brain Cancer Patients
Abstract
1. Introduction
2. Molecular Biomarkers in Brain Cancer
2.1. Glial Fibrillary Acidic Protein (GFAP)
2.2. Methylguanine-DNA Methyltransferase Promoter Methylation (MGMT)
2.3. Isocitrate Dehydrogenase Mutations (IDH1/2)
2.4. Epidermal Growth Factor Receptor (EGFR)
2.5. Telomerase Promoter Mutations (TERT)
2.6. Loss of Heterozygosity
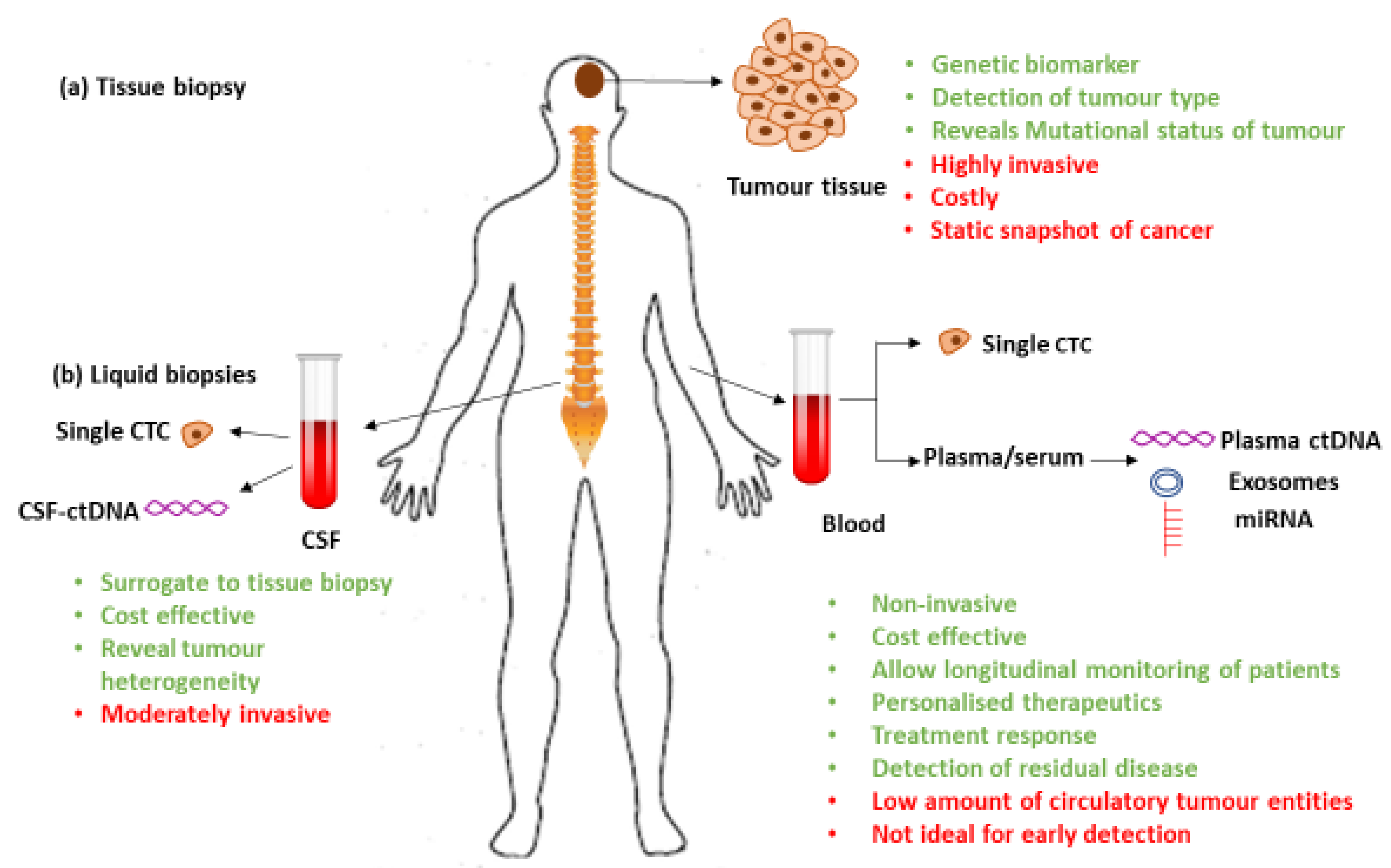
3. Liquid Biopsies
3.1. Circulating Tumor Cells (CTCs)
3.2. Circulating Tumor Nucleic Acids (ctNAs)
3.3. Cerebro-Spinal Fluid
3.4. Exosomes
4. Conclusions
Funding
Conflicts of Interest
References
- Connel, E.; Bartlett, N.; Harvey, J.; Moon, L.; Short, M.; Davis, B. Brain and Other Central Nervous System Cancers; Aihw, S., Ed.; Australian Institute of Health and Welfare: Canberra, Australia, 2017. [Google Scholar]
- Ohgaki, H.; Kleihues, P. Genetic pathways to primary and secondary glioblastoma. Am. J. Pathol. 2007, 170, 1445–1453. [Google Scholar] [CrossRef]
- Reifenberger, G.; Hentschel, B.; Felsberg, J.; Schackert, G.; Simon, M.; Schnell, O.; Westphal, M.; Wick, W.; Pietsch, T.; Loeffler, M.; et al. Predictive impact of MGMT promoter methylation in glioblastoma of the elderly. Int. J. Cancer 2012, 131, 1342–1350. [Google Scholar] [CrossRef]
- Omuro, A.; DeAngelis, L.M. Glioblastoma and other malignant gliomas: A clinical review. Jama 2013, 310, 1842–1850. [Google Scholar] [CrossRef]
- Hygino da Cruz, L.C., Jr.; Rodriguez, I.; Domingues, R.C.; Gasparetto, E.L.; Sorensen, A.G. Pseudoprogression and pseudoresponse: Imaging challenges in the assessment of posttreatment glioma. Am. J. Neuroradiol. 2011, 32, 1978–1985. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabieres, C.; Schwarzenbach, H.; Pantel, K. Circulating tumor cells and circulating tumor DNA. Annu. Rev. Med. 2012, 63, 199–215. [Google Scholar] [CrossRef] [PubMed]
- MacArthur, K.M.; Kao, G.D.; Chandrasekaran, S.; Alonso-Basanta, M.; Chapman, C.; Lustig, R.A.; Wileyto, E.P.; Hahn, S.M.; Dorsey, J.F. Detection of brain tumor cells in the peripheral blood by a telomerase promoter-based assay. Cancer Res. 2014, 74, 2152–2159. [Google Scholar] [CrossRef] [PubMed]
- Lavon, I.; Refael, M.; Zelikovitch, B.; Shalom, E.; Siegal, T. Serum DNA can define tumor-specific genetic and epigenetic markers in gliomas of various grades. Neuro-Oncology 2010, 12, 173–180. [Google Scholar] [CrossRef]
- Siegal, T. Clinical impact of molecular biomarkers in gliomas. J. Clin. Neurosci. 2015, 22, 437–444. [Google Scholar] [CrossRef]
- Jung, C.; Foerch, C.; Schänzer, A.; Heck, A.; Plate, K.; Seifert, V.; Steinmetz, H.; Raabe, A.; Sitzer, M. Serum GFAP is a diagnostic marker for glioblastoma multiforme. Brain 2007, 130, 3336–3341. [Google Scholar] [CrossRef]
- Muller, C.; Holtschmidt, J.; Auer, M.; Heitzer, E.; Lamszus, K.; Schulte, A.; Matschke, J.; Langer-Freitag, S.; Gasch, C.; Stoupiec, M.; et al. Hematogenous dissemination of glioblastoma multiforme. Sci. Transl. Med. 2014, 6, 247ra101. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.-C.; Gorlia, T.; Hamou, M.-F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Fiano, V.; Trevisan, M.; Trevisan, E.; Senetta, R.; Castiglione, A.; Sacerdote, C.; Gillio-Tos, A.; De Marco, L.; Grasso, C.; Magistrello, M.; et al. MGMT promoter methylation in plasma of glioma patients receiving temozolomide. J. Neuro-Oncol. 2014, 117, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Barault, L.; Amatu, A.; Bleeker, F.; Moutinho, C.; Falcomatà, C.; Fiano, V.; Cassingena, A.; Siravegna, G.; Milione, M.; Cassoni, P. Digital PCR quantification of MGMT methylation refines prediction of clinical benefit from alkylating agents in glioblastoma and metastatic colorectal cancer. Ann. Oncol. 2015, 26, 1994–1999. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- Boisselier, B.; Pérez-Larraya, J.G.; Rossetto, M.; Labussière, M.; Ciccarino, P.; Marie, Y.; Delattre, J.-Y.; Sanson, M. Detection of IDH1 mutation in the plasma of patients with glioma. Neurology 2012, 79, 1693–1698. [Google Scholar] [CrossRef] [PubMed]
- Marcucci, G.; Maharry, K.; Wu, Y.-Z.; Radmacher, M.D.; Mrózek, K.; Margeson, D.; Holland, K.B.; Whitman, S.P.; Becker, H.; Schwind, S. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J. Clin. Oncol. 2010, 28, 2348–2355. [Google Scholar] [CrossRef] [PubMed]
- Vengoji, R.; Macha, M.A.; Nimmakayala, R.K.; Rachagani, S.; Siddiqui, J.A.; Mallya, K.; Gorantla, S.; Jain, M.; Ponnusamy, M.P.; Batra, S.K.; et al. Afatinib and Temozolomide combination inhibits tumorigenesis by targeting EGFRvIII-cMet signaling in glioblastoma cells. J. Exp. Clin. Cancer Res. 2019, 38, 266. [Google Scholar] [CrossRef]
- Salkeni, M.A.; Zarzour, A.; Ansay, T.Y.; McPherson, C.M.; Warnick, R.E.; Rixe, O.; Bahassi, E.M. Detection of EGFRvIII mutant DNA in the peripheral blood of brain tumor patients. J. Neuro-Oncol. 2013, 115, 27–35. [Google Scholar] [CrossRef]
- Marchetti, A.; Del Grammastro, M.; Felicioni, L.; Malatesta, S.; Filice, G.; Centi, I.; De Pas, T.; Santoro, A.; Chella, A.; Brandes, A.A. Assessment of EGFR mutations in circulating tumor cell preparations from NSCLC patients by next generation sequencing: Toward a real-time liquid biopsy for treatment. PLoS ONE 2014, 9, e103883. [Google Scholar] [CrossRef]
- Ding, P.N.; Becker, T.; Bray, V.; Chua, W.; Ma, Y.; Xu, B.; Lynch, D.; de Souza, P.; Roberts, T. Plasma next generation sequencing and droplet digital PCR-based detection of epidermal growth factor receptor (EGFR) mutations in patients with advanced lung cancer treated with subsequent-line osimertinib. Thorac. Cancer 2019, 10, 1879–1884. [Google Scholar] [CrossRef]
- Arita, H.; Yamasaki, K.; Matsushita, Y.; Nakamura, T.; Shimokawa, A.; Takami, H.; Tanaka, S.; Mukasa, A.; Shirahata, M.; Shimizu, S.; et al. A combination of TERT promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathol. Commun. 2016, 4, 79. [Google Scholar] [CrossRef]
- Juratli, T.A.; Stasik, S.; Zolal, A.; Schuster, C.; Richter, S.; Daubner, D.; Juratli, M.A.; Thowe, R.; Hennig, S.; Makina, M.; et al. TERT Promoter Mutation Detection in Cell-Free Tumor-Derived DNA in Patients with IDH Wild-Type Glioblastomas: A Pilot Prospective Study. Clin. Cancer Res. 2018, 24, 5282–5291. [Google Scholar] [CrossRef]
- Hayashi, Y.; Fujita, K.; Matsuzaki, K.; Matsushita, M.; Kawamura, N.; Koh, Y.; Nakano, K.; Wang, C.; Ishizuya, Y.; Yamamoto, Y.; et al. Diagnostic potential of TERT promoter and FGFR3 mutations in urinary cell-free DNA in upper tract urothelial carcinoma. Cancer Sci. 2019, 110, 1771–1779. [Google Scholar] [CrossRef]
- Hu, Z.Y.; Xie, N.; Tian, C.; Yang, X.; Liu, L.; Li, J.; Xiao, H.; Wu, H.; Lu, J.; Gao, J.; et al. Identifying Circulating Tumor DNA Mutation Profiles in Metastatic Breast Cancer Patients with Multiline Resistance. EBioMedicine 2018, 32, 111–118. [Google Scholar] [CrossRef]
- Fujisawa, H.; Kurrer, M.; Reis, R.M.; Yonekawa, Y.; Kleihues, P.; Ohgaki, H. Acquisition of the glioblastoma phenotype during astrocytoma progression is associated with loss of heterozygosity on 10q25-qter. Am. J. Pathol. 1999, 155, 387–394. [Google Scholar] [CrossRef]
- Kuhlmann, J.D.; Schwarzenbach, H.; Wimberger, P.; Poetsch, M.; Kimmig, R.; Kasimir-Bauer, S. LOH at 6q and 10q in fractionated circulating DNA of ovarian cancer patients is predictive for tumor cell spread and overall survival. BMC Cancer 2012, 12, 325. [Google Scholar] [CrossRef] [PubMed]
- Tichy, J.; Spechtmeyer, S.; Mittelbronn, M.; Hattingen, E.; Rieger, J.; Senft, C.; Foerch, C. Prospective evaluation of serum glial fibrillary acidic protein (GFAP) as a diagnostic marker for glioblastoma. J. Neuro-Oncol. 2016, 126, 361–369. [Google Scholar] [CrossRef]
- Gállego Pérez-Larraya, J.; Paris, S.; Idbaih, A.; Dehais, C.; Laigle-Donadey, F.; Navarro, S.; Capelle, L.; Mokhtari, K.; Marie, Y.; Sanson, M.; et al. Diagnostic and prognostic value of preoperative combined GFAP, IGFBP-2, and YKL-40 plasma levels in patients with glioblastoma. Cancer 2014, 120, 3972–3980. [Google Scholar] [CrossRef]
- Kiviniemi, A.; Gardberg, M.; Frantzen, J.; Parkkola, R.; Vuorinen, V.; Pesola, M.; Minn, H. Serum levels of GFAP and EGFR in primary and recurrent high-grade gliomas: Correlation to tumor volume, molecular markers, and progression-free survival. J. Neuro-Oncol. 2015, 124, 237–245. [Google Scholar] [CrossRef]
- Hegi, M.E.; Liu, L.; Herman, J.G.; Stupp, R.; Wick, W.; Weller, M.; Mehta, M.P.; Gilbert, M.R. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J. Clin. Oncol. 2008, 26, 4189–4199. [Google Scholar] [CrossRef]
- Szopa, W.; Burley, T.A.; Kramer-Marek, G.; Kaspera, W. Diagnostic and Therapeutic Biomarkers in Glioblastoma: Current Status and Future Perspectives %J BioMed Research International. BioMed Res. Int. 2017, 2017, 13. [Google Scholar] [CrossRef]
- Thon, N.; Kreth, S.; Kreth, F.-W. Personalized treatment strategies in glioblastoma: MGMT promoter methylation status. Oncotargets Ther. 2013, 6, 1363–1372. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Koh, H.J.; Park, D.C.; Song, B.J.; Huh, T.L.; Park, J.W. Cytosolic NADP(+)-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radic. Biol. Med. 2002, 32, 1185–1196. [Google Scholar] [CrossRef]
- Ducray, F.; Idbaih, A.; Wang, X.W.; Cheneau, C.; Labussiere, M.; Sanson, M. Predictive and prognostic factors for gliomas. Expert Rev. Anticancer Ther. 2011, 11, 781–789. [Google Scholar] [CrossRef]
- Lu, Y.; Kwintkiewicz, J.; Liu, Y.; Tech, K.; Frady, L.N.; Su, Y.-T.; Bautista, W.; Moon, S.I.; MacDonald, J.; Ewend, M.G.; et al. Chemosensitivity of IDH1-Mutated Gliomas Due to an Impairment in PARP1-Mediated DNA Repair. Cancer Res. 2017, 77, 1709. [Google Scholar] [CrossRef]
- Shi, J.; Sun, B.; Shi, W.; Zuo, H.; Cui, D.; Ni, L.; Chen, J. Decreasing GSH and increasing ROS in chemosensitivity gliomas with IDH1 mutation. Tumour Biol. 2015, 36, 655–662. [Google Scholar] [CrossRef]
- Agnihotri, S.; Aldape, K.D.; Zadeh, G. Isocitrate dehydrogenase status and molecular subclasses of glioma and glioblastoma. Neurosurg. Focus 2014, 37, E13. [Google Scholar] [CrossRef]
- Beiko, J.; Suki, D.; Hess, K.R.; Fox, B.D.; Cheung, V.; Cabral, M.; Shonka, N.; Gilbert, M.R.; Sawaya, R.; Prabhu, S.S.; et al. IDH1 mutant malignant astrocytomas are more amenable to surgical resection and have a survival benefit associated with maximal surgical resection. Neuro-Oncology 2014, 16, 81–91. [Google Scholar] [CrossRef]
- Li, S.; Chou, A.P.; Chen, W.; Chen, R.; Deng, Y.; Phillips, H.S.; Selfridge, J.; Zurayk, M.; Lou, J.J.; Everson, R.G.; et al. Overexpression of isocitrate dehydrogenase mutant proteins renders glioma cells more sensitive to radiation. Neuro-Oncology 2013, 15, 57–68. [Google Scholar] [CrossRef]
- SongTao, Q.; Lei, Y.; Si, G.; YanQing, D.; HuiXia, H.; XueLin, Z.; LanXiao, W.; Fei, Y. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci. 2012, 103, 269–273. [Google Scholar] [CrossRef]
- Houillier, C.; Wang, X.; Kaloshi, G.; Mokhtari, K.; Guillevin, R.; Laffaire, J.; Paris, S.; Boisselier, B.; Idbaih, A.; Laigle-Donadey, F.; et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology 2010, 75, 1560. [Google Scholar] [CrossRef] [PubMed]
- Sanson, M.; Marie, Y.; Paris, S.; Idbaih, A.; Laffaire, J.; Ducray, F.; El Hallani, S.; Boisselier, B.; Mokhtari, K.; Hoang-Xuan, K.; et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009, 27, 4150–4154. [Google Scholar] [CrossRef] [PubMed]
- Luk, A.; Lynch, D.; Young, F.; Cooper, A.; de Souza, P.; Becker, T. A Sensitive Droplet Digital PCR Assay to Detect the IDH1-R132H Glioma Biomarker. In Proceedings of the Sydney Cancer Conference, Sydney, Australia, 22–23 September 2016. [Google Scholar]
- Normanno, N.; De Luca, A.; Bianco, C.; Strizzi, L.; Mancino, M.; Maiello, M.R.; Carotenuto, A.; De Feo, G.; Caponigro, F.; Salomon, D.S. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 2006, 366, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Aldape, K.D.; Ballman, K.; Furth, A.; Buckner, J.C.; Giannini, C.; Burger, P.C.; Scheithauer, B.W.; Jenkins, R.B.; James, C.D. Immunohistochemical detection of EGFRvIII in high malignancy grade astrocytomas and evaluation of prognostic significance. J. Neuropathol. Exp. Neurol. 2004, 63, 700–707. [Google Scholar] [CrossRef] [PubMed]
- Shinojima, N.; Tada, K.; Shiraishi, S.; Kamiryo, T.; Kochi, M.; Nakamura, H.; Makino, K.; Saya, H.; Hirano, H.; Kuratsu, J.-i. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003, 63, 6962–6970. [Google Scholar] [PubMed]
- Nagane, M.; Levitzki, A.; Gazit, A.; Cavenee, W.K.; Huang, H.J. Drug resistance of human glioblastoma cells conferred by a tumor-specific mutant epidermal growth factor receptor through modulation of Bcl-XL and caspase-3-like proteases. Proc. Natl. Acad. Sci. USA 1998, 95, 5724–5729. [Google Scholar] [CrossRef]
- Heimberger, A.B.; Suki, D.; Yang, D.; Shi, W.; Aldape, K. The natural history of EGFR and EGFRvIII in glioblastoma patients. J. Transl. Med. 2005, 3, 38. [Google Scholar] [CrossRef]
- Montano, N.; Cenci, T.; Martini, M.; D’Alessandris, Q.G.; Pelacchi, F.; Ricci-Vitiani, L.; Maira, G.; Maria, R.D.; Larocca, L.M.; Pallini, R. Expression of EGFRvIII in Glioblastoma: Prognostic Significance Revisited. Neoplasia 2011, 13, 1113-IN6. [Google Scholar] [CrossRef]
- Klingler, S.; Guo, B.; Yao, J.; Yan, H.; Zhang, L.; Vaseva, A.V.; Chen, S.; Canoll, P.; Horner, J.W.; Wang, Y.A. Development of Resistance to EGFR-Targeted Therapy in Malignant Glioma Can Occur through EGFR-Dependent and-Independent Mechanisms. Cancer Res. 2015, 75, 2109–2119. [Google Scholar] [CrossRef]
- Liu, X.; Chen, X.; Shi, L.; Shan, Q.; Cao, Q.; Yue, C.; Li, H.; Li, S.; Wang, J.; Gao, S.; et al. The third-generation EGFR inhibitor AZD9291 overcomes primary resistance by continuously blocking ERK signaling in glioblastoma. J. Exp. Clin. Cancer Res. 2019, 38, 219. [Google Scholar] [CrossRef]
- Ma, Y.; Luk, A.; Young, F.P.; Lynch, D.; Chua, W.; Balakrishnar, B.; de Souza, P.; Becker, T.M. Droplet digital PCR based androgen receptor variant 7 (AR-V7) detection from prostate cancer patient blood biopsies. Int. J. Mol. Sci. 2016, 17, 1264. [Google Scholar] [CrossRef]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef]
- Greider, C.W.; Blackburn, E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 1985, 43, 405–413. [Google Scholar] [CrossRef]
- Horn, S.; Figl, A.; Rachakonda, P.S.; Fischer, C.; Sucker, A.; Gast, A.; Kadel, S.; Moll, I.; Nagore, E.; Hemminki, K. TERT promoter mutations in familial and sporadic melanoma. Science 2013, 339, 959–961. [Google Scholar] [CrossRef]
- Huang, D.-S.; Wang, Z.; He, X.-J.; Diplas, B.H.; Yang, R.; Killela, P.J.; Meng, Q.; Ye, Z.-Y.; Wang, W.; Jiang, X.-T. Recurrent TERT promoter mutations identified in a large-scale study of multiple tumour types are associated with increased TERT expression and telomerase activation. Eur. J. Cancer 2015, 51, 969–976. [Google Scholar] [CrossRef]
- Liu, X.; Bishop, J.; Shan, Y.; Pai, S.; Liu, D.; Murugan, A.K.; Sun, H.; El-Naggar, A.K.; Xing, M. Highly prevalent TERT promoter mutations in aggressive thyroid cancers. Endocr.-Relat. Cancer 2013, 20, 603–610. [Google Scholar] [CrossRef]
- Borah, S.; Xi, L.; Zaug, A.J.; Powell, N.M.; Dancik, G.M.; Cohen, S.B.; Costello, J.C.; Theodorescu, D.; Cech, T.R. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science 2015, 347, 1006–1010. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wu, G.; Shan, Y.; Hartmann, C.; Von Deimling, A.; Xing, M. Highly prevalent TERT promoter mutations in bladder cancer and glioblastoma. Cell Cycle 2013, 12, 1637–1638. [Google Scholar] [CrossRef]
- McEvoy, A.C.; Calapre, L.; Pereira, M.R.; Giardina, T.; Robinson, C.; Khattak, M.A.; Meniawy, T.M.; Pritchard, A.L.; Hayward, N.K.; Amanuel, B.; et al. Sensitive droplet digital PCR method for detection of TERT promoter mutations in cell free DNA from patients with metastatic melanoma. Oncotarget 2017, 8, 78890–78900. [Google Scholar] [CrossRef] [PubMed]
- Mosrati, M.A.; Malmström, A.; Lysiak, M.; Krysztofiak, A.; Hallbeck, M.; Milos, P.; Hallbeck, A.-L.; Bratthäll, C.; Strandéus, M.; Stenmark-Askmalm, M.; et al. TERT promoter mutations and polymorphisms as prognostic factors in primary glioblastoma. Oncotarget 2015, 6, 16663–16673. [Google Scholar] [CrossRef]
- Killela, P.J.; Reitman, Z.J.; Jiao, Y.; Bettegowda, C.; Agrawal, N.; Diaz, L.A.; Friedman, A.H.; Friedman, H.; Gallia, G.L.; Giovanella, B.C. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. USA 2013, 110, 6021–6026. [Google Scholar] [CrossRef]
- Corless, B.C.; Chang, G.A.; Cooper, S.; Syeda, M.M.; Shao, Y.; Osman, I.; Karlin-Neumann, G.; Polsky, D. Development of Novel Mutation-Specific Droplet Digital PCR Assays Detecting TERT Promoter Mutations in Tumor and Plasma Samples. J. Mol. Diagn. 2019, 21, 274–285. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Ishida, E.; Shimada, K.; Kishi, M.; Nakase, H.; Sakaki, T.; Konishi, N. Frequent LOH on 22q12.3 and TIMP-3 inactivation occur in the progression to secondary glioblastomas. Lab. Investig. 2004, 85, 165. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Franceschi, S.; Fukushima, T.; Homma, T.; Ohgaki, H.; Vaccarella, S.; Yonekawa, Y.; Di Patre, P.L. Correlation Among Pathology, Genotype, and Patient Outcomes in Glioblastoma. J. Neuropathol. Exp. Neurol. 2006, 65, 846–854. [Google Scholar] [CrossRef]
- Schmidt, M.C.; Antweiler, S.; Urban, N.; Mueller, W.; Kuklik, A.; Meyer-Puttlitz, B.; Wiestler, O.D.; Louis, D.N.; Fimmers, R.; von Deimling, A. Impact of genotype and morphology on the prognosis of glioblastoma. J. Neuropathol. Exp. Neurol. 2002, 61, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Malara, N.; Guzzi, G.; Mignogna, C.; Trunzo, V.; Camastra, C.; Della Torre, A.; Di Vito, A.; Lavecchia, A.M.; Gliozzi, M.; Ceccotti, C.; et al. Non-invasive real-time biopsy of intracranial lesions using short time expanded circulating tumor cells on glass slide: Report of two cases. BMC Neurol. 2016, 16, 127. [Google Scholar] [CrossRef][Green Version]
- Gao, F.; Cui, Y.; Jiang, H.; Sui, D.; Wang, Y.; Jiang, Z.; Zhao, J.; Lin, S. Circulating tumor cell is a common property of brain glioma and promotes the monitoring system. Oncotarget 2016, 7, 71330–71340. [Google Scholar] [CrossRef]
- Becker, T.M.; Caixeiro, N.J.; Lim, S.H.; Tognela, A.; Kienzle, N.; Scott, K.F.; Spring, K.J.; Souza, P. New frontiers in circulating tumor cell analysis: A reference guide for biomolecular profiling toward translational clinical use. Int. J. Cancer 2014, 134, 2523–2533. [Google Scholar] [CrossRef]
- Touat, M.; Duran-Pena, A.; Alentorn, A.; Lacroix, L.; Massard, C.; Idbaih, A. Emerging circulating biomarkers in glioblastoma: Promises and challenges. Expert Rev. Mol. Diagn. 2015, 15, 1311–1323. [Google Scholar] [CrossRef]
- Sullivan, J.P.; Nahed, B.V.; Madden, M.W.; Oliveira, S.M.; Springer, S.; Bhere, D.; Chi, A.S.; Wakimoto, H.; Rothenberg, S.M.; Sequist, L.V. Brain tumor cells in circulation are enriched for mesenchymal gene expression. Cancer Discov. 2014, 4, 1299–1309. [Google Scholar] [CrossRef]
- Krol, I.; Castro-Giner, F.; Maurer, M.; Gkountela, S.; Szczerba, B.M.; Scherrer, R.; Coleman, N.; Carreira, S.; Bachmann, F.; Anderson, S.; et al. Detection of circulating tumour cell clusters in human glioblastoma. Br. J. Cancer 2018, 119, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Bang-Christensen, S.R.; Pedersen, R.S.; Pereira, M.A.; Clausen, T.M.; Loppke, C.; Sand, N.T.; Ahrens, T.D.; Jorgensen, A.M.; Lim, Y.C.; Goksoyr, L.; et al. Capture and Detection of Circulating Glioma Cells Using the Recombinant VAR2CSA Malaria Protein. Cells 2019, 8, 998. [Google Scholar] [CrossRef] [PubMed]
- Lynch, D.; Powter, B.; Po, J.W.; Cooper, A.; Garrett, C.; Koh, E.-S.; Sheridan, M.; Gelder, J.v.; Darwish, B.; Mckechnie, S. Isolation of Circulating Tumor Cells from Glioblastoma Patients by Direct Immunomagnetic Targeting. Appl. Sci. 2020, 10, 3338. [Google Scholar] [CrossRef]
- Riebensahm, C.; Joosse, S.A.; Mohme, M.; Hanssen, A.; Matschke, J.; Goy, Y.; Witzel, I.; Lamszus, K.; Kropidlowski, J.; Petersen, C.; et al. Clonality of circulating tumor cells in breast cancer brain metastasis patients. Breast Cancer Res. 2019, 21, 101. [Google Scholar] [CrossRef] [PubMed]
- Haber, D.A.; Velculescu, V.E. Blood-based analyses of cancer: Circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014, 4, 650–661. [Google Scholar] [CrossRef] [PubMed]
- Majchrzak-Celińska, A.; Paluszczak, J.; Kleszcz, R.; Magiera, M.; Barciszewska, A.-M.; Nowak, S.; Baer-Dubowska, W. Detection of MGMT, RASSF1A, p15INK4B, and p14ARF promoter methylation in circulating tumor-derived DNA of central nervous system cancer patients. J. Appl. Genet. 2013, 54, 335–344. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra224. [Google Scholar] [CrossRef]
- Schwaederle, M.; Chattopadhyay, R.; Kato, S.; Fanta, P.T.; Banks, K.C.; Choi, I.S.; Piccioni, D.E.; Ikeda, S.; Talasaz, A.; Lanman, R.B.; et al. Genomic Alterations in Circulating Tumor DNA from Diverse Cancer Patients Identified by Next-Generation Sequencing. Cancer Res. 2017, 77, 5419–5427. [Google Scholar] [CrossRef]
- De Mattos-Arruda, L.; Mayor, R.; Ng, C.K.Y.; Weigelt, B.; Martínez-Ricarte, F.; Torrejon, D.; Oliveira, M.; Arias, A.; Raventos, C.; Tang, J.; et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat. Commun. 2015, 6, 8839. [Google Scholar] [CrossRef]
- Miller, A.M.; Shah, R.H.; Pentsova, E.I.; Pourmaleki, M.; Briggs, S.; Distefano, N.; Zheng, Y.; Skakodub, A.; Mehta, S.A.; Campos, C.; et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature 2019, 565, 654–658. [Google Scholar] [CrossRef]
- Ma, C.; Yang, X.; Xing, W.; Yu, H.; Si, T.; Guo, Z. Detection of circulating tumor DNA from non-small cell lung cancer brain metastasis in cerebrospinal fluid samples. Thorac. Cancer 2020. [Google Scholar] [CrossRef]
- Pan, W.; Gu, W.; Nagpal, S.; Gephart, M.H.; Quake, S.R. Brain tumor mutations detected in cerebral spinal fluid. Clin. Chem. 2015, 61, 514–522. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Romero, N.; Carrion-Navarro, J.; Esteban-Rubio, S.; Lazaro-Ibanez, E.; Peris-Celda, M.; Alonso, M.M.; Guzman-De-Villoria, J.; Fernandez-Carballal, C.; de Mendivil, A.O.; Garcia-Duque, S.; et al. DNA sequences within glioma-derived extracellular vesicles can cross the intact blood-brain barrier and be detected in peripheral blood of patients. Oncotarget 2017, 8, 1416–1428. [Google Scholar] [CrossRef] [PubMed]
- Osti, D.; Del Bene, M.; Rappa, G.; Santos, M.; Matafora, V.; Richichi, C.; Faletti, S.; Beznoussenko, G.V.; Mironov, A.; Bachi, A.; et al. Clinical Significance of Extracellular Vesicles in Plasma from Glioblastoma Patients. Clin. Cancer Res. 2019, 25, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Akers, J.C.; Ramakrishnan, V.; Kim, R.; Skog, J.; Nakano, I.; Pingle, S.; Kalinina, J.; Hua, W.; Kesari, S.; Mao, Y.; et al. MiR-21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): A platform for glioblastoma biomarker development. PLoS ONE 2013, 8, e78115. [Google Scholar] [CrossRef]
- Indira Chandran, V.; Welinder, C.; Månsson, A.-S.; Offer, S.; Freyhult, E.; Pernemalm, M.; Lund, S.M.; Pedersen, S.; Lehtiö, J.; Marko-Varga, G.; et al. Ultrasensitive Immunoprofiling of Plasma Extracellular Vesicles Identifies Syndecan-1 as a Potential Tool for Minimally Invasive Diagnosis of Glioma. Clin. Cancer Res. 2019, 25, 3115–3127. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimkhani, S.; Vafaee, F.; Hallal, S.; Wei, H.; Lee, M.Y.T.; Young, P.E.; Satgunaseelan, L.; Beadnall, H.; Barnett, M.H.; Shivalingam, B.; et al. Deep sequencing of circulating exosomal microRNA allows non-invasive glioblastoma diagnosis. Npj Precis. Oncol. 2018, 2, 28. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Chung, J.; Lee, K.; Balaj, L.; Min, C.; Carter, B.S.; Hochberg, F.H.; Breakefield, X.O.; Lee, H.; Weissleder, R. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat. Commun 2015, 6, 6999. [Google Scholar] [CrossRef]
- Zeng, A.; Wei, Z.; Yan, W.; Yin, J.; Huang, X.; Zhou, X.; Li, R.; Shen, F.; Wu, W.; Wang, X.; et al. Exosomal transfer of miR-151a enhances chemosensitivity to temozolomide in drug-resistant glioblastoma. Cancer Lett. 2018, 436, 10–21. [Google Scholar] [CrossRef]
- Lucero, R.; Zappulli, V.; Sammarco, A.; Murillo, O.D.; Cheah, P.S.; Srinivasan, S.; Tai, E.; Ting, D.T.; Wei, Z.; Roth, M.E.; et al. Glioma-Derived miRNA-Containing Extracellular Vesicles Induce Angiogenesis by Reprogramming Brain Endothelial Cells. Cell Rep. 2020, 30, 2065–2074.e4. [Google Scholar] [CrossRef] [PubMed]
| Marker | Clinical Utility in Brain Cancer | Detected in Brain Cancer Circulating Tumor Cells (CTCs) | Detected in Brain Cancer ctNAs | Detected in CTCs or ctNAs # |
|---|---|---|---|---|
| GFAP | Yes [10] | Yes [11] | No | No |
| MGMT * | Yes [12] | No | Yes + [13] | Yes, colorectal cancer [14] |
| IDH1 | Yes [15] | No | Yes + [16] | Yes, leukaemia [17] |
| EGFR ** | Emerging [18] | No | Yes + [19] | Yes, lung cancer [20,21] |
| hTERT | Emerging [22] | Yes [7] | Yes + [23] | Yes, Urothelial cancer [24], Metastatic breast cancer [25] |
| LOH chr10 | Yes [26] | No | Yes + [8] | Yes, Ovarian cancer [27] |
| CTC isolation/ID Method (Reference) | Patient No. | CTC Counts # | Efficiency * | Biomarker Tested | Clinical Utility | Limitations |
|---|---|---|---|---|---|---|
| Lentiviral telomerase reverse transcriptase (TERT)-promoter based detection [7] | 11 | 8.8 (pre-RT) | pre-RT: 72% (8/11) post RT: 8% (1/11) | Epidermal growth factor receptor (EGFR) amplification | Prognostic marker (increased CTC count with recurrence) | Small cohort size, requires viral transduction limited to viable cells, may affect biomarker detection |
| Gradient PBMCs/CTC enrichment, immunocytostainig for glial fibrillary acidic protein (GFAP) [11] | 141 | 0.1–2.2 | 20.6% (29/141) | EGFR amplification | No correlation with OS | GFAP heterogeneous in GBM, use as sole ID marker may under-estimate CTC counts |
| CTC-ichip leucocytes depletion, CTC ID with probing for SOX2, Tubulin b-3, EGFR, A2B5 and c-MET [72] | 33 | 11.8 (progressive disease) 2.1 (stable disease) | 39% (13/33) | N/A | Prognostic marker (progressive disease with greater frequency of CTCs) | Small cohort size |
| FISH CTC detection (chromosome 8 polyploidy) and exclusion of CD45+ cells [69] | 31 | 0.13–1.33 | 71% (24/31) | N/A | CTC count decreases post adjuvant therapy, CTCs may help distinguish radio-necrosis from true tumor progression | Small cohort size |
| Parsortix platform for size based CTC capture, CTC ID testing by EGFR, Ki67, EB1 probing [73] | 13 | 0.3, 1 patient: CTC clusters | 53.8% (7/13) | NGS: APC, XPO1, TFRC, JAK2, BRCA2, ERBB4 and ALK | N/A | Small cohort size |
| VAR2CSA malaria protein based targeted immunomagnetic isolation [74] | 5 (GBM) | 3.5~ | 80% (4/5) | NGS: IDHI, RB1, ALK, LOH 1p/19q, MGMT | N/A | Small cohort size, use of VAR2CSA for both isolation and ID may reduce specificity of CTC detection |
| MCAM, MCSP targeted immunomagnetic isolation, GFAP and GLAST probing [75] | 13 (15 samples) | 1.5 | 60% (9/15) | N/A | No correlation of CTC counts with PFS/OS | Small cohort size |
| ctDNA Source (Reference) | Patients No | Biomarker Tested | Percentage Detection # | Relevance to Disease |
|---|---|---|---|---|
| Plasma ctDNA, CSF ctDNA [81] | 12 | NGS: EGFR, PTEN, ESR1, IDH1, ERBB2, FGFR2 | “higher” sensitivity of mutation detection from CSF ctDNA | Nd |
| Plasma ctDNA, CSF ctDNA [82] | 85 | NGS: TERT, TP53, IDH1, EGFR, and EGFRvIII | 49.4% vs. 15.7% CSF vs. plasma ctDNA | Prognostic: shorter survival of CSF ctDNA positive patients |
| Plasma ctDNA, CSF ctDNA [23] | 38 | pTERT mutation | 92.1% vs. 7.9% CSF vs. plasma ctDNA | pTERT mutation potential poor survival predictor |
| Plasma ctDNA, CSF ctDNA * [83] | 21 | NGS: EGFR, KIT, PIK3CA, TP53, SMAD4, ATM, SMARCB1, PTEN | 95.2% vs. 66.7% CSF vs. plasma ctDNA | Nd |
| Plasma ctDNA, CSF ctDNA [84] | 7 | NGS: NF2, AKT, BRAF, NRAS, EGFR | CSF ctDNA detection “significantly higher” with low systemic disease burden | Nd |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sareen, H.; Garrett, C.; Lynch, D.; Powter, B.; Brungs, D.; Cooper, A.; Po, J.; Koh, E.-S.; Vessey, J.Y.; McKechnie, S.; et al. The Role of Liquid Biopsies in Detecting Molecular Tumor Biomarkers in Brain Cancer Patients. Cancers 2020, 12, 1831. https://doi.org/10.3390/cancers12071831
Sareen H, Garrett C, Lynch D, Powter B, Brungs D, Cooper A, Po J, Koh E-S, Vessey JY, McKechnie S, et al. The Role of Liquid Biopsies in Detecting Molecular Tumor Biomarkers in Brain Cancer Patients. Cancers. 2020; 12(7):1831. https://doi.org/10.3390/cancers12071831
Chicago/Turabian StyleSareen, Heena, Celine Garrett, David Lynch, Branka Powter, Daniel Brungs, Adam Cooper, Joseph Po, Eng-Siew Koh, Joey Yusof Vessey, Simon McKechnie, and et al. 2020. "The Role of Liquid Biopsies in Detecting Molecular Tumor Biomarkers in Brain Cancer Patients" Cancers 12, no. 7: 1831. https://doi.org/10.3390/cancers12071831
APA StyleSareen, H., Garrett, C., Lynch, D., Powter, B., Brungs, D., Cooper, A., Po, J., Koh, E.-S., Vessey, J. Y., McKechnie, S., Bazina, R., Sheridan, M., Gelder, J. v., Darwish, B., Jaeger, M., Roberts, T. L., De Souza, P., & Becker, T. M. (2020). The Role of Liquid Biopsies in Detecting Molecular Tumor Biomarkers in Brain Cancer Patients. Cancers, 12(7), 1831. https://doi.org/10.3390/cancers12071831






