Exosomal MicroRNAs and Organotropism in Breast Cancer Metastasis
Abstract
:1. Introduction
2. Exosomes, MicroRNAs, and Packaging
2.1. Exosome Biogenesis
2.2. MicroRNA Biogenesis
2.3. Mechanisms of Cargo Sorting into Exosomes
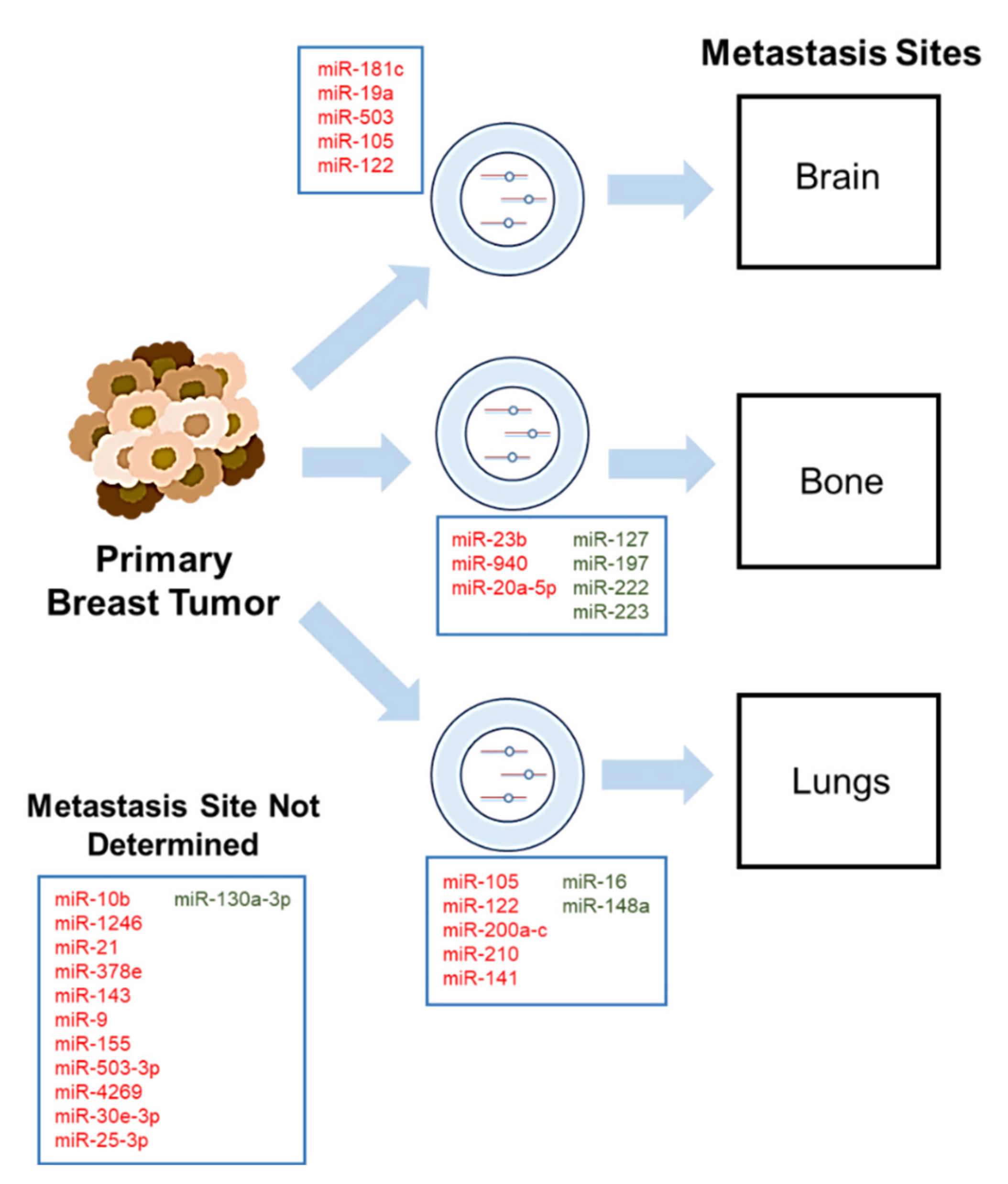
3. Roles of Exosomal MicroRNAs in Breast Cancer Metastasis
3.1. Exosomal MicroRNAs in Breast Cancer Brain Metastasis
3.2. Exosomal MicroRNAs in Breast Cancer Bone Metastasis
3.3. Exosomal MicroRNAs in Breast Cancer Lung Metastasis
3.4. Exosomal MicroRNAs in Breast Cancer Migration, Invasion and Stemness
4. Exosomal MicroRNAs as Cancer Therapeutics
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. ACS J. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast cancer. Nat. Rev. Dis. Primers 2019, 5, 66. [Google Scholar] [CrossRef]
- Wu, Q.; Li, J.; Zhu, S.; Wu, J.; Chen, C.; Liu, Q.; Wei, W.; Zhang, Y.; Sun, S. Breast cancer subtypes predict the preferential site of distant metastases: A SEER based study. Oncotarget 2017, 8, 27990–27996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheang, M.C.; Martin, M.; Nielsen, T.O.; Prat, A.; Voduc, D.; Rodriguez-Lescure, A.; Ruiz, A.; Chia, S.; Shepherd, L.; Ruiz-Borrego, M.; et al. Defining breast cancer intrinsic subtypes by quantitative receptor expression. Oncologist 2015, 20, 474–482. [Google Scholar] [CrossRef] [Green Version]
- Van Mechelen, M.; Van Herck, A.; Punie, K.; Nevelsteen, I.; Smeets, A.; Neven, P.; Weltens, C.; Han, S.; Vanderstichele, A.; Floris, G.; et al. Behavior of metastatic breast cancer according to subtype. Breast Cancer Res. Treat. 2020, 181, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Niikura, N.; Hayashi, N.; Masuda, N.; Takashima, S.; Nakamura, R.; Watanabe, K.; Kanbayashi, C.; Ishida, M.; Hozumi, Y.; Tsuneizumi, M.; et al. Treatment outcomes and prognostic factors for patients with brain metastases from breast cancer of each subtype: A multicenter retrospective analysis. Breast Cancer Res. Treat. 2014, 147, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Waks, A.G.; Winer, E.P. Breast Cancer Treatment: A Review. JAMA 2019, 321, 288–300. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Gao, S.L. Management of breast cancer brain metastases: Focus on human epidermal growth factor receptor 2-positive breast cancer. Chronic. Dis. Transl. Med. 2017, 3, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.V.; Heuser, J.E.; Stahl, P.D. Exosomes: Looking back three decades and into the future. J. Cell Biol. 2013, 200, 367–371. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Shen, M.; Yan, M.; Cui, Y.; Gao, Z.; Meng, X. Exosome-mediated transfer of miR-1290 promotes cell proliferation and invasion in gastric cancer via NKD1. Acta Biochim. Biophys. Sin. 2019, 51, 900–907. [Google Scholar] [CrossRef]
- Osaki, M.; Okada, F. Exosomes and Their Role in Cancer Progression. Yonago Acta Med. 2019, 62, 182–190. [Google Scholar] [CrossRef] [Green Version]
- Peak, T.C.; Praharaj, P.P.; Panigrahi, G.K.; Doyle, M.; Su, Y.; Schlaepfer, I.R.; Singh, R.; Vander Griend, D.J.; Alickson, J.; Hemal, A.; et al. Exosomes secreted by placental stem cells selectively inhibit growth of aggressive prostate cancer cells. Biochem. Biophys. Res. Commun. 2018, 499, 1004–1010. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, S.; Yao, J.; Lowery, F.J.; Zhang, Q.; Huang, W.-C.; Li, P.; Li, M.; Wang, X.; Zhang, C.; et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 2015, 527, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yu, F.; Ding, H.; Wang, Y.; Li, P.; Wang, K. Emerging Function and Clinical Values of Exosomal MicroRNAs in Cancer. Mol. Ther. Nucleic Acids 2019, 16, 791–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kia, V.; Mortazavi, Y.; Paryan, M.; Biglari, A.; Mohammadi-Yeganeh, S. Exosomal miRNAs from highly metastatic cells can induce metastasis in non-metastatic cells. Life Sci. 2019, 220, 162–168. [Google Scholar] [CrossRef]
- Rodrigues, G.; Hoshino, A.; Kenific, C.M.; Matei, I.R.; Steiner, L.; Freitas, D.; Kim, H.S.; Oxley, P.R.; Scandariato, I.; Casanova-Salas, I.; et al. Tumour exosomal CEMIP protein promotes cancer cell colonization in brain metastasis. Nat. Cell Biol. 2019, 21, 1403–1412. [Google Scholar] [CrossRef]
- O‘Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Youngman, E.M.; Claycomb, J.M. From early lessons to new frontiers: The worm as a treasure trove of small RNA biology. Front. Genet. 2014, 5, 416. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Ambros, V. microRNAs: Tiny regulators with great potential. Cell 2001, 107, 823–826. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.C.; Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001, 294, 862–864. [Google Scholar] [CrossRef] [Green Version]
- Nama, S.; Muhuri, M.; Di Pascale, F.; Quah, S.; Aswad, L.; Fullwood, M.; Sampath, P. MicroRNA-138 is a Prognostic Biomarker for Triple-Negative Breast Cancer and Promotes Tumorigenesis via TUSC2 repression. Sci. Rep. 2019, 9, 12718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirjang, S.; Mansoori, B.; Asghari, S.; Duijf, P.H.G.; Mohammadi, A.; Gjerstorff, M.; Baradaran, B. MicroRNAs in cancer cell death pathways: Apoptosis and necroptosis. Free Radic. Biol. Med. 2019, 139, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kogure, A.; Kosaka, N.; Ochiya, T. Cross-talk between cancer cells and their neighbors via miRNA in extracellular vesicles: An emerging player in cancer metastasis. J. Biomed. Sci. 2019, 26, 7. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Pochampally, R.; Watabe, K.; Lu, Z.; Mo, Y.Y. Exosome-mediated transfer of miR-10b promotes cell invasion in breast cancer. Mol. Cancer 2014, 13, 256. [Google Scholar] [CrossRef] [Green Version]
- Harding, C.; Heuser, J.; Stahl, P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 1983, 97, 329–339. [Google Scholar] [CrossRef]
- Pan, B.-T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Ruivo, C.F.; Adem, B.; Silva, M.; Melo, S.A. The Biology of Cancer Exosomes: Insights and New Perspectives. Cancer Res. 2017, 77, 6480–6488. [Google Scholar] [CrossRef] [Green Version]
- Henne, W.M.; Buchkovich, N.J.; Emr, S.D. The ESCRT pathway. Dev. Cell 2011, 21, 77–91. [Google Scholar] [CrossRef] [Green Version]
- Wollert, T.; Wunder, C.; Lippincott-Schwartz, J.; Hurley, J.H. Membrane scission by the ESCRT-III complex. Nature 2009, 458, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Schöneberg, J.; Pavlin, M.R.; Yan, S.; Righini, M.; Lee, I.-H.; Carlson, L.-A.; Bahrami, A.H.; Goldman, D.H.; Ren, X.; Hummer, G.; et al. ATP-dependent force generation and membrane scission by ESCRT-III and Vps4. Science 2018, 362, 1423–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wollert, T.; Hurley, J.H. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 2010, 464, 864–869. [Google Scholar] [CrossRef] [Green Version]
- Guo, E.Z.; Xu, Z. Distinct mechanisms of recognizing endosomal sorting complex required for transport III (ESCRT-III) protein IST1 by different microtubule interacting and trafficking (MIT) domains. J. Biol. Chem. 2015, 290, 8396–8408. [Google Scholar] [CrossRef] [Green Version]
- Odorizzi, G. The multiple personalities of Alix. J. Cell Sci. 2006, 119, 3025–3032. [Google Scholar] [CrossRef] [Green Version]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef]
- Bobrie, A.; Krumeich, S.; Reyal, F.; Recchi, C.; Moita, L.F.; Seabra, M.C.; Ostrowski, M.; Thery, C. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 2012, 72, 4920–4930. [Google Scholar] [CrossRef] [Green Version]
- Temoche-Diaz, M.M.; Shurtleff, M.J.; Nottingham, R.M.; Yao, J.; Fadadu, R.P.; Lambowitz, A.M.; Schekman, R. Distinct mechanisms of microRNA sorting into cancer cell-derived extracellular vesicle subtypes. eLife 2019, 8. [Google Scholar] [CrossRef]
- Sudhof, T.C.; Rothman, J.E. Membrane fusion: Grappling with SNARE and SM proteins. Science 2009, 323, 474–477. [Google Scholar] [CrossRef] [Green Version]
- Montgomery, M.K. RNA interference: Unraveling a mystery. Nat. Struct. Mol. Biol. 2006, 13, 1039–1041. [Google Scholar] [CrossRef] [PubMed]
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Yao, F.; Xiao, Z.; Sun, Y.; Ma, L. MicroRNAs and metastasis: Small RNAs play big roles. Cancer Metastasis Rev. 2018, 37, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An Abundant Class of Tiny RNAs with Probable Regulatory Roles in Caenorhabditis elegans. Science 2001, 294, 858–862. [Google Scholar] [CrossRef] [Green Version]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Carmell, M.A.; Rivas, F.V.; Marsden, C.G.; Thomson, J.M.; Song, J.-J.; Hammond, S.M.; Joshua-Tor, L.; Hannon, G.J. Argonaute2 Is the Catalytic Engine of Mammalian RNAi. Science 2004, 305, 1437–1441. [Google Scholar] [CrossRef] [Green Version]
- Swarts, D.C.; Makarova, K.; Wang, Y.; Nakanishi, K.; Ketting, R.F.; Koonin, E.V.; Patel, D.J.; van der Oost, J. The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol. 2014, 21, 743–753. [Google Scholar] [CrossRef]
- Nakanishi, K.; Weinberg, D.E.; Bartel, D.P.; Patel, D.J. Structure of yeast Argonaute with guide RNA. Nature 2012, 486, 368–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Sheng, G.; Juranek, S.; Tuschl, T.; Patel, D.J. Structure of the guide-strand-containing argonaute silencing complex. Nature 2008, 456, 209–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, B.D.; Claffey, K.P.; White, B.A. Argonaute-2 expression is regulated by epidermal growth factor receptor and mitogen-activated protein kinase signaling and correlates with a transformed phenotype in breast cancer cells. Endocrinology 2009, 150, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef]
- Gregory, R.I.; Yan, K.P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science 2009, 326, 1275–1279. [Google Scholar] [CrossRef]
- Lau, P.W.; Guiley, K.Z.; De, N.; Potter, C.S.; Carragher, B.; MacRae, I.J. The molecular architecture of human Dicer. Nat. Struct. Mol. Biol. 2012, 19, 436–440. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single processing center models for human Dicer and bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef] [Green Version]
- Bernstein, E.; Kim, S.Y.; Carmell, M.A.; Murchison, E.P.; Alcorn, H.; Li, M.Z.; Mills, A.A.; Elledge, S.J.; Anderson, K.V.; Hannon, G.J. Dicer is essential for mouse development. Nat. Genet. 2003, 35, 215–217. [Google Scholar] [CrossRef]
- Chong, M.M.; Zhang, G.; Cheloufi, S.; Neubert, T.A.; Hannon, G.J.; Littman, D.R. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010, 24, 1951–1960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaneda, M.; Tang, F.; O‘Carroll, D.; Lao, K.; Surani, M.A. Essential role for Argonaute2 protein in mouse oogenesis. Epigenetics Chromatin 2009, 2, 9. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.J.; Yang, D.D.; Na, S.; Sandusky, G.E.; Zhang, Q.; Zhao, G. Dicer is required for embryonic angiogenesis during mouse development. J. Biol. Chem. 2005, 280, 9330–9335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Telonis, A.G.; Magee, R.; Loher, P.; Chervoneva, I.; Londin, E.; Rigoutsos, I. Knowledge about the presence or absence of miRNA isoforms (isomiRs) can successfully discriminate amongst 32 TCGA cancer types. Nucleic Acids Res. 2017, 45, 2973–2985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tosar, J.P.; Gambaro, F.; Sanguinetti, J.; Bonilla, B.; Witwer, K.W.; Cayota, A. Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines. Nucleic Acids Res. 2015, 43, 5601–5616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Perez-Hernandez, D.; Vazquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sanchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [Green Version]
- Shurtleff, M.J.; Temoche-Diaz, M.M.; Karfilis, K.V.; Ri, S.; Schekman, R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife 2016, 5. [Google Scholar] [CrossRef]
- Santangelo, L.; Giurato, G.; Cicchini, C.; Montaldo, C.; Mancone, C.; Tarallo, R.; Battistelli, C.; Alonzi, T.; Weisz, A.; Tripodi, M. The RNA-Binding Protein SYNCRIP Is a Component of the Hepatocyte Exosomal Machinery Controlling MicroRNA Sorting. Cell Rep. 2016, 17, 799–808. [Google Scholar] [CrossRef] [Green Version]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Koppers-Lalic, D.; Hackenberg, M.; Bijnsdorp, I.V.; van Eijndhoven, M.A.J.; Sadek, P.; Sie, D.; Zini, N.; Middeldorp, J.M.; Ylstra, B.; de Menezes, R.X.; et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014, 8, 1649–1658. [Google Scholar] [CrossRef] [Green Version]
- Diederichs, S.; Haber, D.A. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 2007, 131, 1097–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cocucci, E.; Racchetti, G.; Meldolesi, J. Shedding microvesicles: Artefacts no more. Trends Cell Biol. 2009, 19, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Harris, D.A.; Patel, S.H.; Gucek, M.; Hendrix, A.; Westbroek, W.; Taraska, J.W. Exosomes released from breast cancer carcinomas stimulate cell movement. PLoS ONE 2015, 10, e0117495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Nedawi, K.; Meehan, B.; Micallef, J.; Lhotak, V.; May, L.; Guha, A.; Rak, J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008, 10, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Donnarumma, E.; Fiore, D.; Nappa, M.; Roscigno, G.; Adamo, A.; Iaboni, M.; Russo, V.; Affinito, A.; Puoti, I.; Quintavalle, C.; et al. Cancer-associated fibroblasts release exosomal microRNAs that dictate an aggressive phenotype in breast cancer. Oncotarget 2017, 8, 19592–19608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, K.; Chen, Y.; Li, A.; Tan, C.; Liu, X. Exosomes play roles in sequential processes of tumor metastasis. Int. J. Cancer 2019, 144, 1486–1495. [Google Scholar] [CrossRef]
- Wang, J.; Wu, Y.; Guo, J.; Fei, X.; Yu, L.; Ma, S. Adipocyte-derived exosomes promote lung cancer metastasis by increasing MMP9 activity via transferring MMP3 to lung cancer cells. Oncotarget 2017, 8, 81880–81891. [Google Scholar] [CrossRef]
- Hanna, J.; Hossain, G.S.; Kocerha, J. The Potential for microRNA Therapeutics and Clinical Research. Front. Genet. 2019, 10, 478. [Google Scholar] [CrossRef] [Green Version]
- Loh, H.Y.; Norman, B.P.; Lai, K.S.; Rahman, N.; Alitheen, N.B.M.; Osman, M.A. The Regulatory Role of MicroRNAs in Breast Cancer. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Qiu, C.; Zhang, H.; Wang, J.; Cui, Q.; Yin, Y. Human microRNA oncogenes and tumor suppressors show significantly different biological patterns: From functions to targets. PLoS ONE 2010, 5. [Google Scholar] [CrossRef]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Agostini, M.; Ganini, C.; Candi, E.; Melino, G. The role of noncoding RNAs in epithelial cancer. Cell Death Discov. 2020, 6, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicoloso, M.S.; Spizzo, R.; Shimizu, M.; Rossi, S.; Calin, G.A. MicroRNAs—The micro steering wheel of tumour metastases. Nat. Rev. Cancer 2009, 9, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Yang, P.; Wang, X.F. Microenvironmental regulation of cancer metastasis by miRNAs. Trends Cell Biol. 2014, 24, 153–160. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Huang, L.F.; Zhao, L.; Zeng, Z.; Wang, X.; Cao, D.; Yang, L.; Ye, Z.; Chen, X.; Liu, B.; et al. Microvesicles (MIVs) secreted from adipose-derived stem cells (ADSCs) contain multiple microRNAs and promote the migration and invasion of endothelial cells. Genes Dis. 2020, 7, 225–234. [Google Scholar] [CrossRef]
- De Palma, M.; Biziato, D.; Petrova, T.V. Microenvironmental regulation of tumour angiogenesis. Nat. Rev. Cancer 2017, 17, 457–474. [Google Scholar] [CrossRef]
- Langley, R.R.; Fidler, I.J. The seed and soil hypothesis revisited--the role of tumor-stroma interactions in metastasis to different organs. Int. J. Cancer 2011, 128, 2527–2535. [Google Scholar] [CrossRef] [Green Version]
- Paget, S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 1989, 8, 98–101. [Google Scholar]
- Le, M.T.; Hamar, P.; Guo, C.; Basar, E.; Perdigao-Henriques, R.; Balaj, L.; Lieberman, J. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J. Clin. Investig. 2014, 124, 5109–5128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Peng, F.; Chen, J. The Role of Exosomal MicroRNAs in the Tumor Microenvironment of Breast Cancer. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Bado, I.; Wang, H.; Zhang, W.; Rosen, J.M.; Zhang, X.H.F. Metastasis Organotropism: Redefining the Congenial Soil. Dev. Cell 2019, 49, 375–391. [Google Scholar] [CrossRef]
- Thulin, A.; Ronnerman, E.; Zhang, C.; De Lara, S.; Chamalidou, C.; Schoenfeldt, A.; Andersson, C.; Kovacs, A.; Enlund, F.; Linderholm, B. Clinical outcome of patients with brain metastases from breast cancer—A population based study over 21 years. Breast 2020, 50, 113–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Younis, M.H.; Fuentes-Rivera, L.; Summers, S.; Pretell-Mazzini, J. Survival in patients with carcinomas presenting with bone metastasis at diagnosis: A SEER population-based cohort study. Arch. Orthop Trauma Surg. 2020. [Google Scholar] [CrossRef] [PubMed]
- Ekici, K.; Temelli, O.; Dikilitas, M.; Halil Dursun, I.; Bozdag Kaplan, N.; Kekilli, E. Survival and prognostic factors in patients with brain metastasis: Single center experience. J. BUON 2016, 21, 958–963. [Google Scholar] [PubMed]
- Suh, J.H.; Kotecha, R.; Chao, S.T.; Ahluwalia, M.S.; Sahgal, A.; Chang, E.L. Current approaches to the management of brain metastases. Nat. Rev. Clin. Oncol. 2020, 17, 279–299. [Google Scholar] [CrossRef]
- Camacho, L.; Guerrero, P.; Marchetti, D. MicroRNA and protein profiling of brain metastasis competent cell-derived exosomes. PLoS ONE 2013, 8, e73790. [Google Scholar] [CrossRef] [Green Version]
- Lo, H.W.; Zhu, H.; Cao, X.; Aldrich, A.; Ali-Osman, F. A novel splice variant of GLI1 that promotes glioblastoma cell migration and invasion. Cancer Res. 2009, 69, 6790–6798. [Google Scholar] [CrossRef] [Green Version]
- Sirkisoon, S.R.; Carpenter, R.L.; Rimkus, T.; Doheny, D.; Zhu, D.; Aguayo, N.R.; Xing, F.; Chan, M.; Ruiz, J.; Metheny-Barlow, L.J.; et al. TGLI1 transcription factor mediates breast cancer brain metastasis via activating metastasis-initiating cancer stem cells and astrocytes in the tumor microenvironment. Oncogene 2020, 39, 64–78. [Google Scholar] [CrossRef]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.; Chin, A.R.; et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arvanitis, C.D.; Ferraro, G.B.; Jain, R.K. The blood-brain barrier and blood-tumour barrier in brain tumours and metastases. Nat. Rev. Cancer 2020, 20, 26–41. [Google Scholar] [CrossRef] [PubMed]
- O’Brown, N.M.; Pfau, S.J.; Gu, C. Bridging barriers: A comparative look at the blood-brain barrier across organisms. Genes Dev. 2018, 32, 466–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, T.H.; Avraham, H.K.; Jiang, S.; Avraham, S. Vascular endothelial growth factor modulates the transendothelial migration of MDA-MB-231 breast cancer cells through regulation of brain microvascular endothelial cell permeability. J. Biol. Chem. 2003, 278, 5277–5284. [Google Scholar] [CrossRef] [Green Version]
- Morad, G.; Carman, C.V.; Hagedorn, E.J.; Perlin, J.R.; Zon, L.I.; Mustafaoglu, N.; Park, T.E.; Ingber, D.E.; Daisy, C.C.; Moses, M.A. Tumor-Derived Extracellular Vesicles Breach the Intact Blood-Brain Barrier via Transcytosis. ACS Nano 2019, 13, 13853–13865. [Google Scholar] [CrossRef]
- Tominaga, N.; Kosaka, N.; Ono, M.; Katsuda, T.; Yoshioka, Y.; Tamura, K.; Lotvall, J.; Nakagama, H.; Ochiya, T. Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood-brain barrier. Nat. Commun. 2015, 6, 6716. [Google Scholar] [CrossRef] [Green Version]
- Xing, F.; Sharma, S.; Liu, Y.; Mo, Y.Y.; Wu, K.; Zhang, Y.Y.; Pochampally, R.; Martinez, L.A.; Lo, H.W.; Watabe, K. miR-509 suppresses brain metastasis of breast cancer cells by modulating RhoC and TNF-alpha. Oncogene 2015, 34, 4890–4900. [Google Scholar] [CrossRef] [Green Version]
- Xing, F.; Liu, Y.; Wu, S.Y.; Wu, K.; Sharma, S.; Mo, Y.Y.; Feng, J.; Sanders, S.; Jin, G.; Singh, R.; et al. Loss of XIST in Breast Cancer Activates MSN-c-Met and Reprograms Microglia via Exosomal miRNA to Promote Brain Metastasis. Cancer Res. 2018, 78, 4316–4330. [Google Scholar] [CrossRef] [Green Version]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.; Li, S.; Chin, A.R.; et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef] [Green Version]
- Lim, P.K.; Bliss, S.A.; Patel, S.A.; Taborga, M.; Dave, M.A.; Gregory, L.A.; Greco, S.J.; Bryan, M.; Patel, P.S.; Rameshwar, P. Gap junction-mediated import of microRNA from bone marrow stromal cells can elicit cell cycle quiescence in breast cancer cells. Cancer Res. 2011, 71, 1550–1560. [Google Scholar] [CrossRef] [Green Version]
- Buonomo, O.C.; Caredda, E.; Portarena, I.; Vanni, G.; Orlandi, A.; Bagni, C.; Petrella, G.; Palombi, L.; Orsaria, P. New insights into the metastatic behavior after breast cancer surgery, according to well-established clinicopathological variables and molecular subtypes. PLoS ONE 2017, 12, e0184680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ono, M.; Kosaka, N.; Tominaga, N.; Yoshioka, Y.; Takeshita, F.; Takahashi, R.U.; Yoshida, M.; Tsuda, H.; Tamura, K.; Ochiya, T. Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Sci. Signal. 2014, 7, ra63. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Ochi, H.; Sunamura, S.; Kosaka, N.; Mabuchi, Y.; Fukuda, T.; Yao, K.; Kanda, H.; Ae, K.; Okawa, A.; et al. Cancer-secreted hsa-miR-940 induces an osteoblastic phenotype in the bone metastatic microenvironment via targeting ARHGAP1 and FAM134A. Proc. Natl. Acad. Sci. USA 2018, 115, 2204–2209. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Zhu, Y.; Li, L.; Zhou, S.; Yin, G.; Yu, G.; Cui, H. Breast cancer cell-derived exosomal miR-20a-5p promotes the proliferation and differentiation of osteoclasts by targeting SRCIN1. Cancer Med. 2019, 8, 5687–5701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, L.; Han, B.; Siegel, E.; Cui, Y.; Giuliano, A.; Cui, X. Breast cancer lung metastasis: Molecular biology and therapeutic implications. Cancer Biol. Ther. 2018, 19, 858–868. [Google Scholar] [CrossRef] [Green Version]
- Gennari, A.; Conte, P.; Rosso, R.; Orlandini, C.; Bruzzi, P. Survival of metastatic breast carcinoma patients over a 20-year period: A retrospective analysis based on individual patient data from six consecutive studies. Cancer 2005, 104, 1742–1750. [Google Scholar] [CrossRef]
- Korpal, M.; Ell, B.J.; Buffa, F.M.; Ibrahim, T.; Blanco, M.A.; Celia-Terrassa, T.; Mercatali, L.; Khan, Z.; Goodarzi, H.; Hua, Y.; et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat. Med. 2011, 17, 1101–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosaka, N.; Iguchi, H.; Hagiwara, K.; Yoshioka, Y.; Takeshita, F.; Ochiya, T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 2013, 288, 10849–10859. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Zhang, W.; Li, B.; Stringer-Reasor, E.; Chu, C.; Sun, L.; Bae, S.; Chen, D.; Wei, S.; Jiao, K.; et al. MicroRNA-200c and microRNA-141 are regulated by a FOXP3-KAT2B axis and associated with tumor metastasis in breast cancer. Breast Cancer Res. 2017, 19, 73. [Google Scholar] [CrossRef]
- Wu, H.J.; Hao, M.; Yeo, S.K.; Guan, J.L. FAK signaling in cancer-associated fibroblasts promotes breast cancer cell migration and metastasis by exosomal miRNAs-mediated intercellular communication. Oncogene 2020, 39, 2539–2549. [Google Scholar] [CrossRef]
- Li, X.J.; Ren, Z.J.; Tang, J.H.; Yu, Q. Exosomal MicroRNA MiR-1246 Promotes Cell Proliferation, Invasion and Drug Resistance by Targeting CCNG2 in Breast Cancer. Cell Physiol. Biochem. 2017, 44, 1741–1748. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; He, F.; Yang, Z.; Cao, X.; Dai, S.; Zou, J.; Xu, P.; Zhou, Z. Exosomal miR-25-3p derived from hypoxia tumor mediates IL-6 secretion and stimulates cell viability and migration in breast cancer. RSC Adv. 2019, 9, 1451–1459. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Mao, J.H.; Wang, B.Y.; Wang, L.X.; Wen, H.Y.; Xu, L.J.; Fu, J.X.; Yang, H. Exosomal miR-1910-3p promotes proliferation, metastasis, and autophagy of breast cancer cells by targeting MTMR3 and activating the NF-κB signaling pathway. Cancer Lett. 2020. [Google Scholar] [CrossRef]
- Kim, K.S.; Park, J.I.; Oh, N.; Cho, H.J.; Park, J.H.; Park, K.S. ELK3 expressed in lymphatic endothelial cells promotes breast cancer progression and metastasis through exosomal miRNAs. Sci. Rep. 2019, 9, 8418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, X.; Zhang, J.; Li, J.; Shao, J.; Fang, L. MiR-130a-3p inhibits migration and invasion by regulating RAB5B in human breast cancer stem cell-like cells. Biochem. Biophys. Res. Commun. 2018, 501, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Kia, V.; Paryan, M.; Mortazavi, Y.; Biglari, A.; Mohammadi-Yeganeh, S. Evaluation of exosomal miR-9 and miR-155 targeting PTEN and DUSP14 in highly metastatic breast cancer and their effect on low metastatic cells. J. Cell. Biochem. 2019, 120, 5666–5676. [Google Scholar] [CrossRef]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Doss, C.G.P.; Lee, S.S. Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine. Mol. Ther. Nucleic Acids 2017, 8, 132–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouchie, A. First microRNA mimic enters clinic. Nat. Biotechnol. 2013, 31, 577. [Google Scholar] [CrossRef] [PubMed]
- Pondé, N.; Aftimos, P.; Piccart, M. Antibody-Drug Conjugates in Breast Cancer: A Comprehensive Review. Curr. Treat. Options Oncol. 2019, 20, 37. [Google Scholar] [CrossRef]
- Wind, N.S.; Holen, I. Multidrug Resistance in Breast Cancer: From In Vitro Models to Clinical Studies. Int. J. Breast Cancer 2011, 2011, 967419. [Google Scholar] [CrossRef]
- Del Re, M.; Bertolini, I.; Crucitta, S.; Fontanelli, L.; Rofi, E.; De Angelis, C.; Diodati, L.; Cavallero, D.; Gianfilippo, G.; Salvadori, B.; et al. Overexpression of TK1 and CDK9 in plasma-derived exosomes is associated with clinical resistance to CDK4/6 inhibitors in metastatic breast cancer patients. Breast Cancer Res. Treat. 2019, 178, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Hu, J.; Lu, P.; Cao, H.; Yu, C.; Li, X.; Qian, X.; Yang, X.; Yang, Y.; Han, N.; et al. Exosome-transmitted miR-567 reverses trastuzumab resistance by inhibiting ATG5 in breast cancer. Cell Death Dis. 2020, 11, 43. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Li, M.; Cui, S.; Wang, D.; Zhang, C.Y.; Zen, K.; Li, L. Shikonin Inhibits the Proliferation of Human Breast Cancer Cells by Reducing Tumor-Derived Exosomes. Molecules 2016, 21. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, Y.; Ye, M.; Wu, J.; Ma, L.; Chen, H. Cisplatin-resistant MDA-MB-231 Cell-derived Exosomes Increase the Resistance of Recipient Cells in an Exosomal miR-423-5p-dependent Manner. Curr. Drug Metab. 2019, 20, 804–814. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.D.; Wu, Y.; Zhang, X.H.; Lv, M.M.; Chen, W.X.; Chen, X.; Yang, S.J.; Shen, H.; Zhong, S.L.; Tang, J.H.; et al. Exosomes from adriamycin-resistant breast cancer cells transmit drug resistance partly by delivering miR-222. Tumour Biol. 2016, 37, 3227–3235. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Lai, X.; Yu, S.; Chen, S.; Ma, Y.; Zhang, Y.; Li, H.; Zhu, X.; Yao, L.; Zhang, J. Exosomal miR-221/222 enhances tamoxifen resistance in recipient ER-positive breast cancer cells. Breast Cancer Res. Treat. 2014, 147, 423–431. [Google Scholar] [CrossRef]
- O‘Brien, K.; Lowry, M.C.; Corcoran, C.; Martinez, V.G.; Daly, M.; Rani, S.; Gallagher, W.M.; Radomski, M.W.; MacLeod, R.A.; O’Driscoll, L. miR-134 in extracellular vesicles reduces triple-negative breast cancer aggression and increases drug sensitivity. Oncotarget 2015, 6, 32774–32789. [Google Scholar] [CrossRef] [Green Version]
- Bovy, N.; Blomme, B.; Frères, P.; Dederen, S.; Nivelles, O.; Lion, M.; Carnet, O.; Martial, J.A.; Noël, A.; Thiry, M.J.O. Endothelial exosomes contribute to the antitumor response during breast cancer neoadjuvant chemotherapy via microRNA transfer. Oncotarget 2015, 6, 10253–10266. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Liang, Y.; Sang, Y.; Song, X.; Zhang, H.; Liu, Y.; Jiang, L.; Yang, Q. MiR-770 suppresses the chemo-resistance and metastasis of triple negative breast cancer via direct targeting of STMN1. Cell Death Dis. 2018, 9, 14. [Google Scholar] [CrossRef]
| Exosome-Secreted miRNAs | Activity | Validated Targets | In Vitro Model(s) | Additional Model(s) | Metastatic Site(s) | Reference |
|---|---|---|---|---|---|---|
| miR-9 | Enriched in TNBC, suppresses PTEN (tumor suppressor) | PTEN* | Human BC cell lines: MDA-MB-231, MCF-7, MCF-10A | N/A | ND | [126] |
| miR-10b | Promotes invasion | HOXD10*, KLF4 | Human BC cell lines: MDA-MB-231, MCF-7, MCF-10A, HMLE | N/A | ND | [26] |
| miR-16 | Tumor suppressive activity (in FAK null-CAFs) | N/A | Human BC cell lines: MDA-MB-231, MCF-7, WI-38 | In vivo (transgenic mice) | Lungs | [120] |
| miR-19a | Increases tumor growth and brain metastasis formation | PTEN* | Human BC cell lines: MDA-MB-231, HCC1954, BT474, MDA-MB-435; mouse cell lines: 4T1, B16BL6 | In vivo (intracarotid, intracranial models) | Brain | [13] |
| miR-20a-5p | Promotes proliferation and migration of BC cells; facilitates osteoclastogenesis | SRCIN1* | Human BC cell lines: MDA-MB-231, MCF-7, MCF-10A; BMMs | Clinical samples | Bone | [114] |
| miR-21 | Increased in CAF-derived exosomes; promotes stemness, EMT, and anchorage-independence | N/A | Human BC cell lines: BT549, MDA-MB-231, T47D | Clinical samples (TCGA) | ND | [75] |
| miR-23b | Promotes dormancy phenotype | MARCKS* | Human BC and BM-MSCs: MDA-MB-231, R14, R36, R37, 4F0218 | In vivo (MFP injections), clinical samples | Bone | [112] |
| miR-25-3p | Promotes proliferation and migration of BC cells in a HIF-1-dependent manner | N/A | Human BC cell line: MDA-MB-231; mouse cell lines: E0771, RAW264.7 (macrophage cell line) | In vivo (MFP injections) | ND | [122] |
| miR-30e-3p | Regulated by ELK3; promote migration and invasion | N/A | Human BC cell lines: MDA-MB-231, Hs-578T, BT-20, MCF-7, MCF-10A; LECs, HUVEC | In vivo (orthotopic inoculation), databases | ND | [124] |
| miR-105 | Promotes metastasis by targeting vascular endothelial barriers | ZO-1 | Human BC cell lines: MDA-MB-231, MCF-10A; HMVECs | In vivo (MFP injection, tumor xenograft model) | Lungs, Brain | [101] |
| miR-122 | Suppress glucose uptake in non-tumorigenic cells to promote metastasis | PKM* | Human BC cell lines: MDA-MB-231, MCF-10A | In vivo (tail vein and intracardiac injections) | Lungs, Brain | [109] |
| miR-127 | Tumor suppressive activity: bone marrow mets transmit miRNAs to BC cells via gap junctions and exosomes | CXCL12* | Human BC cell lines: MDA-MB-231, T47D | N/A | Bone | [110] |
| miR-130a-3p | Tumor suppressive activity: decreased levels associated with lymph node metastasis, O/E inhibited cell proliferation, migration, invasion | RAB5B* | Human BC cell lines: MDA-MB-231, MCF-7, MCF-10A, BCSCs | N/A | ND | [125] |
| miR-141 | Regulates MET (suppresses EMT), which promotes lung metastasis and colonization | SEC23A*, ZEB2, CDH1 | Human BC cell lines: MCF-10A, MDA-MB-231; mouse cell lines: 4T1, 4T07, 67NR | In vivo (tail vein injection) | Lungs | [91] |
| miR-141 | miR-141 is regulated by FOXP3-KAT2B axis, promotes tumor metastasis | N/A | Human BC cell lines: MCF-7, T47D, BT474, MDA-MB-468 | In vivo (transgenic mice), patient samples | Lungs | [119] |
| miR-143 | Increased in CAF-derived exosomes; promotes stemness, EMT, and anchorage-independence | N/A | Human BC cell lines: BT549, MDA-MB-231, T47D | Clinical samples (TCGA) | ND | [75] |
| miR-148a | Tumor suppressive activity (in FAK null-CAFs) | N/A | Human BC cell lines: MDA-MB-231, MCF-7, WI-38 | In vivo (transgenic mice) | Lungs | [120] |
| miR-155 | Enriched in TNBC, suppresses DUSP14 (tumor suppressor) | DUSP14* | Human BC cell lines: MDA-MB-231, MCF-7, MCF-10A | N/A | ND | [126] |
| miR-181c | Promotes breakdown of BBB through abnormal localization of actin; promotes brain metastasis | PDPK1* | Human BC cell lines: MDA-MB-231, BMD1a, BMD2a, BMD2b | In vivo (intracardiac injection and tail vein) | Brain | [106] |
| miR-197 | Tumor suppressive activity: bone marrow mets transmit miRNAs to BC cells via gap junctions and exosomes | CXCL12* | Human BC cell lines: MDA-MB-231, T47D | N/A | Bone | [110] |
| miR-200 (a, b, c) | Regulates MET (suppresses EMT), which promotes lung metastasis and colonization | SEC23A*, ZEB2, CDH1 | Human BC cell lines: MCF-10A, MDA-MB-231; mouse cell lines: 4T1, 4T07, 67NR | In vivo (tail vein injection) | Lungs | [91,117] |
| miR-200c | miR-200c is regulated by FOXP3-KAT2B axis, promotes tumor metastasis | N/A | Human BC cell lines: MCF-7, T47D, BT474, MDA-MB-468 | In vivo (transgenic mice), patient samples | Lungs | [119] |
| miR-210 | Enhances angiogenesis | N/A | Human BC cell lines: MDA-MB-231, MCF-10A; mouse cell line: 4T1 | In vivo (subcutaneous and MFP injections) | Lungs | [118] |
| miR-222 | Tumor suppressive activity: bone marrow mets transmit miRNAs to BC cells via gap junctions and exosomes | CXCL12* | Human BC cell lines: MDA-MB-231, T47D | N/A | Bone | [110] |
| miR-223 | Tumor suppressive activity: bone marrow mets transmit miRs to BC cells via gap junctions and exosomes | CXCL12* | Human BC cell lines: MDA-MB-231, T47D | N/A | Bone | [110] |
| miR-378e | Increased in CAF-derived exosomes; promotes stemness, EMT, and anchorage-independence | N/A | Human BC cell lines: BT549, MDA-MB-231, T47D | Clinical samples (TCGA) | ND | [75] |
| miR-503 | Promotes M1 to M2 polarization of microglia (results in up-regulation of suppressive cytokines that suppress T-cell proliferation) | STAT3 | Human BC cell lines: MCF-7, SKBR3, 231 BRM and SKBRM (brain metastasis cell lines derived from MDA-MB-231 and SKBR3, respectively) | Databases (GEO), in vivo (intracardiac injection) | Brain | [108] |
| miR-503-3p | Regulated by ELK3; promote migration and invasion | N/A | Human BC cell lines: MDA-MB-231, Hs-578T, BT-20, MCF-7, MCF-10A; LECs, HUVEC | In vivo (orthotopic inoculation), databases | ND | [124] |
| miR-940 | Facilitate osteogenic differentiation of host mesenchymal cells (osteoblastic phenotype) | ARHGAP1*, FAM134A* (validated in osteosarcoma line) | Human BC cell line: MDA-MB-231 | In vivo (implant onto calvarial bones/skull or injection into tibia) | Bone | [113] |
| miR-1246 | Promotes proliferation, invasion | CCNG2* | Human BC cell lines: MDA-MB-231, MCF-7, MCF-10A, HMLE | N/A | ND | [121] |
| miR-1910-3p | Promotes proliferation, migration, metastasis, and autophagy | MTMR3* | Human BC cell lines: MDA-MB-231, MCF-7, MCF-10A; 293T (human embryonic kidney cells) | In vivo (subcutaneous xenograft models), clinical samples | ND | [123] |
| miR-4269 | Regulated by ELK3; promote migration and invasion | N/A | Human BC cell lines: MDA-MB-231, Hs-578T, BT-20, MCF-7, MCF-10A; LECs, HUVEC | In vivo (orthotopic inoculation), databases | ND | [124] |
| Exosome-Secreted miRNAs | Drug | Expression Level | Donor Cell | Recipient Cell | Activity | Target Gene(s) | Reference |
|---|---|---|---|---|---|---|---|
| miR-134 | 17-AAG, PU-H71 | Decreased | MCF-7/Resistant | MCF-7/Sensitive | Reduced cell proliferation, invasion, migration and increased cisplatin-induced apoptosis | STAT5B, Hsp90, Bcl-2 | [137] |
| miR-221/222 | Tamoxifen | Increased | MCF-7/Resistant | MCF-7/Sensitive | Increased tamoxifen resistance | p27, ERalpha | [136] |
| miR-222 | Adriamycin | Increased | MCF-7/Resistant | MCF-7/Sensitive | Gained adriamycin-resistance when transfected with miR-222 mimics, lost resistance with miR-222 inhibitors | N/A | [135] |
| miR-423-5p | Cisplatin (DDP) | Increased | MDA-MB-231 | MCF-7, SKBR3 | Increased cell proliferation, migration, and cisplatin resistance | P-gp | [134] |
| miR-503 | Epirubicin, Paclitaxel | Decreased | HUVEC | MDA-MB-231 | Suppressed tumor cell proliferation and invasion | CCND2*, CCND3 | [138] |
| miR-567 | Trastuzumab | Decreased | N/A | SKBR3/R, BT474/R | Inhibits autophagy, reverses chemoresistance | ATG5* | [132] |
| miR-770 | Doxorubicin (DOX) | Decreased | MDA-MB-231, MDA-MB-468 | MDA-MB-231, THP-1 | Increased doxorubicin sensitivity and induced apoptosis; suppressed tumor cell migration and invasion | STMN1* | [139] |
| miR-1246 | Docetaxel (DTX), Epirubicin (EPI), Gemcitabine (GEM) | ND | MDA-MB-231 | HMLE | Increased cell apoptosis after treatment with DTX, EPI, and GEM | CCNG2* | [121] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wong, G.L.; Abu Jalboush, S.; Lo, H.-W. Exosomal MicroRNAs and Organotropism in Breast Cancer Metastasis. Cancers 2020, 12, 1827. https://doi.org/10.3390/cancers12071827
Wong GL, Abu Jalboush S, Lo H-W. Exosomal MicroRNAs and Organotropism in Breast Cancer Metastasis. Cancers. 2020; 12(7):1827. https://doi.org/10.3390/cancers12071827
Chicago/Turabian StyleWong, Grace L., Sara Abu Jalboush, and Hui-Wen Lo. 2020. "Exosomal MicroRNAs and Organotropism in Breast Cancer Metastasis" Cancers 12, no. 7: 1827. https://doi.org/10.3390/cancers12071827





