Circulating Cell-Free DNA-Based Liquid Biopsy Markers for the Non-Invasive Prognosis and Monitoring of Metastatic Pancreatic Cancer
Abstract
1. Introduction
2. Results
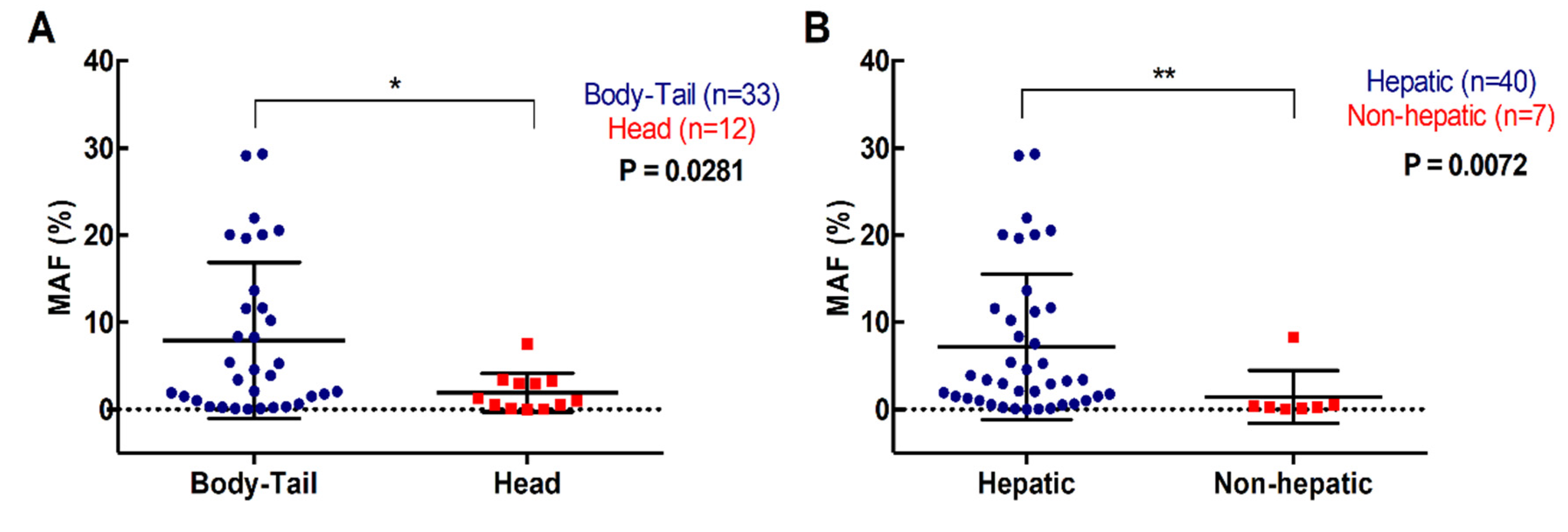
2.1. Clinicopathological Characteristics and RAS Mutation Analysis from Plasma and Tissue
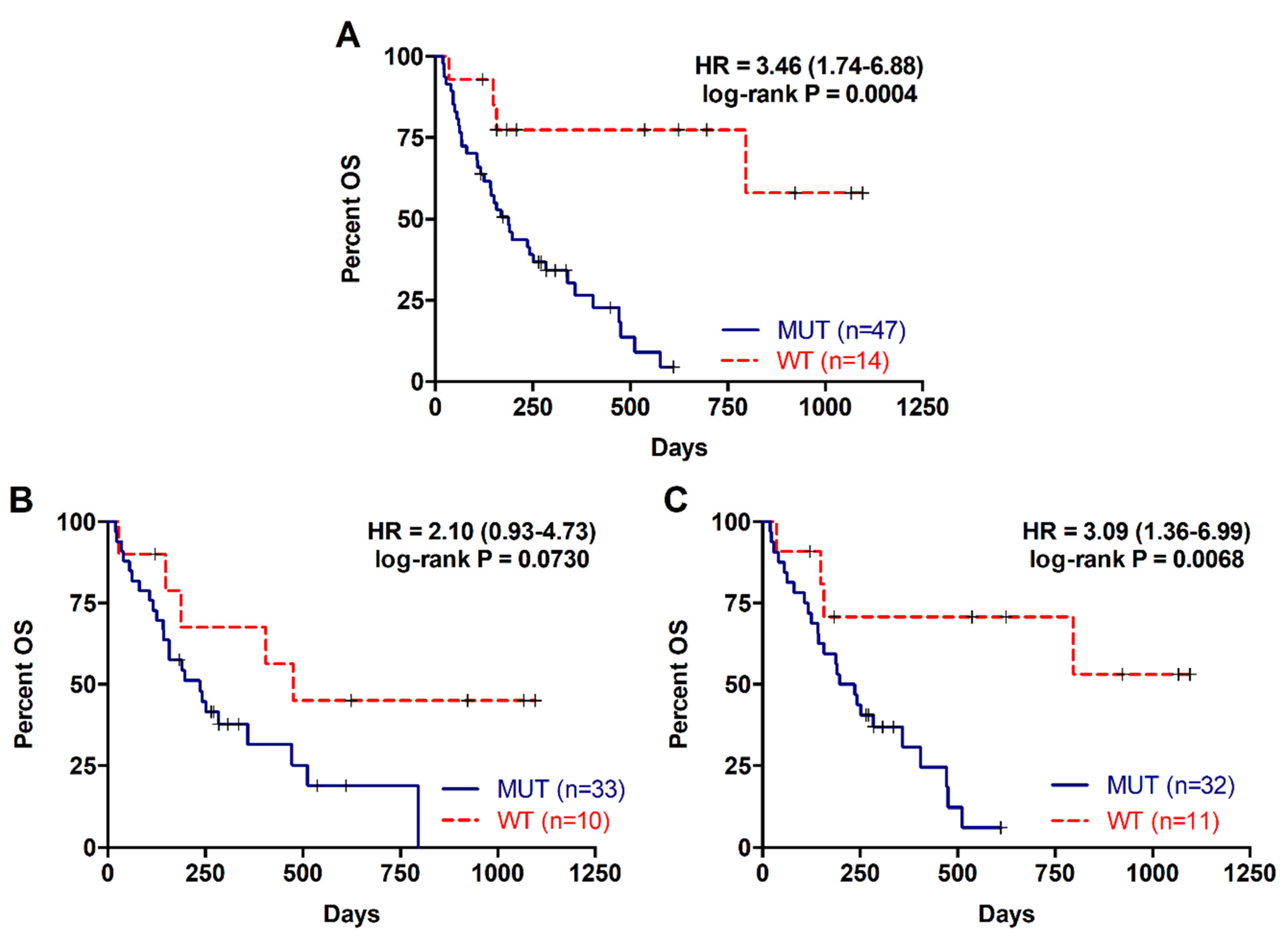
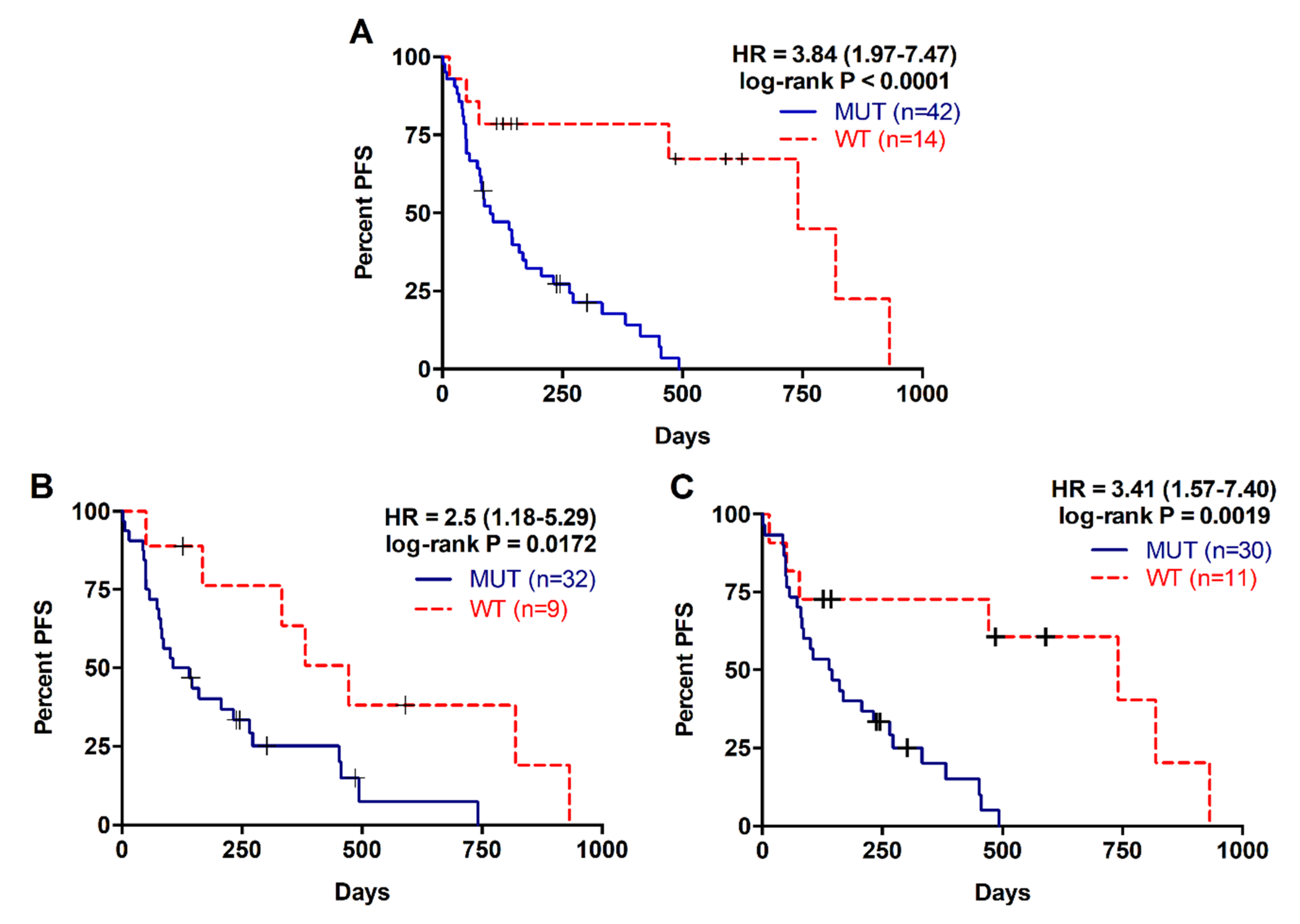
2.2. Detection of RAS Mutations in cfDNA Predicts Poor Prognosis in Metastatic PDAC Patients
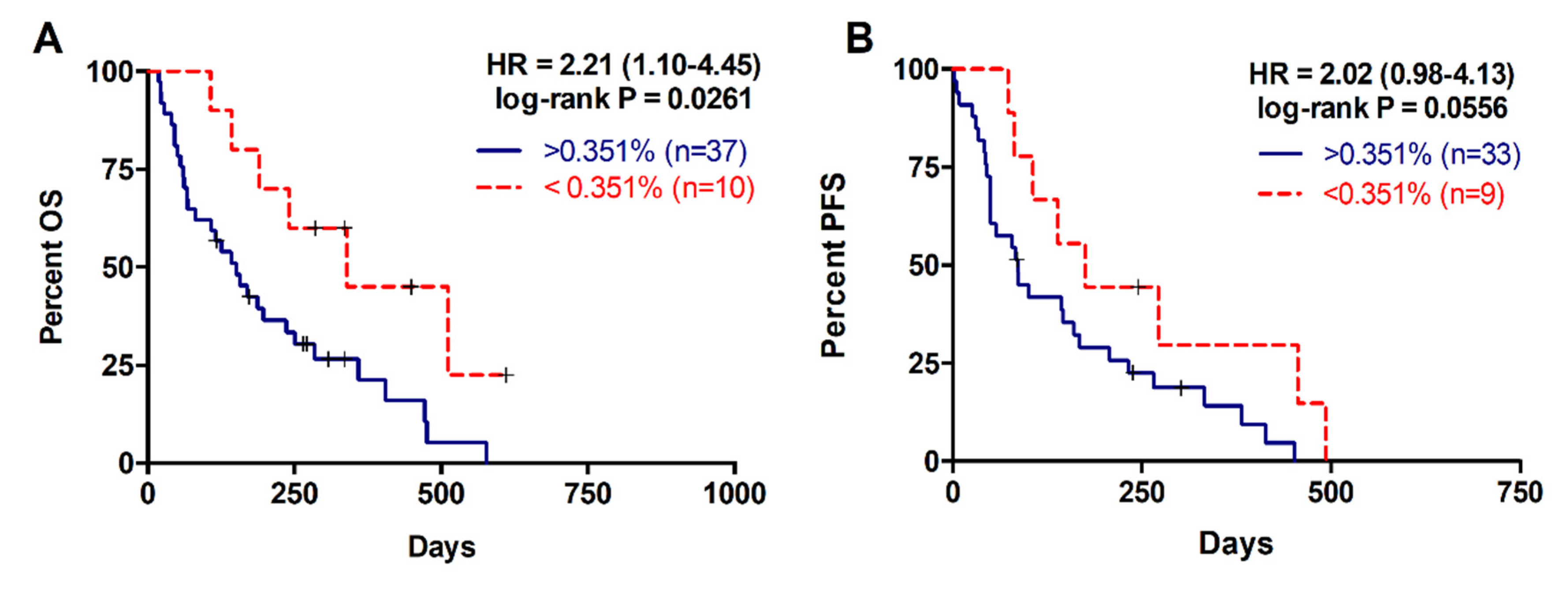
2.3. Higher RAS Mutational Load in cfDNA is Associated with Poor Prognosis in Metastatic PDAC
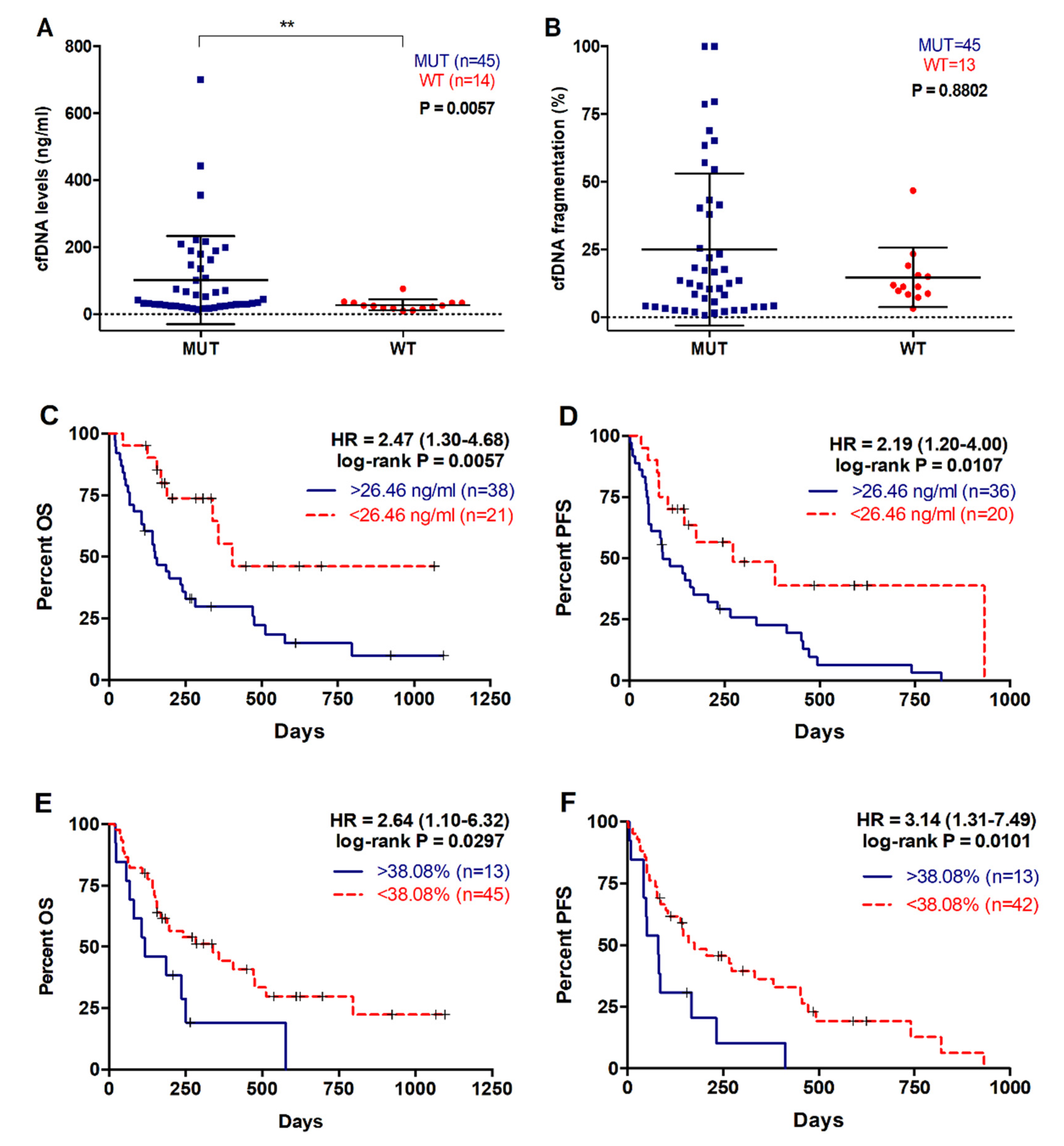
2.4. Higher cfDNA Concentration and Fragmentation Levels Are Associated with Poorer Survival in Metastatic PDAC Patients
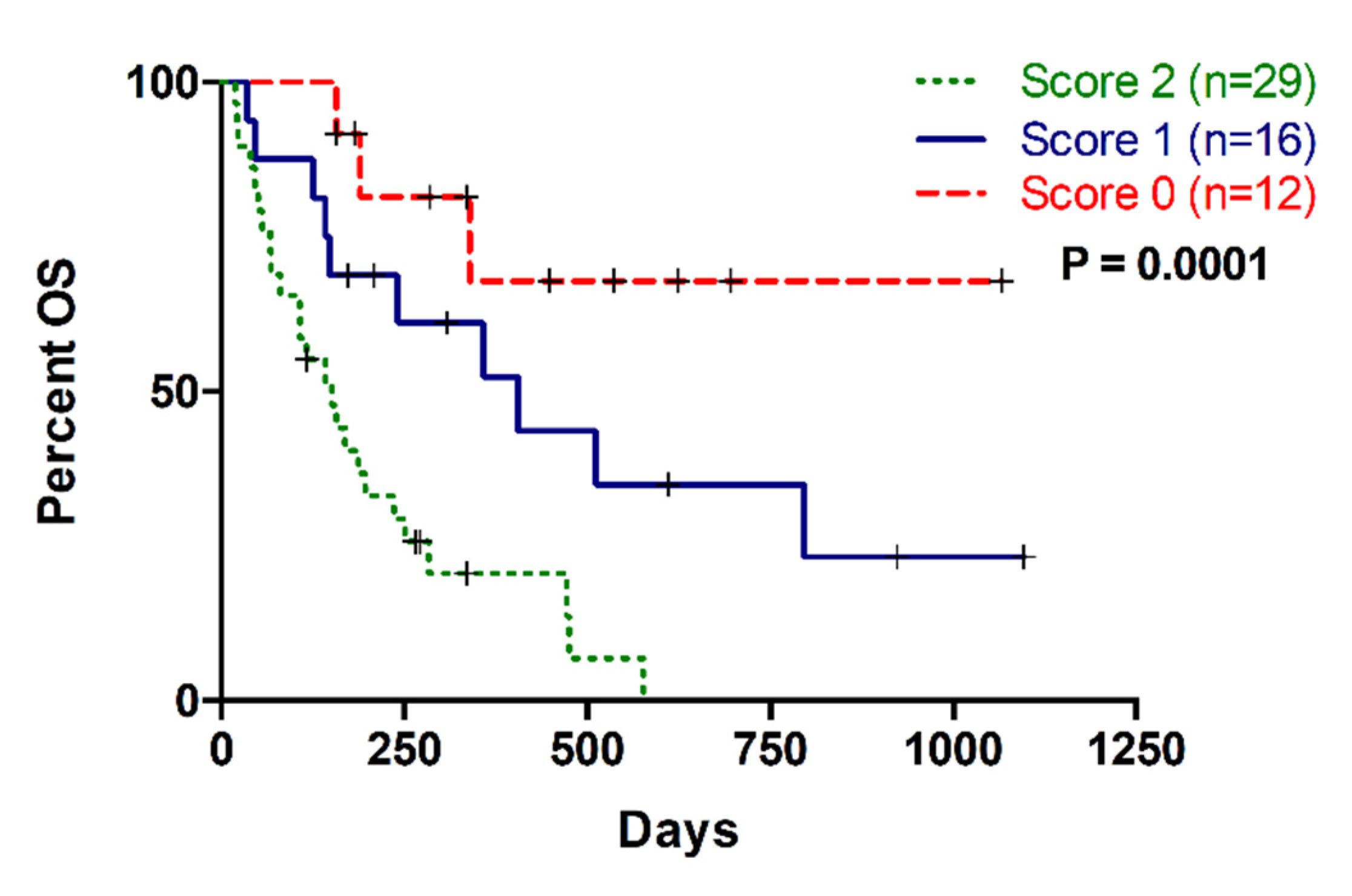
2.5. Multiparameter Liquid Biopsy Refines Prognostic Stratification of Metastatic PDAC Patients
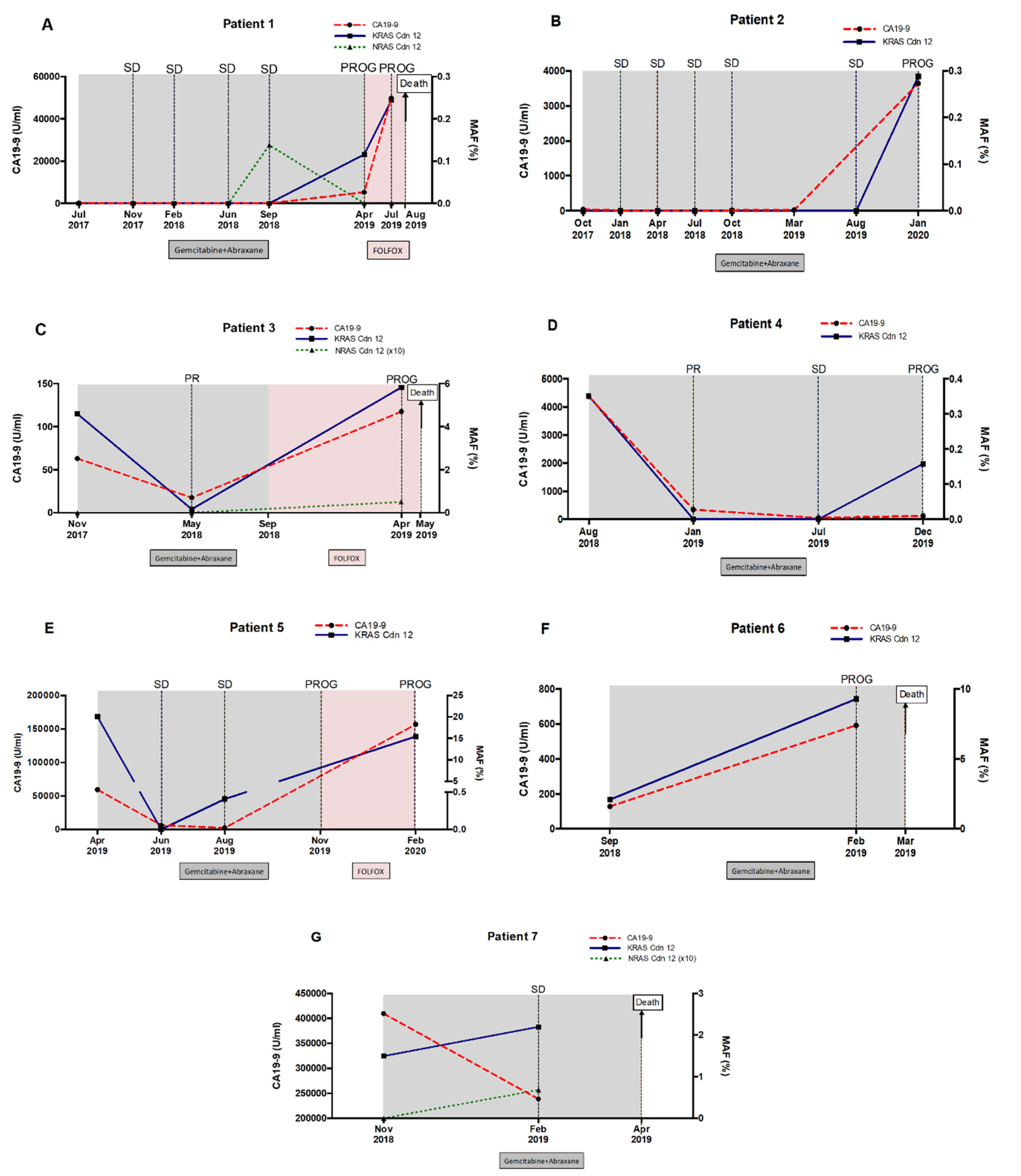
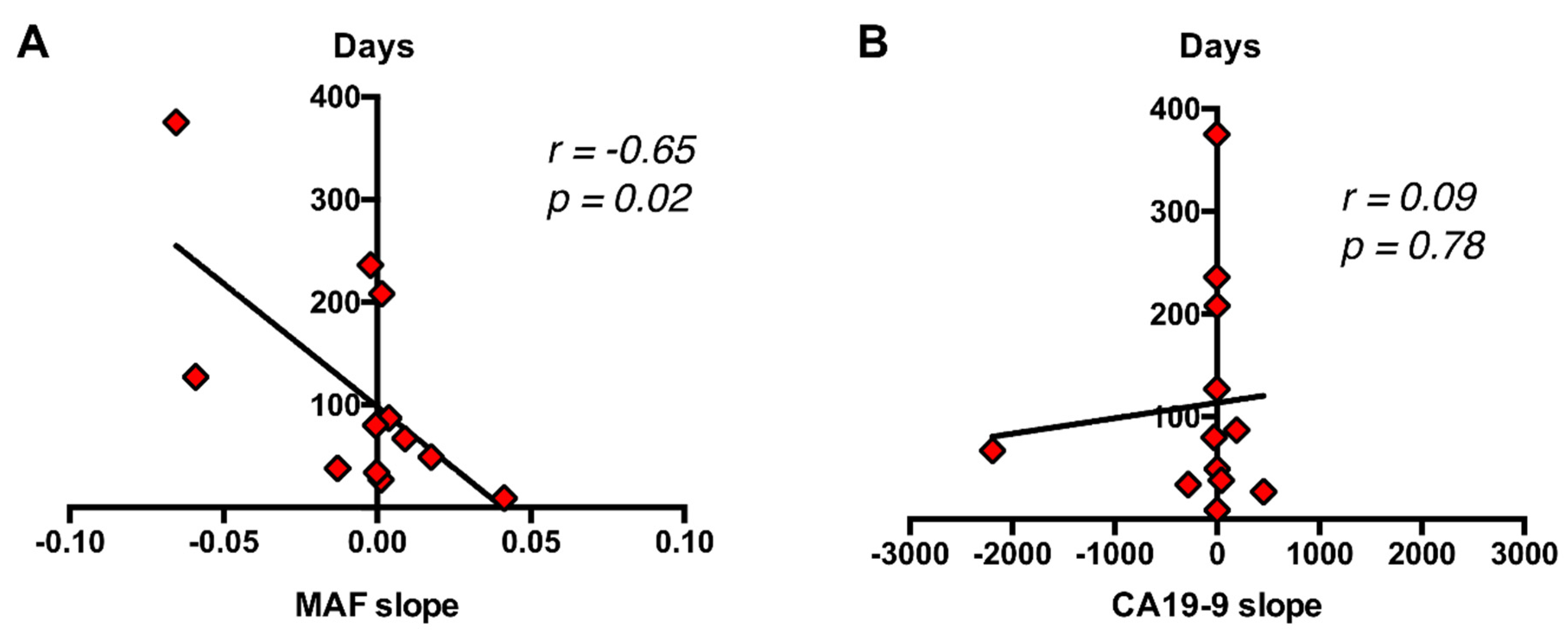
2.6. RAS Mutational Load in cfDNA Enables Monitoring of Disease Progression and Response to Therapy in Metastatic PDAC Patients
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Procedures for Sample Analyses
4.3. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Dyba, T.; Randi, G.; Bettio, M.; Gavin, A.; Visser, O.; Bray, F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur J. Cancer 2018, 103, 356–387. [Google Scholar] [CrossRef]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Ryan, D.P.; Hong, T.S.; Bardeesy, N. Pancreatic adenocarcinoma. N. Engl. J. Med. 2014, 371, 1039–1049. [Google Scholar] [CrossRef]
- Waters, A.M.; Der, C.J. KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb. Perspect. Med. 2018, 8, a031435. [Google Scholar] [CrossRef]
- Baek, H.W.; Park, M.J.; Rhee, Y.Y.; Lee, K.B.; Kim, M.A.; Park, I.A. Diagnostic accuracy of endoscopic ultrasound-guided fine needle aspiration cytology of pancreatic lesions. J. Pathol. Transl. Med. 2015, 49, 52–60. [Google Scholar] [CrossRef]
- Luchini, C.; Veronese, N.; Nottegar, A.; Cappelletti, V.; Daidone, M.G.; Smith, L.; Parris, C.; Brosens, L.A.A.; Caruso, M.G.; Cheng, L.; et al. Liquid Biopsy as Surrogate for Tissue for Molecular Profiling in Pancreatic Cancer: A Meta-Analysis Towards Precision Medicine. Cancers 2019, 11, 1152. [Google Scholar] [CrossRef]
- Tjensvoll, K.; Lapin, M.; Buhl, T.; Oltedal, S.; Steen-Ottosen Berry, K.; Gilje, B.; Soreide, J.A.; Javle, M.; Nordgard, O.; Smaaland, R. Clinical relevance of circulating KRAS mutated DNA in plasma from patients with advanced pancreatic cancer. Mol. Oncol. 2016, 10, 635–643. [Google Scholar] [CrossRef]
- El Messaoudi, S.; Mouliere, F.; Du Manoir, S.; Bascoul-Mollevi, C.; Gillet, B.; Nouaille, M.; Fiess, C.; Crapez, E.; Bibeau, F.; Theillet, C.; et al. Circulating DNA as a Strong Multimarker Prognostic Tool for Metastatic Colorectal Cancer Patient Management Care. Clin. Cancer Res. 2016, 22, 3067–3077. [Google Scholar] [CrossRef] [PubMed]
- Tabernero, J.; Lenz, H.J.; Siena, S.; Sobrero, A.; Falcone, A.; Ychou, M.; Humblet, Y.; Bouche, O.; Mineur, L.; Barone, C.; et al. Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: A retrospective, exploratory analysis of the CORRECT trial. Lancet Oncol. 2015, 16, 937–948. [Google Scholar] [CrossRef]
- Kinugasa, H.; Nouso, K.; Miyahara, K.; Morimoto, Y.; Dohi, C.; Tsutsumi, K.; Kato, H.; Matsubara, T.; Okada, H.; Yamamoto, K. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer 2015, 121, 2271–2280. [Google Scholar] [CrossRef] [PubMed]
- Park, G.; Park, J.K.; Son, D.S.; Shin, S.H.; Kim, Y.J.; Jeon, H.J.; Lee, J.; Park, W.Y.; Lee, K.H.; Park, D. Utility of targeted deep sequencing for detecting circulating tumor DNA in pancreatic cancer patients. Sci. Rep. 2018, 8, 11631. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Ding, X.Q.; Zhu, H.; Wang, R.X.; Pan, X.R.; Tong, J.H. KRAS Mutant Allele Fraction in Circulating Cell-Free DNA Correlates With Clinical Stage in Pancreatic Cancer Patients. Front. Oncol. 2019, 9, 1295. [Google Scholar] [CrossRef]
- Chen, I.; Raymond, V.M.; Geis, J.A.; Collisson, E.A.; Jensen, B.V.; Hermann, K.L.; Erlander, M.G.; Tempero, M.; Johansen, J.S. Ultrasensitive plasma ctDNA KRAS assay for detection, prognosis, and assessment of therapeutic response in patients with unresectable pancreatic ductal adenocarcinoma. Oncotarget 2017, 8, 97769–97786. [Google Scholar] [CrossRef]
- Perets, R.; Greenberg, O.; Shentzer, T.; Semenisty, V.; Epelbaum, R.; Bick, T.; Sarji, S.; Ben-Izhak, O.; Sabo, E.; Hershkovitz, D. Mutant KRAS Circulating Tumor DNA Is an Accurate Tool for Pancreatic Cancer Monitoring. Oncologist 2018, 23, 566–572. [Google Scholar] [CrossRef] [PubMed]
- Hadano, N.; Murakami, Y.; Uemura, K.; Hashimoto, Y.; Kondo, N.; Nakagawa, N.; Sueda, T.; Hiyama, E. Prognostic value of circulating tumour DNA in patients undergoing curative resection for pancreatic cancer. Br. J. Cancer 2016, 115, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Gall, T.M.H.; Belete, S.; Khanderia, E.; Frampton, A.E.; Jiao, L.R. Circulating Tumor Cells and Cell-Free DNA in Pancreatic Ductal Adenocarcinoma. Am. J. Pathol. 2019, 189, 71–81. [Google Scholar] [CrossRef]
- Lennerz, J.K.; Stenzinger, A. Allelic ratio of KRAS mutations in pancreatic cancer. Oncologist 2015, 20, e8. [Google Scholar] [CrossRef]
- Bernard, V.; Kim, D.U.; San Lucas, F.A.; Castillo, J.; Allenson, K.; Mulu, F.C.; Stephens, B.M.; Huang, J.; Semaan, A.; Guerrero, P.A.; et al. Circulating Nucleic Acids Are Associated With Outcomes of Patients With Pancreatic Cancer. Gastroenterology 2019, 156, 108–118. [Google Scholar] [CrossRef]
- Mueller, S.; Engleitner, T.; Maresch, R.; Zukowska, M.; Lange, S.; Kaltenbacher, T.; Konukiewitz, B.; Ollinger, R.; Zwiebel, M.; Strong, A.; et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature 2018, 554, 62–68. [Google Scholar] [CrossRef]
- Birnbaum, D.J.; Bertucci, F.; Finetti, P.; Birnbaum, D.; Mamessier, E. Head and Body/Tail Pancreatic Carcinomas Are Not the Same Tumors. Cancers 2019, 11, 497. [Google Scholar] [CrossRef] [PubMed]
- Dreyer, S.B.; Jamieson, N.B.; Upstill-Goddard, R.; Bailey, P.J.; McKay, C.J.; Biankin, A.V.; Chang, D.K. Defining the molecular pathology of pancreatic body and tail adenocarcinoma. Br. J. Surg. 2018. [Google Scholar] [CrossRef] [PubMed]
- van Erning, F.N.; Mackay, T.M.; van der Geest, L.G.M.; Groot Koerkamp, B.; van Laarhoven, H.W.M.; Bonsing, B.A.; Wilmink, J.W.; van Santvoort, H.C.; de Vos-Geelen, J.; van Eijck, C.H.J.; et al. Association of the location of pancreatic ductal adenocarcinoma (head, body, tail) with tumor stage, treatment, and survival: A population-based analysis. Acta Oncol. 2018, 57, 1655–1662. [Google Scholar] [CrossRef]
- Tomasello, G.; Ghidini, M.; Costanzo, A.; Ghidini, A.; Russo, A.; Barni, S.; Passalacqua, R.; Petrelli, F. Outcome of head compared to body and tail pancreatic cancer: A systematic review and meta-analysis of 93 studies. J. Gastrointest. Oncol. 2019, 10, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Artinyan, A.; Soriano, P.A.; Prendergast, C.; Low, T.; Ellenhorn, J.D.; Kim, J. The anatomic location of pancreatic cancer is a prognostic factor for survival. HPB 2008, 10, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Valpione, S.; Gremel, G.; Mundra, P.; Middlehurst, P.; Galvani, E.; Girotti, M.R.; Lee, R.J.; Garner, G.; Dhomen, N.; Lorigan, P.C.; et al. Plasma total cell-free DNA (cfDNA) is a surrogate biomarker for tumour burden and a prognostic biomarker for survival in metastatic melanoma patients. Eur. J. Cancer 2018, 88, 1–9. [Google Scholar] [CrossRef]
- Lapin, M.; Oltedal, S.; Tjensvoll, K.; Buhl, T.; Smaaland, R.; Garresori, H.; Javle, M.; Glenjen, N.I.; Abelseth, B.K.; Gilje, B.; et al. Fragment size and level of cell-free DNA provide prognostic information in patients with advanced pancreatic cancer. J. Transl. Med. 2018, 16, 300. [Google Scholar] [CrossRef]
- Heitzer, E.; Auinger, L.; Speicher, M.R. Cell-Free DNA and Apoptosis: How Dead Cells Inform About the Living. Trends Mol. Med. 2020, 26, 519–528. [Google Scholar] [CrossRef]
- Zvereva, M.; Roberti, G.; Durand, G.; Voegele, C.; Nguyen, M.D.; Delhomme, T.M.; Chopard, P.; Fabianova, E.; Adamcakova, Z.; Holcatova, I.; et al. Circulating tumour-derived KRAS mutations in pancreatic cancer cases are predominantly carried by very short fragments of cell-free DNA. EBioMedicine 2020, 102462. [Google Scholar] [CrossRef]
- Saad, E.D.; Machado, M.C.; Wajsbrot, D.; Abramoff, R.; Hoff, P.M.; Tabacof, J.; Katz, A.; Simon, S.D.; Gansl, R.C. Pretreatment CA 19-9 level as a prognostic factor in patients with advanced pancreatic cancer treated with gemcitabine. Int. J. Gastrointest. Cancer 2002, 32, 35–41. [Google Scholar] [CrossRef]
- Maisey, N.R.; Norman, A.R.; Hill, A.; Massey, A.; Oates, J.; Cunningham, D. CA19-9 as a prognostic factor in inoperable pancreatic cancer: The implication for clinical trials. Br. J. Cancer 2005, 93, 740–743. [Google Scholar] [CrossRef]
- Scara, S.; Bottoni, P.; Scatena, R. CA 19-9: Biochemical and Clinical Aspects. Adv. Exp. Med. Biol. 2015, 867, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Bauer, T.M.; El-Rayes, B.F.; Li, X.; Hammad, N.; Philip, P.A.; Shields, A.F.; Zalupski, M.M.; Bekaii-Saab, T. Carbohydrate antigen 19-9 is a prognostic and predictive biomarker in patients with advanced pancreatic cancer who receive gemcitabine-containing chemotherapy: A pooled analysis of 6 prospective trials. Cancer 2013, 119, 285–292. [Google Scholar] [CrossRef] [PubMed]
- van der Sijde, F.; Vietsch, E.E.; Mustafa, D.A.M.; Besselink, M.G.; Groot Koerkamp, B.; van Eijck, C.H.J. Circulating Biomarkers for Prediction of Objective Response to Chemotherapy in Pancreatic Cancer Patients. Cancers 2019, 11, 93. [Google Scholar] [CrossRef]
- Vivancos, A.; Aranda, E.; Benavides, M.; Elez, E.; Gomez-Espana, M.A.; Toledano, M.; Alvarez, M.; Parrado, M.R.C.; Garcia-Barberan, V.; Diaz-Rubio, E. Comparison of the Clinical Sensitivity of the Idylla Platform and the OncoBEAM RAS CRC Assay for KRAS Mutation Detection in Liquid Biopsy Samples. Sci. Rep. 2019, 9, 8976. [Google Scholar] [CrossRef]

| Patient Characteristics | Number of Cases (n = 61) | |
|---|---|---|
| Age (median, range) | 65 (40–84) | |
| Sex | Male | 34 (55.7%) |
| Female | 27 (44.3%) | |
| ECOG | 0 | 17 (27.9%) |
| 1 | 31 (50.8%) | |
| 2 | 10 (16.4%) | |
| 3 | 3 (4.9%) | |
| 1st line treatment | Gemcitabine | 3 (4.9%) |
| Gemcitabine/nab-paclitaxel | 39 (63.9%) | |
| Gemcitabine/nab-paclitaxel/FOLFOX | 4 (6.6%) | |
| FOLFIRINOX | 11 (18%) | |
| No treatment | 4 (6.6%) | |
| Survival | Alive | 19 (31.1%) |
| Dead | 42 (68.9%) | |
| Disease progression | Yes | 45 (73.8%) |
| No | 11 (18%) | |
| Not valuable (No treatment or surgery) | 5 (8.2%) | |
| Tissue availability | Yes | 43 (70.5%) |
| No (Cytology) | 18 (29.5%) | |
| Primary tumor location | Tail | 17 (27.9%) |
| Body | 25 (41%) | |
| Head | 18 (29.5%) | |
| Body-Tail | 1 (1.6%) | |
| Number of metastatic lesions | One location | 26 (42.6%) |
| More than one location | 35 (57.4%) | |
| Metastatic lesions location | Hepatic lesions | 48 (78.7%) |
| Non-hepatic lesions | 13 (21.3%) | |
| Tissue Biopsy RAS status 1 | RAS mutated | 33 (76.7%) |
| RAS wild-type | 10 (23.3%) | |
| Liquid Biopsy RAS status | RAS mutated | 47 (77%) |
| RAS wild-type | 14 (23%) |
| Variables | Death Occurrence | Median OS (Days) | HR (95%CI) | p |
|---|---|---|---|---|
| Primary Tumor Location | ||||
| Body/Tail | 28/42 | 187 | 0.818 (0.402–1.667) | p = 0.5802 |
| Head | 12/17 | 173.5 | ||
| Metastatic Location | ||||
| Hepatic | 35/48 | 157 | 2.403 (1.218–4.738) | p = 0.0114 |
| Non-hepatic | 6/13 | 339 | ||
| Number of Metastasis | ||||
| 1 | 16/26 | 197.5 | 0.739 (0.398–1.372) | p = 0.3380 |
| ≥2 | 25/35 | 176 | ||
| KRAS mutation status plasma | ||||
| MUT | 37/47 | 169 | 3.455 (1.736–6.876) | p = 0.0004 |
| WT | 4/14 | 372.5 | ||
| KRAS mutation status tissue | ||||
| MUT | 24/33 | 197 | 2.102 (0.933–4.734) | p = 0.0730 |
| WT | 5/10 | 440 | ||
| KRAS mutation status plasma (with tissue paired sample) | ||||
| MUT | 25/32 | 216.5 | 3.09 (1.364–6.997) | p = 0.0068 |
| WT | 4/11 | 537 | ||
| CA19-9 | ||||
| <45,500 U/mL | 32/50 | 202.5 | 2.272 (1.407–4.930) | p = 0.0408 |
| >45,500 U/mL | 8/10 | 125 | ||
| cfDNA concentration | ||||
| <26.46 ng/mL | 8/21 | 285 | 2.468 (1.302–4.681) | p = 0.0057 |
| >26.46 ng/mL | 31/38 | 149.5 | ||
| MAF | ||||
| <0.351% | 6/10 | 310 | 2.212 (1.099–4.452) | p = 0.0261 |
| >0.351% | 31/37 | 142 | ||
| cfDNA fragmentation | ||||
| <38.08% | 28/45 | 197 | 2.637 (1.1–6.321) | p = 0.0297 |
| >38.08% | 11/13 | 116 |
| Variables | Disease Progression | Median PFS (Days) | HR (95%CI) | p |
|---|---|---|---|---|
| Primary Tumor Location | ||||
| Body/Tail | 33/40 | 152 | 0.783 (0.364–1.685) | p = 0.5318 |
| Head | 10/14 | 81.5 | ||
| Metastatic Location | ||||
| Hepatic | 35/43 | 86 | 2.565 (1.333–4.937) | p = 0.0048 |
| Non-hepatic | 10/13 | 272 | ||
| Number of Metastasis | ||||
| 1 | 19/23 | 127 | 0.86 (0.465–1.591) | p = 0.6304 |
| ≥2 | 26/33 | 139 | ||
| KRAS mutation status plasma | ||||
| MUT | 38/42 | 93.5 | 3.84 (1.974–7.469) | p < 0.0001 |
| WT | 7/14 | 313.5 | ||
| KRAS mutation status tissue | ||||
| MUT | 27/32 | 122.5 | 2.495 (1.176–5.294) | p = 0.0172 |
| WT | 7/9 | 382 | ||
| KRAS mutation status plasma (with tissue paired sample) | ||||
| MUT | 27/30 | 142.5 | 3.41 (1.572–7.395) | p = 0.0019 |
| WT | 7/11 | 472 | ||
| CA19-9 | ||||
| <45,500 U/mL | 35/45 | 143 | 3.013 (1.12–8.103) | p = 0.0289 |
| 45,500 U/mL | 9/10 | 72 | ||
| cfDNA concentration | ||||
| 26.46 ng/mL | 11/20 | 149.5 | 2.190 (1.199–4.001) | p = 0.0107 |
| >26.46 ng/mL | 34/36 | 86.5 | ||
| MAF | ||||
| <0.351% | 8/9 | 175 | 2.015 (0.9834–4.129) | p = 0.0556 |
| >0.351% | 30/33 | 85 | ||
| cfDNA fragmentation | ||||
| <38.08% | 33/42 | 145 | 3.137 (1.313–7.494) | p = 0.0101 |
| >38.08% | 12/13 | 81 |
| Variables | OS | PFS | ||
|---|---|---|---|---|
| p Value | HR (95%CI) | p Value | HR (95%CI) | |
| KRAS mutation status plasma | 0.011 | 5.692 (1.497–21.636) | 0.001 | 8.631 (2.311–32.236) |
| MAF | 0.047 | 1.070 (1.001–1.143) | 0.280 | 1.035 (0.972–1.103) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toledano-Fonseca, M.; Cano, M.T.; Inga, E.; Rodríguez-Alonso, R.; Gómez-España, M.A.; Guil-Luna, S.; Mena-Osuna, R.; de la Haba-Rodríguez, J.R.; Rodríguez-Ariza, A.; Aranda, E. Circulating Cell-Free DNA-Based Liquid Biopsy Markers for the Non-Invasive Prognosis and Monitoring of Metastatic Pancreatic Cancer. Cancers 2020, 12, 1754. https://doi.org/10.3390/cancers12071754
Toledano-Fonseca M, Cano MT, Inga E, Rodríguez-Alonso R, Gómez-España MA, Guil-Luna S, Mena-Osuna R, de la Haba-Rodríguez JR, Rodríguez-Ariza A, Aranda E. Circulating Cell-Free DNA-Based Liquid Biopsy Markers for the Non-Invasive Prognosis and Monitoring of Metastatic Pancreatic Cancer. Cancers. 2020; 12(7):1754. https://doi.org/10.3390/cancers12071754
Chicago/Turabian StyleToledano-Fonseca, Marta, M. Teresa Cano, Elizabeth Inga, Rosa Rodríguez-Alonso, M. Auxiliadora Gómez-España, Silvia Guil-Luna, Rafael Mena-Osuna, Juan R. de la Haba-Rodríguez, Antonio Rodríguez-Ariza, and Enrique Aranda. 2020. "Circulating Cell-Free DNA-Based Liquid Biopsy Markers for the Non-Invasive Prognosis and Monitoring of Metastatic Pancreatic Cancer" Cancers 12, no. 7: 1754. https://doi.org/10.3390/cancers12071754
APA StyleToledano-Fonseca, M., Cano, M. T., Inga, E., Rodríguez-Alonso, R., Gómez-España, M. A., Guil-Luna, S., Mena-Osuna, R., de la Haba-Rodríguez, J. R., Rodríguez-Ariza, A., & Aranda, E. (2020). Circulating Cell-Free DNA-Based Liquid Biopsy Markers for the Non-Invasive Prognosis and Monitoring of Metastatic Pancreatic Cancer. Cancers, 12(7), 1754. https://doi.org/10.3390/cancers12071754






