Targeting Altered Energy Metabolism in Colorectal Cancer: Oncogenic Reprogramming, the Central Role of the TCA Cycle and Therapeutic Opportunities
Abstract
1. Introduction
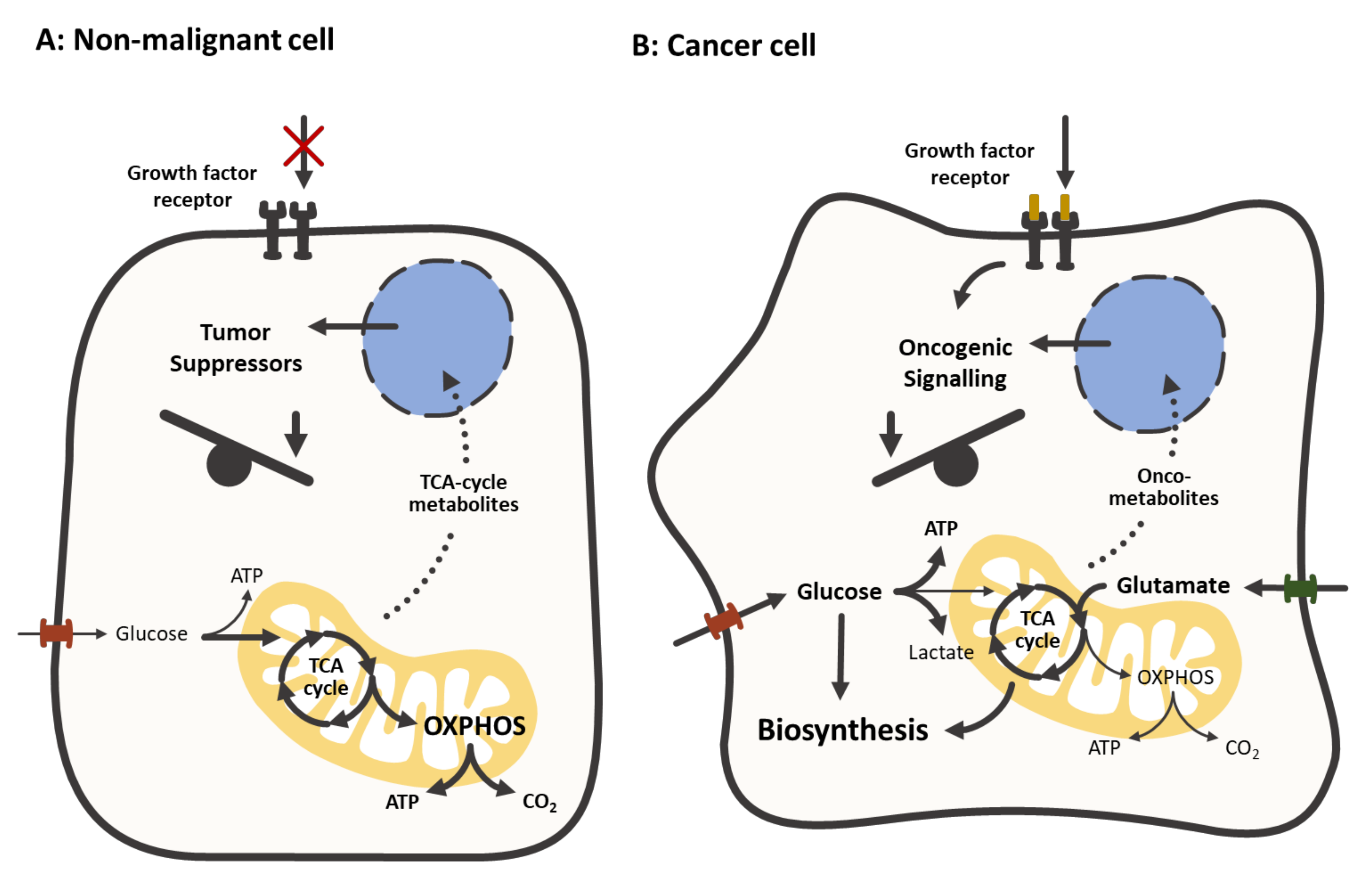
2. Mitochondrial Metabolism of Colorectal Cancer
2.1. The Altered Energy Metabolism in CRC
2.2. The Metabolic Impact of Oncogenes and Tumor Suppressors
2.2.1. KRAS and BRAF
2.2.2. p53
2.2.3. PI3K–AKT–mTOR Network
2.2.4. MYC
2.2.5. HIF
2.3. The Central Role of the Tricarboxylic Acid Cycle
2.4. Metabolic Symbiosis in the Tumor Microenvironment
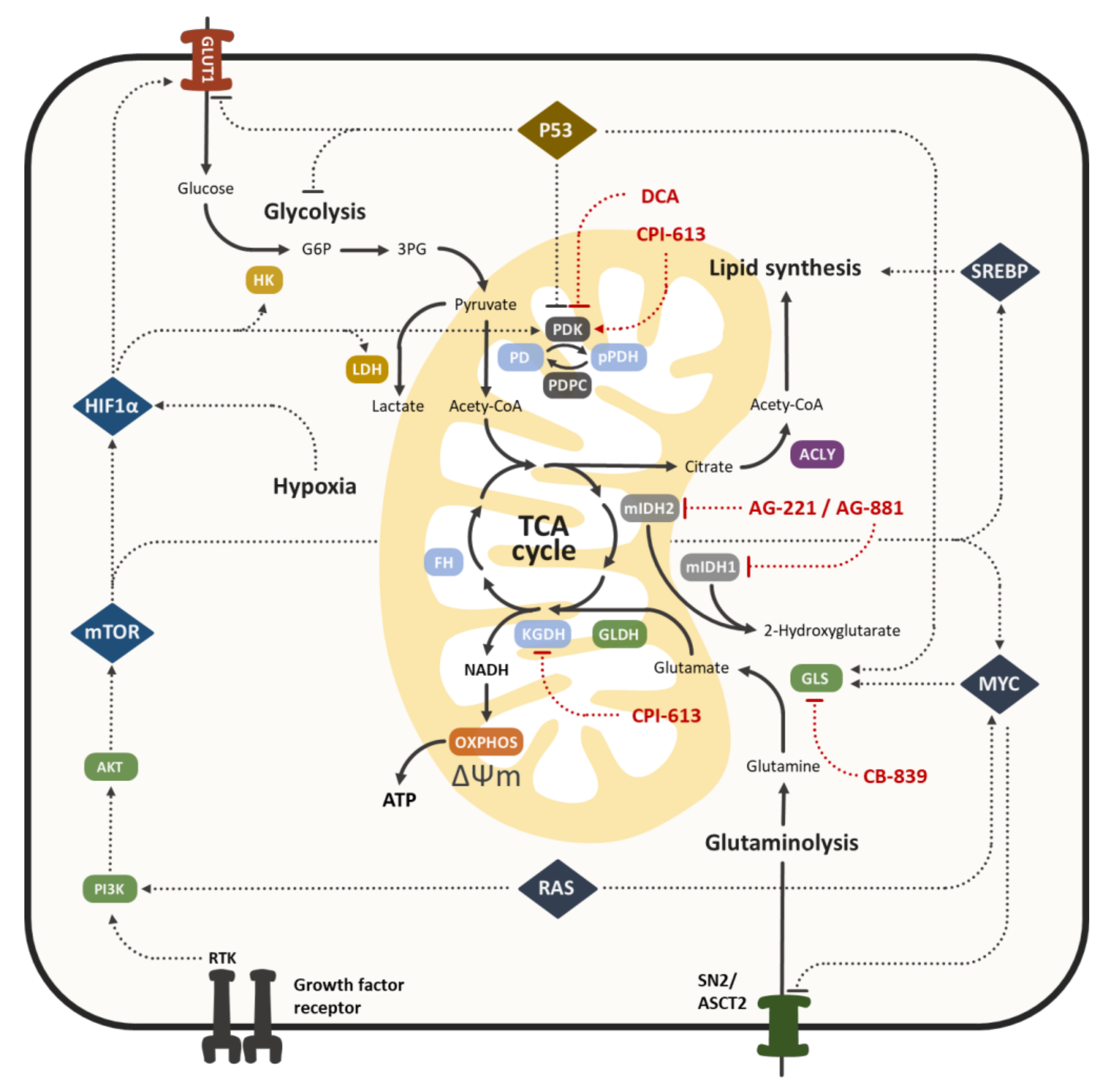
3. Cancer Specific Mitochondrial Metabolism as a Therapeutic Target
4. The Lipoic Acid Derivative CPI-613 in Cancer Therapy
4.1. The Molecular Mechanism of CPI-613 Antitumor Activity
4.2. Cancer Stem Cells as a Preferential Target of CPI-613?
4.3. Application of CPI-613 in Preclinical Studies and Clinical Trials
5. Conclusions and Remarks
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 2-HG | 2-hydroxyglutarate |
| 5-FU | 5-fluorouracil |
| ACC | acetyl CoA carboxylase |
| 2-HG | 2-hydroxyglutarate |
| ADP | adenosine diphosphate |
| αKG | α ketoglutarate |
| AKT | protein kinase B |
| AML | acute myeloid leukemia |
| AMPK | 5’ AMP-activated protein kinase |
| Bax | Bcl-2-associated X protein |
| BW | body weight |
| CAF | cancer-associated fibroblast |
| CD | cluster of differentiation |
| CDK | cyclin-dependent kinase |
| CIMP | CpG island methylator phenotype |
| COX | cyclooxygenase |
| CRC | colorectal cancer |
| CSC | cancer stem cell |
| CYP | cytochrome P450 |
| DCA | dichloroacetate |
| dMMR | DNA mismatch repair system |
| DNA | deoxyribonucleic acid |
| E4BP | eIF4E binding protein |
| ECM | extracellular matrix |
| EGFR | endothelial growth factor receptor |
| EMA | European Medicines Agency |
| EMT | epithelial–mesenchymal transition |
| ERK | extracellular-signal regulated kinases |
| FAD | flavin adenine dinucleotide |
| FDA | Food and Drug Administration |
| FOLFIRI | fluorouracil, leucovorin and irinotecan |
| FOLFIRINOX | fluorouracil, leucovorin, irinotecan and oxaliplatin |
| FOLFOX | fluorouracil, leucovorin and oxaliplatin |
| G6PD | glucose-6-phosphate dehydrogenase |
| GLS | glutaminase |
| GLUD | glutamate dehydrogenase |
| GLUT | glucose transporter |
| GOT | glutamic-oxaloacetic transaminase |
| GTP | glutamate pyruvate transaminase |
| GTP | guanosine-5’-triphosphate |
| HIF | hypoxia-inducible factor |
| HK | hexokinase |
| HRE | hypoxia response element |
| IBD | inflammatory bowel disease |
| IC50 | inhibitory concentration (50%) |
| IDH | isocitrate dehydrogenase |
| IGF | insulin-like growth factor |
| KGDH | α-ketoglutarate dehydrogenase complex |
| LA | α-lipoic acid |
| LC3B | microtubule-associated protein |
| LDHA | lactate dehydrogenase A |
| MAPK | mitogen-activated protein kinase |
| MCT | monocarboxylate transporter |
| MDR | multidrug resistant protein |
| MGMT | O6-methylguanine-DNA methyltransferase |
| MOM | mitochondrial outer membrane |
| MPC | mitochondrial pyruvate carrier |
| MSI | microsatellite instable |
| MSS | microsatellite stable |
| mtDNA | mitochondrial DNA |
| MTD | maximum tolerable dose |
| MTHFD2 | methylenetetrahydrofolate dehydrogenase |
| mTOR | mammalian target of rapamycin |
| MYC | myelocytomatosis |
| NFκB | nuclear factor ‘kappa-light-chain-enhancer’ of activated B-cells |
| Nrf | nuclear factor erythroid-2-related factor |
| OXPHOS | oxidative phosphorylation |
| PARP | poly(ADP-ribose) polymerase |
| PDGF | platelet-derived growth factor |
| PGC | peroxisome proliferator-activated receptor gamma coactivator |
| PDC | pyruvate dehydrogenase complex |
| PDCD4 | programmed cell death protein |
| PDK | pyruvate dehydrogenase kinase |
| PEG2 | prostaglandin E2 |
| PEG | polyethylene glycol |
| PET | positron emission tomography |
| PFK | phosphofructokinase |
| PHD | prolyl hydroxylase domain protein |
| PI3K | phosphoinositide 3-kinases |
| PIP2 | phosphatidylinositol-4,5-bisphosphate |
| PIP3 | phosphatidyl-inositol-3,4,5-trisphosphate |
| PKM | pyruvate kinase |
| PPP | pentose phosphate pathway |
| ROS | reactive oxygen species |
| RTK | receptor tyrosine kinases |
| S6K1 | p70S6 Kinase 1 |
| S9 | supernatant 9000 g,— |
| SCO | synthesis of cytochrome c oxidase |
| SLC | solute carrier |
| SREBP | sterol responsive element binding protein |
| TAM | tumor associated macrophages |
| TCA | tricarboxylic acid cycle |
| Teff cell | effector T cell |
| TGF | transforming growth factor |
| TME | tumor microenvironment |
| TIGAR | TP53-inducible glycolysis and apoptosis regulator |
| Treg cell | regulatory T cell |
| VEGF | vascular endothelial growth factor |
References
- Keum, N.; Giovannucci, E. Global burden of colorectal cancer: Emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 713–732. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Fedewa, S.A.; Anderson, W.F.; Miller, K.D.; Ma, J.; Rosenberg, P.S.; Jemal, A. Colorectal Cancer Incidence Patterns in the United States, 1974–2013. J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef] [PubMed]
- Vuik, F.E.; Nieuwenburg, S.A.; Bardou, M.; Lansdorp-Vogelaar, I.; Dinis-Ribeiro, M.; Bento, M.J.; Zadnik, V.; Pellise, M.; Esteban, L.; Kaminski, M.F.; et al. Increasing incidence of colorectal cancer in young adults in Europe over the last 25 years. Gut 2019, 68, 1820–1826. [Google Scholar] [CrossRef]
- Murphy, N.; Moreno, V.; Hughes, D.J.; Vodicka, L.; Vodicka, P.; Aglago, E.K.; Gunter, M.J.; Jenab, M. Lifestyle and dietary environmental factors in colorectal cancer susceptibility. Mol. Aspects Med. 2019, 69, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Fahrer, J.; Kaina, B. O6-methylguanine-DNA methyltransferase in the defense against N-nitroso compounds and colorectal cancer. Carcinogenesis 2013, 34, 2435–2442. [Google Scholar] [CrossRef]
- Seiwert, N.; Heylmann, D.; Hasselwander, S.; Fahrer, J. Mechanism of colorectal carcinogenesis triggered by heme iron from red meat. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188334. [Google Scholar] [CrossRef]
- Ciccolallo, L.; Capocaccia, R.; Coleman, M.P.; Berrino, F.; Coebergh, J.W.W.; Damhuis, R.A.M.; Faivre, J.; Martinez-Garcia, C.; Møller, H.; De Leon, M.P.; et al. Survival differences between European and US patients with colorectal cancer: Role of stage at diagnosis and surgery. Gut 2005, 54, 268–273. [Google Scholar] [CrossRef]
- Joachim, C.; Macni, J.; Drame, M.; Pomier, A.; Escarmant, P.; Veronique-Baudin, J.; Vinh-Hung, V. Overall survival of colorectal cancer by stage at diagnosis: Data from the Martinique Cancer Registry. Medicine 2019, 98. [Google Scholar] [CrossRef]
- Singh, H.; Shu, E.; Demers, A.; Bernstein, C.N.; Griffith, J.; Fradette, K. Trends in Time to Diagnosis of Colon Cancer and Impact on Clinical Outcomes. Can. J. Gastroenterol. 2012, 26, 877–880. [Google Scholar] [CrossRef]
- Kuipers, E.J.; Grady, W.M.; Lieberman, D.; Seufferlein, T.; Sung, J.J.; Boelens, P.G.; van de Velde, C.J.; Watanabe, T. Colorectal cancer. Nat. Rev. Dis Primers 2015, 1, 15065. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O.; Wind, F.; Negelein, E. The Metabolism of Tumors in the Body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Gatenby, R.A.; Gillies, R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef]
- Jones, R.G.; Thompson, C.B. Tumor suppressors and cell metabolism: A recipe for cancer growth. Genes Dev. 2009, 23, 537–548. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Lum, J.J.; Hatzivassiliou, G.; Thompson, C.B. The Biology of Cancer: Metabolic Reprogramming Fuels Cell Growth and Proliferation. Cell Metab. 2008, 7, 11–20. [Google Scholar] [CrossRef]
- Lyssiotis, C.A.; Kimmelman, A.C. Metabolic Interactions in the Tumor Microenvironment. Trends Cell Biol. 2017, 27, 863–875. [Google Scholar] [CrossRef]
- Weinhouse, S. The Warburg hypothesis fifty years later. Z. Krebsforsch. Klin. Onkol. Cancer Res. Clin. Oncol. 1976, 87, 115–126. [Google Scholar] [CrossRef]
- Wei, T.-T.; Lin, Y.-T.; Tang, S.-P.; Luo, C.-K.; Tsai, C.-T.; Shun, C.-T.; Chen, C.-C. Metabolic targeting of HIF-1α potentiates the therapeutic efficacy of oxaliplatin in colorectal cancer. Oncogene 2020, 39, 414–427. [Google Scholar] [CrossRef]
- La Vecchia, S.; Sebastián, C. Metabolic pathways regulating colorectal cancer initiation and progression. Semin. Cell Dev. Biol. 2020, 98, 63–70. [Google Scholar] [CrossRef]
- Ward, P.S.; Thompson, C.B. Metabolic Reprogramming: A Cancer Hallmark Even Warburg Did Not Anticipate. Cancer Cell 2012, 21, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Reyes, I.; Chandel, N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Marin-Hernandez, A.; Gallardo-Perez, J.C.; Ralph, S.J.; Rodriguez-Enriquez, S.; Moreno-Sanchez, R. HIF-1α Modulates Energy Metabolism in Cancer Cells by Inducing Over-Expression of Specific Glycolytic Isoforms. Mini Rev. Med. Chem. 2009, 9, 1084–1101. [Google Scholar] [CrossRef] [PubMed]
- Papandreou, I.; Cairns, R.A.; Fontana, L.; Lim, A.L.; Denko, N.C. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006, 3, 187–197. [Google Scholar] [CrossRef]
- Nagao, A.; Kobayashi, M.; Koyasu, S.; Chow, C.C.T.; Harada, H. HIF-1-Dependent Reprogramming of Glucose Metabolic Pathway of Cancer Cells and Its Therapeutic Significance. Int. J. Mol. Sci. 2019, 20, 238. [Google Scholar] [CrossRef]
- Manzat Saplacan, R.M.; Balacescu, L.; Gherman, C.; Chira, R.I.; Craiu, A.; Mircea, P.A.; Lisencu, C.; Balacescu, O. The Role of PDGFs and PDGFRs in Colorectal Cancer. Mediat. Inflamm. 2017, 2017. [Google Scholar] [CrossRef]
- Rasheed, S.; Harris, A.L.; Tekkis, P.P.; Turley, H.; Silver, A.; McDonald, P.J.; Talbot, I.C.; Glynne-Jones, R.; Northover, J.M.A.; Guenther, T. Hypoxia-inducible factor-1alpha and -2alpha are expressed in most rectal cancers but only hypoxia-inducible factor-1alpha is associated with prognosis. Br. J. Cancer 2009, 100, 1666–1673. [Google Scholar] [CrossRef]
- Jokilehto, T.; Jaakkola, P.M. The role of HIF prolyl hydroxylases in tumour growth. J. Cell. Mol. Med. 2010, 14, 758–770. [Google Scholar] [CrossRef]
- Bricker, D.K.; Taylor, E.B.; Schell, J.C.; Orsak, T.; Boutron, A.; Chen, Y.-C.; Cox, J.E.; Cardon, C.M.; Van Vranken, J.G.; Dephoure, N.; et al. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science 2012, 337, 96–100. [Google Scholar] [CrossRef]
- Schell, J.C.; Olson, K.A.; Jiang, L.; Hawkins, A.J.; Van Vranken, J.G.; Xie, J.; Egnatchik, R.A.; Earl, E.G.; DeBerardinis, R.J.; Rutter, J. A role for the mitochondrial pyruvate carrier as a repressor of the Warburg effect and colon cancer cell growth. Mol. Cell 2014, 56, 400–413. [Google Scholar] [CrossRef]
- Chekulayev, V.; Mado, K.; Shevchuk, I.; Koit, A.; Kaldma, A.; Klepinin, A.; Timohhina, N.; Tepp, K.; Kandashvili, M.; Ounpuu, L.; et al. Metabolic remodeling in human colorectal cancer and surrounding tissues: Alterations in regulation of mitochondrial respiration and metabolic fluxes. Biochem. Biophys. Rep. 2015, 4, 111–125. [Google Scholar] [CrossRef]
- Sun, X.; Zhan, L.; Chen, Y.; Wang, G.; He, L.; Wang, Q.; Zhou, F.; Yang, F.; Wu, J.; Wu, Y.; et al. Increased mtDNA copy number promotes cancer progression by enhancing mitochondrial oxidative phosphorylation in microsatellite-stable colorectal cancer. Signal. Transduct. Targeted Ther. 2018, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Xiong, L.; Ji, Z.; Cheng, W.; Yang, H. Correlation between increased copy number of mitochondrial DNA and clinicopathological stage in colorectal cancer. Oncol. Lett. 2011, 2, 899–903. [Google Scholar] [CrossRef] [PubMed]
- Vellinga, T.T.; Borovski, T.; de Boer, V.C.J.; Fatrai, S.; van Schelven, S.; Trumpi, K.; Verheem, A.; Snoeren, N.; Emmink, B.L.; Koster, J.; et al. SIRT1/PGC1α-Dependent Increase in Oxidative Phosphorylation Supports Chemotherapy Resistance of Colon Cancer. Clin. Cancer Res. 2015, 21, 2870–2879. [Google Scholar] [CrossRef] [PubMed]
- Denise, C.; Paoli, P.; Calvani, M.; Taddei, M.L.; Giannoni, E.; Kopetz, S.; Kazmi, S.M.A.; Pia, M.M.; Pettazzoni, P.; Sacco, E.; et al. 5-Fluorouracil resistant colon cancer cells are addicted to OXPHOS to survive and enhance stem-like traits. Oncotarget 2015, 6, 41706–41721. [Google Scholar] [CrossRef]
- Hirpara, J.; Eu, J.Q.; Tan, J.K.M.; Wong, A.L.; Clement, M.-V.; Kong, L.R.; Ohi, N.; Tsunoda, T.; Qu, J.; Goh, B.C.; et al. Metabolic reprogramming of oncogene-addicted cancer cells to OXPHOS as a mechanism of drug resistance. Redox Biol. 2019, 25, 101076. [Google Scholar] [CrossRef]
- van Osch, F.H.M.; Voets, A.M.; Schouten, L.J.; Gottschalk, R.W.H.; Simons, C.C.J.M.; van Engeland, M.; Lentjes, M.H.F.M.; van den Brandt, P.A.; Smeets, H.J.M.; Weijenberg, M.P. Mitochondrial DNA copy number in colorectal cancer: Between tissue comparisons, clinicopathological characteristics and survival. Carcinogenesis 2015, 36, 1502–1510. [Google Scholar] [CrossRef][Green Version]
- Rebane-Klemm, E.; Truu, L.; Reinsalu, L.; Puurand, M.; Shevchuk, I.; Chekulayev, V.; Timohhina, N.; Tepp, K.; Bogovskaja, J.; Afanasjev, V.; et al. Mitochondrial Respiration in KRAS and BRAF Mutated Colorectal Tumors and Polyps. Cancers 2020, 12, 815. [Google Scholar] [CrossRef]
- Fritsche-Guenther, R.; Zasada, C.; Mastrobuoni, G.; Royla, N.; Rainer, R.; Roßner, F.; Pietzke, M.; Klipp, E.; Sers, C.; Kempa, S. Alterations of mTOR signaling impact metabolic stress resistance in colorectal carcinomas with BRAF and KRAS mutations. Sci. Rep. 2018, 8, 1–17. [Google Scholar] [CrossRef]
- Vaughn, C.P.; ZoBell, S.D.; Furtado, L.V.; Baker, C.L.; Samowitz, W.S. Frequency of KRAS, BRAF, and NRAS mutations in colorectal cancer. Genes Chromosom. Cancer 2011, 50, 307–312. [Google Scholar] [CrossRef]
- Pylayeva-Gupta, Y.; Grabocka, E.; Bar-Sagi, D. RAS oncogenes: Weaving a tumorigenic web. Nat. Rev. Cancer 2011, 11, 761–774. [Google Scholar] [CrossRef] [PubMed]
- Hutton, J.E.; Wang, X.; Zimmerman, L.J.; Slebos, R.J.C.; Trenary, I.A.; Young, J.D.; Li, M.; Liebler, D.C. Oncogenic KRAS and BRAF Drive Metabolic Reprogramming in Colorectal Cancer. Mol. Cell. Proteomics 2016, 15, 2924–2938. [Google Scholar] [CrossRef]
- Yun, J.; Rago, C.; Cheong, I.; Pagliarini, R.; Angenendt, P.; Rajagopalan, H.; Schmidt, K.; Willson, J.K.V.; Markowitz, S.; Zhou, S.; et al. Glucose Deprivation Contributes to the Development of KRAS Pathway Mutations in Tumor Cells. Science 2009, 325, 1555–1559. [Google Scholar] [CrossRef] [PubMed]
- Lozy, F.; Karantza, V. Autophagy and cancer cell metabolism. Semin. Cell Dev. Biol. 2012, 23, 395–401. [Google Scholar] [CrossRef]
- Guo, J.Y.; Chen, H.-Y.; Mathew, R.; Fan, J.; Strohecker, A.M.; Karsli-Uzunbas, G.; Kamphorst, J.J.; Chen, G.; Lemons, J.M.S.; Karantza, V.; et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011, 25, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Lock, R.; Roy, S.; Kenific, C.M.; Su, J.S.; Salas, E.; Ronen, S.M.; Debnath, J. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol. Biol. Cell 2010, 22, 165–178. [Google Scholar] [CrossRef]
- Kawada, K.; Toda, K.; Sakai, Y. Targeting metabolic reprogramming in KRAS-driven cancers. Int. J. Clin. Oncol. 2017, 22, 651–659. [Google Scholar] [CrossRef]
- Gaglio, D.; Metallo, C.M.; Gameiro, P.A.; Hiller, K.; Danna, L.S.; Balestrieri, C.; Alberghina, L.; Stephanopoulos, G.; Chiaradonna, F. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol. Syst. Biol. 2011, 7, 523. [Google Scholar] [CrossRef]
- Gaglio, D.; Soldati, C.; Vanoni, M.; Alberghina, L.; Chiaradonna, F. Glutamine deprivation induces abortive s-phase rescued by deoxyribonucleotides in k-ras transformed fibroblasts. PLoS ONE 2009, 4, e4715. [Google Scholar] [CrossRef]
- Wong, C.C.; Qian, Y.; Li, X.; Xu, J.; Kang, W.; Tong, J.H.; To, K.-F.; Jin, Y.; Li, W.; Chen, H.; et al. SLC25A22 Promotes Proliferation and Survival of Colorectal Cancer Cells With KRAS Mutations and Xenograft Tumor Progression in Mice via Intracellular Synthesis of Aspartate. Gastroenterology 2016, 151, 945–960.e946. [Google Scholar] [CrossRef]
- Miyo, M.; Konno, M.; Nishida, N.; Sueda, T.; Noguchi, K.; Matsui, H.; Colvin, H.; Kawamoto, K.; Koseki, J.; Haraguchi, N.; et al. Metabolic Adaptation to Nutritional Stress in Human Colorectal Cancer. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Toda, K.; Kawada, K.; Iwamoto, M.; Inamoto, S.; Sasazuki, T.; Shirasawa, S.; Hasegawa, S.; Sakai, Y. Metabolic Alterations Caused by KRAS Mutations in Colorectal Cancer Contribute to Cell Adaptation to Glutamine Depletion by Upregulation of Asparagine Synthetase. Neoplasia 2016, 18, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Chun, S.Y.; Johnson, C.; Washburn, J.G.; Cruz-Correa, M.R.; Dang, D.T.; Dang, L.H. Oncogenic KRAS modulates mitochondrial metabolism in human colon cancer cells by inducing HIF-1α and HIF-2α target genes. Mol. Cancer 2010, 9, 293. [Google Scholar] [CrossRef] [PubMed]
- Olivier, M.; Hollstein, M.; Hainaut, P. TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2010, 2, a001008. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.; Shirole, N.; Tian, R.; Pal, D.; Sordella, R. The Evolution of TP53 Mutations: From Loss-of-Function to Separation-of-Function Mutants. J. Cancer Biol. Res. 2016, 4, 1091. [Google Scholar]
- Wang, L.; Xiong, H.; Wu, F.; Zhang, Y.; Wang, J.; Zhao, L.; Guo, X.; Chang, L.-J.; Zhang, Y.; You, M.J.; et al. Hexokinase 2-Mediated Warburg Effect Is Required for PTEN- and p53-Deficiency-Driven Prostate Cancer Growth. Cell Rep. 2014, 8, 1461–1474. [Google Scholar] [CrossRef]
- Kondoh, H.; Lleonart, M.E.; Gil, J.; Wang, J.; Degan, P.; Peters, G.; Martinez, D.; Carnero, A.; Beach, D. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005, 65, 177–185. [Google Scholar]
- Mathupala, S.P.; Rempel, A.; Pedersen, P.L. Aberrant Glycolytic Metabolism of Cancer Cells: A Remarkable Coordination of Genetic, Transcriptional, Post-translational, and Mutational Events That Lead to a Critical Role for Type II Hexokinase. J. Bioenerg. Biomembr. 1997, 29, 339–343. [Google Scholar] [CrossRef]
- Jiang, P.; Du, W.; Wang, X.; Mancuso, A.; Gao, X.; Wu, M.; Yang, X. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat. Cell Biol. 2011, 13, 310–316. [Google Scholar] [CrossRef]
- Schwartzenberg-Bar-Yoseph, F.; Armoni, M.; Karnieli, E. The Tumor Suppressor p53 Down-Regulates Glucose Transporters GLUT1 and GLUT4 Gene Expression. Cancer Res. 2004, 64, 2627–2633. [Google Scholar] [CrossRef]
- Kawauchi, K.; Araki, K.; Tobiume, K.; Tanaka, N. p53 regulates glucose metabolism through an IKK-NF-κB pathway and inhibits cell transformation. Nat. Cell Biol. 2008, 10, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Zawacka-Pankau, J.; Grinkevich, V.V.; Hünten, S.; Nikulenkov, F.; Gluch, A.; Li, H.; Enge, M.; Kel, A.; Selivanova, G. Inhibition of Glycolytic Enzymes Mediated by Pharmacologically Activated p53 TARGETING WARBURG EFFECT TO FIGHT CANCER. J. Biol. Chem. 2011, 286, 41600–41615. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, J.; Liang, Y.; Wu, R.; Zhao, Y.; Hong, X.; Lin, M.; Yu, H.; Liu, L.; Levine, A.J.; et al. Tumour-associated mutant p53 drives the Warburg effect. Nat. Commun. 2013, 4, 2935. [Google Scholar] [CrossRef] [PubMed]
- Boidot, R.; Végran, F.; Meulle, A.; Breton, A.L.; Dessy, C.; Sonveaux, P.; Lizard-Nacol, S.; Feron, O. Regulation of Monocarboxylate Transporter MCT1 Expression by p53 Mediates Inward and Outward Lactate Fluxes in Tumors. Cancer Res. 2012, 72, 939–948. [Google Scholar] [CrossRef]
- Bensaad, K.; Tsuruta, A.; Selak, M.A.; Vidal, M.N.C.; Nakano, K.; Bartrons, R.; Gottlieb, E.; Vousden, K.H. TIGAR, a p53-Inducible Regulator of Glycolysis and Apoptosis. Cell 2006, 126, 107–120. [Google Scholar] [CrossRef]
- Franklin, D.A.; He, Y.; Leslie, P.L.; Tikunov, A.P.; Fenger, N.; Macdonald, J.M.; Zhang, Y. p53 coordinates DNA repair with nucleotide synthesis by suppressing PFKFB3 expression and promoting the pentose phosphate pathway. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef]
- Ros, S.; Flöter, J.; Kaymak, I.; Da Costa, C.; Houddane, A.; Dubuis, S.; Griffiths, B.; Mitter, R.; Walz, S.; Blake, S.; et al. 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 is essential for p53-null cancer cells. Oncogene 2017, 36, 3287–3299. [Google Scholar] [CrossRef]
- Bao, Y.; Mukai, K.; Hishiki, T.; Kubo, A.; Ohmura, M.; Sugiura, Y.; Matsuura, T.; Nagahata, Y.; Hayakawa, N.; Yamamoto, T.; et al. Energy Management by Enhanced Glycolysis in G1-phase in Human Colon Cancer Cells In Vitro and In Vivo. Mol. Cancer Res. 2013, 11, 973–985. [Google Scholar] [CrossRef]
- Contractor, T.; Harris, C.R. p53 Negatively Regulates Transcription of the Pyruvate Dehydrogenase Kinase Pdk2. Cancer Res. 2012, 72, 560–567. [Google Scholar] [CrossRef]
- Zhang, C.; Lin, M.; Wu, R.; Wang, X.; Yang, B.; Levine, A.J.; Hu, W.; Feng, Z. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc. Natl. Acad. Sci. USA 2011, 108, 16259–16264. [Google Scholar] [CrossRef]
- Matoba, S.; Kang, J.-G.; Patino, W.D.; Wragg, A.; Boehm, M.; Gavrilova, O.; Hurley, P.J.; Bunz, F.; Hwang, P.M. p53 Regulates Mitochondrial Respiration. Science 2006, 312, 1650–1653. [Google Scholar] [CrossRef] [PubMed]
- Okamura, S.; Ng, C.C.; Koyama, K.; Takei, Y.; Arakawa, H.; Monden, M.; Nakamura, Y. Identification of seven genes regulated by wild-type p53 in a colon cancer cell line carrying a well-controlled wild-type p53 expression system. Oncol. Res. 1999, 11, 281–285. [Google Scholar] [PubMed]
- Hu, W.; Zhang, C.; Wu, R.; Sun, Y.; Levine, A.; Feng, Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc. Natl. Acad. Sci. USA 2010, 107, 7455–7460. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Tanaka, T.; Poyurovsky, M.V.; Nagano, H.; Mayama, T.; Ohkubo, S.; Lokshin, M.; Hosokawa, H.; Nakayama, T.; Suzuki, Y.; et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc. Natl. Acad. Sci. USA 2010, 107, 7461–7466. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Hou, L.; Li, L.; Li, L.; Zhu, L.; Wang, Y.; Huang, X.; Hou, Y.; Zhu, D.; Zou, H.; et al. Dichloroacetate restores colorectal cancer chemosensitivity through the p53/miR-149-3p/PDK2-mediated glucose metabolic pathway. Oncogene 2020, 39, 469–485. [Google Scholar] [CrossRef]
- Brown, R.E.; Short, S.P.; Williams, C.S. Colorectal Cancer and Metabolism. Curr. Colorectal Cancer Rep. 2018, 14, 226–241. [Google Scholar] [CrossRef]
- Robey, R.B.; Hay, N. Is Akt the “Warburg kinase”?-Akt-energy metabolism interactions and oncogenesis. Semin. Cancer Biol. 2009, 19, 25–31. [Google Scholar] [CrossRef]
- Fruman, D.A.; Chiu, H.; Hopkins, B.D.; Bagrodia, S.; Cantley, L.C.; Abraham, R.T. The PI3K Pathway in Human Disease. Cell 2017, 170, 605–635. [Google Scholar] [CrossRef]
- Hahn-Windgassen, A.; Nogueira, V.; Chen, C.-C.; Skeen, J.E.; Sonenberg, N.; Hay, N. Akt activates the mammalian target of rapamycin by regulating cellular ATP level and AMPK activity. J. Biol. Chem. 2005, 280, 32081–32089. [Google Scholar] [CrossRef]
- Gottlob, K.; Majewski, N.K.S.; Kandel, E.R.R.B.; Hay, N. Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes Dev. 2001, 1406–1418. [Google Scholar] [CrossRef]
- Roberts, D.J.; Miyamoto, S. Hexokinase II integrates energy metabolism and cellular protection: Akting on mitochondria and TORCing to autophagy. Cell Death Differ. 2015, 22, 248–257. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef]
- Sarbassov, D.D.; Ali, S.M.; Sabatini, D.M. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 2005, 17, 596–603. [Google Scholar] [CrossRef]
- Düvel, K.; Yecies, J.L.; Menon, S.; Raman, P.; Lipovsky, A.I.; Souza, A.L.; Triantafellow, E.; Ma, Q.; Gorski, R.; Cleaver, S.; et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell 2010, 39, 171–183. [Google Scholar] [CrossRef]
- Peterson, T.R.; Sengupta, S.S.; Harris, T.E.; Carmack, A.E.; Kang, S.A.; Balderas, E.; Guertin, D.A.; Madden, K.L.; Carpenter, A.E.; Finck, B.N.; et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 2011, 146, 408–420. [Google Scholar] [CrossRef]
- Muzny, D.M.; Bainbridge, M.N.; Chang, K.; Dinh, H.H.; Drummond, J.A.; Fowler, G.; Kovar, C.L.; Lewis, L.R.; Morgan, M.B.; Newsham, I.F.; et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar]
- Atreya, C.E.; Sangale, Z.; Xu, N.; Matli, M.R.; Tikishvili, E.; Welbourn, W.; Stone, S.; Shokat, K.M.; Warren, R.S. PTEN expression is consistent in colorectal cancer primaries and metastases and associates with patient survival. Cancer Med. 2013, 2, 496–506. [Google Scholar] [CrossRef]
- Kato, S.; Iida, S.; Higuchi, T.; Ishikawa, T.; Takagi, Y.; Yasuno, M.; Enomoto, M.; Uetake, H.; Sugihara, K. PIK3CA mutation is predictive of poor survival in patients with colorectal cancer. Int. J. Cancer 2007, 121, 1771–1778. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Simon, I.; Moreno, V.; Roepman, P.; Tabernero, J.; Snel, M.; van’t Veer, L.; Salazar, R.; Bernards, R.; Capella, G. A combined oncogenic pathway signature of BRAF, KRAS and PI3KCA mutation improves colorectal cancer classification and cetuximab treatment prediction. Gut 2013, 62, 540–549. [Google Scholar] [CrossRef] [PubMed]
- Samuels, Y.; Wang, Z.; Bardelli, A.; Silliman, N.; Ptak, J.; Szabo, S.; Yan, H.; Gazdar, A.; Powell, S.M.; Riggins, G.J.; et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 2004, 304, 554. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Samuels, Y.; Li, Q.; Krokowski, D.; Guan, B.-J.; Wang, C.; Jin, Z.; Dong, B.; Cao, B.; Feng, X.; et al. Oncogenic PIK3CA mutations reprogram glutamine metabolism in colorectal cancer. Nat. Commun. 2016, 7, 11971. [Google Scholar] [CrossRef] [PubMed]
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V.; O’Donnell, K.A.; Zeller, K.I.; Nguyen, T.; Osthus, R.C.; Li, F. The c-Myc target gene network. Semin. Cancer Biol. 2006, 16, 253–264. [Google Scholar] [CrossRef] [PubMed]
- He, T.C.; Sparks, A.B.; Rago, C.; Hermeking, H.; Zawel, L.; da Costa, L.T.; Morin, P.J.; Vogelstein, B.; Kinzler, K.W. Identification of c-MYC as a target of the APC pathway. Science 1998, 281, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Satoh, K.; Yachida, S.; Sugimoto, M.; Oshima, M.; Nakagawa, T.; Akamoto, S.; Tabata, S.; Saitoh, K.; Kato, K.; Sato, S.; et al. Global metabolic reprogramming of colorectal cancer occurs at adenoma stage and is induced by MYC. Proc. Natl. Acad. Sci. USA 2017, 114, E7697–E7706. [Google Scholar] [CrossRef]
- Tang, J.; Yan, T.; Bao, Y.; Shen, C.; Yu, C.; Zhu, X.; Tian, X.; Guo, F.; Liang, Q.; Liu, Q.; et al. LncRNA GLCC1 promotes colorectal carcinogenesis and glucose metabolism by stabilizing c-Myc. Nat. Commun. 2019, 10, 3499. [Google Scholar] [CrossRef]
- Guo, X.; Zhu, Y.; Hong, X.; Zhang, M.; Qiu, X.; Wang, Z.; Qi, Z.; Hong, X. miR-181d and c-myc-mediated inhibition of CRY2 and FBXL3 reprograms metabolism in colorectal cancer. Cell Death Dis. 2017, 8, e2958. [Google Scholar] [CrossRef]
- Brenner, C.; Deplus, R.; Didelot, C.; Loriot, A.; Viré, E.; Smet, C.; Gutierrez, A.; Danovi, D.; Bernard, D.; Boon, T.; et al. Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J. 2005, 24, 336–346. [Google Scholar] [CrossRef]
- Dang, C.V. MYC on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef]
- Jang, M.; Kim, S.S.; Lee, J. Cancer cell metabolism: Implications for therapeutic targets. Exp. Mol. Med. 2013, 45, e45. [Google Scholar] [CrossRef]
- Kim, T.; Jeon, Y.-J.; Cui, R.; Lee, J.-H.; Peng, Y.; Kim, S.-H.; Tili, E.; Alder, H.; Croce, C.M. Role of MYC-regulated long noncoding RNAs in cell cycle regulation and tumorigenesis. J. Natl. Cancer Inst. 2015, 107. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, Y.; Zeller, K.I.; Potter, J.J.; Wonsey, D.R.; O’Donnell, K.A.; Kim, J.-W.; Yustein, J.T.; Lee, L.A.; Dang, C.V. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol. Cell. Biol. 2005, 25, 6225–6234. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Tchernyshyov, I.; Chang, T.-C.; Lee, Y.-S.; Kita, K.; Ochi, T.; Zeller, K.I.; Marzo, A.M.; van Eyk, J.E.; Mendell, J.T.; et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 2009, 458, 762–765. [Google Scholar] [CrossRef]
- Xu, X.; Li, J.; Sun, X.; Guo, Y.; Chu, D.W.L.; Li, X.; Yang, G.; Liu, X.; Yao, L.; Zhang, J.; et al. Tumor suppressor NDRG2 inhibits glycolysis and glutaminolysis in colorectal cancer cells by repressing c-Myc expression. Oncotarget 2015, 6, 26161–26176. [Google Scholar] [CrossRef] [PubMed]
- Locasale, J.W. Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat. Rev. Cancer 2013, 13, 572–583. [Google Scholar] [CrossRef]
- Higashi, K.; Yamagishi, T.; Ueda, Y.; Ishigaki, Y.; Shimasaki, M.; Nakamura, Y.; Oguchi, M.; Takegami, T.; Sagawa, M.; Tonami, H. Correlation of HIF-1α/HIF-2α expression with FDG uptake in lung adenocarcinoma. Ann. Nucl. Med. 2016, 30, 708–715. [Google Scholar] [CrossRef] [PubMed]
- Nagaraju, G.P.; Bramhachari, P.V.; Raghu, G.; El-Rayes, B.F. Hypoxia inducible factor-1α: Its role in colorectal carcinogenesis and metastasis. Cancer Lett. 2015, 366, 11–18. [Google Scholar] [CrossRef]
- Vadde, R.; Vemula, S.; Jinka, R.; Merchant, N.; Bramhachari, P.V.; Nagaraju, G.P. Role of hypoxia-inducible factors (HIF) in the maintenance of stemness and malignancy of colorectal cancer. Crit. Rev. Oncol. Hematol. 2017, 113, 22–27. [Google Scholar] [CrossRef]
- Semenza, G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 2003, 3, 721–732. [Google Scholar] [CrossRef]
- Cao, D.; Hou, M.; Guan, Y.-S.; Jiang, M.; Yang, Y.; Gou, H.-F. Expression of HIF-1alpha and VEGF in colorectal cancer: Association with clinical outcomes and prognostic implications. BMC Cancer 2009, 9, 432. [Google Scholar] [CrossRef]
- Tsujii, M.; Kamano, S.; Tsuji, S.; Sawaoka, H.; H´ori, M.; DuBois, R.N. Cyclooxygenase Regulates Angiogenesis Induced by Colon Cancer Cells. Cell 1998, 93, 705–716. [Google Scholar] [CrossRef]
- Forsythe, J.A.; Jiang, B.-H.; Iyer, N.V.; Agani, F.; Leung, S.W.; Koos, R.D.; Semenza, G.L. Activation of Vascular Endothelial Growth Factor Gene Transcription by Hypoxia-Inducible Factor 1. Mol. Cell. Biol. 1996, 16, 4604–4613. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Yang, L.; Xie, X.; Xie, F.; Pan, F.; Li, J.; He, J.; Liang, H. Expression and significance of hypoxia-inducible factor-1 alpha and MDR1/P-glycoprotein in human colon carcinoma tissue and cells. J. Cancer Res. Clin. Oncol. 2010, 136, 1697–1707. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ding, Z.; Peng, Y.; Pan, F.; Li, J.; Zou, L.; Zhang, Y.; Liang, H. HIF-1α inhibition reverses multidrug resistance in colon cancer cells via downregulation of MDR1/P-glycoprotein. PLoS ONE 2014, 9, e98882. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Carbonneau, M.; Gagné, M.L.; Lalonde, M.-E.; Germain, M.-A.; Motorina, A.; Guiot, M.-C.; Secco, B.; Vincent, E.E.; Tumber, A.; Hulea, L.; et al. The oncometabolite 2-hydroxyglutarate activates the mTOR signalling pathway. Nat. Commun. 2016, 7, 12700. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, X.; Chen, V.; Feng, Y.; Wang, L.; Croniger, C.; Conlon, R.A.; Markowitz, S.; Fearon, E.; Puchowicz, M.; et al. Colorectal cancers utilize glutamine as an anaplerotic substrate of the TCA cycle in vivo. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Owen, O.E.; Kalhan, S.C.; Hanson, R.W. The Key Role of Anaplerosis and Cataplerosis for Citric Acid Cycle Function. J. Biol. Chem. 2002, 277, 30409–30412. [Google Scholar] [CrossRef]
- Song, Z.; Wei, B.; Lu, C.; Li, P.; Chen, L. Glutaminase sustains cell survival via the regulation of glycolysis and glutaminolysis in colorectal cancer. Oncol. Lett. 2017, 14, 3117–3123. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Zhang, Q.; Ma, H.; Lv, Q.; Zhang, T. Expression of glutaminase is upregulated in colorectal cancer and of clinical significance. Int. J. Clin. Exp. Pathol. 2014, 7, 1093–1100. [Google Scholar]
- Liu, G.; Zhu, J.; Yu, M.; Cai, C.; Zhou, Y.; Yu, M.; Fu, Z.; Nguyen, C.; Yang, B.; Li, Y.; et al. Glutamate dehydrogenase is a novel prognostic marker and predicts metastases in colorectal cancer patients. J. Transl. Med. 2015, 13, 144. [Google Scholar] [CrossRef] [PubMed]
- Marin-Valencia, I.; Yang, C.; Mashimo, T.; Cho, S.; Baek, H.; Yang, X.-L.; Rajagopalan, K.N.; Maddie, M.; Vemireddy, V.; Zhao, Z.; et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012, 15, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Budczies, J.; Weichert, W.; Wohlgemuth, G.; Scholz, M.; Kind, T.; Niesporek, S.; Noske, A.; Buckendahl, A.; Dietel, M.; et al. Metabolite profiling of human colon carcinoma—Deregulation of TCA cycle and amino acid turnover. Mol. Cancer 2008, 7, 72. [Google Scholar] [CrossRef]
- Mal, M.; Koh, P.K.; Cheah, P.Y.; Chan, E.C.Y. Metabotyping of human colorectal cancer using two-dimensional gas chromatography mass spectrometry. Anal. Bioanal. Chem. 2012, 403, 483–493. [Google Scholar] [CrossRef]
- Ong, E.S.; Zou, L.; Li, S.; Cheah, P.Y.; Eu, K.W.; Ong, C.N. Metabolic profiling in colorectal cancer reveals signature metabolic shifts during tumorigenesis. Mol. Cell. Proteomics 2010. [Google Scholar] [CrossRef] [PubMed]
- Shiratori, R.; Furuichi, K.; Yamaguchi, M.; Miyazaki, N.; Aoki, H.; Chibana, H.; Ito, K.; Aoki, S. Glycolytic suppression dramatically changes the intracellular metabolic profile of multiple cancer cell lines in a mitochondrial metabolism-dependent manner. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Cairns, R.A.; Mak, T.W. Oncogenic isocitrate dehydrogenase mutations: Mechanisms, models, and clinical opportunities. Cancer Discov. 2013, 3, 730–741. [Google Scholar] [CrossRef] [PubMed]
- Marquez, J.; Flores, J.; Kim, A.H.; Nyamaa, B.; Nguyen, A.T.T.; Park, N.; Han, J. Rescue of TCA Cycle Dysfunction for Cancer Therapy. J. Clin. Med. 2019, 8, 2161. [Google Scholar] [CrossRef]
- Waitkus, M.S.; Diplas, B.H.; Yan, H. Biological Role and Therapeutic Potential of IDH Mutations in Cancer. Cancer Cell 2018, 34, 186–195. [Google Scholar] [CrossRef]
- Tougeron, D.; Guilloteau, K.; Karayan-Tapon, L. Absence of IDH mutation in colorectal cancers with microsatellite instability. Dig. Liver Dis. 2016, 48, 681–683. [Google Scholar] [CrossRef]
- Whitehall, V.L.J.; Dumenil, T.D.; McKeone, D.M.; Bond, C.E.; Bettington, M.L.; Buttenshaw, R.L.; Bowdler, L.; Montgomery, G.W.; Wockner, L.F.; Leggett, B.A. Isocitrate dehydrogenase 1 R132C mutation occurs exclusively in microsatellite stable colorectal cancers with the CpG island methylator phenotype. Epigenetics 2014, 9, 1454–1460. [Google Scholar] [CrossRef] [PubMed]
- de Vogel, S.; Weijenberg, M.P.; Herman, J.G.; Wouters, K.A.; de Goeij, A.F.; van den Brandt, P.A.; de Bruine, A.P.; van Engeland, M. MGMT and MLH1 promoter methylation versus APC, KRAS and BRAF gene mutations in colorectal cancer: Indications for distinct pathways and sequence of events. Ann. Oncol. 2009, 20, 1216–1222. [Google Scholar] [CrossRef]
- Fallah-Rad, N.; Bedard, P.L.; Siu, L.L.; Kamel-Reid, S.; Chow, H.; Weijiang, Z.; Jang, R.; Krzyzanowska, M.K.; Razak, A.R.A.; Chen, E.X. Association of isocitrate dehydorgenase-1 (IDH-1) mutations with elevated oncometabolite 2-hydroxyglutarate (2HG) in advanced colorectal cancer. J. Clin. Oncol. 2016, 34, 627. [Google Scholar] [CrossRef]
- Alpert, L.; Yassan, L.; Poon, R.; Kadri, S.; Niu, N.; Patil, S.A.; Mujacic, I.; Montes, D.; Galbo, F.; Wurst, M.N.; et al. Targeted mutational analysis of inflammatory bowel disease-associated colorectal cancers. Hum. Pathol. 2019, 89, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Yaeger, R.; Shah, M.A.; Miller, V.A.; Kelsen, J.R.; Wang, K.; Heins, Z.J.; Ross, J.S.; He, Y.; Sanford, E.; Yantiss, R.K.; et al. Genomic Alterations Observed in Colitis-Associated Cancers Are Distinct From Those Found in Sporadic Colorectal Cancers and Vary by Type of Inflammatory Bowel Disease. Gastroenterology 2016, 151, 278–287. [Google Scholar] [CrossRef]
- Colvin, H.; Nishida, N.; Konno, M.; Haraguchi, N.; Takahashi, H.; Nishimura, J.; Hata, T.; Kawamoto, K.; Asai, A.; Tsunekuni, K.; et al. Oncometabolite D-2-Hydroxyglurate Directly Induces Epithelial-Mesenchymal Transition and is Associated with Distant Metastasis in Colorectal Cancer. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.Q.; Hanse, E.A.; Habowski, A.N.; Li, H.; Ishak Gabra, M.B.; Yang, Y.; Lowman, X.H.; Ooi, A.M.; Liao, S.Y.; Edwards, R.A.; et al. α-Ketoglutarate attenuates Wnt signaling and drives differentiation in colorectal cancer. Nat. Cancer 2020, 1, 345–358. [Google Scholar] [CrossRef]
- Lawson, D.A.; Kessenbrock, K.; Davis, R.T.; Pervolarakis, N.; Werb, Z. Tumour heterogeneity and metastasis at single-cell resolution. Nat. Cell Biol. 2018, 20, 1349–1360. [Google Scholar] [CrossRef]
- Pavlides, S.; Whitaker-Menezes, D.; Castello-Cros, R.; Flomenberg, N.; Witkiewicz, A.K.; Frank, P.G.; Casimiro, M.C.; Wang, C.; Fortina, P.; Addya, S.; et al. The reverse Warburg effect: Aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle 2009, 8, 3984–4001. [Google Scholar] [CrossRef]
- Zhou, W.; Xu, G.; Wang, Y.; Xu, Z.; Liu, X.; Xu, X.; Ren, G.; Tian, K. Oxidative stress induced autophagy in cancer associated fibroblast enhances proliferation and metabolism of colorectal cancer cells. Cell Cycle 2017, 16, 73–81. [Google Scholar] [CrossRef]
- Koukourakis, M.I.; Giatromanolaki, A.; Harris, A.L.; Sivridis, E. Comparison of metabolic pathways between cancer cells and stromal cells in colorectal carcinomas: A metabolic survival role for tumor-associated stroma. Cancer Res. 2006, 66, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Dalerba, P.; Dylla, S.J.; Park, I.-K.; Liu, R.; Wang, X.; Cho, R.W.; Hoey, T.; Gurney, A.; Huang, E.H.; Simeone, D.M.; et al. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 10158–10163. [Google Scholar] [CrossRef] [PubMed]
- Munro, M.J.; Wickremesekera, S.K.; Peng, L.; Tan, S.T.; Itinteang, T. Cancer stem cells in colorectal cancer: A review. J. Clin. Pathol. 2018, 71, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J. Cancer stem cells and chemoresistance: The smartest survives the raid. Pharmacol. Ther. 2016, 160, 145–158. [Google Scholar] [CrossRef]
- Sancho, P.; Barneda, D.; Heeschen, C. Hallmarks of cancer stem cell metabolism. Br. J. Cancer 2016, 114, 1305–1312. [Google Scholar] [CrossRef]
- Vincent, Z.; Urakami, K.; Maruyama, K.; Yamaguchi, K.; Kusuhara, M. CD133-positive cancer stem cells from Colo205 human colon adenocarcinoma cell line show resistance to chemotherapy and display a specific metabolomic profile. Genes Cancer 2014, 5, 250–260. [Google Scholar] [CrossRef]
- Song, I.-S.; Jeong, Y.J.; Jeong, S.H.; Heo, H.J.; Kim, H.K.; Bae, K.B.; Park, Y.-H.; Kim, S.U.; Kim, J.-M.; Kim, N.; et al. FOXM1-Induced PRX3 Regulates Stemness and Survival of Colon Cancer Cells via Maintenance of Mitochondrial Function. Gastroenterology 2015, 149, 1006–1016.e1009. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, K.J.; Seo, Y.; Kwon, J.-H.; Yoon, J.P.; Kang, J.Y.; Lee, H.J.; Park, S.J.; Hong, S.P.; Cheon, J.H.; et al. Effects of metformin on colorectal cancer stem cells depend on alterations in glutamine metabolism. Sci. Rep. 2018, 8, 409. [Google Scholar] [CrossRef]
- Kahlert, U.D.; Mooney, S.M.; Natsumeda, M.; Steiger, H.J.; Maciaczyk, J. Targeting cancer stem-like cells in glioblastoma and colorectal cancer through metabolic pathways. Int. J. Cancer 2017, 140, 10–22. [Google Scholar] [CrossRef]
- Cassim, S.; Pouyssegur, J. Tumor Microenvironment: A Metabolic Player that Shapes the Immune Response. Int. J. Mol. Sci. 2019, 21, 157. [Google Scholar] [CrossRef]
- Herbel, C.; Patsoukis, N.; Bardhan, K.; Seth, P.; Weaver, J.D.; Boussiotis, V.A. Clinical significance of T cell metabolic reprogramming in cancer. Clin. Transl. Med. 2016, 5. [Google Scholar] [CrossRef]
- Wang, R.; Dillon, C.P.; Shi, L.Z.; Milasta, S.; Carter, R.; Finkelstein, D.; McCormick, L.L.; Fitzgerald, P.; Chi, H.; Munger, J.; et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 2011, 35, 871–882. [Google Scholar] [CrossRef]
- Chou, C.; Pinto, A.K.; Curtis, J.D.; Persaud, S.P.; Cella, M.; Lin, C.-C.; Edelson, B.T.; Allen, P.M.; Colonna, M.; Pearce, E.L.; et al. c-Myc-induced transcription factor AP4 is required for host protection mediated by CD8+ T cells. Nat. Immunol. 2014, 15, 884–893. [Google Scholar] [CrossRef]
- Jacobs, S.R.; Herman, C.E.; MacIver, N.J.; Wofford, J.A.; Wieman, H.L.; Hammen, J.J.; Rathmell, J.C. Glucose Uptake Is Limiting in T Cell Activation and Requires CD28-Mediated Akt-Dependent and Independent Pathways. J. Immunol. 2008, 180, 4476–4486. [Google Scholar] [CrossRef] [PubMed]
- Carr, E.L.; Kelman, A.; Wu, G.S.; Gopaul, R.; Senkevitch, E.; Aghvanyan, A.; Turay, A.M.; Frauwirth, K.A. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J. Immunol. 2010, 185, 1037–1044. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-H.; Qiu, J.; O’Sullivan, D.; Buck, M.D.; Noguchi, T.; Curtis, J.D.; Chen, Q.; Gindin, M.; Gubin, M.M.; van der Windt, G.J.W.; et al. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell 2015, 162, 1229–1241. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.-C.; Bihuniak, J.D.; Macintyre, A.N.; Staron, M.; Liu, X.; Amezquita, R.; Tsui, Y.-C.; Cui, G.; Micevic, G.; Perales, J.C.; et al. Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell 2015, 162, 1217–1228. [Google Scholar] [CrossRef]
- Nakaya, M.; Xiao, Y.; Zhou, X.; Chang, J.-H.; Chang, M.; Cheng, X.; Blonska, M.; Lin, X.; Sun, S.-C. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity 2014, 40, 692–705. [Google Scholar] [CrossRef]
- Patsoukis, N.; Bardhan, K.; Chatterjee, P.; Sari, D.; Liu, B.; Bell, L.N.; Karoly, E.D.; Freeman, G.J.; Petkova, V.; Seth, P.; et al. PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat. Commun. 2015, 6, 6692. [Google Scholar] [CrossRef]
- Gerriets, V.A.; Rathmell, J.C. Metabolic pathways in T cell fate and function. Trends Immunol. 2012, 33, 168–173. [Google Scholar] [CrossRef]
- Michalek, R.D.; Gerriets, V.A.; Jacobs, S.R.; Macintyre, A.N.; MacIver, N.J.; Mason, E.F.; Sullivan, S.A.; Nichols, A.G.; Rathmell, J.C. Cutting Edge: Distinct Glycolytic and Lipid Oxidative Metabolic Programs Are Essential for Effector and Regulatory CD4+ T Cell Subsets. J. Immunol. 2011, 186, 3299–3303. [Google Scholar] [CrossRef]
- Gerriets, V.A.; Kishton, R.J.; Johnson, M.O.; Cohen, S.; Siska, P.J.; Nichols, A.G.; Warmoes, M.O.; de Cubas, A.A.; MacIver, N.J.; Locasale, J.W.; et al. Foxp3 and Toll-like receptor signaling balance Treg cell anabolic metabolism for suppression. Nat. Immunol. 2016, 17, 1459–1466. [Google Scholar] [CrossRef] [PubMed]
- Pollard, J.W. Tumour-educated macrophages promote tumour progression and metastasis. Nat. Rev. Cancer 2004, 4, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.-Z.; Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Penny, H.L.; Sieow, J.L.; Adriani, G.; Yeap, W.H.; See Chi Ee, P.; San Luis, B.; Lee, B.; Lee, T.; Mak, S.Y.; Ho, Y.S.; et al. Warburg metabolism in tumor-conditioned macrophages promotes metastasis in human pancreatic ductal adenocarcinoma. Oncoimmunology 2016, 5, e1191731. [Google Scholar] [CrossRef] [PubMed]
- Tannahill, G.M.; Curtis, A.M.; Adamik, J.; Palsson-McDermott, E.M.; McGettrick, A.F.; Goel, G.; Frezza, C.; Bernard, N.J.; Kelly, B.; Foley, N.H.; et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 2013, 496, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.-Y.; Huang, T.-W.; Hsieh, Y.-T.; Wang, Y.-F.; Yen, C.-C.; Lee, G.-L.; Yeh, C.-C.; Peng, Y.-J.; Kuo, Y.-Y.; Wen, H.-T.; et al. Cancer-Derived Succinate Promotes Macrophage Polarization and Cancer Metastasis via Succinate Receptor. Mol. Cell 2020, 77, 213–227.e215. [Google Scholar] [CrossRef] [PubMed]
- Parks, S.K.; Mueller-Klieser, W.; Pouysségur, J. Lactate and Acidity in the Cancer Microenvironment. Annu. Rev. Cancer Biol. 2020, 4, 141–158. [Google Scholar] [CrossRef]
- Colegio, O.R.; Chu, N.-Q.; Szabo, A.L.; Chu, T.; Rhebergen, A.M.; Jairam, V.; Cyrus, N.; Brokowski, C.E.; Eisenbarth, S.C.; Phillips, G.M.; et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 2014, 513, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Tang, Z.; Huang, H.; Zhou, G.; Cui, C.; Weng, Y.; Liu, W.; Kim, S.; Lee, S.; Perez-Neut, M.; et al. Metabolic regulation of gene expression by histone lactylation. Nature 2019, 574, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Fischer, B.; Müller, B.; Fisch, P.; Kreutz, W. An acidic microenvironment inhibits antitumoral non-major histocompatibility complex-restricted cytotoxicity: Implications for cancer immunotherapy. J. Immunother. 2000, 23, 196–207. [Google Scholar] [CrossRef]
- Calcinotto, A.; Filipazzi, P.; Grioni, M.; Iero, M.; De Milito, A.; Ricupito, A.; Cova, A.; Canese, R.; Jachetti, E.; Rossetti, M.; et al. Modulation of microenvironment acidity reverses anergy in human and murine tumor-infiltrating T lymphocytes. Cancer Res. 2012, 72, 2746–2756. [Google Scholar] [CrossRef] [PubMed]
- Bosticardo, M.; Ariotti, S.; Losana, G.; Bernabei, P.; Forni, G.; Novelli, F. Biased activation of human T lymphocytes due to low extracellular pH is antagonized by B7/CD28 costimulation. Eur. J. Immunol. 2001, 31, 2829–2838. [Google Scholar] [CrossRef]
- Fulda, S.; Galluzzi, L.; Kroemer, G. Targeting mitochondria for cancer therapy. Nat. Rev. Drug Discov. 2010, 9, 447–464. [Google Scholar] [CrossRef] [PubMed]
- Tennant, D.A.; Durán, R.V.; Gottlieb, E. Targeting metabolic transformation for cancer therapy. Nat. Rev. Cancer 2010, 10, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Wilson, W.R.; Hay, M.P. Targeting hypoxia in cancer therapy. Nat. Rev. Cancer 2011, 11, 393–410. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G. Targeting cancer metabolism: A therapeutic window opens. Nat. Rev. Drug Discov. 2011, 10, 671–684. [Google Scholar] [CrossRef]
- Anderson, R.G.; Ghiraldeli, L.P.; Pardee, T.S. Mitochondria in cancer metabolism, an organelle whose time has come? Biochim. Biophys. Acta Rev. Cancer 2018, 1870, 96–102. [Google Scholar] [CrossRef]
- Jones, N.P.; Schulze, A. Targeting cancer metabolism—Aiming at a tumour’s sweet-spot. Drug Discov. Today 2012, 17, 232–241. [Google Scholar] [CrossRef]
- Luengo, A.; Gui, D.Y.; Heiden, M.G.V. Targeting Metabolism for Cancer Therapy. Cell Chem. Biol. 2017, 24, 1161–1180. [Google Scholar] [CrossRef]
- Sośnicki, S.; Kapral, M.; Węglarz, L. Molecular targets of metformin antitumor action. Pharmacol. Rep. 2016, 68, 918–925. [Google Scholar] [CrossRef] [PubMed]
- Jara, J.A.; López-Muñoz, R. Metformin and cancer: Between the bioenergetic disturbances and the antifolate activity. Pharmacol. Res. 2015, 101, 102–108. [Google Scholar] [CrossRef]
- Andrzejewski, S.; Gravel, S.-P.; Pollak, M.; St-Pierre, J. Metformin directly acts on mitochondria to alter cellular bioenergetics. Cancer Metabol. 2014, 2, 12. [Google Scholar] [CrossRef] [PubMed]
- Kamarudin, M.N.A.; Sarker, M.M.R.; Zhou, J.-R.; Parhar, I. Metformin in colorectal cancer: Molecular mechanism, preclinical and clinical aspects. J. Exp. Clin. Cancer Res. 2019, 38, 491. [Google Scholar] [CrossRef]
- Ciccarone, F.; Vegliante, R.; Di Leo, L.; Ciriolo, M.R. The TCA cycle as a bridge between oncometabolism and DNA transactions in cancer. Semin. Cancer Biol. 2017, 47, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.M.; Mucka, P.; Kern, J.G.; Feng, H. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell 2018, 9, 216–237. [Google Scholar] [CrossRef] [PubMed]
- Michelakis, E.D.; Sutendra, G.; Dromparis, P.; Webster, L.; Haromy, A.; Niven, E.; Maguire, C.; Gammer, T.L.; Mackey, J.R.; Fulton, D.; et al. Metabolic modulation of glioblastoma with dichloroacetate. Sci. Transl. Med. 2010, 2, 31ra34. [Google Scholar] [CrossRef]
- Liu, X.; Gong, Y. Isocitrate dehydrogenase inhibitors in acute myeloid leukemia. Biomark. Res. 2019, 7, 22. [Google Scholar] [CrossRef]
- Gross, M.I.; Demo, S.D.; Dennison, J.B.; Chen, L.; Chernov-Rogan, T.; Goyal, B.; Janes, J.R.; Laidig, G.J.; Lewis, E.R.; Li, J.; et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol. Cancer Ther. 2014, 13, 890–901. [Google Scholar] [CrossRef]
- Reckzeh, E.S.; Karageorgis, G.; Schwalfenberg, M.; Ceballos, J.; Nowacki, J.; Stroet, M.C.M.; Binici, A.; Knauer, L.; Brand, S.; Choidas, A.; et al. Inhibition of Glucose Transporters and Glutaminase Synergistically Impairs Tumor Cell Growth. Cell Chem. Biol. 2019, 26, 1214–1228.e1225. [Google Scholar] [CrossRef]
- Li, L.; Meng, Y.; Li, Z.; Dai, W.; Xu, X.; Bi, X.; Bian, J. Discovery and development of small molecule modulators targeting glutamine metabolism. Eur. J. Med. Chem. 2019, 163, 215–242. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, D.; Xu, N.; Fang, J.; Yu, Y.; Hou, W.; Ruan, H.; Zhu, P.; Ma, R.; Lu, S.; et al. Novel 1,3,4-Selenadiazole-Containing Kidney-Type Glutaminase Inhibitors Showed Improved Cellular Uptake and Antitumor Activity. J. Med. Chem. 2019, 62, 589–603. [Google Scholar] [CrossRef] [PubMed]
- Stuart, S.D.; Schauble, A.; Gupta, S.; Kennedy, A.D.; Keppler, B.R.; Bingham, P.M.; Zachar, Z. A strategically designed small molecule attacks alpha-ketoglutarate dehydrogenase in tumor cells through a redox process. Cancer Metab. 2014, 2, 4. [Google Scholar] [CrossRef] [PubMed]
- Zachar, Z.; Marecek, J.; Maturo, C.; Gupta, S.; Stuart, S.D.; Howell, K.; Schauble, A.; Lem, J.; Piramzadian, A.; Karnik, S.; et al. Non-redox-active lipoate derivates disrupt cancer cell mitochondrial metabolism and are potent anticancer agents in vivo. J. Mol. Med. 2011, 89, 1137–1148. [Google Scholar] [CrossRef]
- Dörsam, B.; Fahrer, J. The disulfide compound α-lipoic acid and its derivatives: A novel class of anticancer agents targeting mitochondria. Cancer Lett. 2016, 371, 12–19. [Google Scholar] [CrossRef]
- Farhat, D.; Lincet, H. Lipoic acid a multi-level molecular inhibitor of tumorigenesis. Biochim. Biophys. Acta (BBA) Rev. Cancer 2020, 1873, 188317. [Google Scholar] [CrossRef]
- Dörsam, B.; Göder, A.; Seiwert, N.; Kaina, B.; Fahrer, J. Lipoic acid induces p53-independent cell death in colorectal cancer cells and potentiates the cytotoxicity of 5-fluorouracil. Arch. Toxicol. 2015, 89, 1829–1846. [Google Scholar] [CrossRef]
- Wenzel, U.; Nickel, A.; Daniel, H. α-lipoic acid induces apoptosis in human colon cancer cells by increasing mitochondrial respiration with a concomitant O2-*- generation. Apoptosis 2005, 10, 359–368. [Google Scholar] [CrossRef]
- Neitzel, C.; Seiwert, N.; Göder, A.; Diehl, E.; Weber, C.; Nagel, G.; Stroh, S.; Rasenberger, B.; Christmann, M.; Fahrer, J. Lipoic Acid Synergizes with Antineoplastic Drugs in Colorectal Cancer by Targeting p53 for Proteasomal Degradation. Cells 2019, 8, 794. [Google Scholar] [CrossRef]
- Göder, A.; Nagel, G.; Kraus, A.; Dörsam, B.; Seiwert, N.; Kaina, B.; Fahrer, J. Lipoic acid inhibits the DNA repair protein O 6-methylguanine-DNA methyltransferase (MGMT) and triggers its depletion in colorectal cancer cells with concomitant autophagy induction. Carcinogenesis 2015, 36, 817–831. [Google Scholar] [CrossRef]
- Bunik, V.I. 2-Oxo acid dehydrogenase complexes in redox regulation. Eur. J. Biochem. 2003, 270, 1036–1042. [Google Scholar] [CrossRef]
- Pardee, T.S.; Lee, K.; Luddy, J.; Maturo, C.; Rodriguez, R.; Isom, S.; Miller, L.D.; Stadelman, K.M.; Levitan, D.; Hurd, D.; et al. A phase I study of the first-in-class antimitochondrial metabolism agent, CPI-613, in patients with advanced hematologic malignancies. Clin. Cancer Res. 2014, 20, 5255–5264. [Google Scholar] [CrossRef] [PubMed]
- Oppermann, H.; Schnabel, L.; Meixensberger, J.; Gaunitz, F. Pyruvate attenuates the anti-neoplastic effect of carnosine independently from oxidative phosphorylation. Oncotarget 2016, 7, 85848–85860. [Google Scholar] [CrossRef]
- O’Brien, M.; Chalker, J.; Slade, L.; Gardiner, D.; Mailloux, R.J. Protein S-glutathionylation alters superoxide/hydrogen peroxide emission from pyruvate dehydrogenase complex. Free Radic. Biol. Med. 2017, 106, 302–314. [Google Scholar] [CrossRef] [PubMed]
- Slade, L.; Chalker, J.; Kuksal, N.; Young, A.; Gardiner, D.; Mailloux, R.J. Examination of the superoxide/hydrogen peroxide forming and quenching potential of mouse liver mitochondria. Biochim. Biophys. Acta (BBA) Gen. Subj. 2017, 1861, 1960–1969. [Google Scholar] [CrossRef] [PubMed]
- Dogar, I.; Dixon, S.; Gill, R.; Young, A.; Mallay, S.; Oldford, C.; Mailloux, R.J. C57BL/6J mice upregulate catalase to maintain the hydrogen peroxide buffering capacity of liver mitochondria. Free Radic. Biol. Med. 2020, 146, 59–69. [Google Scholar] [CrossRef]
- Young, A.; Gardiner, D.; Brosnan, M.E.; Brosnan, J.T.; Mailloux, R.J. Physiological levels of formate activate mitochondrial superoxide/hydrogen peroxide release from mouse liver mitochondria. FEBS Lett. 2017, 591, 2426–2438. [Google Scholar] [CrossRef]
- Young, A.; Oldford, C.; Mailloux, R.J. Lactate dehydrogenase supports lactate oxidation in mitochondria isolated from different mouse tissues. Redox Biol. 2020, 28, 101339. [Google Scholar] [CrossRef]
- Meyer, A.P.; Carvalho, R.A.; Lorenz, H.M.; Souto-Carneiro, M. 05.15 Altering peripheral cd8+ t-cell function in ra through metabolic modulation with small molecule agents 3-bromopyruvate, fx-11 and cpi-613. Ann. Rheum. Dis. 2017, 76, A58. [Google Scholar] [CrossRef]
- Arai, S.; Chen, S.; Xie, L.; Balk, S.P. Ubiquitin Ligase MARCH5 Regulates Apoptosis through Mediation of Stress-Induced and NOXA-Dependent MCL1 Degradation. bioRxiv 2020. [Google Scholar] [CrossRef]
- Gao, L.; Xu, Z.; Huang, Z.; Tang, Y.; Yang, D.; Huang, J.; He, L.; Liu, M.; Chen, Z.; Teng, Y. CPI-613 rewires lipid metabolism to enhance pancreatic cancer apoptosis via the AMPK-ACC signaling. J. Exp. Clin. Cancer Res. 2020, 39, 73. [Google Scholar] [CrossRef] [PubMed]
- Pardee, T.S.; Anderson, R.G.; Pladna, K.M.; Isom, S.; Ghiraldeli, L.P.; Miller, L.D.; Chou, J.W.; Jin, G.; Zhang, W.; Ellis, L.R.; et al. A Phase I Study of CPI-613 in Combination with High-Dose Cytarabine and Mitoxantrone for Relapsed or Refractory Acute Myeloid Leukemia. Clin. Cancer Res. 2018, 24, 2060–2073. [Google Scholar] [CrossRef] [PubMed]
- Pardee, T.S.; Miller, L.D.; Pladna, K.; Isom, S.; Ellis, L.R.; Berenzon, D.; Howard, D.; Manuel, M.; Dralle, S.; Lyerly, S.; et al. TCA Cycle Inhibition By Cpi-613 Increases Sensitivity to Chemotherapy in Older and Poor Risk Acute Myeloid Leukemia (AML). Blood 2016, 128, 4062. [Google Scholar] [CrossRef]
- Lee, K.C.; Maturo, C.; Shorr, R.; Rodriguez, R. Uncommon Toxicologic Profile at Toxic Doses of CPI-613 (an Agent Selectively Alters Tumor Energy Metabolism) in Rats and Minipigs Reflects Novel Mechanism. Am. J. Pharmacol. Toxicol. 2011, 5, 183–208. [Google Scholar] [CrossRef]
- Lee, K.C.; Shorr, R.; Rodriguez, R.; Maturo, C.; Boteju, L.W.; Sheldon, A. Formation and anti-tumor activity of uncommon in vitro and in vivo metabolites of CPI-613, a novel anti-tumor compound that selectively alters tumor energy metabolism. Drug Metab. Lett. 2011, 5, 163–182. [Google Scholar] [CrossRef] [PubMed]
- Lycan, T.W.; Pardee, T.S.; Petty, W.J.; Bonomi, M.; Alistar, A.; Lamar, Z.S.; Isom, S.; Chan, M.D.; Miller, A.A.; Ruiz, J. A Phase II Clinical Trial of CPI-613 in Patients with Relapsed or Refractory Small Cell Lung Carcinoma. PLoS ONE 2016, 11, e0164244. [Google Scholar] [CrossRef]
- Lee, K.C.; Maturo, C.; Perera, C.N.; Luddy, J.; Rodriguez, R.; Shorr, R. Translational assessment of mitochondrial dysfunction of pancreatic cancer from in vitro gene microarray and animal efficacy studies, to early clinical studies, via the novel tumor-specific anti-mitochondrial agent, CPI-613. Ann. Transl. Med. 2014, 2, 91. [Google Scholar] [CrossRef] [PubMed]
- Perera, C.N.; Rodriguez, R.; Shorr, R. Abstract 3807: Regulation of pancreatic, gliosarcoma and non-small cell lung cancer via CPI-613, a novel anticancer therapeutic agent. Cancer Res. 2012, 72, 3807. [Google Scholar] [CrossRef]
- Egawa, Y.; Saigo, C.; Kito, Y.; Moriki, T.; Takeuchi, T. Therapeutic potential of CPI-613 for targeting tumorous mitochondrial energy metabolism and inhibiting autophagy in clear cell sarcoma. PLoS ONE 2018, 13, e0198940. [Google Scholar] [CrossRef]
- Mordhorst, B.R.; Kerns, K.C.; Schauflinger, M.; Zigo, M.; Murphy, S.L.; Ross, R.M.; Wells, K.D.; Green, J.A.; Sutovsky, P.; Prather, R.S. Pharmacologic treatment with CPI-613 and PS48 decreases mitochondrial membrane potential and increases quantity of autolysosomes in porcine fibroblasts. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Wang, X.; Li, X.; Fu, X.; Bai, M.; Li, X.; Mei, Q.; Nie, J.; Wu, Z.; Han, W. Eliminating ovarian cancer stem cells: A potential therapeutic target for ovarian cancer chemoresistance. Curr. Protein Peptide Sci. 2015, 16, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Shin, S.S.; Nam, J.-H.; Kim, Y.-T.; Kim, Y.-M.; Kim, J.H.; Choi, E.K. Prospective randomized comparison of monthly fluorouracil and cisplatin versus weekly cisplatin concurrent with pelvic radiotherapy and high-dose rate brachytherapy for locally advanced cervical cancer. Gynecol. Oncol. 2008, 108, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Bellio, C.; DiGloria, C.; Spriggs, D.R.; Foster, R.; Growdon, W.B.; Rueda, B.R. The Metabolic Inhibitor CPI-613 Negates Treatment Enrichment of Ovarian Cancer Stem Cells. Cancers 2019, 11, 1678. [Google Scholar] [CrossRef] [PubMed]
- Pastò, A.; Bellio, C.; Pilotto, G.; Ciminale, V.; Silic-Benussi, M.; Guzzo, G.; Rasola, A.; Frasson, C.; Nardo, G.; Zulato, E.; et al. Cancer stem cells from epithelial ovarian cancer patients privilege oxidative phosphorylation, and resist glucose deprivation. Oncotarget 2014, 5, 4305–4319. [Google Scholar] [CrossRef] [PubMed]
- Perera, C.N.; Rodriguez, R.; Shorr, R. Abstract A65: Heterogeneous response of H460 non-small lung carcinoma cells to CPI-613, a novel compound that selectively alters tumor energy metabolism. Cancer Res. 2015, 75, A65. [Google Scholar] [CrossRef]
- Sotgia, F.; Martinez-Outschoorn, U.E.; Howell, A.; Pestell, R.G.; Pavlides, S.; Lisanti, M.P. Caveolin-1 and Cancer Metabolism in the Tumor Microenvironment: Markers, Models, and Mechanisms. Annu. Rev. Pathol. Mech. Dis. 2012, 7, 423–467. [Google Scholar] [CrossRef]
- Capparelli, C.; Guido, C.; Whitaker-Menezes, D.; Bonuccelli, G.; Balliet, R.; Pestell, T.G.; Goldberg, A.F.; Pestell, R.G.; Howell, A.; Sneddon, S.; et al. Autophagy and senescence in cancer-associated fibroblasts metabolically supports tumor growth and metastasis, via glycolysis and ketone production. Cell Cycle 2012, 11, 2285–2302. [Google Scholar] [CrossRef]
- Fiori, M.E.; Di Franco, S.; Villanova, L.; Bianca, P.; Stassi, G.; De Maria, R. Cancer-associated fibroblasts as abettors of tumor progression at the crossroads of EMT and therapy resistance. Mol. Cancer 2019, 18, 70. [Google Scholar] [CrossRef]
- Mordhorst, B.R.; Murphy, S.L.; Ross, R.M.; Samuel, M.S.; Rojas Salazar, S.; Ji, T.; Behura, S.K.; Wells, K.D.; Green, J.A.; Prather, R.S. Pharmacologic Reprogramming Designed to Induce a Warburg Effect in Porcine Fetal Fibroblasts Alters Gene Expression and Quantities of Metabolites from Conditioned Media Without Increased Cell Proliferation. Cell. Reprogram. 2018, 20, 38–48. [Google Scholar] [CrossRef]
- Sai, K.S.; Zachar, Z.; Almaguel, F.; Stuart, S.D.; Dahan, M.S.; Guardado, M.; Rideout, S.; Wang, M.; Sattiraju, A.; Bingham, P.M.; et al. Abstract 2867: FDG-PET imaging as a potential biomarker of mitochondrial targeting by CPI-613, a novel inhibitor of mitochondrial metabolism. Cancer Res. 2017, 77, 2867. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, Z.; Liu, H.; Fetse, J.P.; Jain, A.; Lin, C.-Y.; Cheng, K. Development of a Tumor-Responsive Nanopolyplex Targeting Pancreatic Cancer Cells and Stroma. ACS Appl. Mater. Interfaces 2019, 11, 45390–45403. [Google Scholar] [CrossRef] [PubMed]
- Alistar, A.; Morris, B.B.; Desnoyer, R.; Klepin, H.D.; Hosseinzadeh, K.; Clark, C.; Cameron, A.; Leyendecker, J.; D’Agostino, R.; Topaloglu, U.; et al. Safety and tolerability of the first-in-class agent CPI-613 in combination with modified FOLFIRINOX in patients with metastatic pancreatic cancer: A single-centre, open-label, dose-escalation, phase 1 trial. Lancet Oncol. 2017, 18, 770–778. [Google Scholar] [CrossRef]
- Lamar, Z.S.; Isom, S.; Vaidya, R.; Beaven, A.W.; McIver, Z.A. Phase I Dose-Escalation Study of Cpi-613, in Combination with Bendamustine, in Relapsed or Refractory T-Cell Non-Hodgkin Lymphoma. Blood 2016, 128, 4163. [Google Scholar] [CrossRef]
- Pardee, T.S.; Luther, S.; Buyse, M.; Powell, B.L.; Cortes, J. Devimistat in combination with high dose cytarabine and mitoxantrone compared with high dose cytarabine and mitoxantrone in older patients with relapsed/refractory acute myeloid leukemia: ARMADA 2000 Phase III study. Future Oncol. 2019, 15, 3197–3208. [Google Scholar] [CrossRef] [PubMed]
- Philip, P.A.; Buyse, M.E.; Alistar, A.T.; Rocha Lima, C.M.; Luther, S.; Pardee, T.S.; Van Cutsem, E. A Phase III open-label trial to evaluate efficacy and safety of CPI-613 plus modified FOLFIRINOX (mFFX) versus FOLFIRINOX (FFX) in patients with metastatic adenocarcinoma of the pancreas. Future Oncol. 2019, 15, 3189–3196. [Google Scholar] [CrossRef]
- Pardee, T.S.; DeFord-Watts, L.M.; Peronto, E.; Levitan, D.A.; Hurd, D.D.; Kridel, S.; Harrelson, R.; Manuel, M.; Lyerly, S.; Powell, B.L. Evaluation of the first-in-class antimitochondrial metabolism agent CPI-613 in hematologic malignancies. J. Clin. Oncol. 2012, 30, 6524. [Google Scholar] [CrossRef]
- Anderson, E.M.; Zhang, J.; Russell, G.; Bowline, I.G.; Thyagarajan, B.; Li, D.; Ma, L.; Anderson, E.R.; Murea, M. A Single-Center Retrospective Study of Acute Kidney Injury Incidence in Patients With Advanced Malignancies Treated With Antimitochondrial Targeted Drug. Kidney Int. Rep. 2019, 4, 310–320. [Google Scholar] [CrossRef]
- Retter, A.S. Translational assessment of the efficacy of CPI-613 against pancreatic cancer in animal models versus patients with stage IV disease. J. Clin. Oncol. 2012, 30, 3075. [Google Scholar] [CrossRef]
- Weiner, I.D.; Verlander, J.W. Renal Ammonia Metabolism and Transport. In Comprehensive Physiology; American Cancer Society: Atlanta, GA, USA, 2013; pp. 201–220. [Google Scholar]
- Lord, C.J.; Ashworth, A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017, 355, 1152–1158. [Google Scholar] [CrossRef]
- Singh, N.; Pay, S.L.; Bhandare, S.B.; Arimpur, U.; Motea, E.A. Therapeutic Strategies and Biomarkers to Modulate PARP Activity for Targeted Cancer Therapy. Cancers 2020, 12, 972. [Google Scholar] [CrossRef]
- Dörsam, B.; Seiwert, N.; Foersch, S.; Stroh, S.; Nagel, G.; Begaliew, D.; Diehl, E.; Kraus, A.; McKeague, M.; Minneker, V.; et al. PARP-1 protects against colorectal tumor induction, but promotes inflammation-driven colorectal tumor progression. Proc. Natl. Acad. Sci. USA 2018, 115, E4061–E4070. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, K.; Stadler, Z.K.; Cercek, A.; Mendelsohn, R.B.; Shia, J.; Segal, N.H.; Diaz, L.A., Jr. Immunotherapy in colorectal cancer: Rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 361–375. [Google Scholar] [CrossRef] [PubMed]
| Drug | Phase | Condition(s) | Combination Therapy | Identification No. |
|---|---|---|---|---|
| CPI-613 | I | Advanced or Metastatic Cancer, Metastatic Cancer, Lymphoma, Solid Tumors, Advanced Malignancies | - | NCT00741403 |
| I | (Advanced or Metastatic) CRC | 5-FU | NCT02232152 | |
| I/II | Cancer, Pancreatic Cancer | Gemcitabine | NCT00907166 | |
| II | Cancer | - | NCT01832857 | |
| DCA | I | Recurrent or Metastatic Solid Tumors | - | NCT00566410 |
| AG-221 | I/II | Solid Tumor, Glioma, Angioimmunoblastic T-cell Lymphoma, Intrahepatic Cholangiocarcinoma, Chondrosarcoma | - | NCT02273739 |
| I/II | Refractory or Recurrent Malignancies | - | NCT02813135 | |
| CB-839 | I/II | Solid Tumor, Clear Cell Renal Cell Carcinoma, (Triple-Negative) Breast Cancer, CRC, (Clear Cell) Renal Cell Carcinoma | Talazoparib | NCT03875313 |
| I/II | CRC, Solid Tumor | Capecitabine | NCT02861300 | |
| I/II | (Metastatic or Refractory) CRC | Panitumumab + Irinotecan | NCT03263429 | |
| I/II | Solid Tumors, Non-Small Cell Lung Cancer, CRC | Palbociclib | NCT03965845 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neitzel, C.; Demuth, P.; Wittmann, S.; Fahrer, J. Targeting Altered Energy Metabolism in Colorectal Cancer: Oncogenic Reprogramming, the Central Role of the TCA Cycle and Therapeutic Opportunities. Cancers 2020, 12, 1731. https://doi.org/10.3390/cancers12071731
Neitzel C, Demuth P, Wittmann S, Fahrer J. Targeting Altered Energy Metabolism in Colorectal Cancer: Oncogenic Reprogramming, the Central Role of the TCA Cycle and Therapeutic Opportunities. Cancers. 2020; 12(7):1731. https://doi.org/10.3390/cancers12071731
Chicago/Turabian StyleNeitzel, Carina, Philipp Demuth, Simon Wittmann, and Jörg Fahrer. 2020. "Targeting Altered Energy Metabolism in Colorectal Cancer: Oncogenic Reprogramming, the Central Role of the TCA Cycle and Therapeutic Opportunities" Cancers 12, no. 7: 1731. https://doi.org/10.3390/cancers12071731
APA StyleNeitzel, C., Demuth, P., Wittmann, S., & Fahrer, J. (2020). Targeting Altered Energy Metabolism in Colorectal Cancer: Oncogenic Reprogramming, the Central Role of the TCA Cycle and Therapeutic Opportunities. Cancers, 12(7), 1731. https://doi.org/10.3390/cancers12071731





