Protein Conformational Changes in Breast Cancer Sera Using Infrared Spectroscopic Analysis
Abstract
1. Introduction
1.1. Biomedical Application of FTIR Spectroscopy
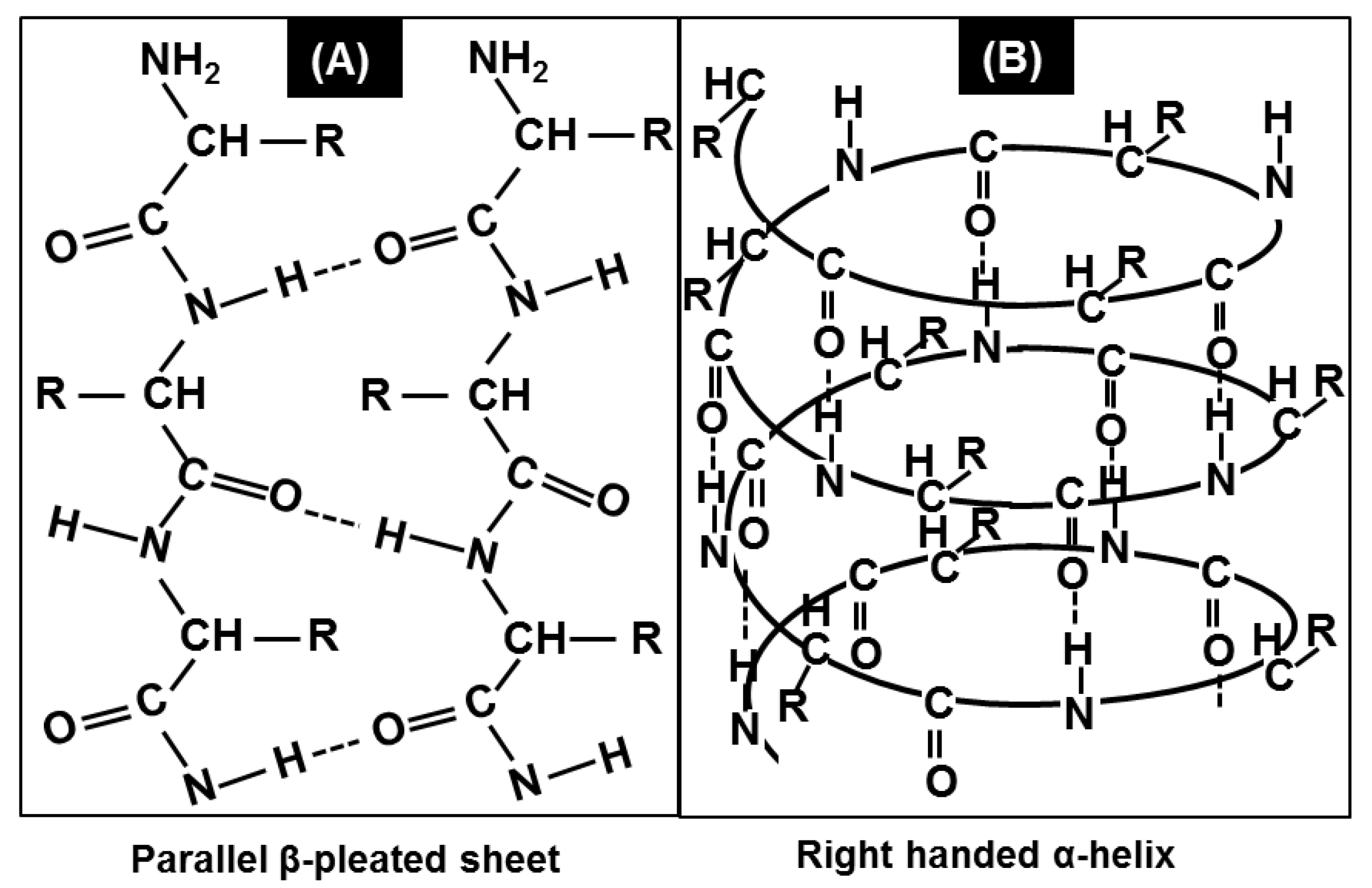
1.2. Protein Conformational Studies Using FTIR Spectroscopy
2. Results
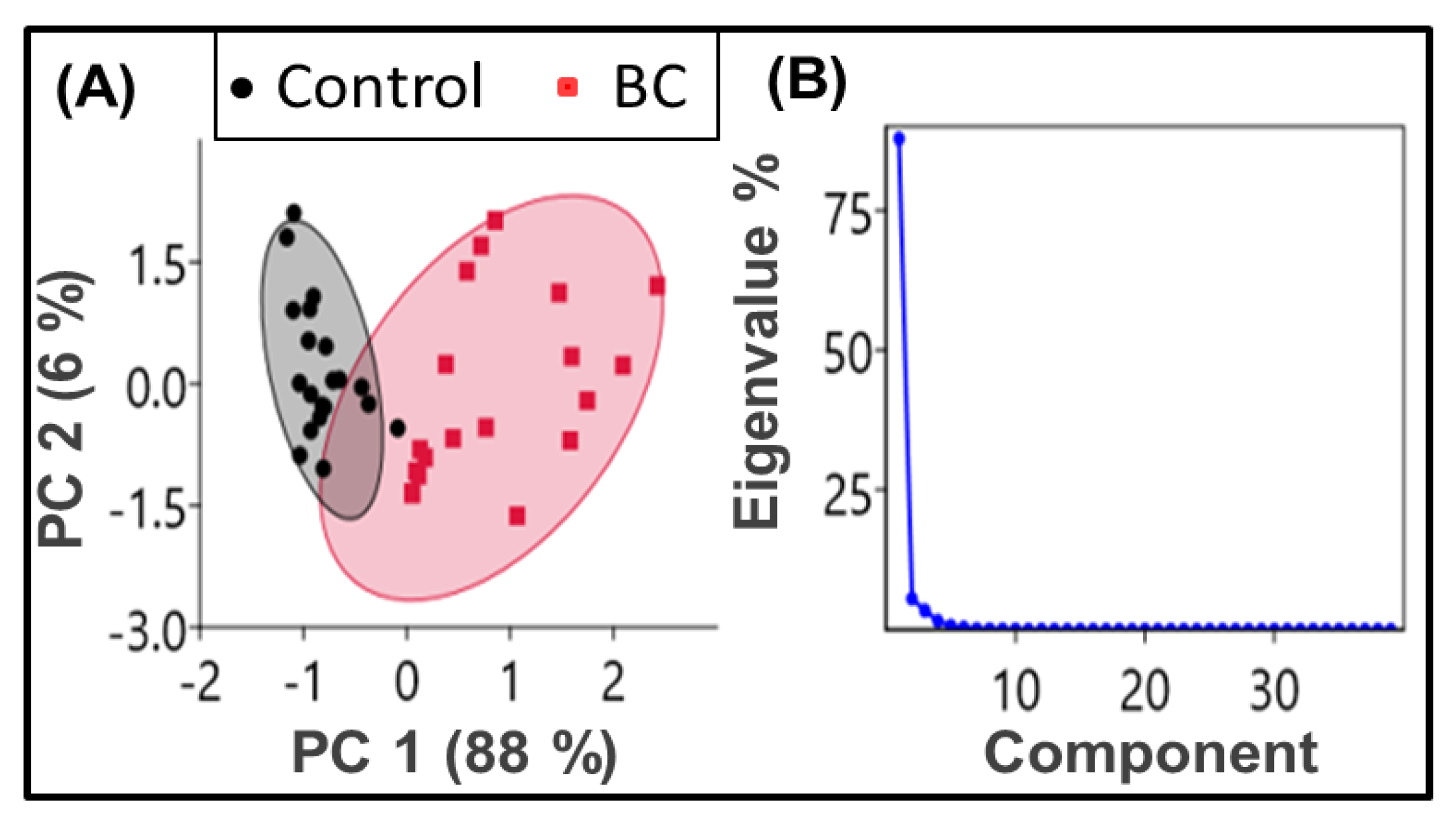
2.1. Principal Component Analysis (PCA)
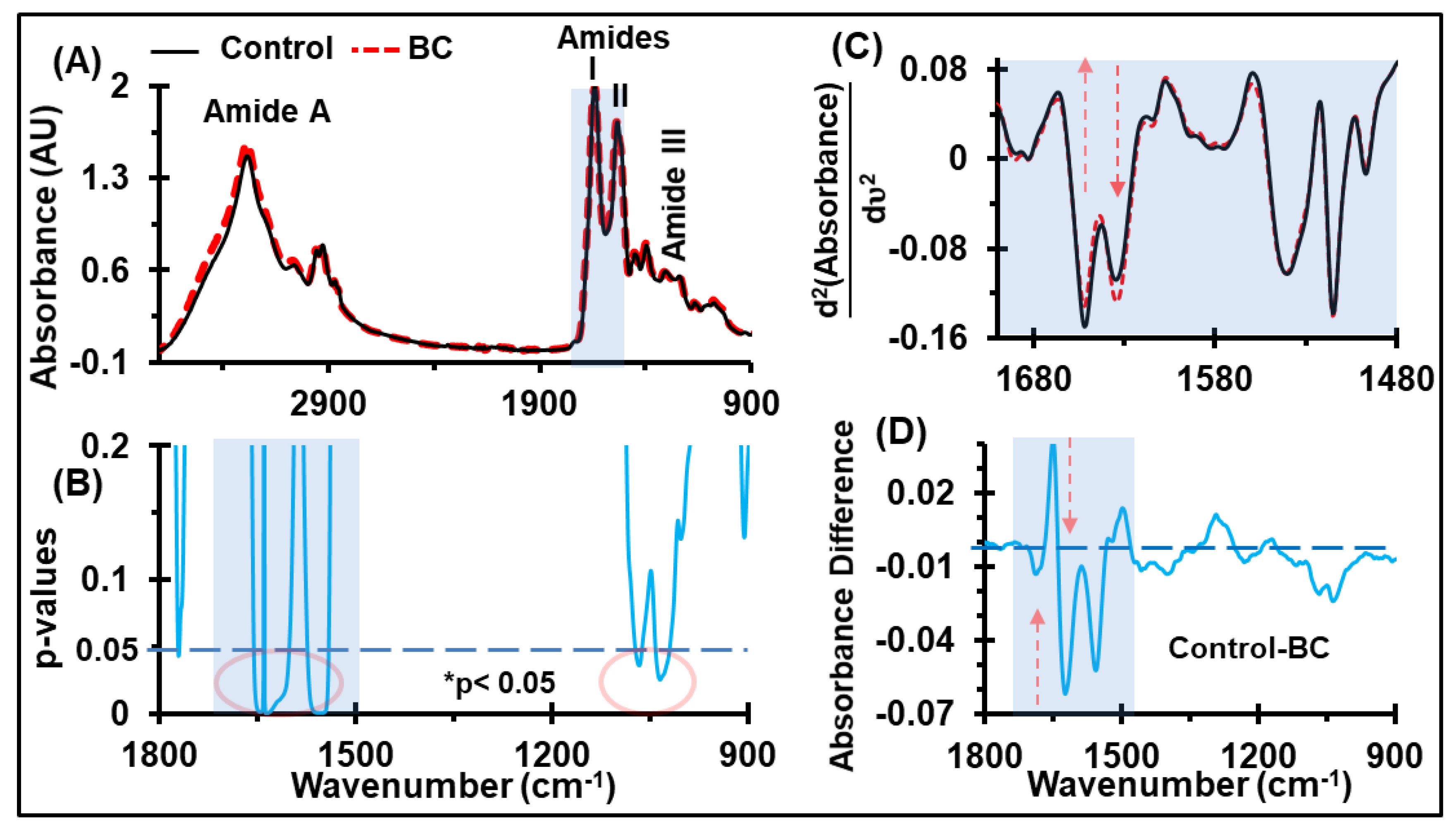
2.2. Discrimination of Average Absorbance
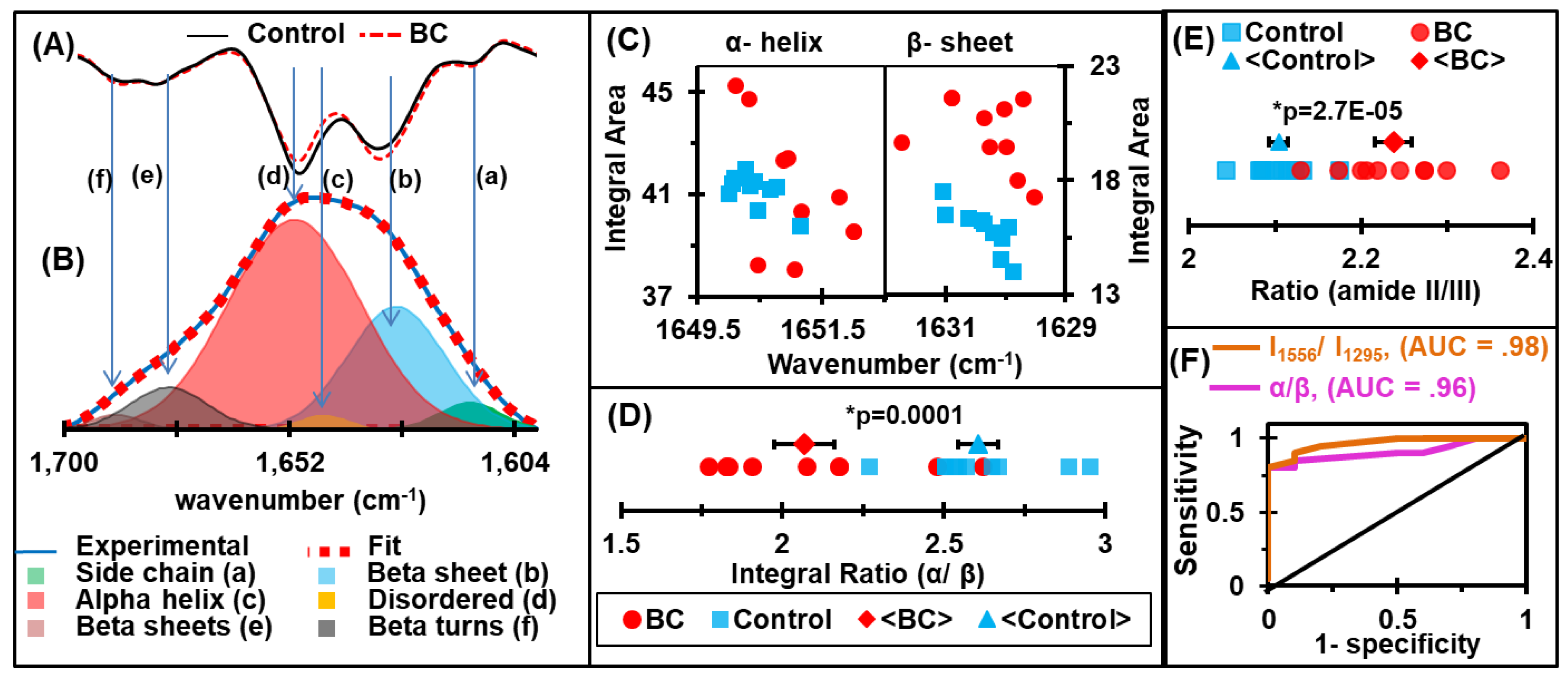
2.3. Discrimination of Protein Secondary Structures
2.4. Receiver Operating Characteristic (ROC) Curves and Area Under the Curve (AUC) Values
3. Discussion
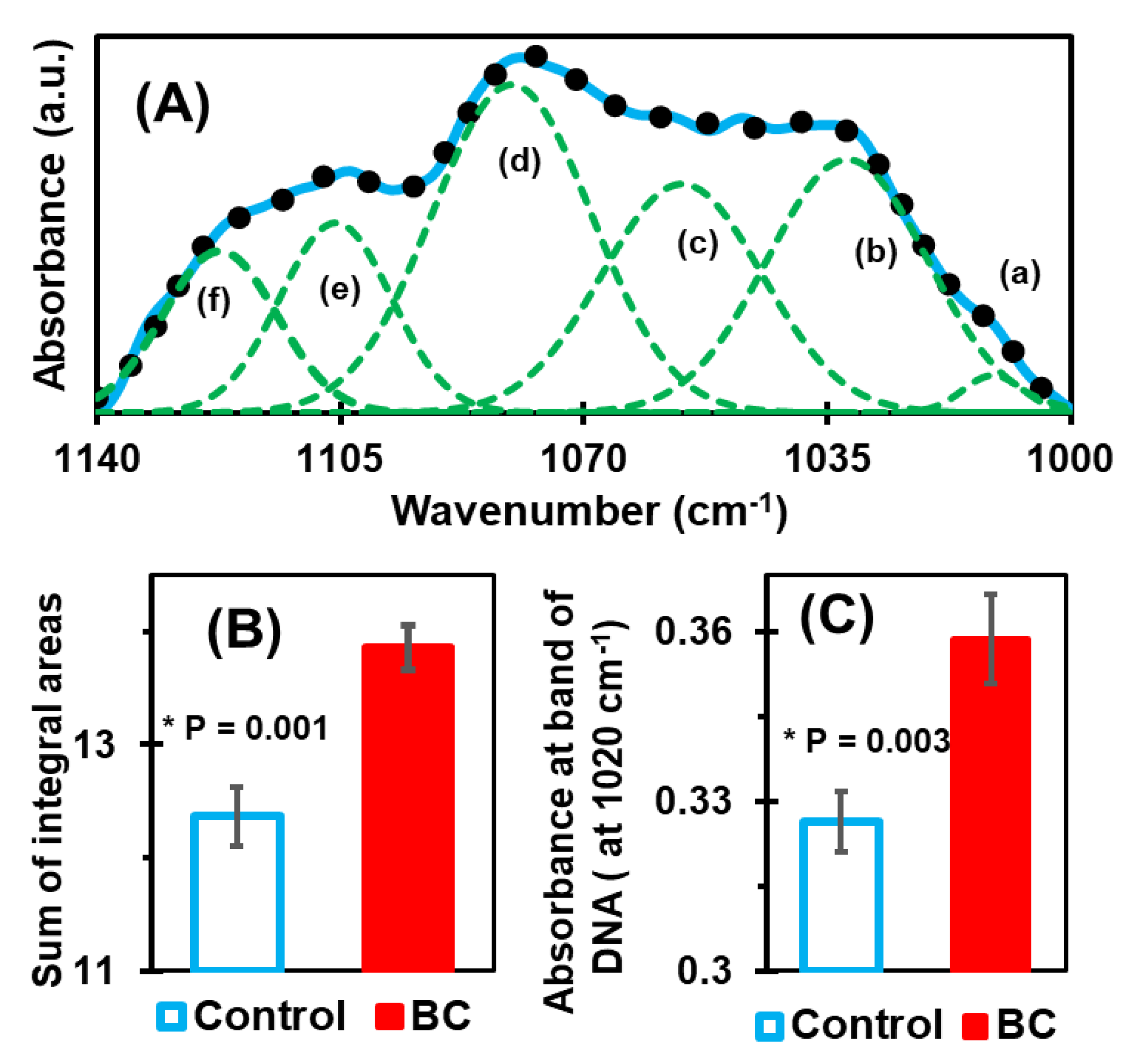
3.1. Deconvolution of Spectral Range 1140–1000 cm−1
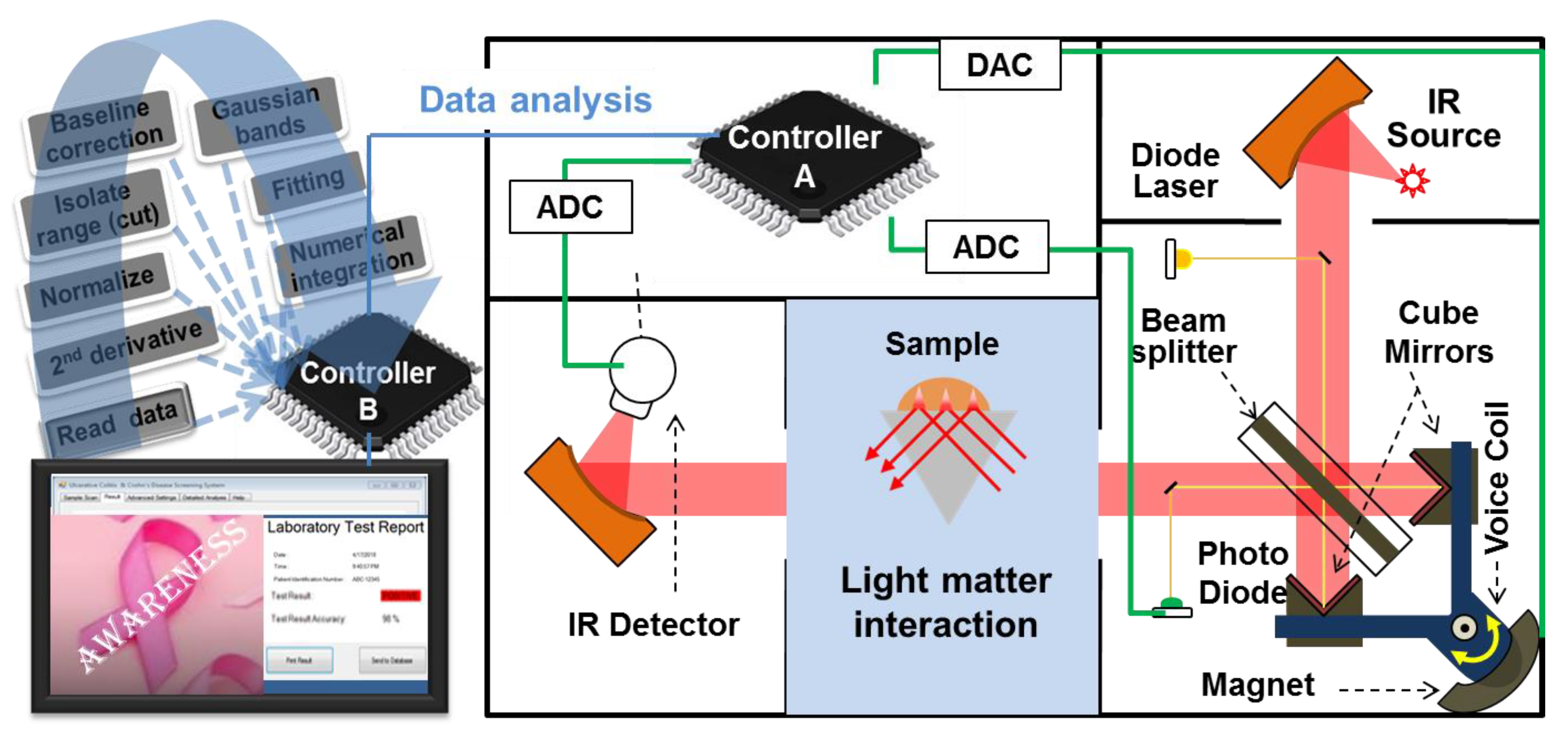
3.2. Potential Prototype for Clinical Application
4. Materials and Methods
4.1. Human Sera
4.2. FTIR Spectrometer
4.3. ATR-FTIR Spectral Measurements
4.4. Spectral Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stewart, B.; Wild, C.P. World cancer report 2014. Public Health 2019; International Agency for Research on Cancer: Lyon, France, 2019. [Google Scholar]
- McGuire, S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015; Oxford University Press: New York, NY, USA, 2016. [Google Scholar]
- Society, A.C. Breast Cancer Facts & Figures 2017–2018; American Cancer Society: Atlanta, GA, USA, 2017. [Google Scholar]
- Yankaskas, B.C.; Haneuse, S.; Kapp, J.M.; Kerlikowske, K.; Geller, B.; Buist, D.S. Performance of first mammography examination in women younger than 40 years. J. Natl. Cancer Inst. 2010, 102, 692–701. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.B.; Wall, C.; Baines, C.J.; Sun, P.; To, T.; Narod, S.A.J.B. Twenty five year follow-up for breast cancer incidence and mortality of the Canadian National Breast Screening Study: Randomised screening trial. BMJ 2014, 348. [Google Scholar] [CrossRef] [PubMed]
- Misek, D.E.; Kim, E.H. Protein biomarkers for the early detection of breast cancer. Int. J. Proteom. 2011, 2011, 343582. [Google Scholar] [CrossRef] [PubMed]
- Kazarian, A.; Blyuss, O.; Metodieva, G.; Gentry-Maharaj, A.; Ryan, A.; Kiseleva, E.M.; Prytomanova, O.M.; Jacobs, I.J.; Widschwendter, M.; Menon, U.; et al. Testing breast cancer serum biomarkers for early detection and prognosis in pre-diagnosis samples. Br. J. Cancer 2017, 116, 501–508. [Google Scholar] [CrossRef]
- Blanco, M.; Villarroya, I. NIR spectroscopy: A rapid-response analytical tool. TrAC Trends Anal. Chem. 2002, 21, 240–250. [Google Scholar] [CrossRef]
- Kendall, C.; Isabelle, M.; Bazant-Hegemark, F.; Hutchings, J.; Orr, L.; Babrah, J.; Baker, R.; Stone, N. Vibrational spectroscopy: A clinical tool for cancer diagnostics. Analyst 2009, 134, 1029–1045. [Google Scholar] [CrossRef]
- Dubois, J.; Shaw, R.A. Peer Reviewed: IR Spectroscopy in Clinical and Diagnostic Applications; ACS Publications: Washington, DC, USA, 2004. [Google Scholar]
- Parker, F. Applications of Infrared Spectroscopy in Biochemistry, Biology, and Medicine; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Movasaghi, Z.; Rehman, S.; ur Rehman, D.I. Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2008, 43, 134–179. [Google Scholar] [CrossRef]
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771. [Google Scholar] [CrossRef]
- Yang, H.; Yang, S.; Kong, J.; Dong, A.; Yu, S. Obtaining information about protein secondary structures in aqueous solution using Fourier transform IR spectroscopy. Nat. Protoc. 2015, 10, 382. [Google Scholar] [CrossRef]
- Trevisan, J.; Angelov, P.P.; Carmichael, P.L.; Scott, A.D.; Martin, F.L. Extracting biological information with computational analysis of Fourier-transform infrared (FTIR) biospectroscopy datasets: Current practices to future perspectives. Analyst 2012, 137, 3202–3215. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.G.; Angelov, P.P.; Trevisan, J.; Vlachopoulou, A.; Paraskevaidis, E.; Martin-Hirsch, P.L.; Martin, F.L. Robust classification of low-grade cervical cytology following analysis with ATR-FTIR spectroscopy and subsequent application of self-learning classifier eClass. Anal. Bioanal. Chem. 2010, 398, 2191–2201. [Google Scholar] [CrossRef] [PubMed]
- Eckel, R.; Huo, H.; Guan, H.W.; Hu, X.; Che, X.; Huang, W. Characteristic infrared spectroscopic patterns in the protein bands of human breast cancer tissue. Vib. Spectrosc. 2001, 27, 165–173. [Google Scholar] [CrossRef]
- Blobe, G.C.; Obeid, L.M.; Hannun, Y.A. Regulation of protein kinase C and role in cancer biology. Cancer Metastasis Rev. 1994, 13, 411–431. [Google Scholar] [CrossRef] [PubMed]
- Bartkova, J.; Lukas, J.; Müller, H.; Lützhøt, D.; Strauss, M.; Bartek, J. Cyclin D1 protein expression and function in human breast cancer. Int. J. Cancer 1994, 57, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Jacquemier, J.; Ginestier, C.; Rougemont, J.; Bardou, V.J.; Charafe-Jauffret, E.; Geneix, J.; Adélaïde, J.; Koki, A.; Houvenaeghel, G.; Hassoun, J. Protein expression profiling identifies subclasses of breast cancer and predicts prognosis. Cancer Res. 2005, 65, 767–779. [Google Scholar]
- Li, J.; Zhang, Z.; Rosenzweig, J.; Wang, Y.Y.; Chan, D.W. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin. Chem. 2002, 48, 1296–1304. [Google Scholar] [CrossRef]
- Elmi, F.; Movaghar, A.F.; Elmi, M.M.; Alinezhad, H.; Nikbakhsh, N. Application of FT-IR spectroscopy on breast cancer serum analysis. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 187, 87–91. [Google Scholar] [CrossRef]
- Zelig, U.; Barlev, E.; Bar, O.; Gross, I.; Flomen, F.; Mordechai, S.; Kapelushnik, J.; Nathan, I.; Kashtan, H.; Wasserberg, N.; et al. Early detection of breast cancer using total biochemical analysis of peripheral blood components: A preliminary study. BMC Cancer 2015, 15, 408. [Google Scholar] [CrossRef]
- Ostrovsky, E.; Zelig, U.; Gusakova, I.; Ariad, S.; Mordechai, S.; Nisky, I.; Kapilushnik, J. Detection of cancer using advanced computerized analysis of infrared spectra of peripheral blood. IEEE Trans. Biomed. Eng. 2012, 60, 343–353. [Google Scholar] [CrossRef]
- Gao, T.; Feng, J.; Ci, Y. Human breast carcinomal tissues display distinctive FTIR spectra: Implication for the histological characterization of carcinomas. Anal. Cell. Pathol. 1999, 18, 87–93. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lyman, D.J.; Murray-Wijelath, J. Fourier transform infrared attenuated total reflection analysis of human hair: Comparison of hair from breast cancer patients with hair from healthy subjects. Appl. Spectrosc. 2005, 59, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Han, S.M.; Chikawa, J.I.; Jeon, J.K.; Hwang, M.Y.; Lim, J.; Jeong, Y.J.; Park, S.H.; Kim, H.T.; Jheon, S.; Kim, J.K. Synchrotron nanoscopy imaging study of scalp hair in breast cancer patients and healthy individuals: Difference in medulla loss and cortical membrane enhancements. Microsc. Res. Tech. 2016, 79, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Malins, D.C.; Polissar, N.L.; Schaefer, S.; Su, Y.; Vinson, M. A unified theory of carcinogenesis based on order–disorder transitions in DNA structure as studied in the human ovary and breast. Proc. Natl. Acad. Sci. USA 1998, 95, 7637–7642. [Google Scholar] [CrossRef]
- Kotkova, M.; Sitnikova, V.; Nosenko, T.; Kotkova, T.; Uspenskaya, M.; Olekhnovich, R. Spectroscopic Study of Blood Serum of Patients with Breast Cancer. In Proceedings of the 2018 IEEE-EMBS Conference on Biomedical Engineering and Sciences (IECBES), Sarawak, Malaysia, 3−6 December 2018; pp. 657–660. [Google Scholar]
- Mitchell, A.L.; Gajjar, K.B.; Theophilou, G.; Martin, F.L.; Martin-Hirsch, P.L. Vibrational spectroscopy of biofluids for disease screening or diagnosis: Translation from the laboratory to a clinical setting. J. Biophotonics 2014, 7, 153–165. [Google Scholar] [CrossRef]
- Aaboe, M.; Offersen, B.V.; Christensen, A.; Andreasen, P.A. Vitronectin in human breast carcinomas. Biochim. Biophys. Acta Mol. Basis Dis. 2003, 1638, 72–82. [Google Scholar] [CrossRef]
- Karplus, M.; McCammon, J.A. Molecular dynamics simulations of biomolecules. Nat. Struct. Mol. Biol. 2002, 9, 646. [Google Scholar] [CrossRef]
- Drenth, J. Principles of Protein X-Ray Crystallography; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Wüthrich, K. NMR with proteins and nucleic acids. Europhys. News 1986, 17, 11–13. [Google Scholar] [CrossRef]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta 2007, 1767, 1073–1101. [Google Scholar] [CrossRef]
- Jabs, A.J. Determination of Secondary Structure in Proteins by Fourier Transform Infrared Spectroscopy (FTIR); Leibniz Institute on Aging-Fritz Lipmann Institute: Jena, Germany, 2005. [Google Scholar]
- Titus, J.; Ghimire, H.; Viennois, E.; Merlin, D.; Perera, A.G. Protein secondary structure analysis of dried blood serum using infrared spectroscopy to identify markers for colitis screening. J. Biophotonics 2018, 11, e201700057. [Google Scholar] [CrossRef]
- Hering, J.A.; Haris, P.I. FTIR spectroscopy for analysis of protein secondary structure. Biol. Biomed. Infrared Spectrosc. 2009, 2, 129–167. [Google Scholar]
- Surewicz, W.K.; Mantsch, H.H.; Chapman, D. Determination of protein secondary structure by Fourier transform infrared spectroscopy: A critical assessment. Biochemistry 1993, 32, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.C.; Haris, P.I.; Chapman, D.; Mitchell, R.C. Determination of protein secondary structure using factor analysis of infrared spectra. Biochemistry 1990, 29, 9185–9193. [Google Scholar] [CrossRef] [PubMed]
- Pribic, R.; Vanstokkum, I.; Chapman, D.; Haris, P.I.; Bloemendal, M. Protein secondary structure from Fourier transform infrared and/or circular dichroism spectra. Anal. Biochem. 1993, 214, 366–378. [Google Scholar] [CrossRef] [PubMed]
- Miller, L.M.; Bourassa, M.W.; Smith, R.J. FTIR spectroscopic imaging of protein aggregation in living cells. Biochim. Biophys. Acta Biomembr. 2013, 1828, 2339–2346. [Google Scholar] [CrossRef]
- Haris, P.I.; Severcan, F. FTIR spectroscopic characterization of protein structure in aqueous and non-aqueous media. J. Mol. Catal. B Enzym. 1999, 7, 207–221. [Google Scholar] [CrossRef]
- Garczarek, F.; Gerwert, K. Functional waters in intraprotein proton transfer monitored by FTIR difference spectroscopy. Nature 2006, 439, 109. [Google Scholar] [CrossRef]
- Grdadolnik, J.; Maréchal, Y. Hydrogen–Deuterium Exchange in Bovine Serum Albumin Protein Monitored by Fourier Transform Infrared Spectroscopy, Part I: Structural Studies. Appl. Spectrosc. 2005, 59, 1347–1356. [Google Scholar] [CrossRef]
- Prestrelski, S.J.; Tedeschi, N.; Arakawa, T.; Carpenter, J.F. Dehydration-induced conformational transitions in proteins and their inhibition by stabilizers. Biophys. J. 1993, 65, 661–671. [Google Scholar] [CrossRef]
- Lu, R.; Li, W.W.; Katzir, A.; Raichlin, Y.; Yu, H.Q.; Mizaikoff, B. Probing the secondary structure of bovine serum albumin during heat-induced denaturation using mid-infrared fiberoptic sensors. Analyst 2015, 140, 765–770. [Google Scholar] [CrossRef]
- Fabian, H.; Schultz, C.; Naumann, D.; Landt, O.; Hahn, U.; Saenger, W. Secondary structure and temperature-induced unfolding and refolding of ribonuclease T1 in aqueous solution: A Fourier transform infrared spectroscopic study. J. Mol. Biol. 1993, 232, 967–981. [Google Scholar] [CrossRef]
- Baker, M.J.; Gazi, E.; Brown, M.D.; Shanks, J.H.; Gardner, P.; Clarke, N.W. FTIR-based spectroscopic analysis in the identification of clinically aggressive prostate cancer. Br. J. Cancer 2008, 99, 1859. [Google Scholar] [CrossRef] [PubMed]
- Ghimire, H.; Venkataramani, M.; Bian, Z.; Liu, Y.; Perera, A.U. ATR-FTIR spectral discrimination between normal and tumorous mouse models of lymphoma and melanoma from serum samples. Sci. Rep. 2017, 7, 16993. [Google Scholar] [CrossRef] [PubMed]
- Choo, L.P.I.; Wetzel, D.L.; Halliday, W.C.; Jackson, M.; LeVine, S.M.; Mantsch, H.H. In situ characterization of beta-amyloid in Alzheimer’s diseased tissue by synchrotron Fourier transform infrared microspectroscopy. Biophys. J. 1996, 71, 1672–1679. [Google Scholar] [CrossRef]
- Szczerbowska-Boruchowska, M.; Dumas, P.; Kastyak, M.Z.; Chwiej, J.; Lankosz, M.; Adamek, D.; Krygowska-Wajs, A. Biomolecular investigation of human substantia nigra in Parkinson’s disease by synchrotron radiation Fourier transform infrared microspectroscopy. Arch. Biochem. Biophys. 2007, 459, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Kretlow, A.; Wang, Q.; Beekes, M.; Naumann, D.; Miller, L.M. Changes in protein structure and distribution observed at pre-clinical stages of scrapie pathogenesis. Biochim. Biophys. Acta Mol. Basis Dis. 2008, 1782, 559–565. [Google Scholar] [CrossRef]
- Haris, P.I. Probing protein–protein interaction in biomembranes using Fourier transform infrared spectroscopy. Biochim. Biophys. Acta Biomembr. 2013, 1828, 2265–2271. [Google Scholar] [CrossRef]
- Jackson, M.; Haris, P.I.; Chapman, D. Fourier transform infrared spectroscopic studies of calcium-binding proteins. Biochemistry 1991, 30, 9681–9686. [Google Scholar] [CrossRef]
- Jackson, M.; Mantsch, H.H. The use and misuse of FTIR spectroscopy in the determination of protein structure. Crit. Rev. Biochem. Mol. Biol. 1995, 30, 95–120. [Google Scholar] [CrossRef]
- Surewicz, W.K.; Mantsch, H.H. New insight into protein secondary structure from resolution-enhanced infrared spectra. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1988, 952, 115–130. [Google Scholar] [CrossRef]
- Fabian, H.; Naumann, D. Methods to study protein folding by stopped-flow FT-IR. Methods 2004, 34, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Rich, A. Direct conversion of an oligopeptide from a β-sheet to an α-helix: A model for amyloid formation. Proc. Natl. Acad. Sci. USA 1997, 94, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Yu, S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophys. Sin. 2007, 39, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Dovbeshko, G.I.; Gridina, N.Y.; Kruglova, E.B.; Pashchuk, O.P. FTIR spectroscopy studies of nucleic acid damage. Talanta 2000, 53, 233–246. [Google Scholar] [CrossRef]
- Sahu, R.; Argov, S.; Salman, A.; Huleihel, M.; Grossman, N.; Hammody, Z.; Kapelushnik, J.; Mordechai, S. Characteristic absorbance of nucleic acids in the Mid-IR region as possible common biomarkers for diagnosis of malignancy. Technol. Cancer Res. Treat. 2004, 3, 629–638. [Google Scholar] [CrossRef]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Ollesch, J.; Drees, S.L.; Heise, H.M.; Behrens, T.; Brüning, T.; Gerwert, K. FTIR spectroscopy of biofluids revisited: An automated approach to spectral biomarker identification. Analyst 2013, 138, 4092–4102. [Google Scholar] [CrossRef]
- Lovergne, L.; Clemens, G.; Untereiner, V.; Lukaszweski, R.A.; Sockalingum, G.D.; Baker, M.J. Investigating optimum sample preparation for infrared spectroscopic serum diagnostics. Anal. Methods 2015, 7, 7140–7149. [Google Scholar] [CrossRef][Green Version]
- Ghazarian, H.; Idoni, B.; Oppenheimer, S.B. A glycobiology review: Carbohydrates, lectins and implications in cancer therapeutics. Acta Histochem. 2011, 113, 236–247. [Google Scholar] [CrossRef]
- Heitzer, E.; Ulz, P.; Geigl, J.B. Circulating tumor DNA as a liquid biopsy for cancer. Clin. Chem. 2015, 61, 112–123. [Google Scholar] [CrossRef]
- Meurens, M.; Wallon, J.; Tong, J.; Noel, H.; Haot, J. Breast cancer detection by Fourier transform infrared spectrometry. Vib. Spectrosc. 1996, 10, 341–346. [Google Scholar] [CrossRef]
- Baker, M.J.; Hussain, S.R.; Lovergne, L.; Untereiner, V.; Hughes, C.; Lukaszewski, R.A.; Thiéfin, G.; Sockalingum, G.D. Developing and understanding biofluid vibrational spectroscopy: A critical review. Chem. Soc. Rev. 2016, 45, 1803–1818. [Google Scholar] [CrossRef] [PubMed]
- Gazi, E.; Baker, M.; Dwyer, J.; Lockyer, N.P.; Gardner, P.; Shanks, J.H.; Reeve, R.S.; Hart, C.A.; Clarke, N.W.; Brown, M.D. A correlation of FTIR spectra derived from prostate cancer biopsies with Gleason grade and tumour stage. Eur. Urol. 2006, 50, 750–761. [Google Scholar] [CrossRef] [PubMed]
- Gajjar, K.; Heppenstall, L.D.; Pang, W.; Ashton, K.M.; Trevisan, J.; Patel, I.I.; Llabjani, V.; Stringfellow, H.F.; Martin-Hirsch, P.L.; Dawson, T. Diagnostic segregation of human brain tumours using Fourier-transform infrared and/or Raman spectroscopy coupled with discriminant analysis. Anal. Methods 2013, 5, 89–102. [Google Scholar] [CrossRef]
- Hands, J.R.; Clemens, G.; Stables, R.; Ashton, K.; Brodbelt, A.; Davis, C.; Dawson, T.P.; Jenkinson, M.D.; Lea, R.W.; Walker, C. Brain tumour differentiation: Rapid stratified serum diagnostics via attenuated total reflection Fourier-transform infrared spectroscopy. J. Neuro-Oncol. 2016, 127, 463–472. [Google Scholar] [CrossRef]
- Hands, J.R.; Dorling, K.M.; Abel, P.; Ashton, K.M.; Brodbelt, A.; Davis, C.; Dawson, T.; Jenkinson, M.D.; Lea, R.W.; Walker, C. Attenuated total reflection Fourier transform infrared (ATR-FTIR) spectral discrimination of brain tumour severity from serum samples. J. Biophotonics 2014, 7, 189–199. [Google Scholar] [CrossRef]
- Krimm, S.; Bandekar, J. Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. In Advances in Protein Chemistry; Elsevier: Amsterdam, The Netherlands, 1986; Volume 38. [Google Scholar]
- Goormaghtigh, E.; Ruysschaert, J.M.; Raussens, V. Evaluation of the information content in infrared spectra for protein secondary structure determination. Biophys. J. 2006, 90, 2946–2957. [Google Scholar] [CrossRef]
- Swinehart, D.J. The beer-lambert law. J. Chem. Educ. 1962, 39, 333. [Google Scholar] [CrossRef]
- Schweizer, K.S.; Chandler, D. Vibrational dephasing and frequency shifts of polyatomic molecules in solution. J. Chem. Phys. 1982, 76, 2296–2314. [Google Scholar] [CrossRef]
- Šimundić, A.M. Measures of diagnostic accuracy: Basic definitions. Ejifcc 2009, 19, 203. [Google Scholar]
- Gast, M.C.W.; Van Gils, C.H.; Wessels, L.F.; Harris, N.; Bonfrer, J.M.; Rutgers, E.J.; Schellens, J.H.; Beijnen, J.H. Serum protein profiling for diagnosis of breast cancer using SELDI-TOF MS. Oncol. Rep. 2009, 22, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Backhaus, J.; Mueller, R.; Formanski, N.; Szlama, N.; Meerpohl, H.G.; Eidt, M.; Bugert, P. Diagnosis of breast cancer with infrared spectroscopy from serum samples. Vib. Spectrosc. 2010, 52, 173–177. [Google Scholar] [CrossRef]
- Andrei, A.B.; Fleschin, Ş.; Aboul-Enein, H.Y. Cancer diagnosis by FT-IR Spectrophotometry. Rev. Roum. Chim. 2015, 60, 415–426. [Google Scholar]
- Schwarzenbach, H.; Pantel, K. Circulating DNA as biomarker in breast cancer. Breast Cancer Res. 2015, 17, 136. [Google Scholar] [CrossRef] [PubMed]
- Kriege, M.; Brekelmans, C.T.; Obdeijn, I.M.; Boetes, C.; Zonderland, H.M.; Muller, S.H.; Kok, T.; Manoliu, R.A.; Besnard, A.P.E.; Tilanus-Linthorst, M.M.; et al. Factors affecting sensitivity and specificity of screening mammography and MRI in women with an inherited risk for breast cancer. Breast Cancer Res. Treat. 2006, 100, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Kriege, M.; Brekelmans, C.T.M.; Boetes, C.; Besnard, P.E.; Zonderland, H.M.; Obdeijn, I.M.; Manoliu, R.A.; Kok, T.; Peterse, H.; Tilanuslinthorst, M.M.A.; et al. Efficacy of MRI and mammography for breast-cancer screening in women with a familial or genetic predisposition. N. Engl. J. Med. 2004, 351, 427–437. [Google Scholar] [CrossRef]
- Leach, M.O.; Boggis, C.; Dixon, A.K.; Easton, D.F.; Eeles, R.A.; Evans, D.G.R.; Gilbert, F.F.; Griebsch, I.; Hoff, R.; Kessar, P.; et al. Screening with magnetic resonance imaging and mammography of a UK population at high familial risk of breast cancer: A prospective multicentre cohort study (MARIBS). Lancet 2005, 365, 1769–1778. [Google Scholar]
- Naylor, D.A.; Tahic, M.K. Apodizing functions for Fourier transform spectroscopy. J. Opt. Soc. Am. A 2007, 24, 3644–3648. [Google Scholar] [CrossRef]
| Wavenumber (cm−1) | Biomolecular Assignments |
|---|---|
| 1700–1600 | Amide I: sensitive to protein secondary structures of proteins, which arises mainly due to C=O stretching vibrations and the C-N groups. |
| 1580–1480 | Amide II: sensitive for protein conformation, originates mainly from the in-plane N-H bending mode along with C-N and C-C stretching vibrations. |
| 1140–1000 | Carbohydrates: sensitive to C-O, C-C stretching, C-H bending, and nucleic acids: sensitive to deoxyribose/ribose DNA, RNA, νs(PO2−). |
| Signatures | Average ± st. Error | Range of Values | Cutoff Value | AUC | Sensitivity % | Specificity % | p-value | ||
|---|---|---|---|---|---|---|---|---|---|
| Control | BC | Control | BC | ||||||
| α/β | 2.61 ± 0.06 | 2.07 ± 0.09 | 2.27–2.95 | 1.77–2.62 | 2.25 | 0.96 | 90 | 90 | 1.4 × 10−4 |
| I1556/ I1295 | 2.10 ± 0.01 | 2.24 ± 0.02 | 2.04–2.17 | 2.13–2.36 | 2.12 | 0.98 | 100 | 80 | 2.7× 10−5 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghimire, H.; Garlapati, C.; Janssen, E.A.M.; Krishnamurti, U.; Qin, G.; Aneja, R.; Perera, A.G.U. Protein Conformational Changes in Breast Cancer Sera Using Infrared Spectroscopic Analysis. Cancers 2020, 12, 1708. https://doi.org/10.3390/cancers12071708
Ghimire H, Garlapati C, Janssen EAM, Krishnamurti U, Qin G, Aneja R, Perera AGU. Protein Conformational Changes in Breast Cancer Sera Using Infrared Spectroscopic Analysis. Cancers. 2020; 12(7):1708. https://doi.org/10.3390/cancers12071708
Chicago/Turabian StyleGhimire, Hemendra, Chakravarthy Garlapati, Emiel A. M. Janssen, Uma Krishnamurti, Gengsheng Qin, Ritu Aneja, and A. G. Unil Perera. 2020. "Protein Conformational Changes in Breast Cancer Sera Using Infrared Spectroscopic Analysis" Cancers 12, no. 7: 1708. https://doi.org/10.3390/cancers12071708
APA StyleGhimire, H., Garlapati, C., Janssen, E. A. M., Krishnamurti, U., Qin, G., Aneja, R., & Perera, A. G. U. (2020). Protein Conformational Changes in Breast Cancer Sera Using Infrared Spectroscopic Analysis. Cancers, 12(7), 1708. https://doi.org/10.3390/cancers12071708






