Rac1 Promotes Cell Motility by Controlling Cell Mechanics in Human Glioblastoma
Abstract
1. Introduction
2. Results
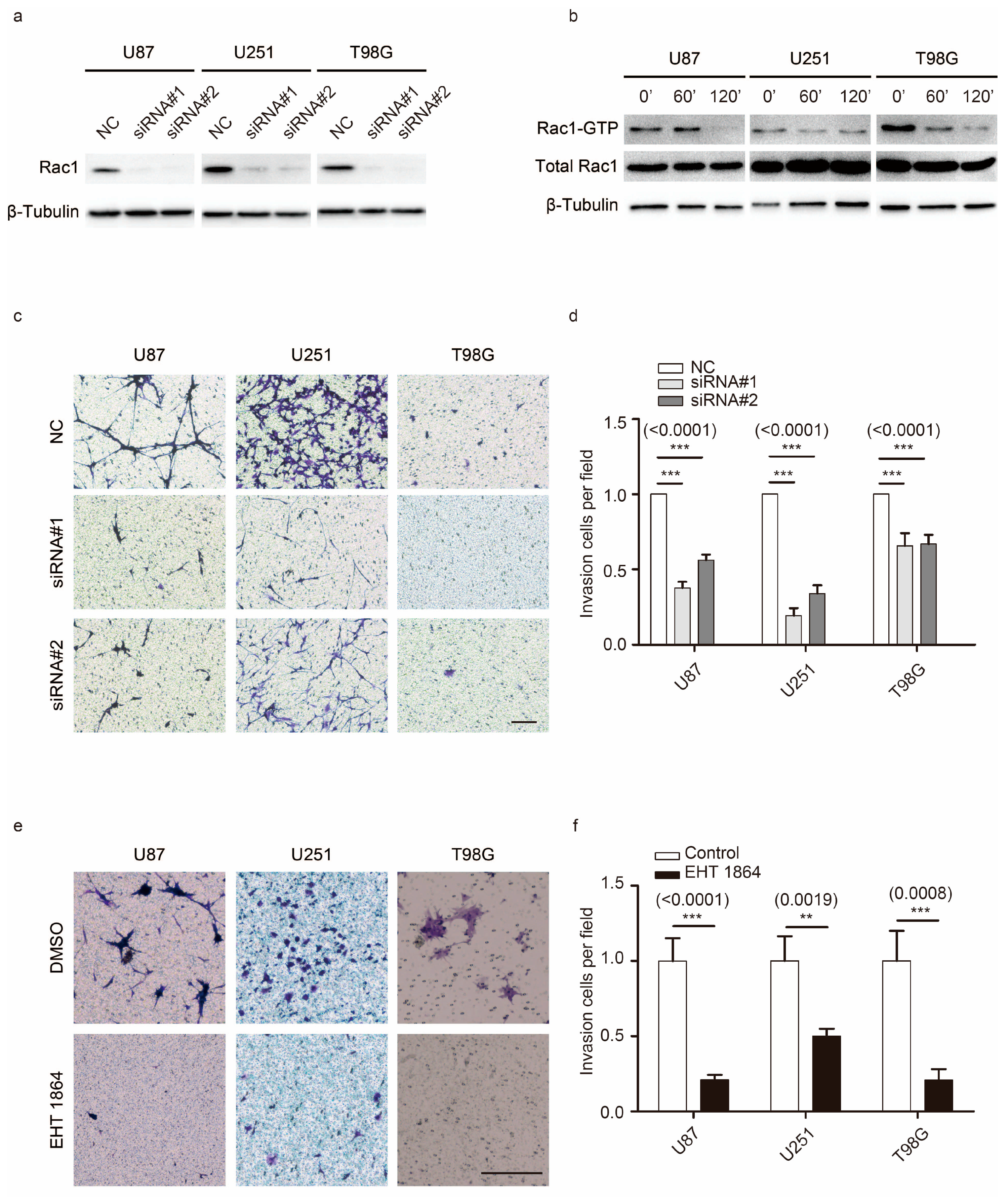
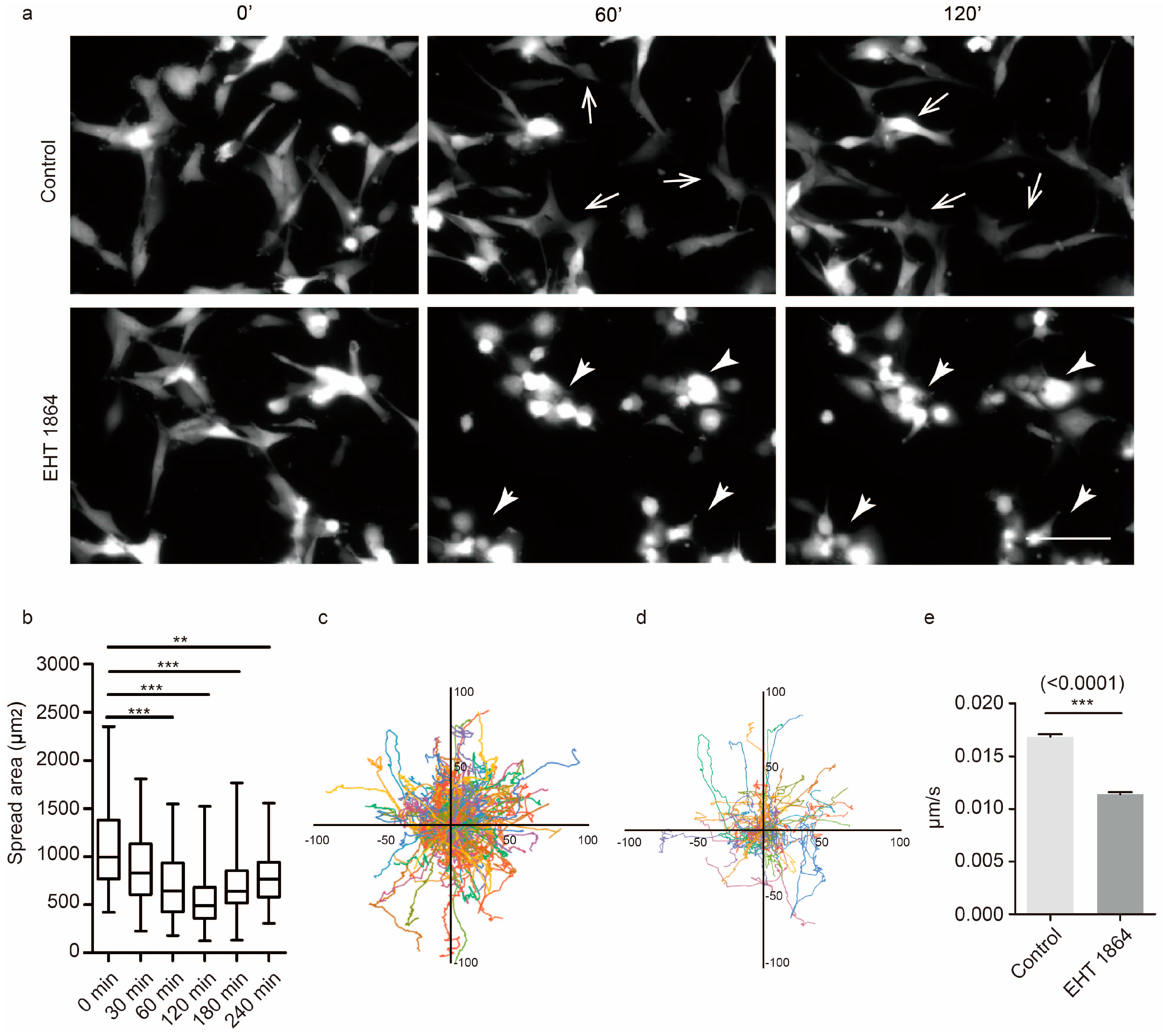
2.1. Rac1 Is Essential for GBM Motility
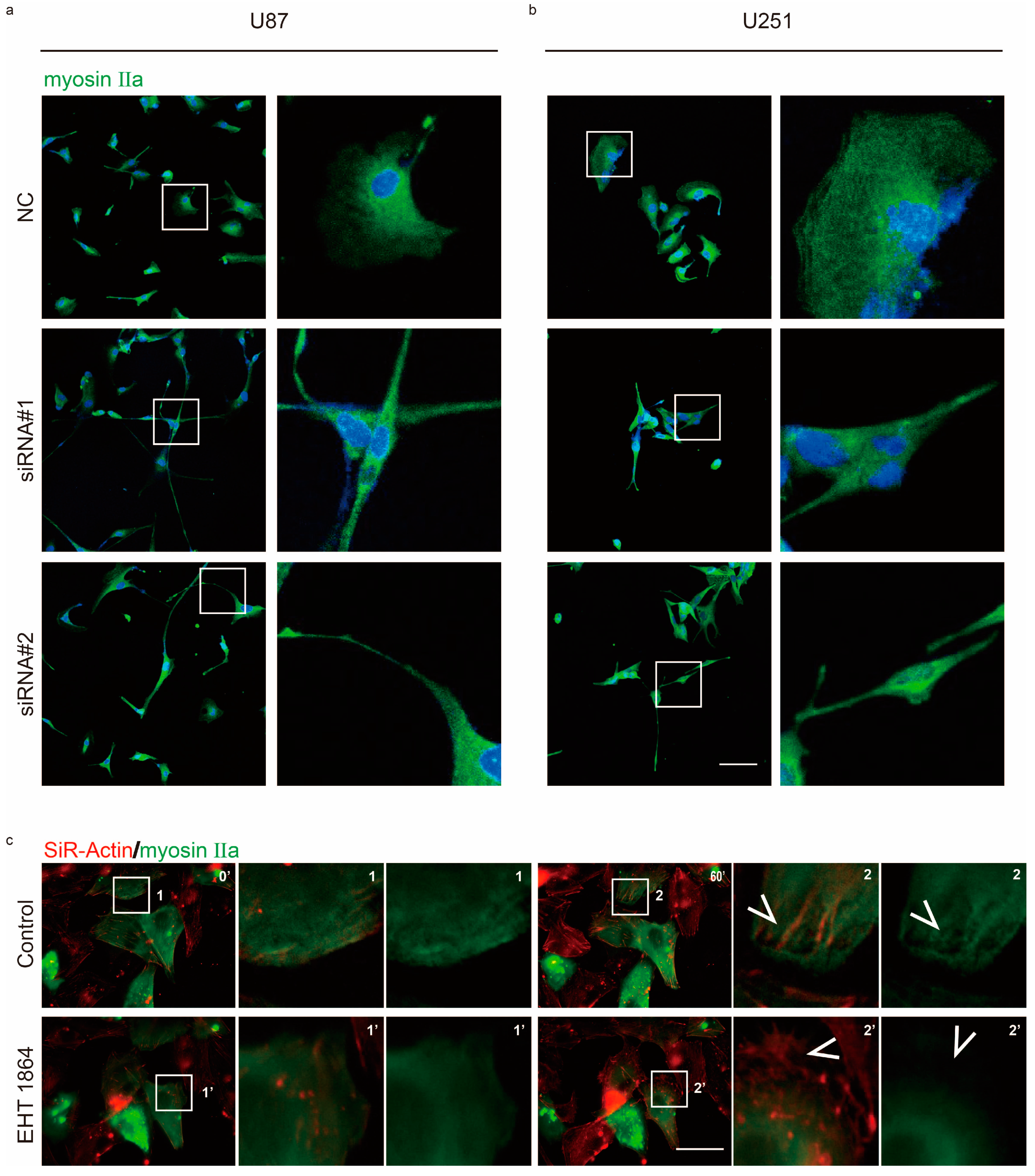
2.2. Rac1 Signaling Regulates Myosin IIa Localization
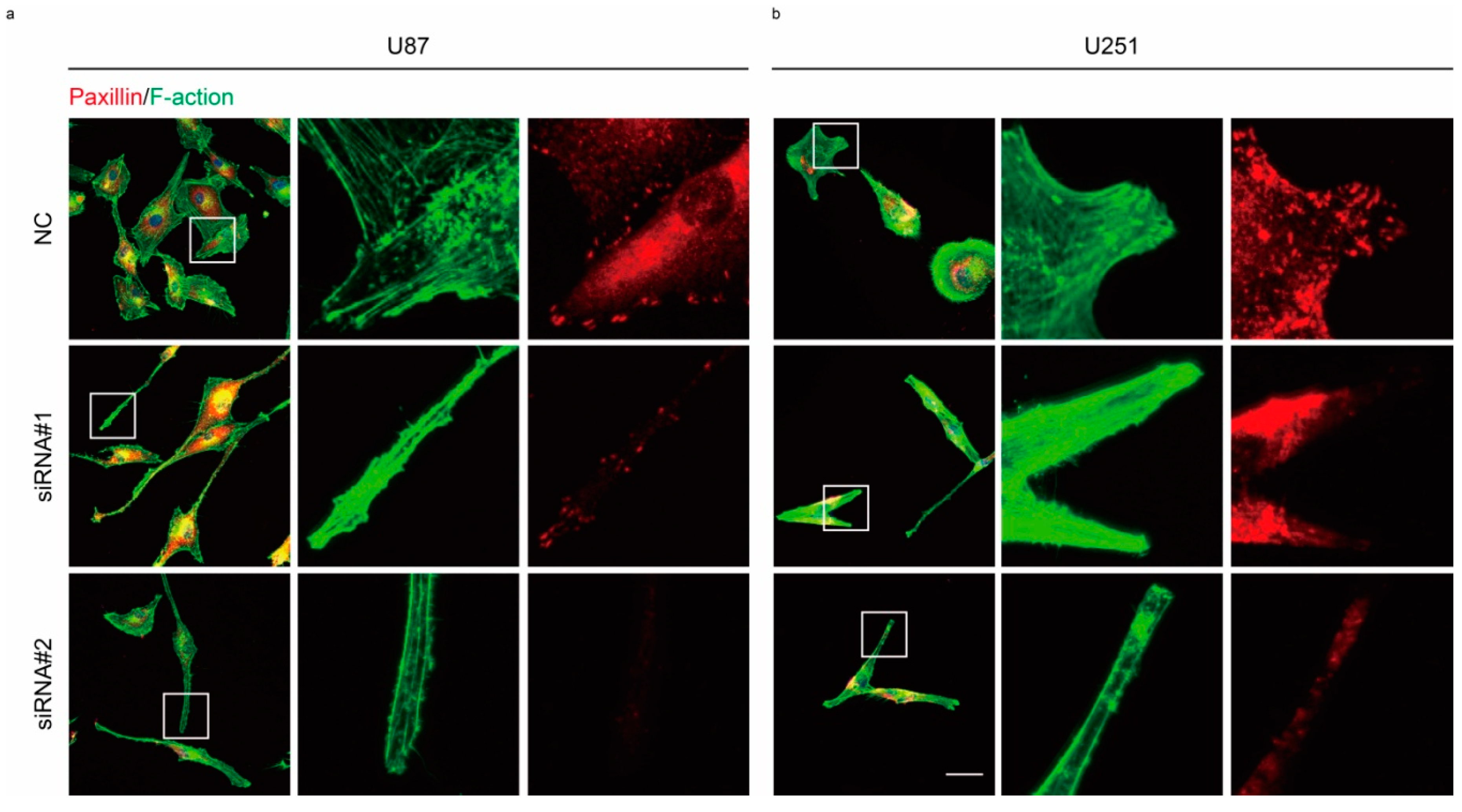
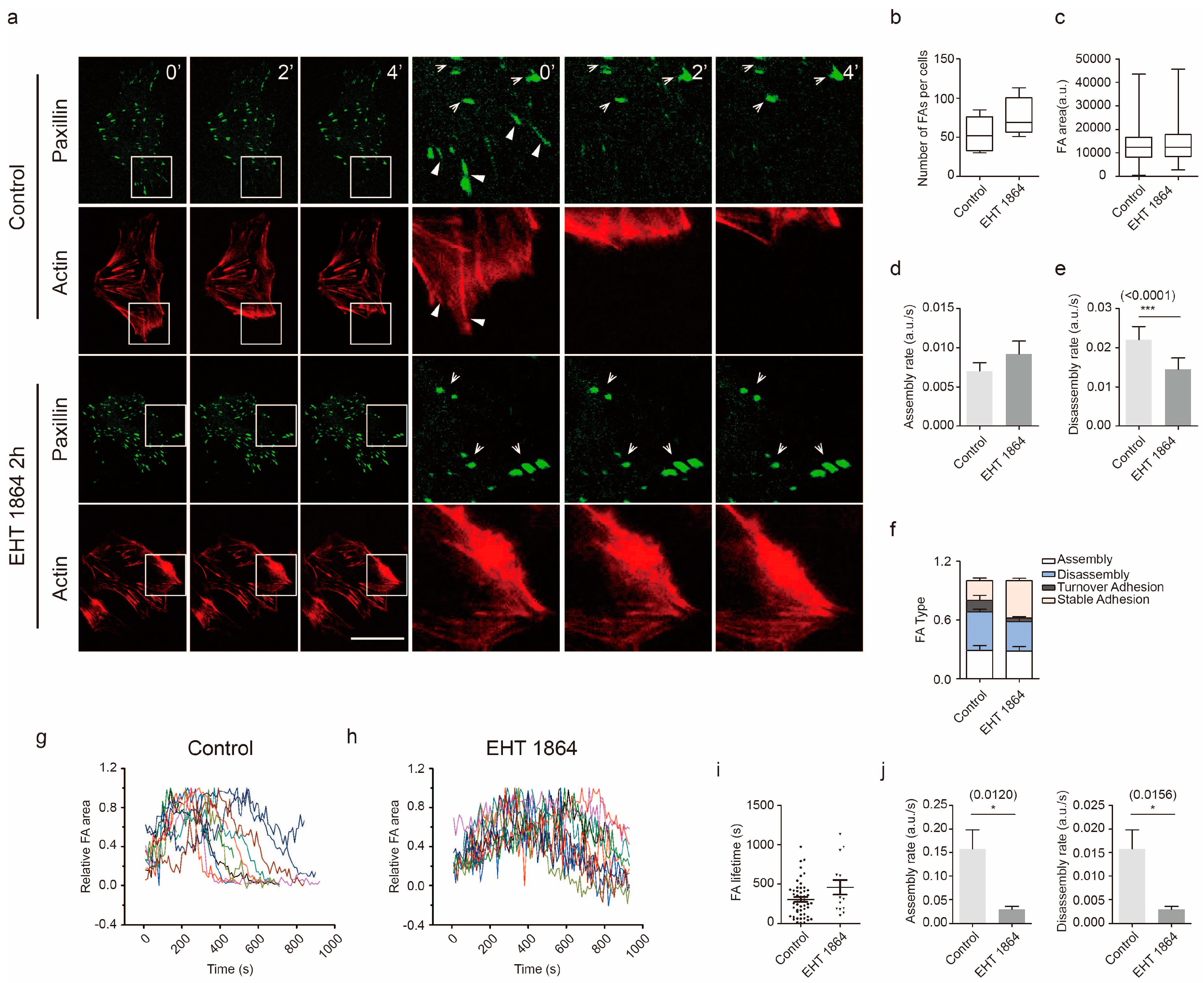
2.3. Rac1 Signaling in Cell Adhesion Formation
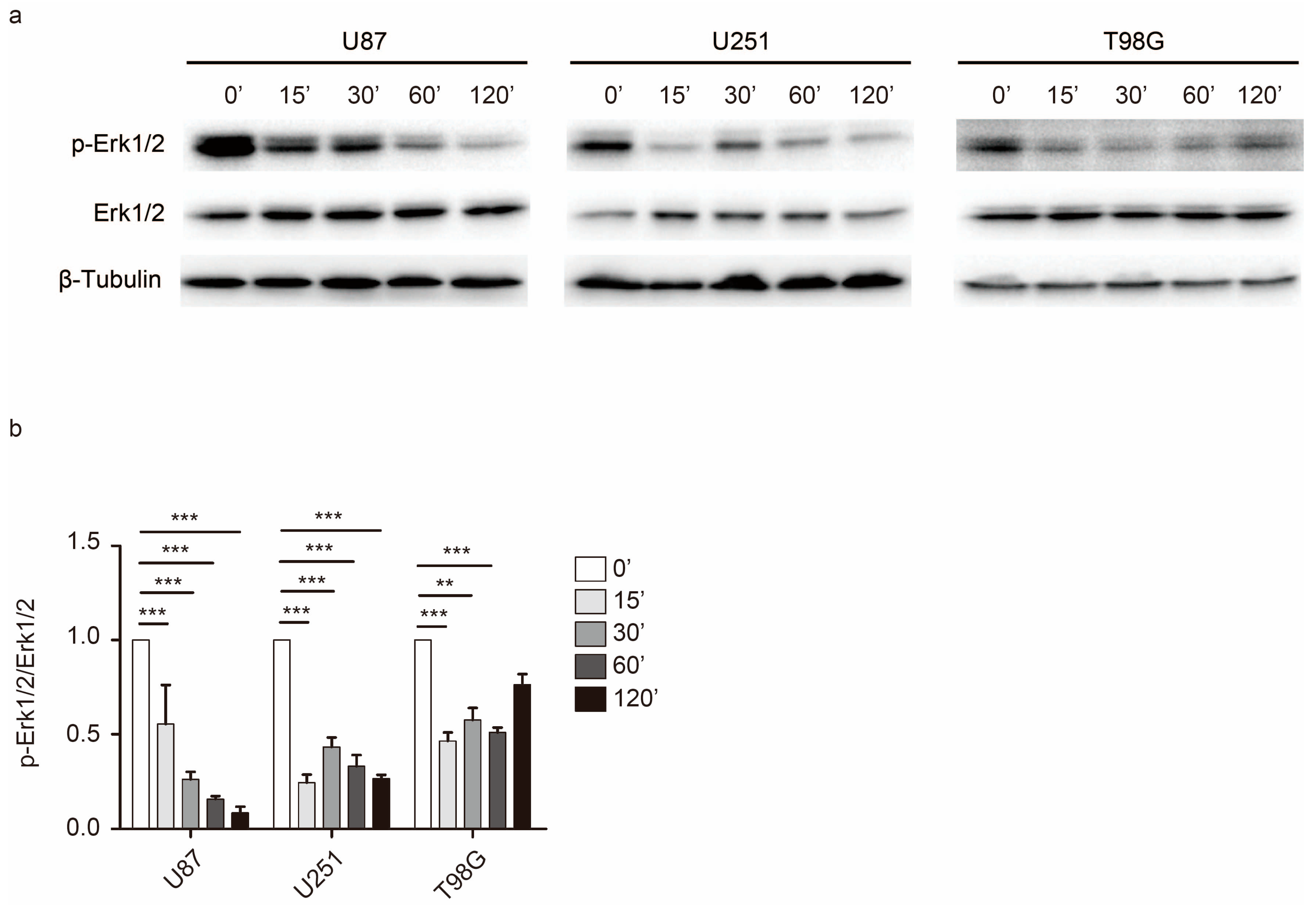
2.4. Rac1 Activates Erk to Mediate Cell Adhesion Dynamics
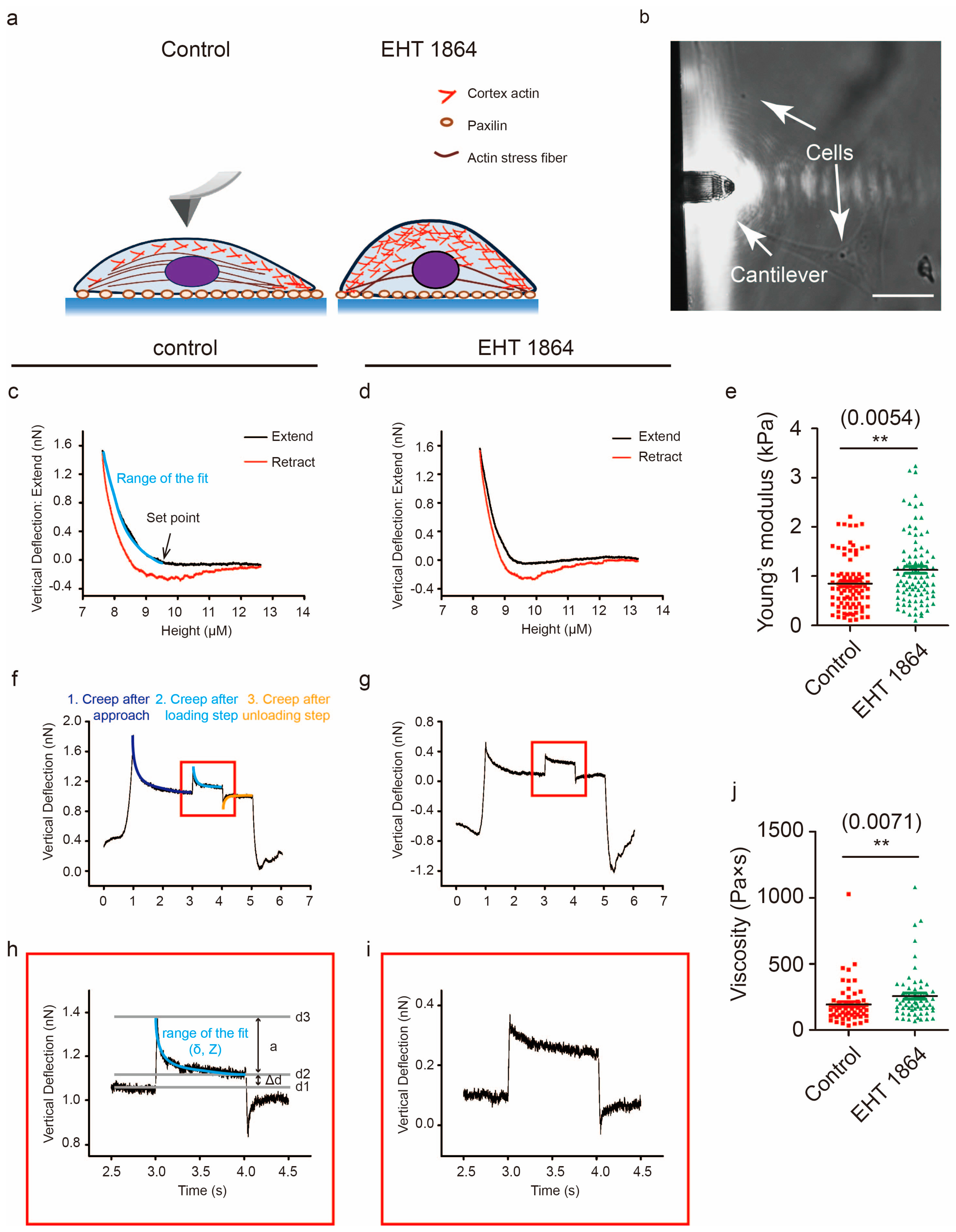
2.5. Characterization of the Mechanical Properties in Response to Rac1 Inhibition
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Gene Silencing
4.3. Immunofluorescent Staining and RT-PCR
4.4. Cell Proliferation Assay
4.5. Cell Transfection and Live Cell Imaging
4.6. Tracking Assays and Data Analysis
4.7. Active Rac1 Pull-Down Assay and Western Blot
4.8. Transwell Assay
4.9. Force Spectroscopy of Living Cells and Data Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy Plus Concomitant and Adjuvant Temozolomide for Glioblastoma. New Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Hegi, M.E.; Mason, W.P.; van den Bent, M.J.; Taphoorn, M.J.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of Radiotherapy with Concomitant and Adjuvant Temozolomide Versus Radiotherapy Alone on Survival in Glioblastoma in a Randomised Phase Iii Study: 5-Year Analysis of the Eortc-Ncic Trial. Lancet Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef]
- Chan, A.Y.; Coniglio, S.J.; Chuang, Y.Y.; Michaelson, D.; Knaus, U.G.; Philips, M.R.; Symons, M. Roles of the Rac1 and Rac3 Gtpases in Human Tumor Cell Invasion. Oncogene 2005, 24, 7821–7829. [Google Scholar] [CrossRef] [PubMed]
- Parri, M.; Chiarugi, P. Rac and Rho Gtpases in Cancer Cell Motility Control. Cell Commun. Signal 2010, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Chi, X.; Wang, S.; Huang, Y.; Stamnes, M.; Chen, J.L. Roles of Rho Gtpases in Intracellular Transport and Cellular Transformation. Int. J. Mol. Sci. 2013, 14, 7089–7108. [Google Scholar] [CrossRef] [PubMed]
- Ridley, A.J.; Paterson, H.F.; Johnston, C.L.; Diekmann, D.; Hall, A. The Small Gtp-Binding Protein Rac Regulates Growth Factor-Induced Membrane Ruffling. Cell 1992, 70, 401–410. [Google Scholar] [CrossRef]
- Blanchoin, L.; Amann, K.J.; Higgs, H.N.; Marchand, J.B.; Kaiser, D.A.; Pollard, T.D. Direct Observation of Dendritic Actin Filament Networks Nucleated by Arp2/3 Complex and Wasp/Scar Proteins. Nature 2000, 404, 1007–1011. [Google Scholar] [CrossRef]
- Goley, E.D.; Welch, M.D. The Arp2/3 Complex: An Actin Nucleator Comes of Age. Nat. Rev. Mol. CellBiol. 2006, 7, 713–726. [Google Scholar] [CrossRef]
- Takenawa, T.; Miki, H. Wasp and Wave Family Proteins: Key Molecules for Rapid Rearrangement of Cortical Actin Filaments and Cell Movement. J. Cell Sci. 2001, 114, 1801–1809. [Google Scholar]
- Ridley, A.J. Rho Gtpases and Actin Dynamics in Membrane Protrusions and Vesicle Trafficking. Trends Cell Biol. 2006, 16, 522–529. [Google Scholar] [CrossRef]
- Malliri, A.; van der Kammen, R.A.; Clark, K.; van der Valk, M.; Michiels, F.; Collard, J.G. Mice Deficient in the Rac Activator Tiam1 Are Resistant to Ras-Induced Skin Tumours. Nature 2002, 417, 867–871. [Google Scholar] [CrossRef] [PubMed]
- Kissil, J.L.; Walmsley, M.J.; Hanlon, L.; Haigis, K.M.; Bender Kim, C.F.; Sweet-Cordero, A.; Eckman, M.S.; Tuveson, D.A.; Capobianco, A.J.; Tybulewicz, V.L.; et al. Requirement for Rac1 in a K-Ras Induced Lung Cancer in the Mouse. Cancer Res. 2007, 67, 8089–8094. [Google Scholar] [CrossRef] [PubMed]
- Kazanietz, M.G.; Caloca, M.J. The RacGtpase in Cancer: From Old Concepts to New Paradigms. Cancer Res. 2017, 77, 5445–5451. [Google Scholar] [CrossRef] [PubMed]
- Gastonguay, A.; Berg, T.; Hauser, A.D.; Schuld, N.; Lorimer, E.; Williams, C.L. The Role of Rac1 in the Regulation of Nf-Kappab Activity, Cell Proliferation, and Cell Migration in Non-Small Cell Lung Carcinoma. Cancer Biol. Ther. 2012, 13, 647–656. [Google Scholar] [CrossRef]
- Yang, W.H.; Lan, H.Y.; Huang, C.H.; Tai, S.K.; Tzeng, C.H.; Kao, S.Y.; Wu, K.J.; Hung, M.C.; Yang, M.H. Rac1 Activation Mediates Twist1-Induced Cancer Cell Migration. Nat. Cell Biol. 2012, 14, 366–374. [Google Scholar] [CrossRef]
- Etienne-Manneville, S.; Hall, A. Rho Gtpases in Cell Biology. Nature 2002, 420, 629–635. [Google Scholar] [CrossRef]
- Shutes, A.; Onesto, C.; Picard, V.; Leblond, B.; Schweighoffer, F.; Der, C.J. Specificity and Mechanism of Action of Eht 1864, a Novel Small Molecule Inhibitor of Rac Family Small Gtpases. J. Biol. Chem. 2007, 282, 35666–35678. [Google Scholar] [CrossRef]
- Cross, S.E.; Jin, Y.S.; Rao, J.; Gimzewski, J.K. Nanomechanical Analysis of Cells from Cancer Patients. Nat. Nanotechnol. 2007, 2, 780–783. [Google Scholar] [CrossRef]
- Remmerbach, T.W.; Wottawah, F.; Dietrich, J.; Lincoln, B.; Wittekind, C.; Guck, J. Oral Cancer Diagnosis by Mechanical Phenotyping. Cancer Res. 2009, 69, 1728–1732. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, D.; Luo, W.; Yu, Y.; Yu, J.; Li, J.; Zhang, X.; Zhang, B.; Chen, J.; Wu, X.R.; et al. X-Linked Inhibitor of Apoptosis Protein (Xiap) Mediates Cancer Cell Motility Via Rho Gdp Dissociation Inhibitor (Rhogdi)-Dependent Regulation of the Cytoskeleton. J. Biol. Chem. 2011, 286, 15630–15640. [Google Scholar] [CrossRef]
- Olson, M.F.; Sahai, E. The Actin Cytoskeleton in Cancer Cell Motility. Clin. Exp. Metastasis 2009, 26, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, D.; Kurisu, S.; Takenawa, T. Regulation of Cancer Cell Motility through Actin Reorganization. Cancer Sci. 2005, 96, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Jordan, M.A.; Wilson, L. Microtubules and Actin Filaments: Dynamic Targets for Cancer Chemotherapy. Curr. Opin. Cell Biol. 1998, 10, 123–130. [Google Scholar] [CrossRef]
- Kavallaris, M. Microtubules and Resistance to Tubulin-Binding Agents. Nat. Rev. Cancer 2010, 10, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Dia, V.P.; Pangloli, P. Epithelial-to-Mesenchymal Transition in Paclitaxel-Resistant Ovarian Cancer Cells Is Downregulated by Luteolin. J. Cell Physiol. 2017, 232, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Darling, E.M.; Di Carlo, D. High-Throughput Assessment of Cellular Mechanical Properties. Annu. Rev. Biomed. Eng. 2015, 17, 35–62. [Google Scholar] [CrossRef]
- Taatjes, D.J.; Quinn, A.S.; Rand, J.H.; Jena, B.P. Atomic force microscopy: High resolution dynamic imaging of cellular and molecular structure in health and disease. J. Cell Physiol. 2013, 228, 1949–1955. [Google Scholar] [CrossRef]
- Fletcher, D.A.; Mullins, R.D. Cell Mechanics and the Cytoskeleton. Nature 2010, 463, 485–492. [Google Scholar] [CrossRef]
- Zhang, T. Cytoplasmic Motion Induced by Cytoskeleton Stretching and Its Effect on Cell Mechanics. Mol. Cell Biomech. 2011, 8, 169–193. [Google Scholar]
- Yango, A.; Schape, J.; Rianna, C.; Doschke, H.; Radmacher, M. Measuring the Viscoelastic Creep of Soft Samples by Step Response Afm. Soft Matter 2016, 12, 8297–8306. [Google Scholar] [CrossRef]
- Xiao, M.; Li, X.; Song, Q.; Zhang, Q.; Lazzarino, M.; Cheng, G.; Ulloa Severino, F.P.; Torre, V. A Fully 3D Interconnected Graphene-Carbon Nanotube Web Allows the Study of Glioma Infiltration in Bioengineered 3D Cortex-Like Networks. Adv. Mater 2018, 30, e1806132. [Google Scholar] [CrossRef]
- Nobes, C.D.; Hall, A. Rho, Rac, and Cdc42 Gtpases Regulate the Assembly of Multimolecular Focal Complexes Associated with Actin Stress Fibers, Lamellipodia, and Filopodia. Cell 1995, 81, 53–62. [Google Scholar] [CrossRef]
- Eden, S.; Rohatgi, R.; Podtelejnikov, A.V.; Mann, M.; Kirschner, M.W. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 2002, 418, 790–793. [Google Scholar] [CrossRef] [PubMed]
- Even-Ram, S.; Doyle, A.D.; Conti, M.A.; Matsumoto, K.; Adelstein, R.S.; Yamada, K.M. Myosin Iia Regulates Cell Motility and Actomyosin-Microtubule Crosstalk. Nat. Cell Biol. 2007, 9, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Vicente-Manzanares, M.; Zareno, J.; Whitmore, L.; Choi, C.K.; Horwitz, A.F. Regulation of Protrusion, Adhesion Dynamics, and Polarity by MyosinsIia and Iib in Migrating Cells. J. Cell Biol. 2007, 176, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Webb, N.J.; Donais, K.; Whitmore, L.A.; Thomas, S.M.; Turner, C.E.; Parsons, J.T.; Horwitz, A.F. Fak-SrcSignalling through Paxillin, Erk and Mlck Regulates Adhesion Disassembly. Nature 2004, 6, 154–161. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Galvanetto, N.; Nie, J.; Yang, Y.; Torre, V. Rac1 Promotes Cell Motility by Controlling Cell Mechanics in Human Glioblastoma. Cancers 2020, 12, 1667. https://doi.org/10.3390/cancers12061667
Xu J, Galvanetto N, Nie J, Yang Y, Torre V. Rac1 Promotes Cell Motility by Controlling Cell Mechanics in Human Glioblastoma. Cancers. 2020; 12(6):1667. https://doi.org/10.3390/cancers12061667
Chicago/Turabian StyleXu, Jing, Nicola Galvanetto, Jihua Nie, Yili Yang, and Vincent Torre. 2020. "Rac1 Promotes Cell Motility by Controlling Cell Mechanics in Human Glioblastoma" Cancers 12, no. 6: 1667. https://doi.org/10.3390/cancers12061667
APA StyleXu, J., Galvanetto, N., Nie, J., Yang, Y., & Torre, V. (2020). Rac1 Promotes Cell Motility by Controlling Cell Mechanics in Human Glioblastoma. Cancers, 12(6), 1667. https://doi.org/10.3390/cancers12061667



