1. Introduction
Hepatocellular carcinoma (HCC) is the most common form of liver cancer and the second deadliest cancer, affecting over 500,000 people worldwide every year [
1]. One well-established risk factor for HCC is chronic hepatitis B (HBV) infection, which accounts for approximately 50% of all cases [
2]. In the absence of diagnostic markers for the early detection of the disease, HBV-related HCC exhibits extremely poor prognosis, with a median survival of less than 16 months [
3]. Another well-established risk factor is excessive alcohol use. Studies have shown that alcohol consumption significantly increases the risk of HCC in HBV-related patients [
4]. Multiple epidemiologic and pathologic studies have reported the synergism between alcohol and HBV infection in the context of HCC [
5,
6,
7]. Despite the recent advancements in the knowledge of HCC and cancer in general, the molecular effects of ethanol exposure on the microenvironment and on HCC pathogenesis and progression, and its specific effects in HCC with a chronic viral hepatitis B background, remain poorly characterized.
The microbiome is defined as the genetic material of all the microbes—bacteria, fungi, protozoa and viruses—that live on and inside the human body. Since the microbiome is located primarily in the gut, it is not surprising that the gut microbiome has been the most studied and has been implicated in a wide variety of human diseases, including Alzheimer’s disease, cardiovascular disease, diabetes, arthritis, and cancer [
8]. In addition, there is an overwhelming amount of evidence on how the gut microbiome regulates liver function, and consequently, liver diseases, including nonalcoholic fatty liver disease (NAFLD), cirrhosis, and hepatocellular carcinoma [
9,
10]. Since the liver receives 70% of its blood supply from the intestine, it acts as the first line of defense against gut-derived foreign entities. The gut bacteria play a critical role in the maintenance of gut-liver axis health, and breakdown in the homeostasis between bacteria and the host can lead to a disruption of the intestinal barrier function and may allow the migration of the bacteria from the intestinal lumen to other extraintestinal organs or sites. In fact, the derangement of the gut flora occurs in 20–75% of patients with chronic liver disease [
10]. In contrast to our understanding of the gut microbiome, there have been very limited reports on the intratumor liver microbiome, especially in HCC. To date, there have been no etiology-specific studies focusing on the role of the liver microbiome in HCC pathogenesis and progression. Recent studies have shown that the microbiome is able to modulate the immune system and thereby alter homeostasis [
11]. The altered abundance of specific microbes may increase or decrease immune activity [
12]. Additionally, microbial metabolites may alter cell signaling pathways, thereby affecting cancer cells as well [
13]. Through these mechanisms, microbes within the intratumoral liver microbiome may affect carcinogenesis.
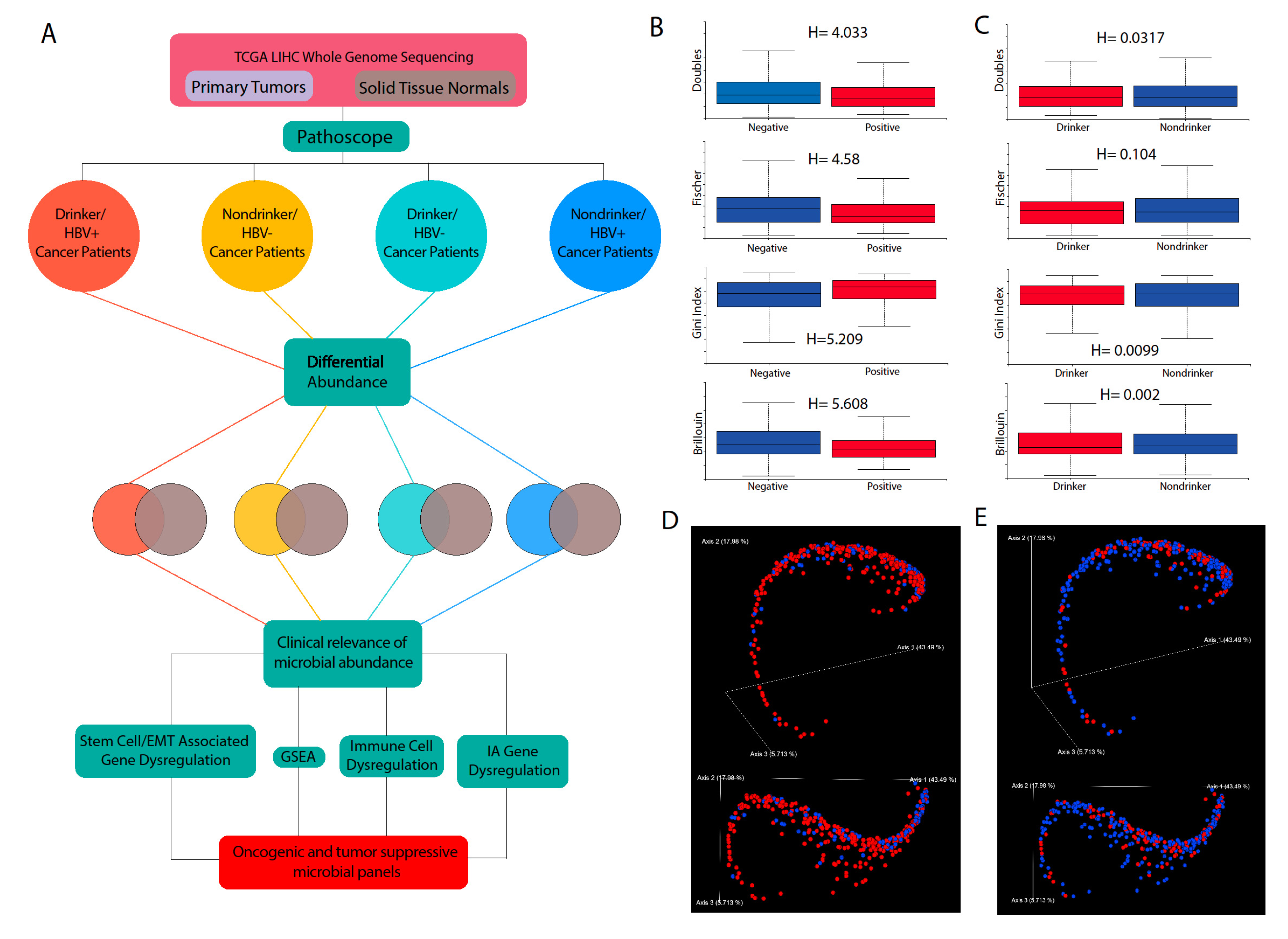
In this study, we sought to expand the current understanding of the interplay among alcohol consumption, HBV, and HCC, using next-generation RNA-sequencing data from HBV+ HCC patients and adjacent normal liver tissues. We identified the most abundant bacterial microbiome based on etiology and divided our subjects into four cohorts: HBV− nondrinkers, HBV+ nondrinkers, HBV− drinkers, and HBV+ drinkers. We investigated the association between microbial diversity and abundance and their correlation with clinical variables, including survival, tumor stage, histologic grade, fibrosis Ishak score, and Pugh classification grade. We assessed whether alcohol consumption synergizes with HBV to regulate the microbiome and how intratumor HCC microbial dysbiosis in turn correlates with the clinical variables. In addition, we studied the correlation between microbial abundance and transcriptomic alterations. The results of this study suggest that both heavy alcohol use and HBV infection may utilize the intratumor microbiome to promote cancer development and that the microbiome likely plays previously uncharacterized roles in the HBV and alcohol-associated pathogenesis and progression of HCC.
3. Discussion
The microbiome has been significantly implicated in cancer development in recent years. Recent research has illustrated the microbiome as a powerful factor in influencing drug metabolism, inflammation, cancer progression, and cancer treatment outcomes [
16]. However, the precise role of the microbiome in cancer is just beginning to be elucidated. Determining how microbes are involved in cancer is often a significant challenge, because they can either be harmful or beneficial [
17]. A potential factor that determines whether a microbe is oncogenic or tumor suppressive is thought to be its effects on modulating the immune system, which could lead to chronic inflammation and an increased risk of cancer, but could also lead to immune activation, which is critical for eliminating cancerous cells when they arise.
In this study, we present provocative and unprecedented evidence that the liver microbiome is dysregulated in patients who are drinkers and who are infected with HBV, two major risk factors that are known to cause liver cancer development. Our study is among the first to demonstrate the existence of an intratumor microbiome in the liver. While multiple studies have investigated the gut microbiome-liver axis, no established evidence exists that proves that the liver harbors a microbiome in humans. However, it is likely that the liver could, because of its proximity to the gut. The liver receives drainage through the portal vein from the intestines, which accounts for 70% of blood influx into the liver [
18]. While the portal vein should not be permeable to bacteria because of the gastrointestinal barrier, a number of diseases and conditions are associated with the disruption of this barrier, including inflammatory bowel disease, celiac disease, bowel obstruction, and gastrointestinal infection [
19]. Breaching of this barrier has been associated with the entrance of microbes into the portal vein [
19]. Other routes of origin of bacteria into the liver could also be conceived. A recent publication found that microbes migrate into the pancreas from the intestines through the pancreatic duct [
20]. Through a similar mechanism, bacteria can theoretically enter the liver from the gut through the bile duct. There are also multiple reports of a biliary microbiome [
21,
22].
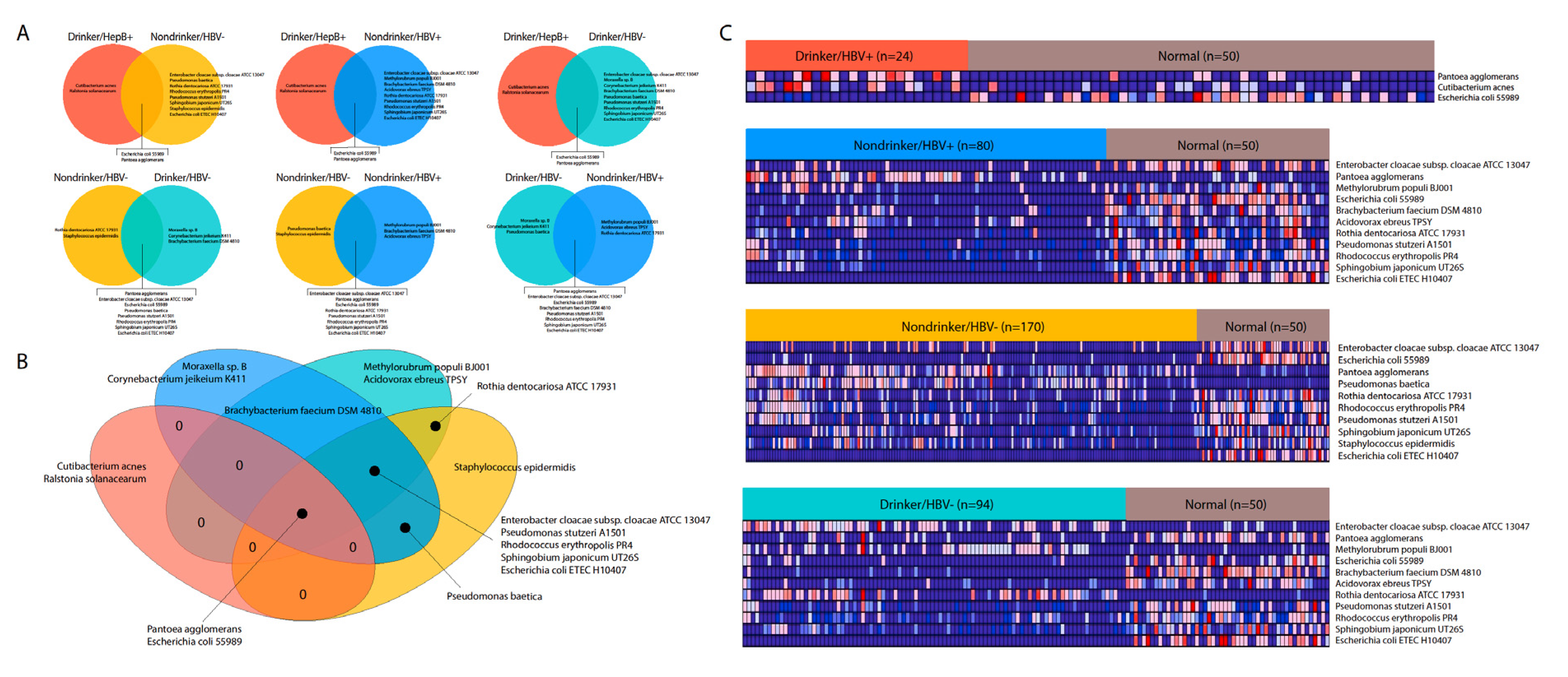
Exploring the association between the liver microbiome and two of the main etiological factors for LIHC, viral hepatitis B and heavy alcohol drinking, we discovered a significant microbiome dysregulation landscape, mediated by exposure to alcohol and HBV. Interestingly, we found that most microbes affected by alcohol and HBV exposure were downregulated compared to adjacent normal tissue, although there are a few exceptions. A relatively larger number of microbes was dysregulated after exposure to either HBV or alcohol than after exposure to both factors, and the microbes dysregulated by HBV or alcohol alone are also not similar to those dysregulated by the combined effects of HBV and alcohol. Therefore, HBV and alcohol combined may result in unique synergistic or antagonistic effects on the liver microbiome that have yet to be elucidated.
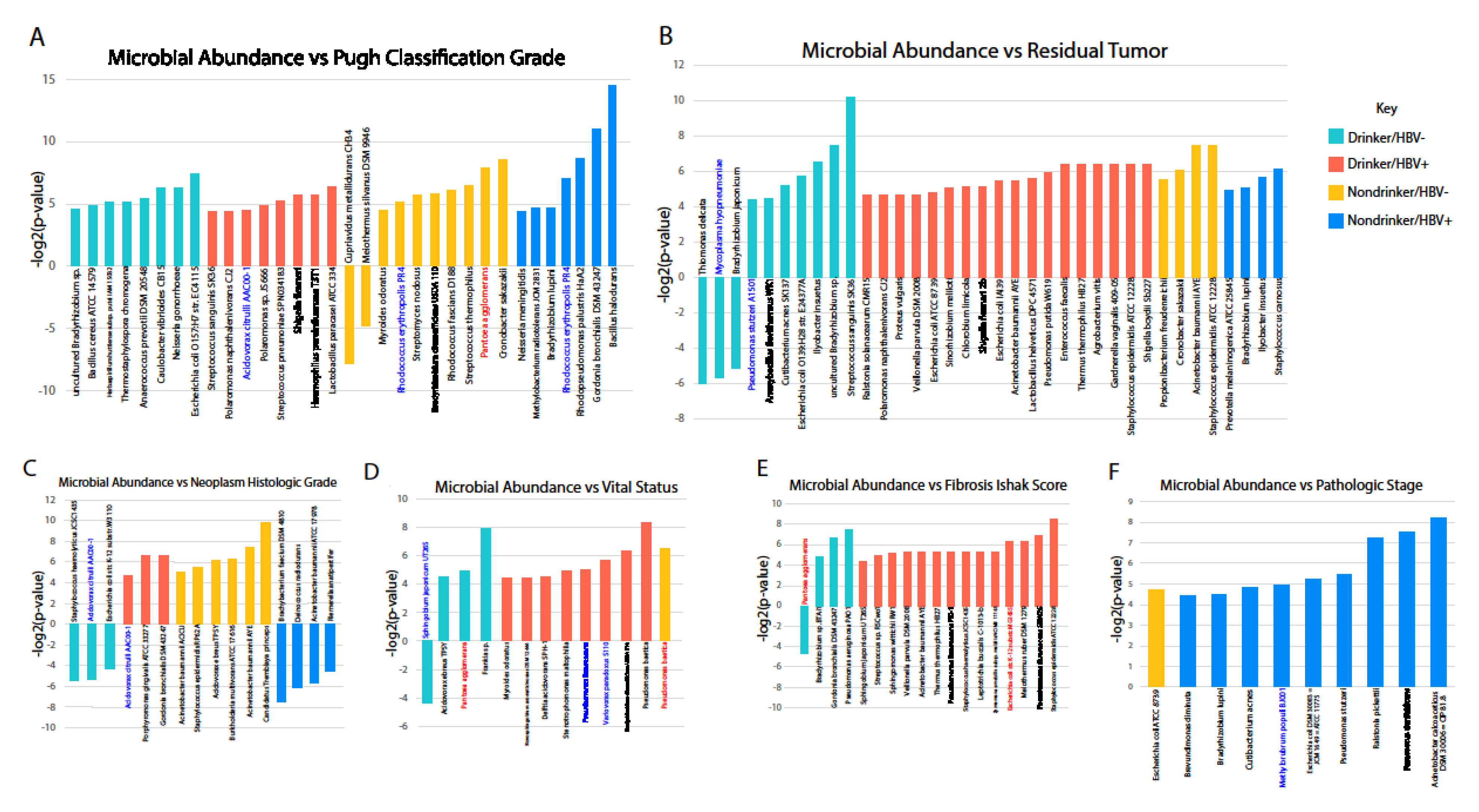
We also correlated the presence of different bacterial species with various clinical variables, including the Pugh classification grade, histologic grade, vital status, residual tumor, Ishak score for fibrosis, and pathologic stage. We discovered that, with few exceptions, most correlations with clinical variables were positive with poor prognosis. In other words, the increased abundance of most microbes was associated with poorer prognosis. Most microbes exhibiting strong correlation with poor prognosis are not dysregulated in LIHC or by either of the etiologies we examined. However, many microbes downregulated by alcohol or HBV also exhibited the same positive correlation with poor prognosis, raising the possibility that the dysregulation of these microbes is tumor suppressive. Since many microbes that are not dysregulated in alcohol and HBV-associated LIHC correlate with poor prognosis, we may infer that microbes that are already present in the liver environment could be contributing to tumor development. Indeed, many of these microbes’ abundance correlated with the increased activity of cancer-associated pathways, including the AKT, ERK, EZH2, and ATF2 pathways.
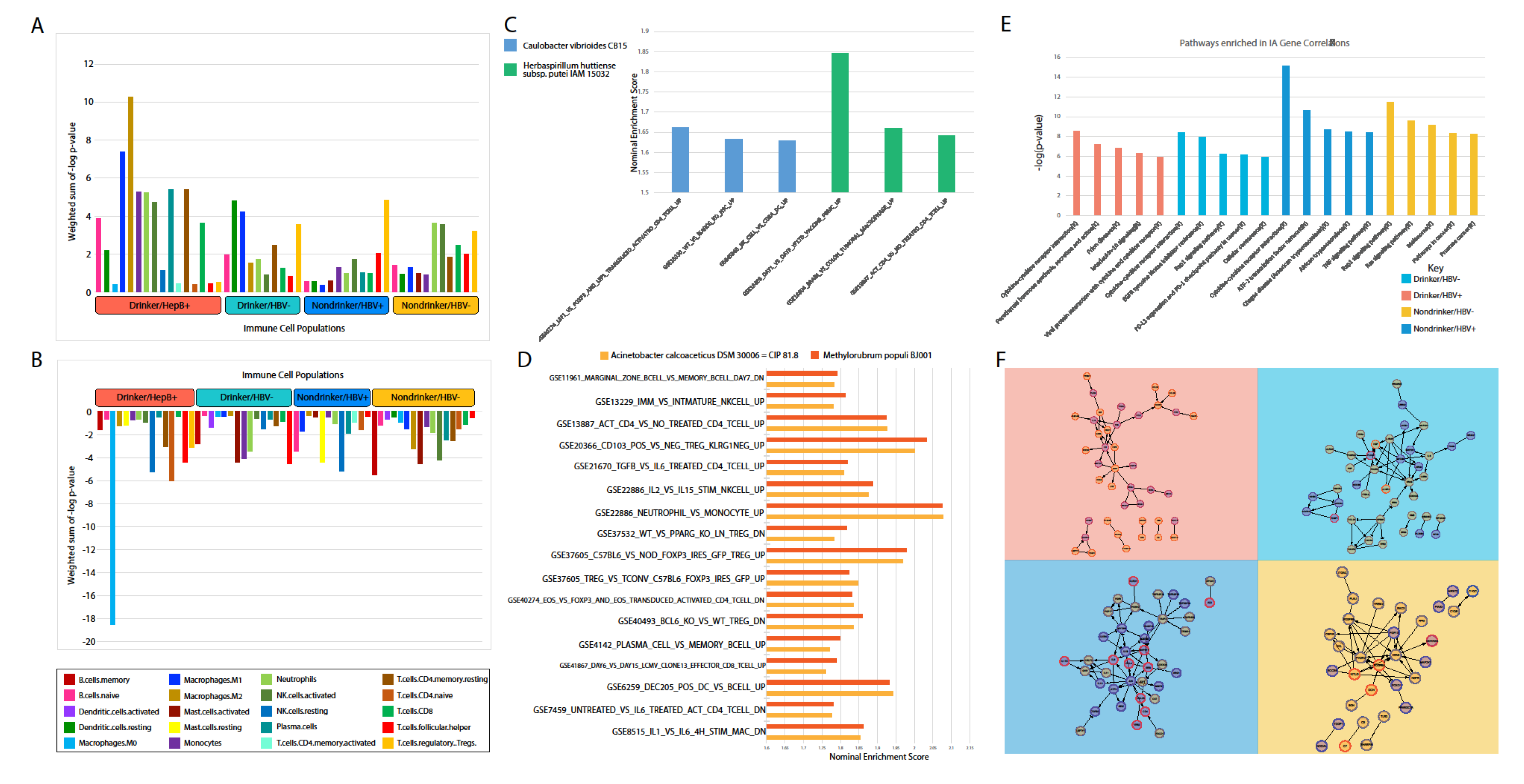
In addition to correlations with clinical variables and cancer pathways, we also examined the correlations between microbes and stem cell-related genes and microbes. In the cancer stem cell theory, a population of cells in the tumor exhibits stem cell features, and could self-renew, produce heterogeneous cancer cells, and orchestrate tumor growth, contributing to treatment resistance, metastasis, and relapse [
23]. We found that, in HBV-exposed patients who do not drink, many microbes’ abundance positively correlated with increased activities of stem cell-related signatures. For the few microbes that are downregulated by HBV, their dysregulation associated strongly with decreased stem cell activity. Our results thus suggest that HBV-induced LIHC may downregulate cancer stem cell activity through downregulating microbe levels. However, the liver microbiome itself could contribute to increased cancer stem cells activity, thus contributing to tumor development and progression.
Based on our results, an important outstanding question is what determines if a tumor-promoting microbe can be downregulated by an etiological agent. The answer may be dependent on both the identity of the microbe and the etiological agent. For example,
Pantoea agglomerans, which is known to cause infection in cancer patients [
24], appears to be upregulated in LIHC samples from HBV+/nondrinkers and HBV−/nondrinkers. However, in samples from HBV−/drinkers, the same microbe is highly downregulated. Understanding how and why alcohol might induce the opposite trend of dysregulation for this microbe as that being induced by HBV may elucidate why certain microbes are downregulated in LIHC.
In order to elucidate the potential mechanism through which microbial abundance may be pro-tumor, we correlated microbial abundance to the expression of immune-associated genes in each of our cohorts. We discovered that cytokine signaling was strongly correlated with the abundance of microbes. For example, in the HBV+/drinker cohort, the upregulation of microbes correlated with the increased expression of cytokines, including CCL28, CCL26, CSF3, and SOCS3. However, several cytokines, like IL6 and IL10, were suppressed by microbe presence. Overall, a complex landscape of cytokine regulation might be conducted by the microbiome, as evidenced by the large number of microbial abundance correlations to cytokine expression.
In conclusion, we found that most microbes in the liver microbiome are tumor promoting in the context of HBV and alcohol exposure, but both factors could also downregulate microbes that are pro-tumor. However, only HBV, not alcohol, could downregulate microbes that may promote stem-cell function. These conclusions suggest different approaches to the diagnosis and management of HCC, depending on patient lifestyle and medical history. Recent findings have demonstrated the efficacy of blood serum diagnosis of cancer presence and risk using microbial signatures [
25]. Additionally, future cancer treatments may aim to target the bioproducts of certain microbes to augment the immune system. Recent data may support the use of metabolites as markers for HCC, as HCC metabolism has been shown to have a unique signature. Specifically, glucose and acetate usage are significantly altered in HCC versus normal liver metabolism [
26,
27,
28]. Our analysis has identified strains of
Escherichia coli to be potentially important to HCC progression. Glucose and acetate concentration and metabolism play a key role in
E. coli growth. When utilizing glucose to anaerobically grow,
E. coli secretes acetate. The subsequent increase in concentration inhibits further
E. coli growth without significantly changing
E. coli metabolism [
29]. This mechanism, called overflow metabolism, is hypothesized to be a common mechanism amongst fast-growing bacteria [
30,
31]. The growth-inhibiting role of acetate, coupled with anaerobic respiration, suggests a few possible mechanisms of liver dysbiosis. Changes to the gut microbiome may alter concentrations of acetate and glucose at the liver, thereby causing oncogenic dysbiosis. Alternatively, liver microenvironment changes in cancer may cause changes to bacteria levels. Dysbiotic bacteria may then alter concentration of metabolites and thereby affect other microbes and the tumor microenvironment at large. A further analysis of the metabolism of microbes identified in this study could possibly contribute to our understanding of HCC metabolism and therefore provide diagnostic and therapeutic opportunities. Further analysis of other gut disorders and their effect on the cancer microbiome may be important, as bacteria in the liver potentially originated from the gut. The effects of the microbiome on HCC could be attributed to manipulation by the gut microbiome, as is the case for breast cancer [
32].
Several limitations of our study exist. First, the microbiome of healthy, normal livers may be hard to characterize because of the lack of sample availability. We were restricted to comparing microbes within the tumor to those in adjacent normal tissue, but it is conceivable that microbes in both regions might be the same once cancer develops. Second, direct alignment does not provide as high a resolution of microbe detection as 16s rRNA sequencing. Direct alignment is also restricted by already sequenced bacteria genomes, which are a fraction of the total microbiome. Finally, we would not be able to differentiate between live microbes in the liver and nucleic acid fragments from microbes in other parts of the body. Further in vitro and in vivo experimentations are needed to validate our findings and elucidate the mechanisms by which HBV and alcohol may mediate the microbiome composition.
4. Materials and Methods
4.1. Data Acquisition from TCGA
Raw whole-transcriptome RNA-sequencing data for tumor tissue were downloaded from the TCGA legacy archive [
33] on 5 August 2018, for 373 LIHC samples and 50 adjacent normal samples. Level 3 normalized mRNA expression read counts for the above samples were downloaded from the GDC portal [
34]. Clinical information for all patients were downloaded from the Broad GDAC Firehose [
35].
4.2. Extraction of Microbial Reads and Calculation of Microbial Abundance
Using the Pathoscope 2.0 program [
36], RNA-sequencing data were filtered for bacterial reads via direct alignment through a wrapper for Bowtie2. Bacterial sequences deposited at the NCBI nucleotide database [
37] were used. Pathoscope generates two output measures quantifying the amount of bacterial species present in samples. One measure, best guess, quantifies the relative abundance of each species, expressed as a percentage. The other measure, best hit, signifies the absolute integer count of each species in the sequencing data.
4.3. Determination of Microbiome Diversity in Patient Samples
Using the Qiime2 framework [
14], the best guess data output from Pathoscope were used to calculate alpha diversity and beta diversity using the
qiime diversity alpha and
qimme diversity beta modules respectively. A principle component analysis of the beta diversity results was done via the
qiime diversity pcoa module and visualized using the
qiime emperor plot module, the latter of which uses the EMPeror tool [
38].
4.4. Differential Microbial Abundance between Cancer and Normal Patients
Differential abundance analysis was performed to compare microbe abundance (percentage abundance) in cancer tissues to microbe abundance in normal tissues of the same body site. Microbes that are present in less than ten percent of the patients in a cancer cohort were excluded. The Kruskal–Wallis analysis test was then applied to determine differential abundance (p < 0.05).
4.5. Correlation of Microbial Abundance to Survival and Clinical Variables
Survival analyses were performed while using the Kaplan–Meier model, with microbe expression being designated as a binary variable based on presence or absence of microbe in tumor samples. A univariate Cox regression analysis was used to identify candidates that were significantly associated with patient survival (p < 0.05). A clinical variable analysis was performed using the Kruskal–Wallis test, as described above.
4.6. Association between Microbial Abundance and IA Gene Expression
Using edgeR, a differential expression analysis was performed between mRNA expression of cancer and normal tissues to identify significantly dysregulated IA genes for each cancer (FDR < 0.05 and |log fold change| > 1). The Kruskal–Wallis test was used to correlate the abundance of significantly dysregulated microbes to significantly dysregulated IA genes (p < 0.05). Microbe abundance was modeled as a binary variable of presence and absence.
4.7. Identifying Connections between Significantly Dysregulated IA Genes
For each cohort, we filtered the significantly dysregulated IA genes identified from the previous analysis by p-value. The top 100 or so IA genes that had the most significant p-values when correlated to microbial abundance and were associated to microbes that were significantly dysregulated in the respective cancer cohort vs. normal were used. These IA genes were inputted into ReactomeFIViz which shows interactions between genes, to visualize the connections between the IA genes, in addition to finding pathways that contained these IA genes for each cohort.
4.8. Association between Microbial Abundance and Stem Cell and EMT-Associated Gene Expression
A panel of stem cell and EMT-associated genes were compiled by researching the literature. Using edgeR, a differential expression analysis was performed between mRNA expression of cancer and normal tissues, to identify significantly dysregulated stem cell and EMT-associated genes for each cohort (FDR < 0.05 and |log fold change| > 1). The Kruskal–Wallis test was used to correlate the abundance of significantly dysregulated microbes to significantly dysregulated stem cell and EMT-associated genes (p < 0.05). Microbe abundance was modeled as a binary variable of presence and absence.
4.9. Correlation of Microbial Abundance to Immune Infiltration
Estimated relative immune cell infiltration levels for 22 cell types were computed using the software CibersortX [
39]. Microbe abundance was then correlated with immune cell infiltration levels for each microbe using the Kruskal–Wallis test (
p < 0.05). Microbe abundance was modeled as a binary variable of presence and absence. The immune cell types examined include naïve B-cells, memory B-cells, plasma cells, CD8 T-cells, CD4 naïve T-cells, CD4 memory resting T-cells, CD4 memory activated T-cells, follicular helper T-cells, regulatory T-cells, gamma-delta T-cells, resting NK cells, activated NK cells, monocytes, M0-M2 macrophages, resting dendritic cells, activated dendritic cells, resting mast cells, activated mast cells, eosinophils, and neutrophils.
To determine the overall correlation of the microbiota of each cancer cohort with the infiltration levels of each immune cell type, the negative logged p-values for all correlations to each immune cell type were normalized as a fraction of the maximum negative logged p-value within each cancer cohort. For each immune cell population in each cancer cohort, the normalized log p-value were separately added based on whether they were correlatively negatively or positively to immune cell populations, in order to calculate the weighted sum.
4.10. Correlation of Microbial Abundance to Cancer and Immune-Associates Signatures
Signature enrichment corresponding to microbial abundance was measured using the gene set enrichment analysis (GSEA). Cancer and stem cell-associated signatures were chosen from the C6 set of signatures from the Molecular Signatures Database (MSigDB) [
15]. Immune-associated signatures were chosen from the C7 set of signatures. Significantly enriched signatures were identified by a nominal enrichment score > 1 and a nominal
p-value < 0.05. The direction of pathway enrichment was filtered to match the direction of clinical variable correlations per microbe.
4.11. Evaluation of Contamination Using Date of Sequencing
We applied a heuristic algorithm to extract sequencing dates where this overexpression occurs, which allowed us to determine potential contaminants’ relationship with the sequencing date. We visualized the microbial abundance of cancer patients in the form of a heat map and removed any microbe where stretches of dates with high microbial abundance exist, which we identified as contamination. In other words, contaminants are marked by marked non-uniform abundance across sequencing dates. For all the following analyses, we removed all microbes that were identified as contaminants.
4.12. Evaluation of Contamination Based on Plates
The abundance values of microbes were associated with plates on which the samples were stored prior to sequencing using the Kruskal–Wallis test (p < 0.05) and the visual examination of abundance differences between different plates using a boxplot.
4.13. Evaluation of Contamination Using Microbial Abundance Counts
The abundance of individual microbes in each patient is plotted against total microbe reads in the same patient, to determine if any microbe is likely a contaminant. Best hit results from Pathoscope are used for this analysis because absolute counts are required. In the resulting scatterplots, if a positive slope exists, it is likely that the microbe was biologically relevant and physically present in the sample, since the counts per microbe increased with the number of microbes sequenced. If the scatterplot has a slope of close to zero, and the counts of all the microbes are substantially above zero, it is likely that the microbe was a contaminant. This reasoning follows from the assumption that similar amounts of microbes will be present, regardless of how many microbes are present in the tissue sample if the microbe is an environmental contaminant. The Spearman correlation test and the correlation coefficient (R2) was used to calculate significance of a linear trendline and the slope of that trendline, respectively.