Molecular Interplay between Dormant Bone Marrow-Resident Cells (BMRCs) and CTCs in Breast Cancer
Abstract
1. Introduction
2. Results
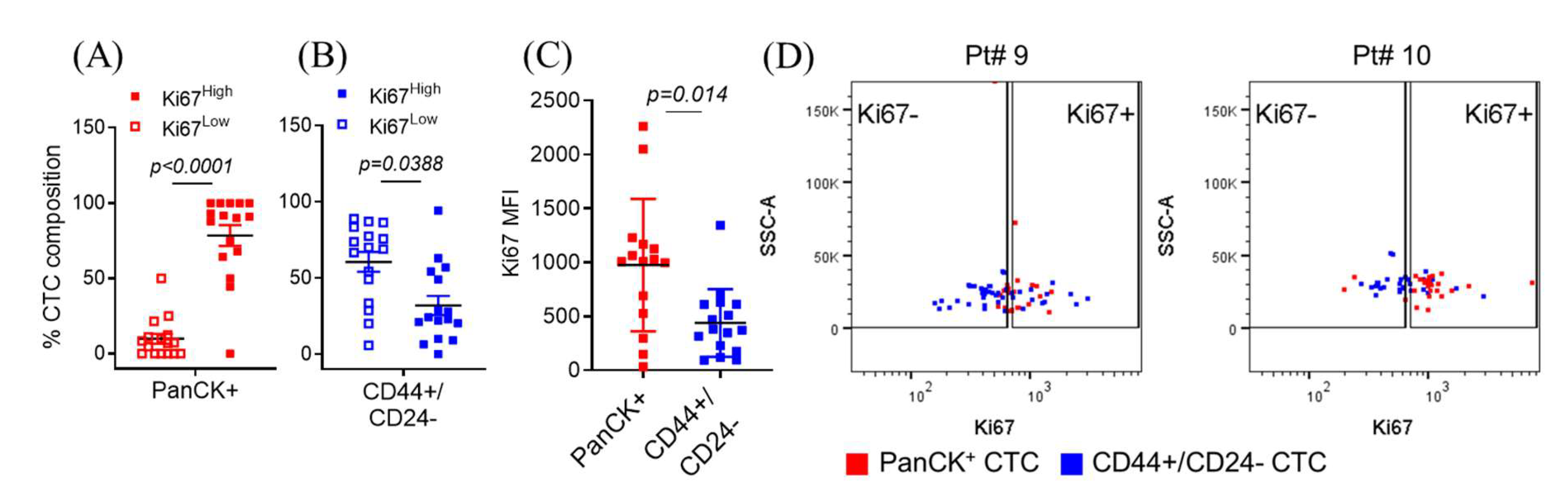
2.1. Isolated Patient CTCs Are Characterized by Proliferative/Epithelial Markers or Quiescent/Stem-Like Markers
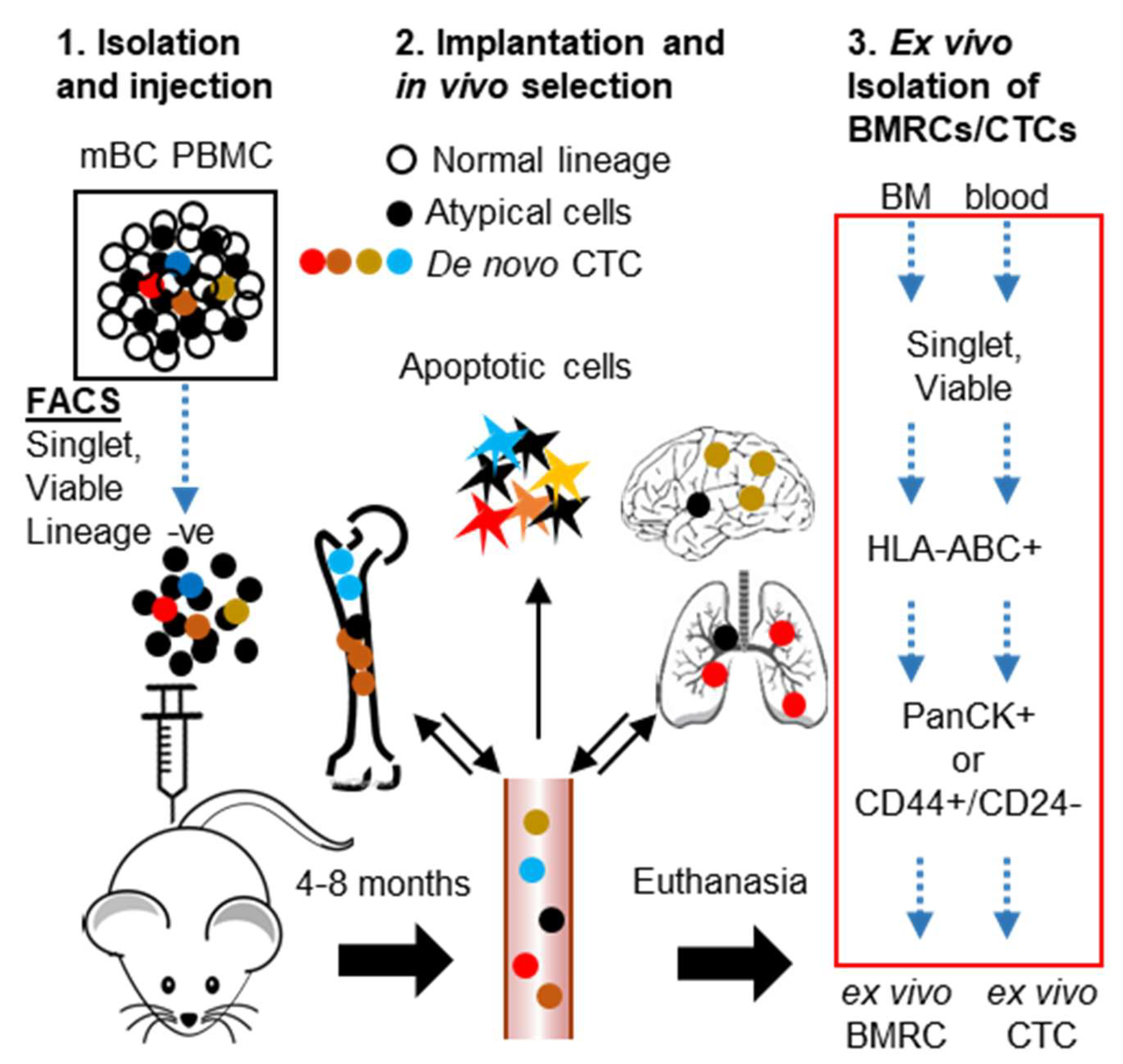
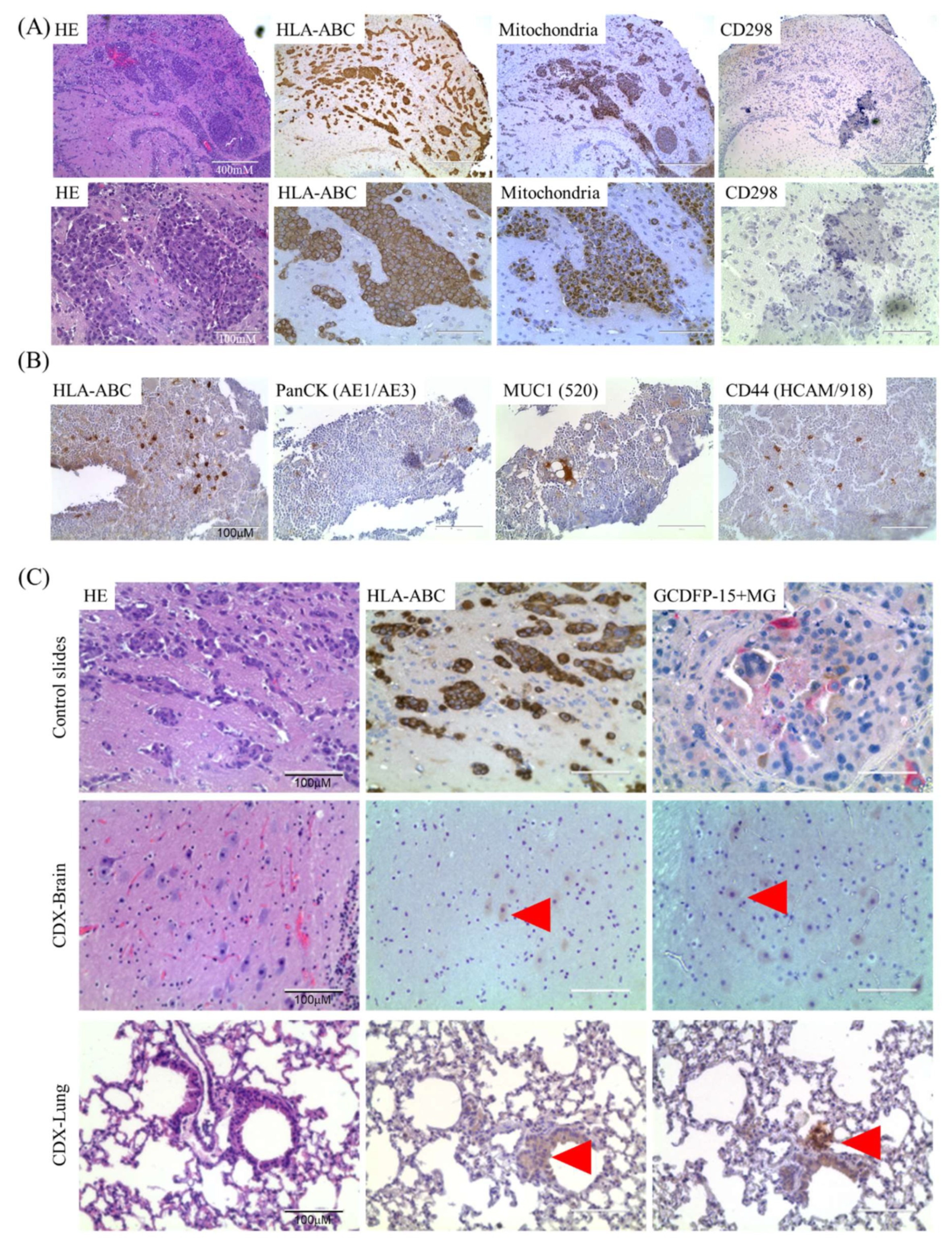
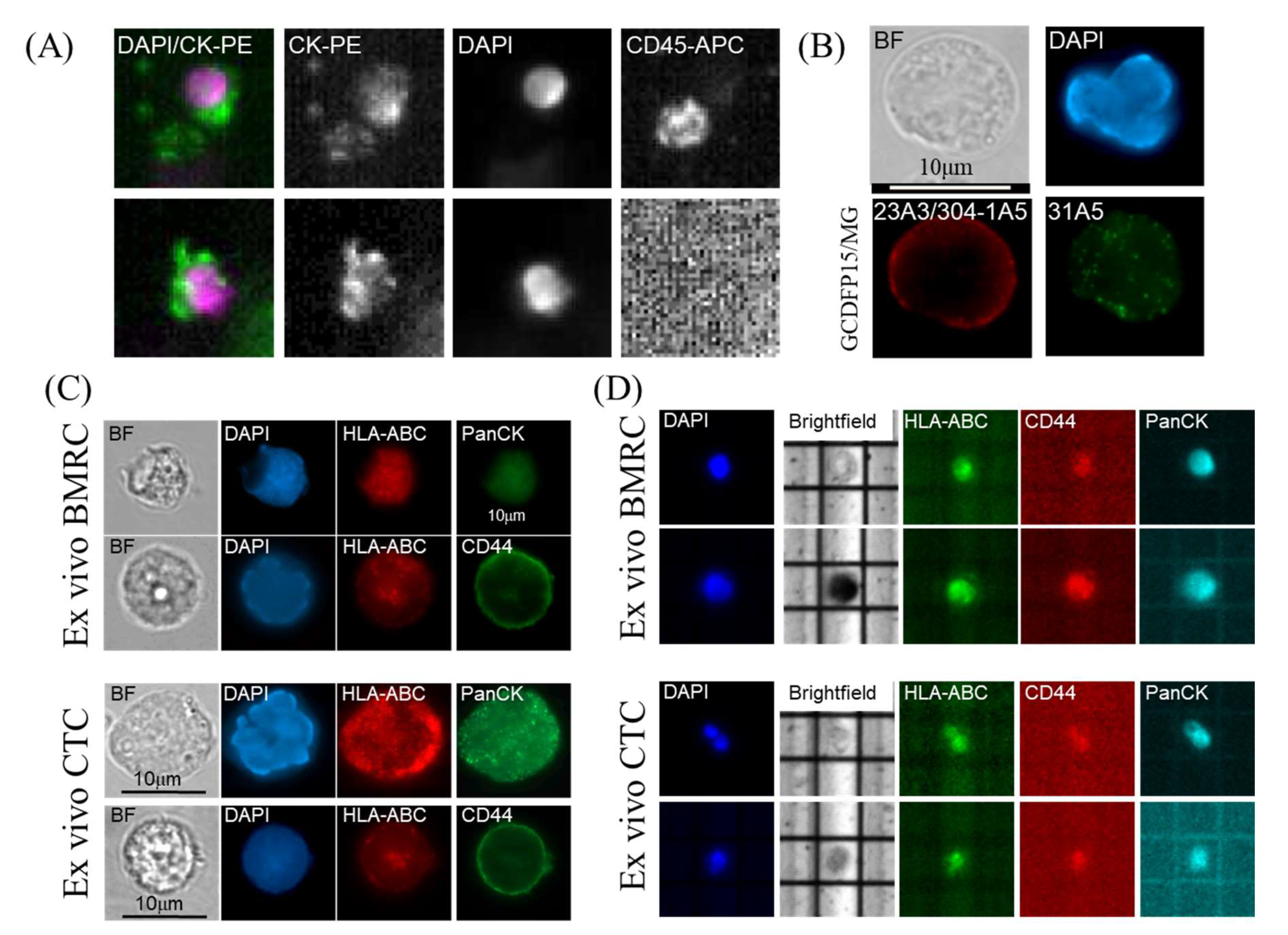
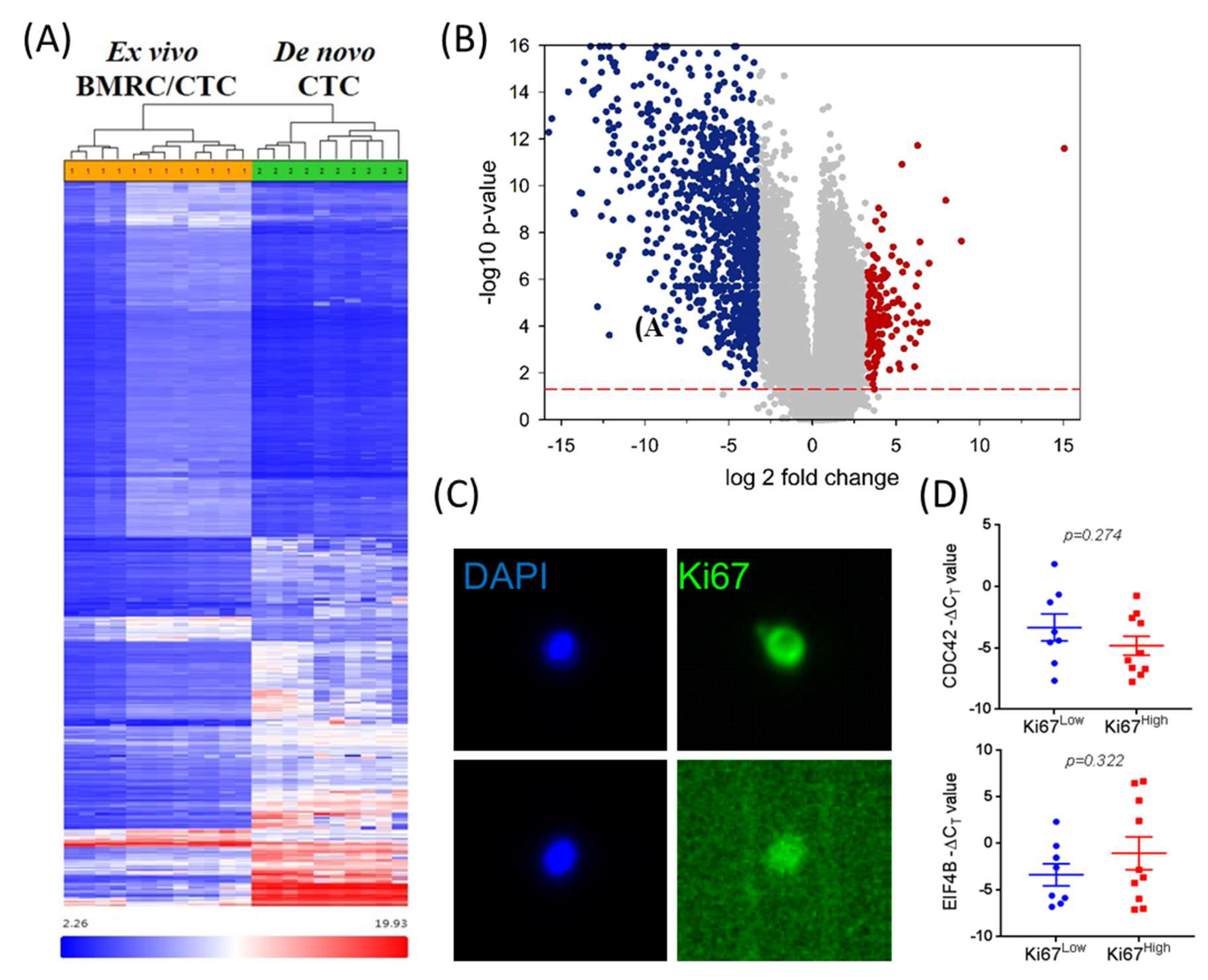
2.2. Isolation and Characterization of Ex Vivo BMRCs and CTCs
2.3. mTOR Signaling Is Upregulated in Ex Vivo CTCs
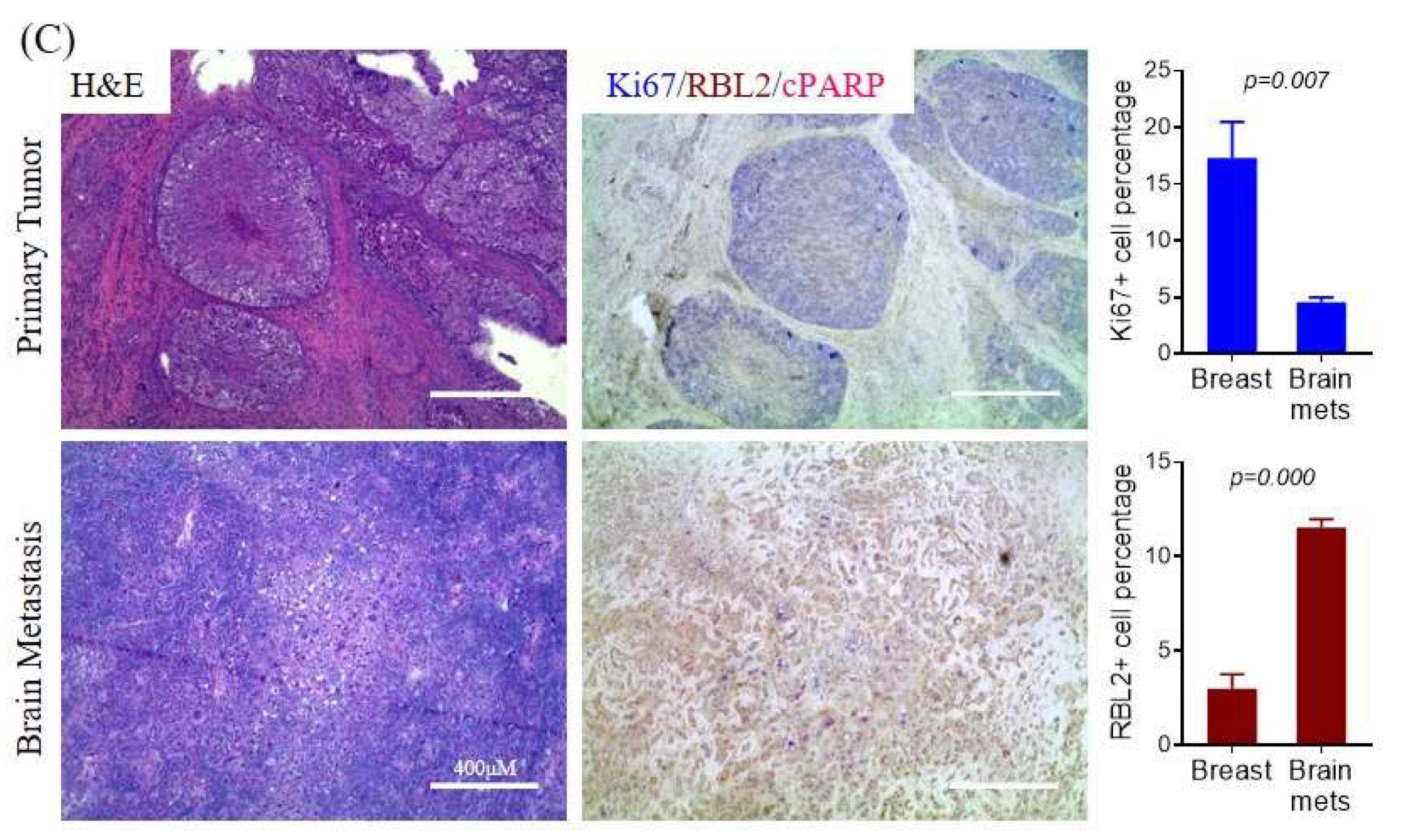
2.4. Differential mTORC Activities in Quiescence vs Proliferation Properties of CTCs and Metastatic Tumors
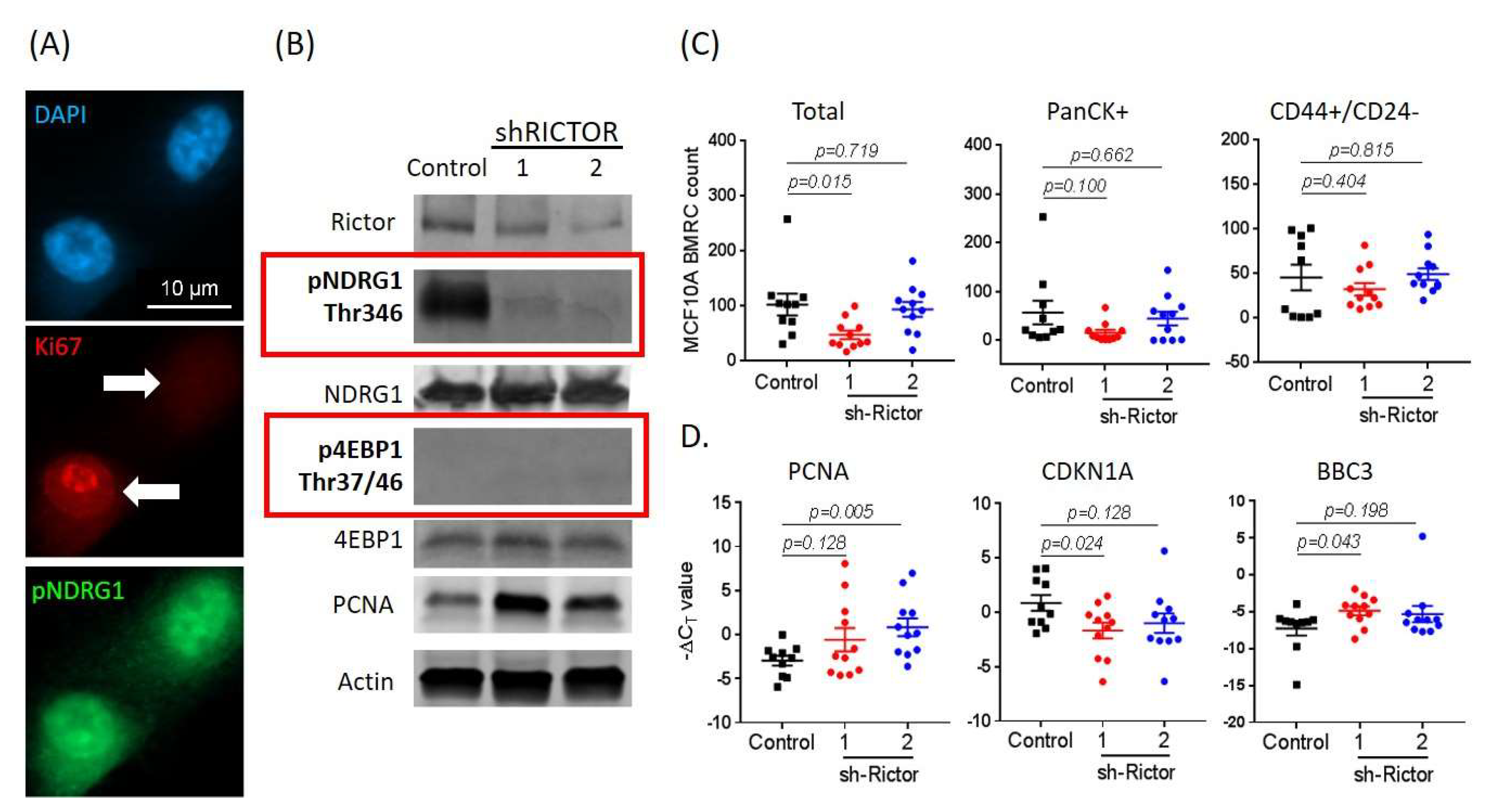
2.5. mTORC2 Inhibition Affects BMRC Survival/Proliferation
3. Discussion
4. Materials and Methods
4.1. Patient Blood Collection and Samples
4.2. Cell Lines and Tissue Culture
4.3. Flow Cytometry
4.4. CellSearch Analyses and CTC Enumeration
4.5. CTC-Derived Xenografts
4.6. RNA Microarrays and Pathways Analysis
4.7. Immunofluorescence, Immunohistochemistry and DEPArray
4.8. ShRNA, qPCR, and Western Blotting Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Howlader, N.; Noone, A.M.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; et al. SEER Cancer Statistics Review, 1975–2016; National Cancer Institute: Bethesda, MD, USA, 2020; Available online: https://seer.cancer.gov/statfacts/html/breast.html (accessed on 16 June 2020).
- Schumacher, T.N.; Schreiber, R.D. Neoantigens in cancer immunotherapy. Science 2015, 348, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, H.; Obradović, M.M.S.; Hoffmann, M.; Harper, K.L.; Sosa, M.S.; Werner-Klein, M.; Nanduri, S.L.K.; Werno, C.; Ehrl, C.; Maneck, M.; et al. Early dissemination seeds metastasis in breast cancer. Nature 2016, 540, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Rhim, A.D.; Mirek, E.T.; Aiello, N.M.; Maitra, A.; Bailey, J.M.; McAllister, F.; Reichert, M.; Beatty, G.L.; Rustgi, A.K.; Vonderheide, R.H.; et al. EMT and Dissemination Precede Pancreatic Tumor Formation. Cell 2012, 148, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Rack, B.; Schindlbeck, C.; Jückstock, J.; Andergassen, U.; Hepp, P.; Zwingers, T.; Friedl, T.W.P.; Lorenz, R.; Tesch, H.; Fasching, P.A.; et al. Circulating tumor cells predict survival in early average-to-high risk breast cancer patients. J. Natl. Cancer Inst. 2014, 106, 106. [Google Scholar] [CrossRef]
- Luzzi, K.J.; Macdonald, I.C.; Schmidt, E.E.; Kerkvliet, N.; Morris, V.L.; Chambers, A.F.; Groom, A.C. Multistep Nature of Metastatic Inefficiency: Dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am. J. Pathol. 1998, 153, 865–873. [Google Scholar] [CrossRef]
- Klein, C.A. Selection and adaptation during metastatic cancer progression. Nature 2013, 501, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Pantel, K.; Alix-Panabieres, C.; Riethdorf, S. Cancer micrometastases. Nat. Rev. Clin. Oncol. 2009, 6, 339–351. [Google Scholar] [CrossRef]
- Jenkins, D.E.; Hornig, Y.S.; Oei, Y.; Dusich, J.; Purchio, T. Bioluminescent human breast cancer cell lines that permit rapid and sensitive in vivo detection of mammary tumors and multiple metastases in immune deficient mice. Breast Cancer Res. 2005, 7, R444–R454. [Google Scholar] [CrossRef]
- Naumov, G.N.; Macdonald, I.C.; Weinmeister, P.M.; Kerkvliet, N.; Nadkarni, K.V.; Wilson, S.M.; Morris, V.L.; Groom, A.C.; Chambers, A.F. Persistence of solitary mammary carcinoma cells in a secondary site: A possible contributor to dormancy. Cancer Res. 2002, 62, 2162–2168. [Google Scholar]
- Bidard, F.-C.; Vincent-Salomon, A.; Gomme, S.; Nos, C.; De Rycke, Y.; Thiery, J.P.; Sigal-Zafrani, B.; Mignot, L.; Sastre-Garau, X.; Pierga, J.-Y. Disseminated Tumor Cells of Breast Cancer Patients: A Strong Prognostic Factor for Distant and Local Relapse. Clin. Cancer Res. 2008, 14, 3306–3311. [Google Scholar] [CrossRef]
- Stefanovic, S.; Diel, I.; Sinn, P.; Englert, S.; Hennigs, A.; Mayer, C.; Schott, S.; Wallwiener, M.; Blumenstein, M.; Golatta, M.; et al. Disseminated Tumor Cells in the Bone Marrow of Patients with Operable Primary Breast Cancer: Prognostic Impact in Immunophenotypic Subgroups and Clinical Implication for Bisphosphonate Treatment. Ann. Surg. Oncol. 2015, 23, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Janni, W.; Vogl, F.D.; Wiedswang, G.; Synnestvedt, M.; Fehm, D.M.T.; Jückstock, J.; Borgen, E.; Rack, B.; Braun, S.; Sommer, H.; et al. Persistence of Disseminated Tumor Cells in the Bone Marrow of Breast Cancer Patients Predicts Increased Risk for Relapse—A European Pooled Analysis. Clin. Cancer Res. 2011, 17, 2967–2976. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ridgway, L.D.; Wetzel, M.D.; Ngo, J.; Yin, W.; Kumar, D.; Goodman, J.C.; Groves, M.D.; Marchetti, D. The Identification and Characterization of Breast Cancer CTCs Competent for Brain Metastasis. Sci. Transl. Med. 2013, 5, 180ra48. [Google Scholar] [CrossRef] [PubMed]
- Khoo, B.L.; Grenci, G.; Jing, T.; Lim, Y.B.; Lee, S.C.; Thiery, J.P.; Han, J.; Lim, C.T. Liquid biopsy and therapeutic response: Circulating tumor cell cultures for evaluation of anticancer treatment. Sci. Adv. 2016, 2, e1600274. [Google Scholar] [CrossRef] [PubMed]
- Grillet, F.; Bayet, E.; Villeronce, O.; Zappia, L.; Lagerqvist, E.L.; Lunke, S.; Charafe-Jauffret, E.; Pham, K.; Molck, C.; Rolland, N.; et al. Circulating tumour cells from patients with colorectal cancer have cancer stem cell hallmarks in ex vivo culture. Gut 2017, 66, 1802–1810. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov. 2016, 6, 479–491. [Google Scholar] [CrossRef]
- Markou, A.; Lazaridou, M.; Paraskevopoulos, P.; Chen, S.; Świerczewska, M.; Budna-Tukan, J.; Kuske, A.; Gorges, T.; Joosse, S.A.; Kroneis, T.; et al. Multiplex Gene Expression Profiling of In Vivo Isolated Circulating Tumor Cells in High-Risk Prostate Cancer Patients. Clin. Chem. 2018, 64, 297–306. [Google Scholar] [CrossRef]
- Bankó, P.; Lee, S.Y.; Nagygyörgy, V.; Zrinyi, M.; Chae, C.H.; Cho, D.H.; Telekes, A. Technologies for circulating tumor cell separation from whole blood. J. Hematol. Oncol. 2019, 12, 48. [Google Scholar] [CrossRef]
- Vishnoi, M.; Boral, D.; Liu, H.; Sprouse, M.L.; Yin, W.; Goswami-Sewell, D.; Tetzlaff, M.; Davies, M.A.; Glitza, I.C.; Marchetti, D. Targeting USP7 Identifies a Metastasis-Competent State within Bone Marrow–Resident Melanoma CTCs. Cancer Res. 2018, 78, 5349–5362. [Google Scholar] [CrossRef]
- Boral, D.; Vishnoi, M.; Liu, H.N.; Yin, W.; Sprouse, M.L.; Scamardo, A.; Hong, D.S.; Tan, T.Z.; Thiery, J.P.; Chang, J.C.; et al. Molecular characterization of breast cancer CTCs associated with brain metastasis. Nat. Commun. 2017, 8, 196. [Google Scholar] [CrossRef]
- Ghiso, J.A.A.; Kovalski, K.; Ossowski, L. Tumor Dormancy Induced by Downregulation of Urokinase Receptor in Human Carcinoma Involves Integrin and MAPK Signaling. J. Cell Biol. 1999, 147, 89–104. [Google Scholar] [CrossRef] [PubMed]
- Vishnoi, M.; Peddibhotla, S.; Yin, W.; Scamardo, A.T.; George, G.C.; Hong, D.S.; Marchetti, D. The isolation and characterization of CTC subsets related to breast cancer dormancy. Sci. Rep. 2015, 5, 17533. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Kang, Y. Cradle of evil: Osteogenic niche for early bone metastasis. Cancer Cell 2015, 27, 153–155. [Google Scholar] [CrossRef]
- Meng, S.; Tripathy, D.; Frenkel, E.P.; Shete, S.; Naftalis, E.Z.; Huth, J.F.; Beitsch, P.D.; Leitch, M.; Hoover, S.; Euhus, D.M.; et al. Circulating Tumor Cells in Patients with Breast Cancer Dormancy. Clin. Cancer Res. 2004, 10, 8152–8162. [Google Scholar] [CrossRef]
- Zhang, X.H.-F.; Wang, Q.; Gerald, W.; Hudis, C.A.; Norton, L.; Smid, M.; Foekens, J.A.; Massagué, J. Latent Bone Metastasis in Breast Cancer Tied to Src-Dependent Survival Signals. Cancer Cell 2009, 16, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Pantel, K. Tumor cell dissemination: Emerging biological insights from animal models and cancer patients. Cancer Cell 2013, 23, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Sprouse, M.L.; Welte, T.; Boral, D.; Liu, H.N.; Yin, W.; Vishnoi, M.; Goswami-Sewell, D.; Li, L.; Pei, G.; Jia, P.; et al. PMN-MDSCs Enhance CTC Metastatic Properties through Reciprocal Interactions via ROS/Notch/Nodal Signaling. Int. J. Mol. Sci. 2019, 20, 1916. [Google Scholar] [CrossRef]
- Vishnoi, M.; Liu, N.H.; Yin, W.; Boral, D.; Scamardo, A.; Hong, D.; Marchetti, D. The identification of a TNBC liver metastasis gene signature by sequential CTC-xenograft modeling. Mol. Oncol. 2019, 13, 1913–1926. [Google Scholar] [CrossRef]
- Magbanua, M.J.M.; Rugo, H.S.; Hauranieh, L.; Roy, R.; Scott, J.H.; Lee, J.C.; Hsiao, F.; Sosa, E.V.; Veer, L.V.; Esserman, L.J.; et al. Genomic and expression profiling reveal molecular heterogeneity of disseminated tumor cells in bone marrow of early breast cancer. NPJ Breast Cancer 2018, 4, 31. [Google Scholar] [CrossRef]
- Chaffer, C.L.; Juan, B.P.S.; Lim, E.; Weinberg, R.A. EMT, cell plasticity and metastasis. Cancer Metastasis Rev. 2016, 35, 645–654. [Google Scholar] [CrossRef]
- Kim, L.C.; Cook, R.S.; Chen, J. mTORC1 and mTORC2 in cancer and the tumor microenvironment. Oncogene 2016, 36, 2191–2201. [Google Scholar] [CrossRef] [PubMed]
- Schewe, D.M.; Aguirre-Ghiso, J. ATF6α-Rheb-mTOR signaling promotes survival of dormant tumor cells in vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 10519–10524. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Frias, M.A.; Chatterjee, A.; Yellen, P.; Foster, D.A. The Enigma of Rapamycin Dosage. Mol. Cancer Ther. 2016, 15, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Dai, Y.; Vethanayagam, R.R.; Hebert, M.F.; Thummel, K.E.; Unadkat, J.D.; Ross, D.D.; Mao, Q. Cyclosporin A, tacrolimus and sirolimus are potent inhibitors of the human breast cancer resistance protein (ABCG2) and reverse resistance to mitoxantrone and topotecan. Cancer Chemother. Pharmacol. 2006, 58, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Sarbassov, D.D.; Ali, S.M.; Kim, D.-H.; Guertin, D.A.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. Rictor, a Novel Binding Partner of mTOR, Defines a Rapamycin-Insensitive and Raptor-Independent Pathway that Regulates the Cytoskeleton. Curr. Biol. 2004, 14, 1296–1302. [Google Scholar] [CrossRef]
- Pullen, N.A.; Thomas, G. The modular phosphorylation and activation of p70s6k. FEBS Lett. 1997, 410, 78–82. [Google Scholar] [CrossRef]
- Fingar, D.C.; Salama, S.; Tsou, C.; Harlow, E.; Blenis, J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002, 16, 1472–1487. [Google Scholar] [CrossRef]
- Murray, J.; Campbell, D.G.; Morrice, N.; Auld, G.C.; Shpiro, N.; Marquez, R.; Peggie, M.; Bain, J.; Bloomberg, G.B.; Grahammer, F.; et al. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem. J. 2004, 384, 477–488. [Google Scholar] [CrossRef]
- García-Martínez, J.M.; Alessi, D.R. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1). Biochem. J. 2008, 416, 375–385. [Google Scholar] [CrossRef]
- Sarbassov, D.D.; Guertin, D.A.; Ali, S.M.; Sabatini, D.M. Phosphorylation and Regulation of Akt/PKB by the Rictor-mTOR Complex. Science 2005, 307, 1098–1101. [Google Scholar] [CrossRef]
- Yerushalmi, R.; Woods, R.; Ravdin, P.M.; Hayes, M.M.; Gelmon, K.A. Ki67 in breast cancer: Prognostic and predictive potential. Lancet Oncol. 2010, 11, 174–183. [Google Scholar] [CrossRef]
- Lawson, D.A.; Bhakta, N.R.; Kessenbrock, K.; Prummel, K.D.; Yu, Y.; Takai, K.; Zhou, A.; Eyob, H.; Balakrishnan, S.; Wang, C.-Y.; et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature 2015, 526, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Thibaudeau, L.; Taubenberger, A.V.; Holzapfel, B.M.; Quent, V.M.; Führmann, T.; Hesami, P.; Brown, T.D.; Dalton, P.D.; Power, C.A.; Hollier, B.G.; et al. A tissue-engineered humanized xenograft model of human breast cancer metastasis to bone. Dis. Model. Mech. 2014, 7, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Eyre, R.; Alférez, D.G.; Spence, K.; Kamal, M.; Shaw, F.L.; Simões, B.M.; Santiago-Gómez, A.; Sarmiento-Castro, A.; Bramley, M.; Absar, M.; et al. Patient-derived Mammosphere and Xenograft Tumour Initiation Correlates with Progression to Metastasis. J. Mammary Gland. Biol. Neoplasia 2016, 21, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Kanwar, N.; Hu, P.; Bedard, P.L.; Clemons, M.; McCready, D.; Done, S.J. Identification of genomic signatures in circulating tumor cells from breast cancer. Int. J. Cancer 2015, 137, 332–344. [Google Scholar] [CrossRef]
- Schardt, J.A.; Meyer, M.; Hartmann, C.H.; Schubert, F.; Schmidt-Kittler, O.; Fuhrmann, C.; Polzer, B.; Petronio, M.; Eils, R.; Klein, C.A. Genomic analysis of single cytokeratin-positive cells from bone marrow reveals early mutational events in breast cancer. Cancer Cell 2005, 8, 227–239. [Google Scholar] [CrossRef]
- Boral, D.; Marchetti, D. Liquid Biopsy in Prostate Cancer: A Case for Comprehensive Genomic Characterization of Circulating Tumor Cells. Clin. Chem. 2018, 64, 251–253. [Google Scholar] [CrossRef]
- Linde, N.; Fluegen, G.; Aguirre-Ghiso, J.A. The Relationship Between Dormant Cancer Cells and Their Microenvironment. Adv. Cancer Res. 2016, 132, 45–71. [Google Scholar] [CrossRef]
- Klein, C.A. Parallel progression of primary tumours and metastases. Nat. Rev. Cancer 2009, 9, 302–312. [Google Scholar] [CrossRef]
- Price, T.T.; Burness, M.L.; Sivan, A.; Warner, M.J.; Cheng, R.; Lee, C.H.; Olivere, L.; Comatas, K.; Magnani, J.; Lyerly, H.K.; et al. Dormant breast cancer micrometastases reside in specific bone marrow niches that regulate their transit to and from bone. Sci. Transl. Med. 2016, 8, 340ra73. [Google Scholar] [CrossRef]
- Zaytseva, Y.Y.; Valentino, J.D.; Gulhati, P.; Evers, B.M. mTOR inhibitors in cancer therapy. Cancer Lett. 2012, 319, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Rodrik-Outmezguine, V.S.; Okaniwa, M.; Yao, Z.; Novotny, C.J.; McWhirter, C.; Banaji, A.; Won, H.; Wong, W.; Berger, M.; De Stanchina, E.; et al. Overcoming mTOR resistance mutations with a new-generation mTOR inhibitor. Nature 2016, 534, 272–276. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Hendrickson, A.E.; Yun, S.S.; Han, J.J.; Schneider, P.A.; Koh, B.D.; Stenson, M.J.; Wellik, L.E.; Shing, J.C.; Peterson, K.L.; et al. Dual mTORC1/mTORC2 inhibition diminishes Akt activation and induces Puma-dependent apoptosis in lymphoid malignancies. Blood 2012, 119, 476–487. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Spandidos, A.; Wang, H.; Seed, B. PrimerBank: A PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 2011, 40, D1144–D1149. [Google Scholar] [CrossRef]


| mBC Pt # | Tumor Subtype | Length of in vivo Selection (days) | BMRCs | CTCs | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CD44+/CD24− | PanCK+ | CD44+/CD24− and PanCK+ | Total | CD44+/CD24− | PanCK+ | CD44+/CD24− and PanCK+ | Total | |||
| 4 | ER+/PR+ | 264 | 60 | 101 | 583 | 744 | 7 | 1272 | 138 | 1417 |
| 17 | ER+/PR+ | 160 | 40 | 323 | 97 | 460 | 38 | 240 | 22 | 300 |
| 18 | HER2+ | 72 | 3 | 111 | 106 | 220 | 10 | 39 | 1 | 50 |
| 19 | HER2+ | 140 | 40 | 96 | 53 | 189 | 40 | 417 | 31 | 488 |
| 8 | TNBC | 97 | 0 | 50 | 176 | 226 | 163 | 48 | 14 | 225 |
| 20 | TNBC | 64 | 64 | 96 | 478 | 638 | 7 | 1345 | 86 | 1438 |
| Canonical Pathways | −log (p-value) | z-score | Functions Annotation | p-value | z-score |
|---|---|---|---|---|---|
| mTOR Signaling | 8.06 | 0.632 | cancer cell death | 1.59 × 10−10 | 2.771 |
| Wnt/β-catenin Signaling | 2.91 | 1.897 | osteosarcoma cell death | 4.08 × 10−9 | 4.082 |
| Cardiac β-adrenergic Signaling | 2.28 | 0.905 | tumor necrosis | 1.17 × 10−8 | 3.001 |
| EIF2 Signaling | 16.4 | −4.025 | metastatic solid tumor | 1.37 × 10−8 | −2.567 |
| CD28 Signaling in T Helper Cells | 4.25 | −1.265 | cell invasion | 1.92 × 10−8 | −2.448 |
| Cdc42 Signaling | 3.14 | −0.816 | tumor cell line invasion | 5.24 × 10−8 | −2.368 |
| Regulator | Molecule Type | z-score | p-value of Overlap |
|---|---|---|---|
| Rapamycin | chemical drug | 4.849 | 8.74 × 10−25 |
| 5-fluorouracil | chemical drug | 3.64 | 8.06 × 10−18 |
| CD 437 | chemical drug | 5.34 | 3.82 × 10−17 |
| ST1926 | chemical drug | 4.904 | 1.4 × 10−14 |
| RICTOR | other | 4.842 | 1.61 × 10−13 |
| MYCN | transcription factor | −3.69 | 5.81 × 10−15 |
| LPS | chemical drug | −3.058 | 8.38 × 10−13 |
| IFNG | cytokine | −3.229 | 1.01 × 10−7 |
| poly rI:rC-RNA | biologic drug | −3.535 | 2.17 × 10−7 |
| TCR | complex | −2.331 | 2.81 × 10−7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boral, D.; Liu, H.N.; Kenney, S.R.; Marchetti, D. Molecular Interplay between Dormant Bone Marrow-Resident Cells (BMRCs) and CTCs in Breast Cancer. Cancers 2020, 12, 1626. https://doi.org/10.3390/cancers12061626
Boral D, Liu HN, Kenney SR, Marchetti D. Molecular Interplay between Dormant Bone Marrow-Resident Cells (BMRCs) and CTCs in Breast Cancer. Cancers. 2020; 12(6):1626. https://doi.org/10.3390/cancers12061626
Chicago/Turabian StyleBoral, Debasish, Haowen N. Liu, S. Ray Kenney, and Dario Marchetti. 2020. "Molecular Interplay between Dormant Bone Marrow-Resident Cells (BMRCs) and CTCs in Breast Cancer" Cancers 12, no. 6: 1626. https://doi.org/10.3390/cancers12061626
APA StyleBoral, D., Liu, H. N., Kenney, S. R., & Marchetti, D. (2020). Molecular Interplay between Dormant Bone Marrow-Resident Cells (BMRCs) and CTCs in Breast Cancer. Cancers, 12(6), 1626. https://doi.org/10.3390/cancers12061626






