Integrated Analysis of Key Differentially Expressed Genes Identifies DBN1 as a Predictive Marker of Response to Endocrine Therapy in Luminal Breast Cancer
Abstract
1. Introduction
2. Results
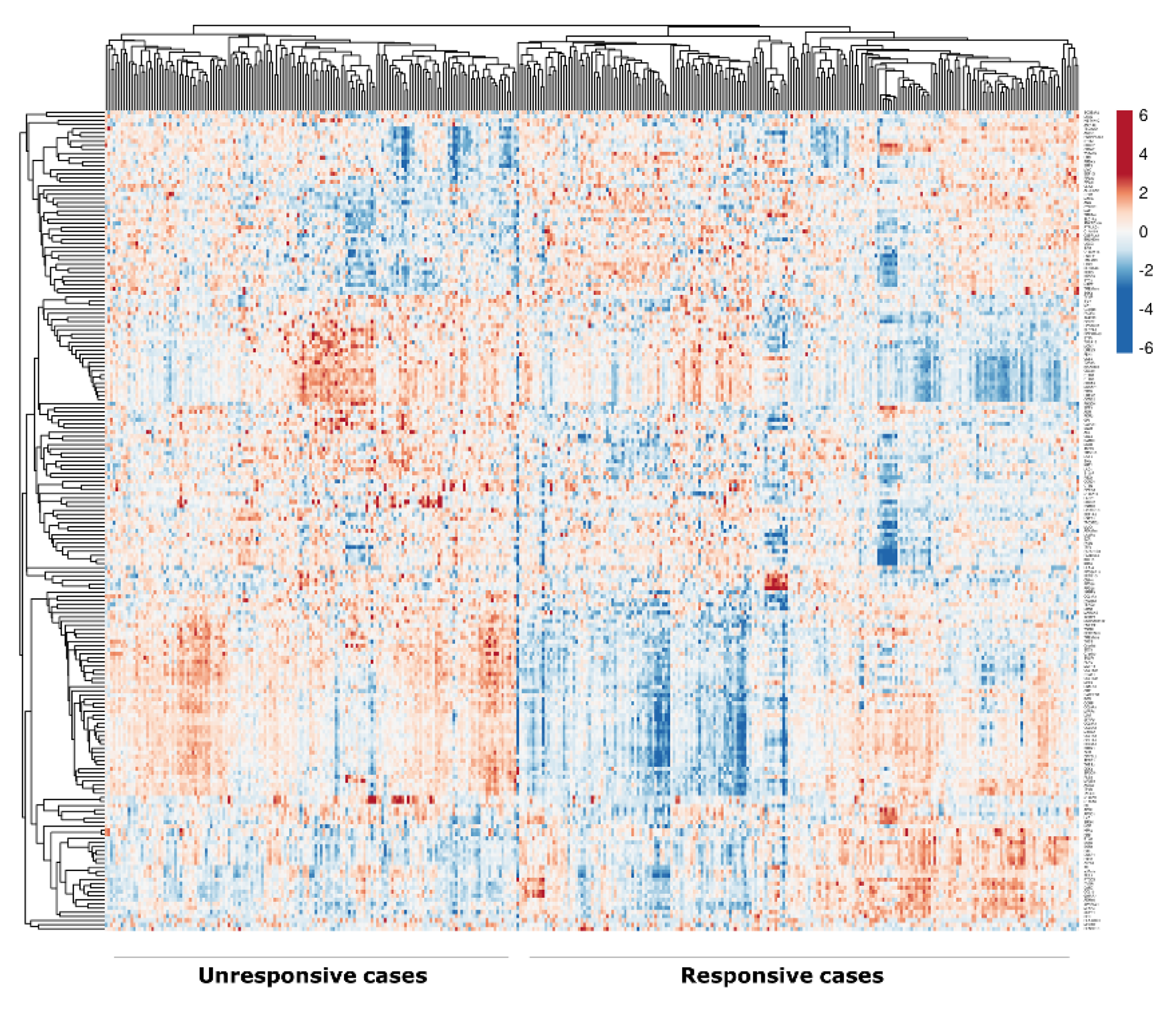
2.1. Identification of DEGs
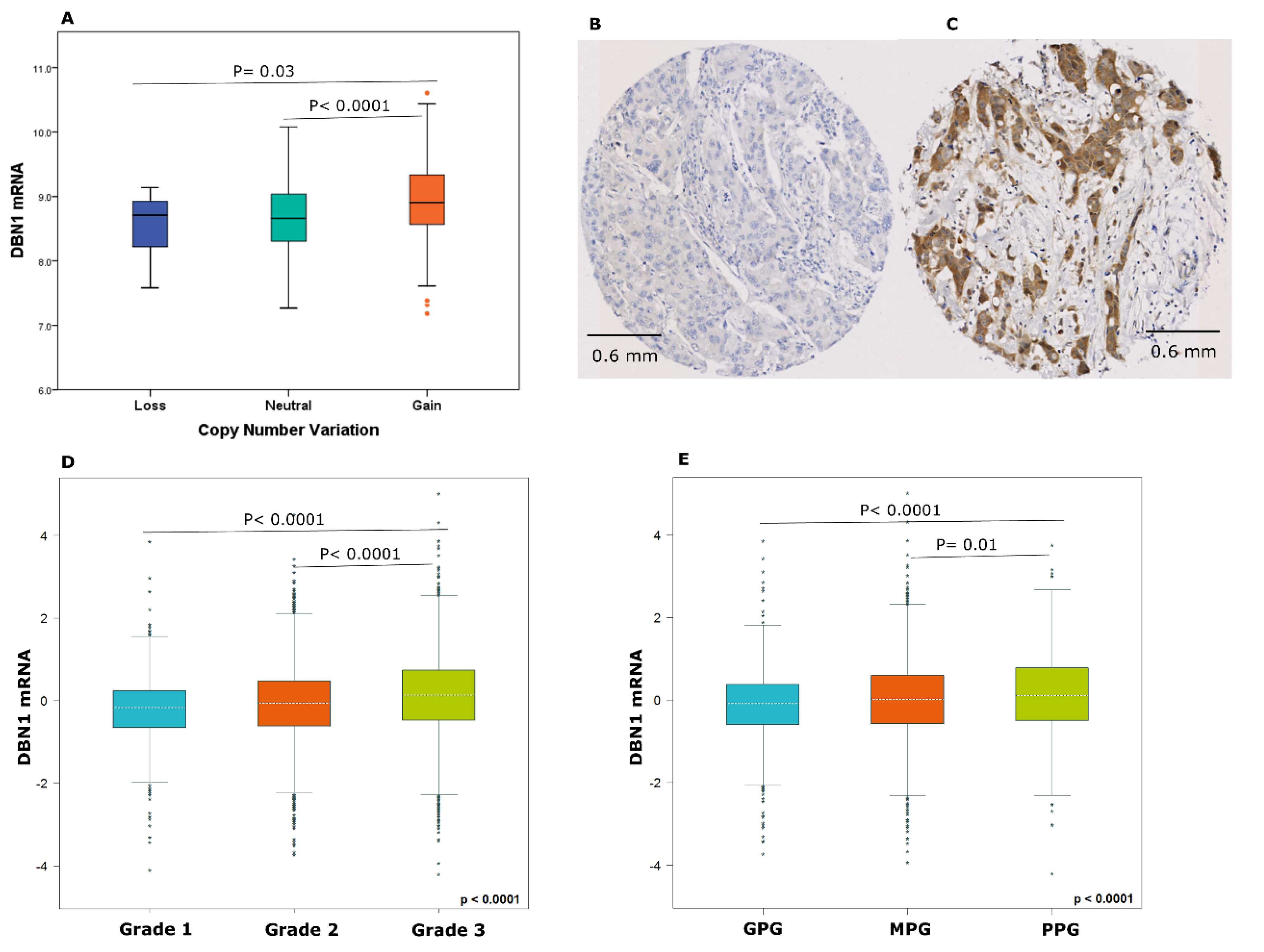
2.2. DBN1 Expression in Luminal Breast Cancer
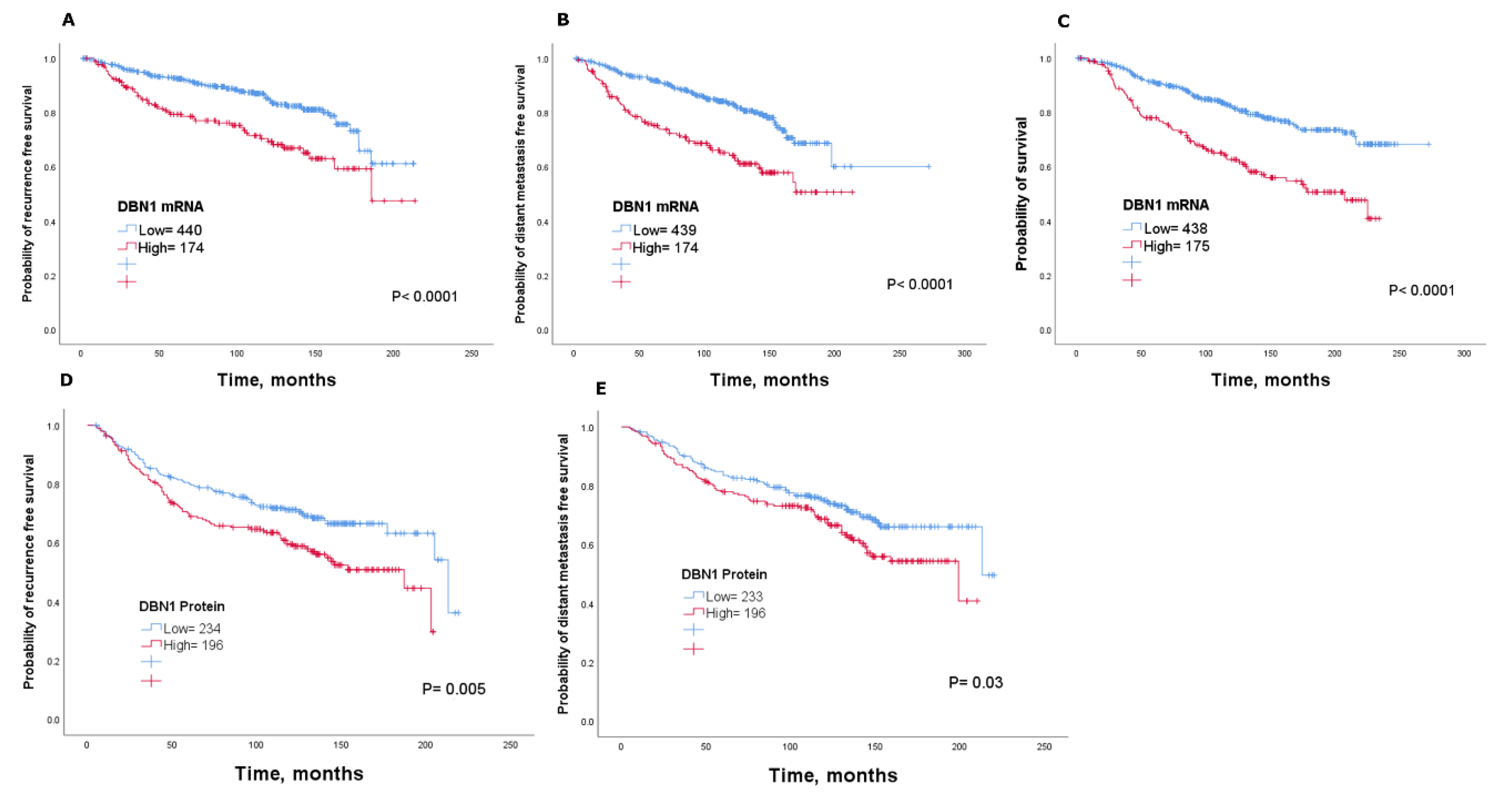
2.3. Clinical Significance of DBN1
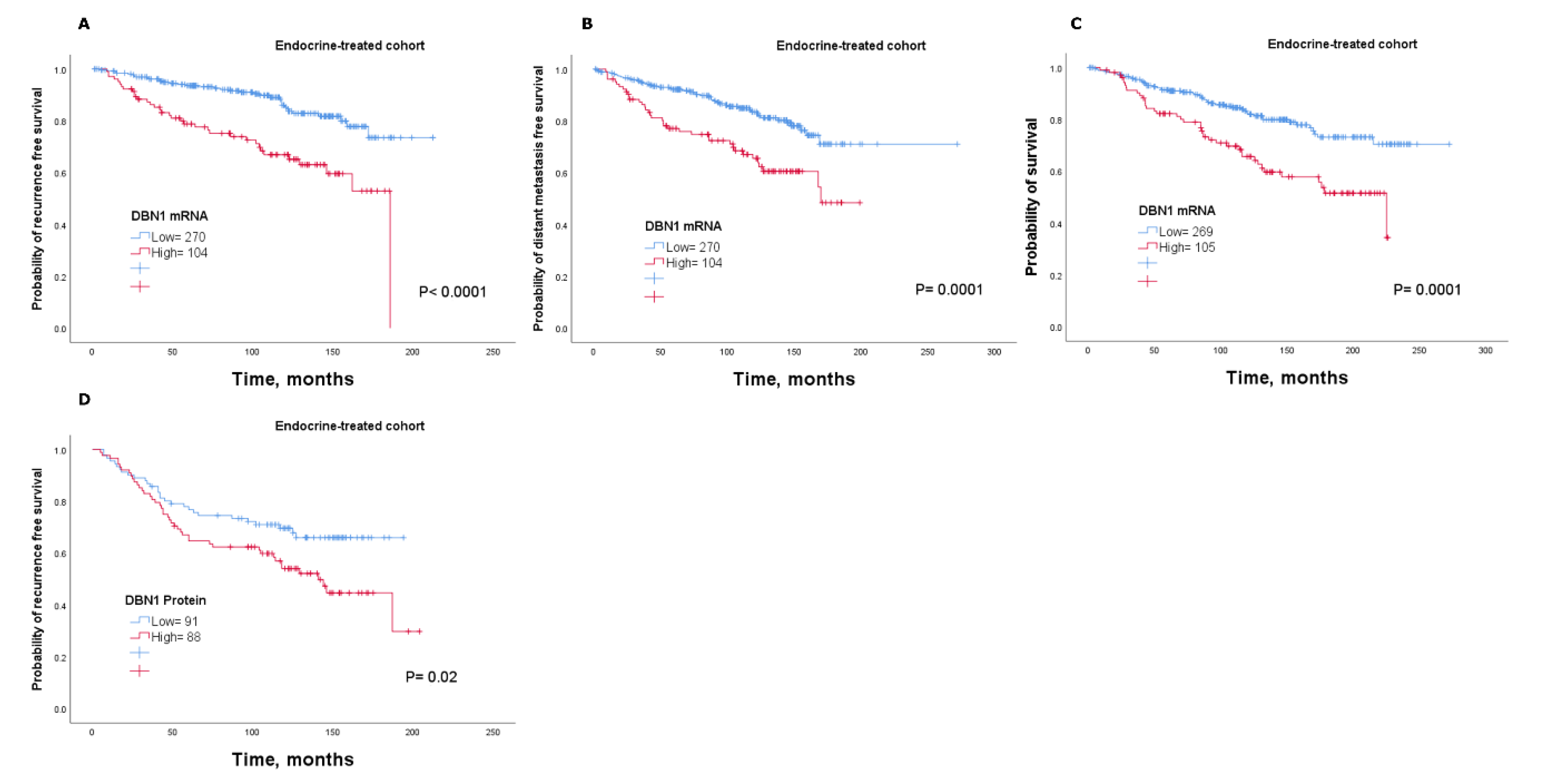
2.4. DBN1 Expression Predicts Poor Response in Endocrine-Treated Patients
3. Discussion
4. Materials and Methods
4.1. Ethical Approval
4.2. DEGs Analysis
4.3. mRNA Expression Cohorts
4.4. Protein Expression Analysis
4.5. Clinical Outcomes Data
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Early Breast Cancer Trialists’ Collaborative Group (EBCTCG); Davies, K.E.; Godwin, J.; Gray, R.; Clarke, M.; Cutter, D.; Darby, S.; McGale, P.; Pan, H.C.; Taylor, C.; et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: Patient-level meta-analysis of randomised trials. Lancet 2011, 378, 771–784. [Google Scholar] [CrossRef]
- Szostakowska-Rodzos, M.; Trebinska-Stryjewska, A.; Grzybowska, E.A.; Fabisiewicz, A. Resistance to endocrine therapy in breast cancer: Molecular mechanisms and future goals. Breast Cancer Res. Treat. 2018, 173, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Park, C.C.; Zhang, H.; Pallavicini, M.; Gray, J.W.; Baehner, F.; Park, C.J.; Bissell, M.J. Beta1 integrin inhibitory antibody induces apoptosis of breast cancer cells, inhibits growth, and distinguishes malignant from normal phenotype in three dimensional cultures and in vivo. Cancer Res. 2006, 66, 1526–1535. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Liu, M.; Yang, L.; Tu, G.; Zhu, Q.; Chen, M.; Cheng, H.; Luo, H.; Fu, W.; Li, Z.; et al. Acquisition of epithelial-mesenchymal transition phenotype in the tamoxifen-resistant breast cancer cell: A new role for G protein-coupled estrogen receptor in mediating tamoxifen resistance through cancer-associated fibroblast-derived fibronectin and β1-integrin signaling pathway in tumor cells. Breast Cancer Res. 2015, 17, 69. [Google Scholar] [CrossRef] [PubMed]
- Pontiggia, O.; Sampayo, R.; Raffo, D.; Motter, A.; Xu, R.; Bissell, M.J.; Joffe, E.B.D.K.; Simian, M. The tumor microenvironment modulates tamoxifen resistance in breast cancer: A role for soluble stromal factors and fibronectin through β1 integrin. Breast Cancer Res. Treat. 2011, 133, 459–471. [Google Scholar] [CrossRef]
- Shajahan-Haq, A.; Cook, K.; Schwartz-Roberts, J.L.; Eltayeb, A.E.; Demas, D.M.; Warri, A.M.; Facey, C.O.B.; Halakivi-Clarke, L.; Clarke, R.B. MYC regulates the unfolded protein response and glucose and glutamine uptake in endocrine resistant breast cancer. Mol. Cancer 2014, 13, 239. [Google Scholar] [CrossRef]
- Stendahl, M.; Kronblad, A.; Rydén, L.; Emdin, S.; Bengtsson, N.O.; Landberg, G. Cyclin D1 overexpression is a negative predictive factor for tamoxifen response in postmenopausal breast cancer patients. Br. J. Cancer 2004, 90, 1942–1948. [Google Scholar] [CrossRef]
- Osborne, C.K.; Schiff, R. Mechanisms of endocrine resistance in breast cancer. Annu. Rev. Med. 2011, 62, 233–247. [Google Scholar] [CrossRef]
- Alfarsi, L.H.; El-Ansari, R.; Craze, M.L.; Masisi, B.K.; Mohammed, O.J.; Ellis, I.O. Co-Expression Effect of SLC7A5/SLC3A2 to Predict Response to Endocrine Therapy in Oestrogen-Receptor-Positive Breast Cancer. Int. J. Mol. Sci. 2020, 21, 1407. [Google Scholar] [CrossRef]
- El Ansari, R.; Craze, M.; Miligy, I.; Diez-Rodriguez, M.; Nolan, C.C.; Ellis, I.; Rakha, E.A.; Green, A. The amino acid transporter SLC7A5 confers a poor prognosis in the highly proliferative breast cancer subtypes and is a key therapeutic target in luminal B tumours. Breast Cancer Res. 2018, 20, 21. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Li, L.; Coyaud, É; Luna, A.; Sander, C.; Raught, B.; Asara, J.M.; Brown, M.; Muthuswamy, S. LLGL2 rescues nutrient stress by promoting leucine uptake in ER+ breast cancer. Nature 2019, 569, 275–279. [Google Scholar] [CrossRef]
- Dun, X.-P.; Chilton, J.K. Control of cell shape and plasticity during development and disease by the actin-binding protein Drebrin. Histol. Histopathol. 2010, 25, 533–540. [Google Scholar]
- Peitsch, W. Drebrin particles: Components in the ensemble of proteins regulating actin dynamics of lamellipodia and filopodia. Eur. J. Cell Biol. 2001, 80, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Mitra, R.; Lee, J.; Jo, J.; Milani, M.; McClintick, J.N.; Edenberg, H.J.; Kesler, K.A.; Rieger, K.M.; Badve, S.S.; Cummings, O.W.; et al. Prediction of Postoperative Recurrence-Free Survival in Non-small Cell Lung Cancer by Using an Internationally Validated Gene Expression Model. Clin. Cancer Res. 2011, 17, 2934–2946. [Google Scholar] [CrossRef] [PubMed]
- Terakawa, Y.; Agnihotri, S.; Golbourn, B.; Nadi, M.; Sabha, N.; Smith, C.A.; Croul, S.E.; Rutka, J.T. The role of drebrin in glioma migration and invasion. Exp. Cell Res. 2013, 319, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Tan, H.T.; Lim, T.K.; Khoo, A.; Lim, K.H.; Chung, M.C.M. iTRAQ analysis of colorectal cancer cell lines suggests Drebrin (DBN1) is overexpressed during liver metastasis. Proteomics 2014, 14, 1434–1443. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.-Q.; Buraschi, S.; Morcavallo, A.; Genua, M.; Shirao, T.; Peiper, S.C.; Gomella, L.G.; Birbe, R.; Belfiore, A.; Iozzo, R.V.; et al. A novel role for drebrin in regulating progranulin bioactivity in bladder cancer. Oncotarget 2015, 6, 10825–10839. [Google Scholar] [CrossRef]
- Peitsch, W.K.; Hofmann, I.; Bulkescher, J.; Hergt, M.; Spring, H.; Bleyl, U.; Goerdt, S.; Franke, W. Drebrin, an Actin-Binding, Cell-Type Characteristic Protein: Induction and Localization in Epithelial Skin Tumors and Cultured Keratinocytes. J. Investig. Dermatol. 2005, 125, 761–774. [Google Scholar] [CrossRef]
- Dart, A.E.; Worth, D.C.; Muir, G.; Chandra, A.; Morris, J.D.; McKee, C.; Verrill, C.; Bryant, R.J.; Gordon-Weeks, P. The drebrin/EB3 pathway drives invasive activity in prostate cancer. Oncogene 2017, 36, 4111–4123. [Google Scholar] [CrossRef]
- Pérez-Martínez, M.; Gordon, M.; Cabrero, J.R.; Barrero-Villar, M.; Rey, M.; Mittelbrunn, M.; Lamana, A.; Morlino, G.; Calabia, C.; Yamazaki, H.; et al. F-actin-binding protein drebrin regulates CXCR4 recruitment to the immune synapse. J. Cell Sci. 2010, 123, 1160–1170. [Google Scholar] [CrossRef]
- Sun, Y.; Mao, X.; Fan, C.; Liu, C.; Guo, A.; Guan, S.; Jin, Q.; Li, B.; Yao, F.; Jin, F. CXCL12-CXCR4 axis promotes the natural selection of breast cancer cell metastasis. Tumor Biol. 2014, 35, 7765–7773. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Yoon, Y.; Votaw, J.; Goodman, M.M.; Williams, L.; Shim, H. Silencing of CXCR4 blocks breast cancer metastasis. Cancer Res. 2005, 65, 967–971. [Google Scholar] [PubMed]
- Hernandez, L.; Magalhaes, M.; Coniglio, S.J.; Condeelis, J.S.; Segall, J.E. Opposing roles of CXCR4 and CXCR7 in breast cancer metastasis. Breast Cancer Res. 2011, 13, R128. [Google Scholar] [CrossRef] [PubMed]
- Iyama, S.; Ono, M.; Kawai-Nakahara, H.; Husni, R.E.; Dai, T.; Shiozawa, T.; Sakata, A.; Kohrogi, H.; Noguchi, M. Drebrin: A new oncofetal biomarker associated with prognosis of lung adenocarcinoma. Lung Cancer 2016, 102, 74–81. [Google Scholar] [CrossRef]
- Sparano, J.; Gray, R.J.; Ravdin, P.M.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E.; Dees, E.C.; Goetz, M.P.; et al. Clinical and Genomic Risk to Guide the Use of Adjuvant Therapy for Breast Cancer. N. Engl. J. Med. 2019, 380, 2395–2405. [Google Scholar] [CrossRef]
- Alfarsi, L.; Johnston, S.; Liu, N.-X.; Rakha, E.A.; Green, A. Current issues with luminal subtype classification in terms of prediction of benefit from endocrine therapy in early breast cancer. Histopathology 2018, 73, 545–558. [Google Scholar] [CrossRef]
- Sauerbrei, W.; Taube, S.E.; McShane, L.M.; Cavenagh, M.M.; Altman, U.G. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): An Abridged Explanation and Elaboration. J. Natl. Cancer Inst. 2018, 110, 803–811. [Google Scholar] [CrossRef]
- Kadota, K.; Nakai, Y.; Shimizu, K. A weighted average difference method for detecting differentially expressed genes from microarray data. Algorithms Mol. Biol. 2008, 3, 8. [Google Scholar] [CrossRef]
- Curtis, C.; Shah, S.P.; Chin, S.-F.; Turashvili, G.; Rueda, O.M.; Dunning, M.; Speed, D.; Lynch, A.G.; Samarajiwa, S.A.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [PubMed]
- Győrffy, B.; Lanczky, A.; Eklund, A.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res. Treat. 2009, 123, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Jézéquel, P.; Campone, M.; Gouraud, W.; Guérin-Charbonnel, C.; Leux, C.; Ricolleau, G.; Campion, L. bc-GenExMiner: An easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res. Treat. 2011, 131, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Alfarsi, L.H.; El Ansari, R.; Craze, M.L.; Toss, M.S.; Masisi, B.; Ellis, I.O.; Rakha, E.A.; Green, A. CDC20 expression in oestrogen receptor positive breast cancer predicts poor prognosis and lack of response to endocrine therapy. Breast Cancer Res. Treat. 2019, 178, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Mccarty, K.S. Histochemical approaches to steroid receptor analyses. Semin. Diagn. Pathol. 1984, 1, 297–308. [Google Scholar] [PubMed]
| METABRIC Cohort | ||||
| Parameters | Recurrence-Free Survival | |||
| HR (95% CI) | p | p * | ||
| DBN1 Tumour size Tumour grade Nodal stage | 2.2 (1.4–3.5) 1.3 (0.8–2.1) 1.2 (0.9–1.8) 1.1 (0.7–1.5) | 0.0003 0.17 0.14 0.5 | 0.001 0.2 0.3 0.6 | |
| Parameters | Distant Metastasis-Free Survival | |||
| HR (95% CI) | p | p * | ||
| DBN1 Tumour size Tumour grade Nodal stage | 2.9 (1.8–4.8) 2.0 (1.1–3.3) 1.3 (0.9–2.0) 1.3 (0.9–2.0) | 0.00001 0.008 0.1 0.09 | 0.0001 0.02 0.12 0.1 | |
| Parameters | Breast Cancer Specific Survival | |||
| HR (95% CI) | p | p * | ||
| DBN1 Tumour size Tumour grade Nodal stage | 3.3 (1.9–5.8) 2.0 (1.1–3.5) 1.8 (1.1–2.9) 1.4 (0.9–2.1) | 0.00001 0.01 0.008 0.09 | 0.0001 0.016 0.02 0.1 | |
| Nottingham Cohort | ||||
| Parameters | Recurrence-Free Survival | |||
| HR (95% CI) | p | p * | ||
| DBN1 Tumour size Tumour grade Nodal stage | 1.5 (1.1–2.0) 1.4 (1.0–2.0) 1.1 (0.9–1.5) 1.6 (1.2–2.0) | 0.009 0.02 0.1 0.00005 | 0.02 0.03 0.12 0.0003 | |
| Parameters | Distant Metastasis-Free Survival | |||
| HR (95% CI) | p | p * | ||
| DBN1 Tumour size Tumour grade Nodal stage | 1.4 (1.0–1.9) 1.8 (1.3–2.7) 1.3 (1.0–1.7) 1.6 (1.3–2.1) | 0.04 0.0004 0.01 0.00003 | 0.05 0.001 0.016 0.0002 | |
| METABRIC Cohort | ||||
| Parameters | Recurrence-Free Survival | |||
| HR (95% CI) | p | p * | ||
| DBN1 Tumour size Tumour grade Nodal stage | 2.5 (1.4–4.3) 1.1 (0.6–1.9) 1.1 (0.7–1.7) 1.0 (0.6–1.6) | 0.001 0.6 0.5 0.8 | 0.005 1.0 1.2 1.0 | |
| Parameters | Distant Metastasis-Free Survival | |||
| HR (95% CI) | p | p * | ||
| DBN1 Tumour size Tumour grade Nodal stage | 2.5 (1.4–4.6) 1.4 (0.8–2.7) 1.3 (0.7–2.8) 1.4 (0.8–2.2) | 0.001 0.1 0.2 0.1 | 0.005 0.25 0.25 0.16 | |
| Parameters | Breast Cancer Specific Survival | |||
| HR (95% CI) | p | p * | ||
| DBN1 Tumour size Tumour grade Nodal stage | 3.2 (1.6–6.1) 1.8 (0.9–3.7) 1.6 (0.9–3.0) 1.2 (0.7–2.1) | 0.001 0.08 0.09 0.3 | 0.005 0.2 0.15 0.37 | |
| Nottingham Cohort | ||||
| Parameters | Recurrence-Free Survival | |||
| HR (95% CI) | p | p * | ||
| DBN1 Tumour size Tumour grade Nodal stage | 1.9 (1.1–3.1) 1.6 (1.0–2.6) 1.3 (0.9–2.0) 1.7 (1.2–2.4) | 0.007 0.05 0.1 0.002 | 0.01 0.08 0.12 0.01 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alfarsi, L.H.; El Ansari, R.; Masisi, B.K.; Parks, R.; Mohammed, O.J.; Ellis, I.O.; Rakha, E.A.; Green, A.R. Integrated Analysis of Key Differentially Expressed Genes Identifies DBN1 as a Predictive Marker of Response to Endocrine Therapy in Luminal Breast Cancer. Cancers 2020, 12, 1549. https://doi.org/10.3390/cancers12061549
Alfarsi LH, El Ansari R, Masisi BK, Parks R, Mohammed OJ, Ellis IO, Rakha EA, Green AR. Integrated Analysis of Key Differentially Expressed Genes Identifies DBN1 as a Predictive Marker of Response to Endocrine Therapy in Luminal Breast Cancer. Cancers. 2020; 12(6):1549. https://doi.org/10.3390/cancers12061549
Chicago/Turabian StyleAlfarsi, Lutfi H., Rokaya El Ansari, Brendah K. Masisi, Ruth Parks, Omar J Mohammed, Ian O. Ellis, Emad A. Rakha, and Andrew R. Green. 2020. "Integrated Analysis of Key Differentially Expressed Genes Identifies DBN1 as a Predictive Marker of Response to Endocrine Therapy in Luminal Breast Cancer" Cancers 12, no. 6: 1549. https://doi.org/10.3390/cancers12061549
APA StyleAlfarsi, L. H., El Ansari, R., Masisi, B. K., Parks, R., Mohammed, O. J., Ellis, I. O., Rakha, E. A., & Green, A. R. (2020). Integrated Analysis of Key Differentially Expressed Genes Identifies DBN1 as a Predictive Marker of Response to Endocrine Therapy in Luminal Breast Cancer. Cancers, 12(6), 1549. https://doi.org/10.3390/cancers12061549







