ESM-1 Overexpression is Involved in Increased Tumorigenesis of Radiotherapy-Resistant Breast Cancer Cells
Abstract
1. Introduction
2. Results
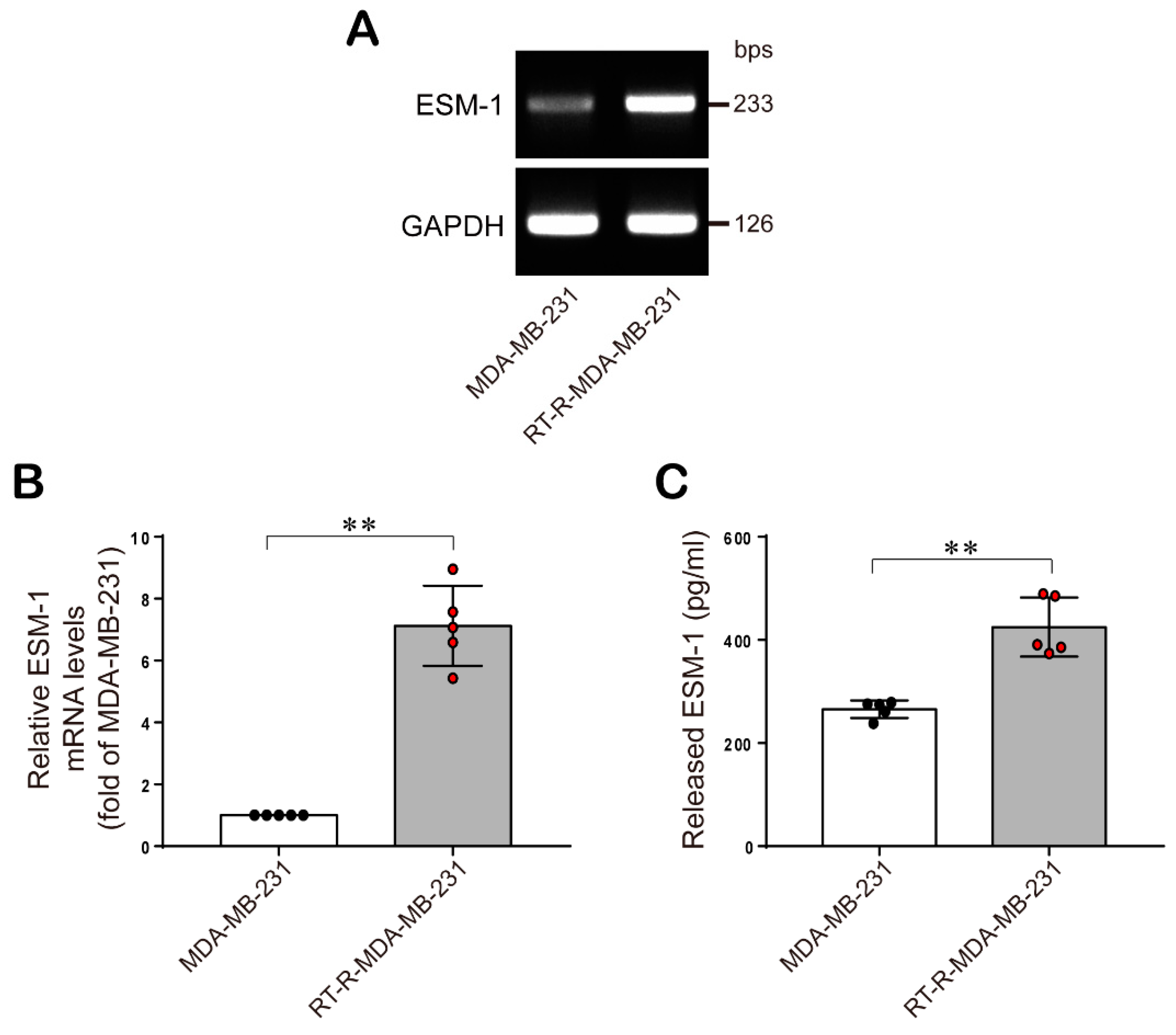
2.1. ESM-1 is Significantly Overexpressed in RT-R-MDA-MB-231 Cells Compared with MDA-MB-231 Cells
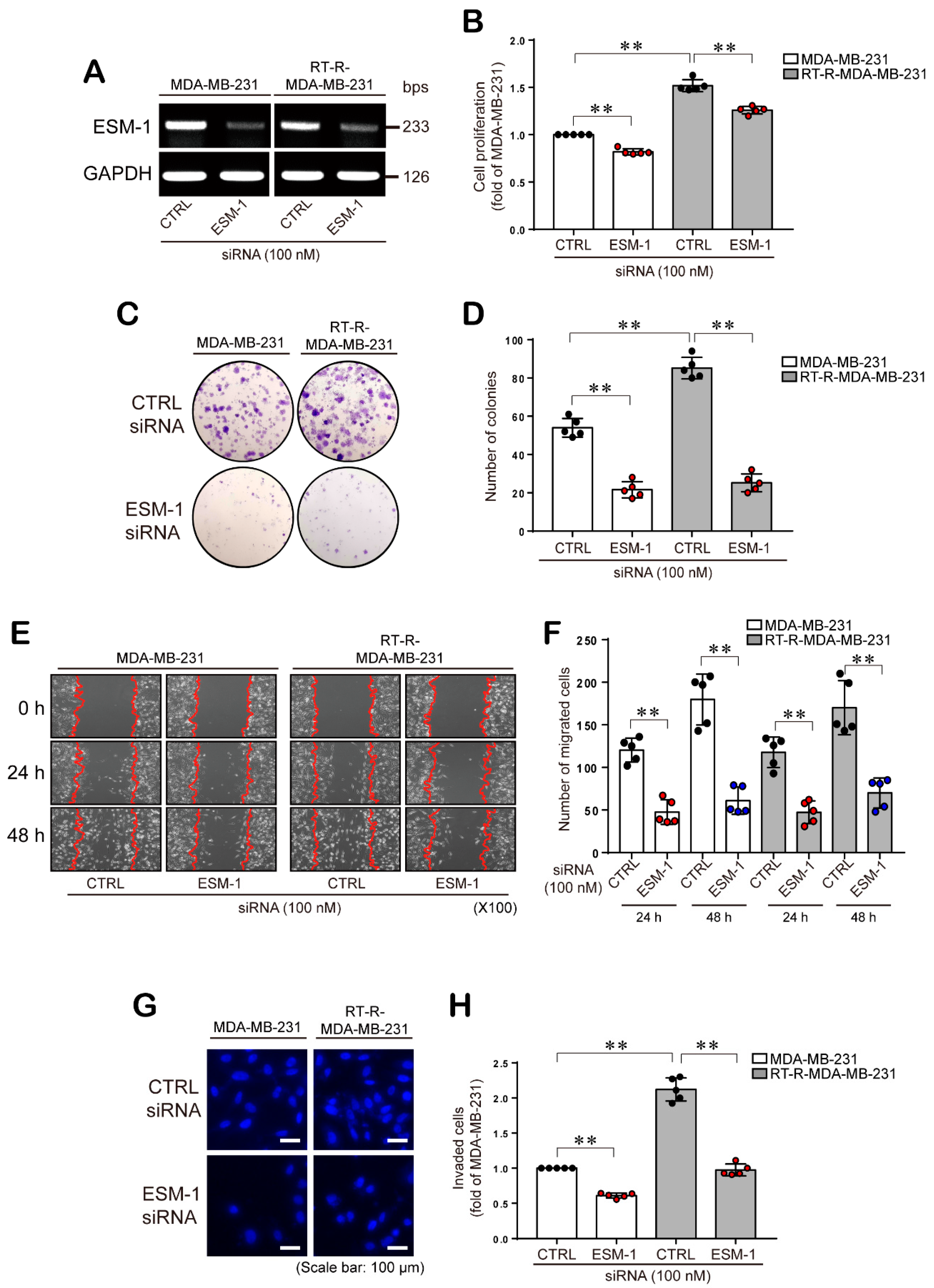
2.2. ESM-1 Knockdown Inhibits the Proliferation, Colony Formation Ability, Migration, and Invasion of both MDA-MB-231 and RT-R-MDA-MB-231 Cells
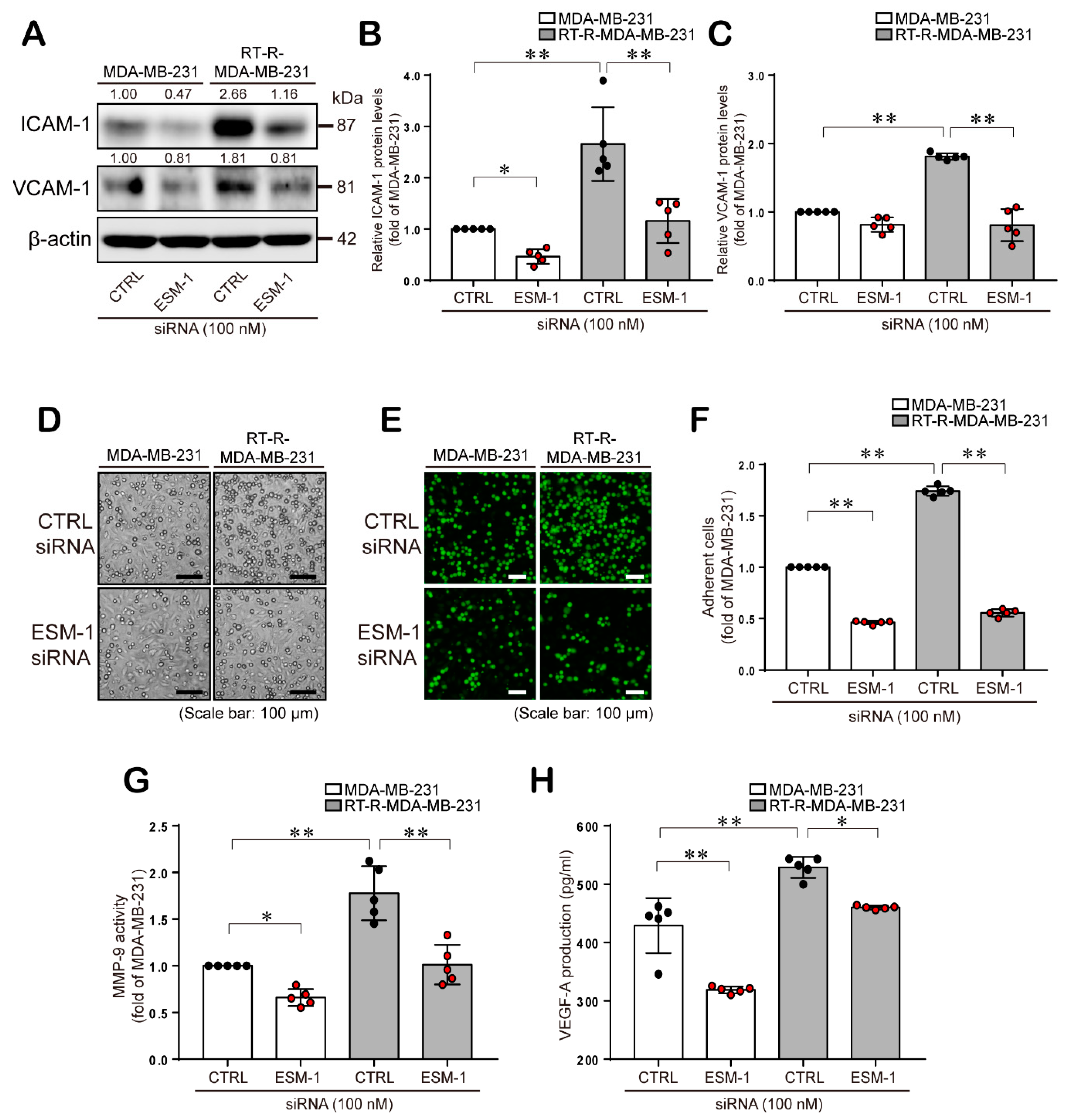
2.3. ESM-1 Plays a Role in Tumorigenesis in MDA-MB-231 and RT-R-MDA-MB-231 Cells through Induction of Adhesion Molecules, Resulting in Adhesion of these Cells to ECs, MMP-9 Activity, and VEGF-A Production
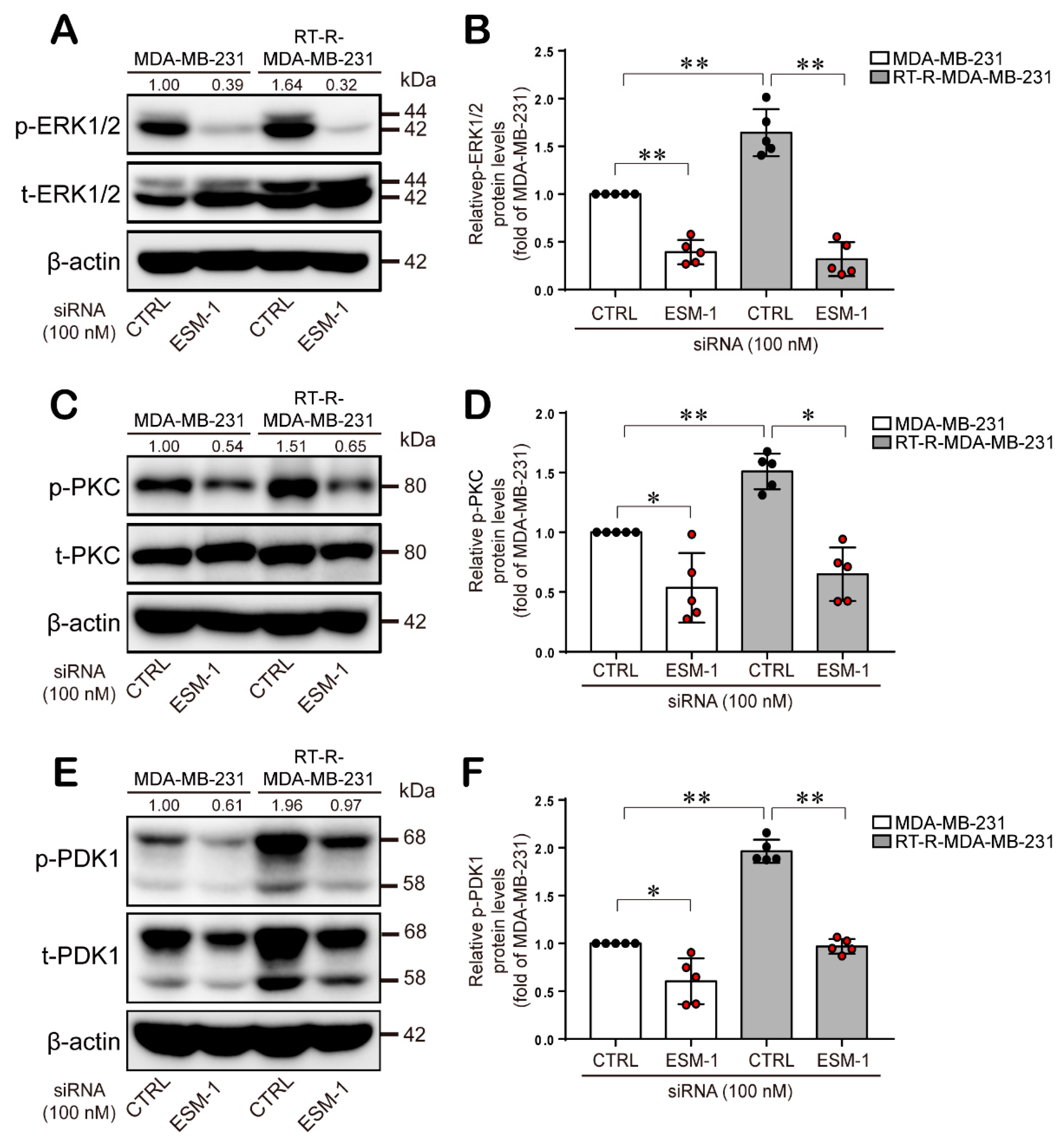
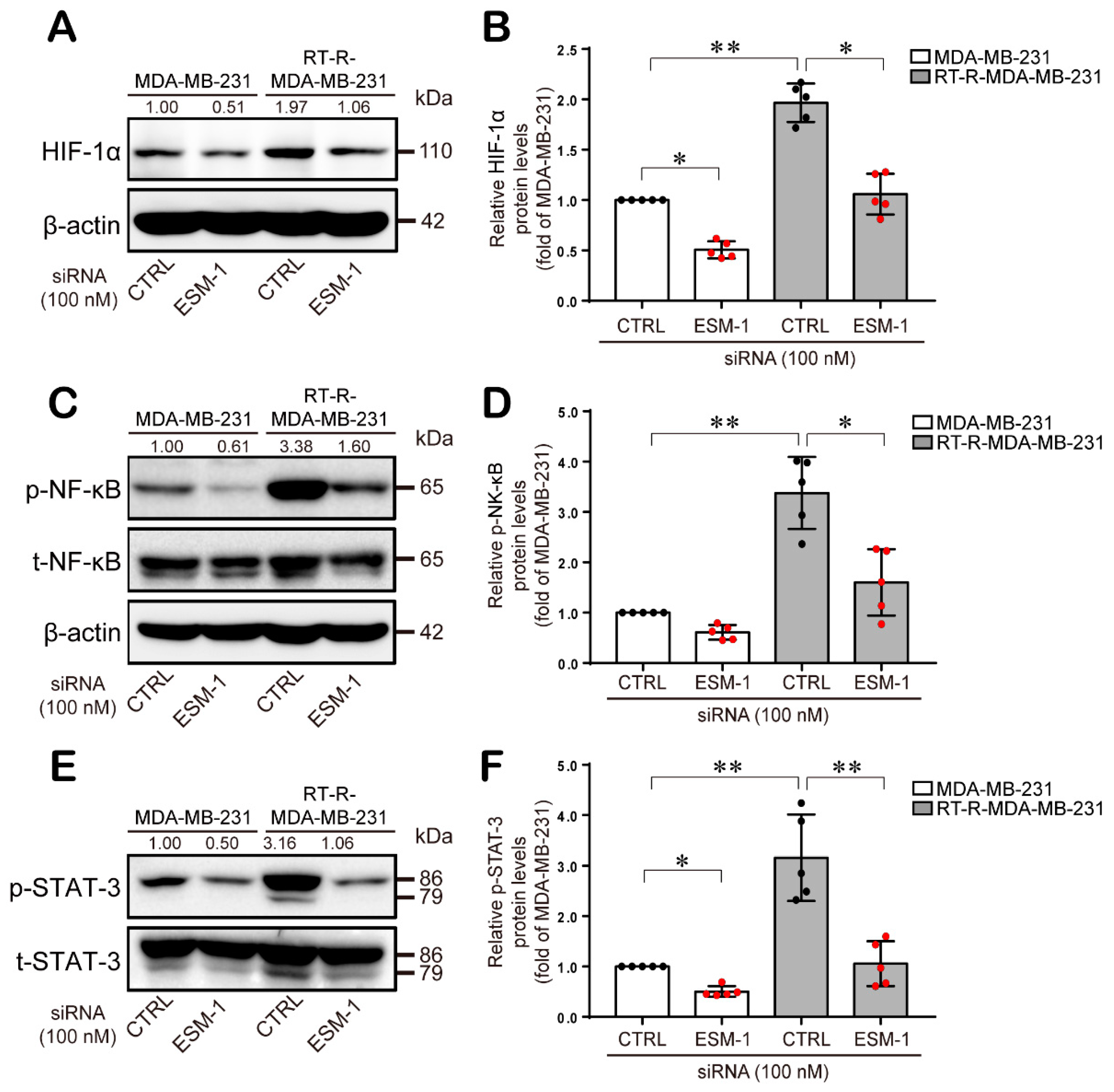
2.4. ESM-1-Overexpressing RT-R-MDA-MB-231 Cells Increase ERK1/2, PKC, and PDK1 Activation and then Transcription Factor Hypoxia-Inducible Factor-1α (HIF-1α) Induction and NF-κB and STAT-3 Activation
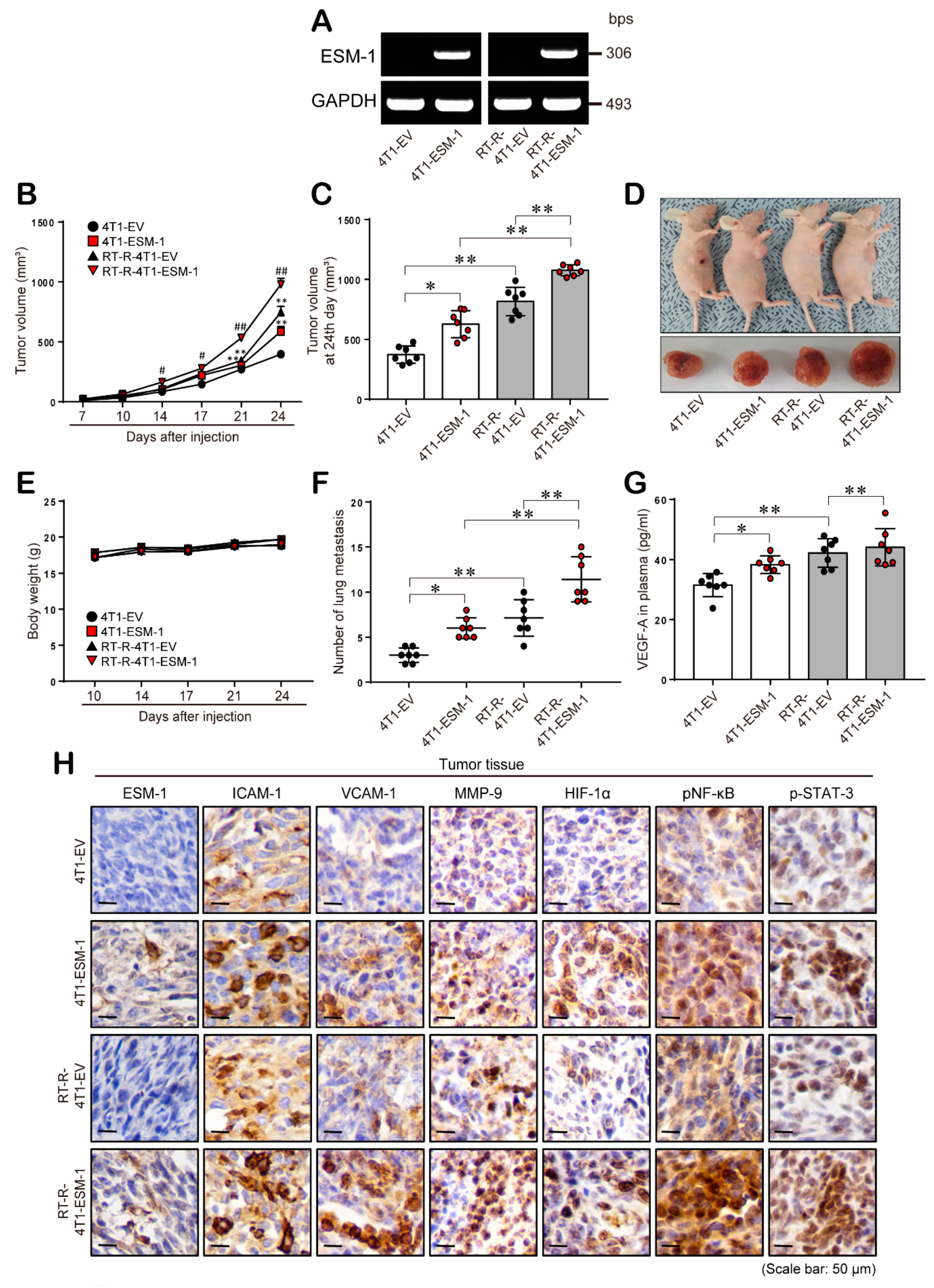
2.5. ESM-1 Overexpression Increases Tumor Growth and Metastasis in an In Vivo Mouse Model
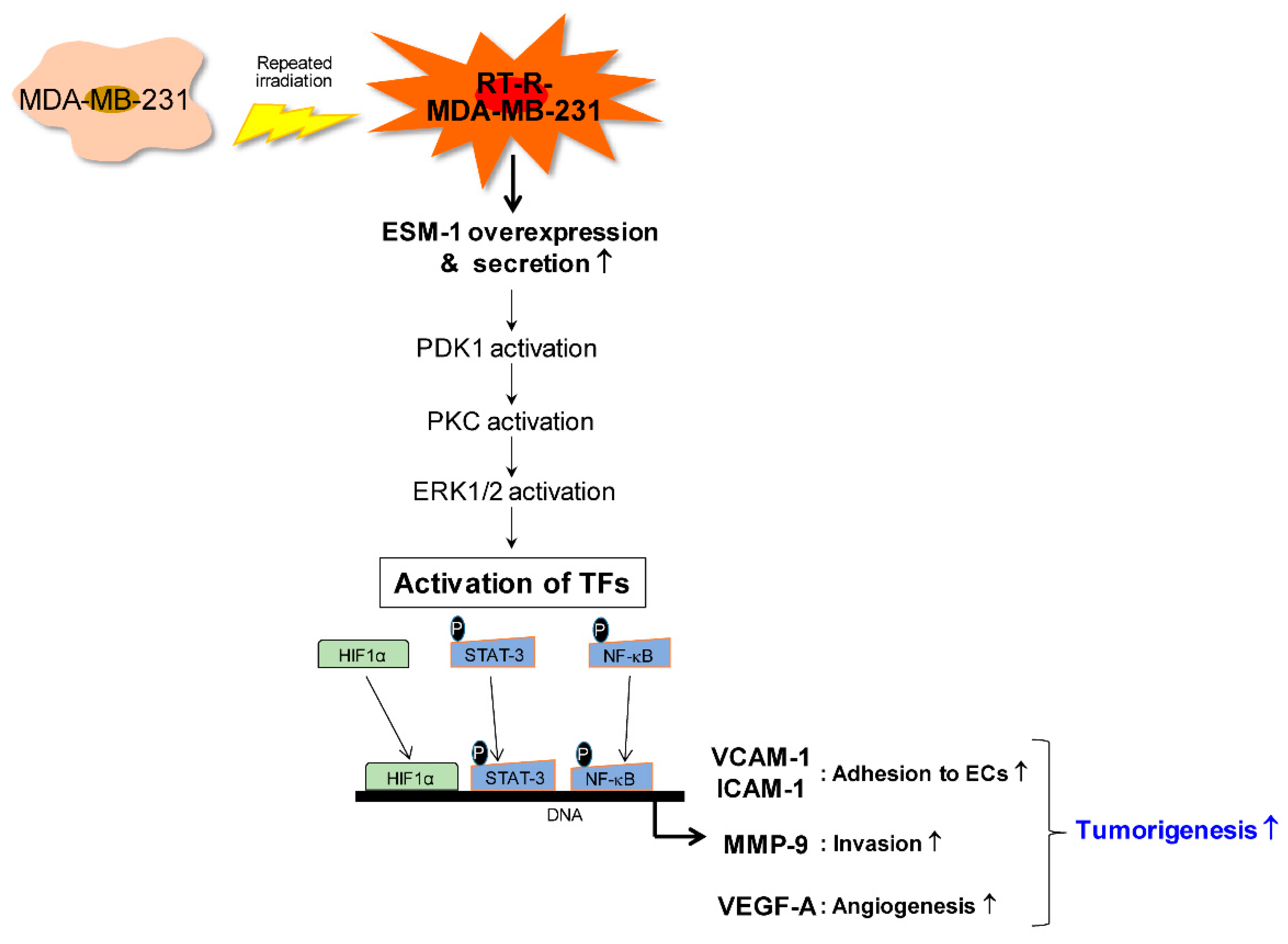
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Establishment of RT-R-Breast Cancer Cells
4.3. Gene Expression Array Analysis
4.4. Gene Silencing with siRNA
4.5. RT-PCR
4.6. Cell Proliferation Assay
4.7. Colony Formation Assay
4.8. Wound Healing Assay
4.9. Adhesion Assay
4.10. Matrigel Invasion Assay
4.11. Gelatin Zymography
4.12. Quantitative VEGF-A Immunoassay
4.13. Quantification of ESM-1 Secretion
4.14. Western Blot Analysis
4.15. ESM-1 Overexpression in 4T1 and RT-R-4T1 cells
4.16. Animal Experiments
4.17. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| BC | breast cancer |
| CCK-8 | cell counting kit-8 |
| CTRL | control |
| DAB | 3,3′-diaminobenzidine tetrahydrochloride |
| DAPI | 4′,6-diamidino-2-phenyindole, dilactate |
| DMEM | Dulbecco’s modification of Eagle medium |
| EC | endothelial cell |
| ECL | enhanced chemiluminescence |
| ER | estrogen receptors |
| ERK | extracellular-signal-regulated kinase |
| ESM-1 | endothelial cell-specific molecule 1 |
| EV | empty vector |
| FBS | fetal bovine serum |
| HER2 | human epidermal growth factor receptor 2 |
| HIF-1α | hypoxia-inducible factor 1 alpha |
| ICAM-1 | intercellular adhesion molecule-1 |
| IFNγ | interferon gamma |
| IHC | immunohistochemistry |
| IL-1β | interleukin 1 beta |
| MDA-MB-231 BR | MDA-MB-231 cells with a brain metastatic phenotype |
| MMP | matrix metalloproteinase |
| NF-κB | nuclear factor kappa B |
| PDK1 | phosphoinoisitide dependent kinase 1 |
| PI3K | phosphoinositide 3-kinase |
| PKC | protein kinase C |
| PR | progesterone receptors |
| P2Y2R | P2Y purinergic receptor 2 |
| RT-PCR | reverse transcription-polymerase chain reaction |
| RT | radiotherapy |
| RT-R | radiotherapy resistant |
| SD | standard deviation |
| SDS-PAGE | sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| siRNA | small interfering RNA |
| STAT-3 | signal transducer and activator of transcription 3 |
| TNBC | triple negative breast cancer |
| TNFα | tumor necrosis factor alpha |
| VCAM-1 | vascular cell adhesion molecule-1 |
| VEGF | vascular endothelial growth factor |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Gnant, M. Breast cancer. Lancet 2017, 389, 1134–1150. [Google Scholar] [CrossRef]
- Nguyen, Y.T.K.; Moon, J.Y.; Ediriweera, M.K.; Cho, S.K. Phenethyl isothiocyanate suppresses stemness in the chemo-and radio-resistant triple-negative breast cancer cell line MDA-MB-231/IR via downregulation of metadherin. Cancers 2020, 12, 268. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.; Turnbull, A.K.; Ward, C.; Meehan, J.; Martínez-Pérez, C.; Bonello, M.; Pang, L.Y.; Langdon, S.P.; Kunkler, I.H.; Murray, A.; et al. Development and characterisation of acquired radioresistant breast cancer cell lines. Radiat. Oncol. 2019, 14, 64. [Google Scholar] [CrossRef]
- Ko, Y.S.; Jin, H.; Lee, J.S.; Park, S.W.; Chang, K.C.; Kang, K.M.; Jeong, B.K.; Kim, H.J. Radioresistant breast cancer cells exhibit increased resistance to chemotherapy and enhanced invasive properties due to cancer stem cells. Oncol. Rep. 2018, 40, 3752–3762. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Ko, Y.W.; Kim, H.J. P2Y2R-mediated inflammasome activation is involved in tumor progression in breast cancer cells and in radiotherapy-resistant breast cancer. Int. J. Oncol. 2018, 53, 1953–1966. [Google Scholar] [CrossRef]
- Maurage, C.A.; Adam, E.; Minéo, J.F.; Sarrazin, S.; Debunne, M.; Siminski, R.M.; Baroncini, M.; Lassalle, P.; Blond, S.; Delehedde, M. Endocan expression and localization in human glioblastomas. J. Neuropathol. Exp. Neurol. 2009, 68, 633–641. [Google Scholar] [CrossRef]
- Béchard, D.; Gentina, T.; Delehedde, M.; Scherpereel, A.; Lyon, M.; Aumercier, M.; Vazeux, R.; Richet, C.; Degand, P.; Jude, B.; et al. Endocan is a novel chondroitin sulfate/dermatan sulfate proteoglycan that promotes hepatocyte growth factor/scatter factor mitogenic activity. J. Biol. Chem. 2001, 276, 48341–48349. [Google Scholar] [CrossRef]
- Bechard, D.; Meignin, V.; Scherpereel, A.; Oudin, S.; Kervoaze, G.; Bertheau, P.; Janin, A.; Tonnel, A.; Lassalle, P. Characterization of the secreted form of endothelial-cell-specific molecule 1 by specific monoclonal antibodies. J. Vasc. Res. 2000, 37, 417–425. [Google Scholar] [CrossRef]
- Lassalle, P.; Molet, S.; Janin, A.; Heyden, J.V.; Tavernier, J.; Fiers, W.; Devos, R.; Tonnel, A.B. ESM-1 is a novel human endothelial cell-specific molecule expressed in lung and regulated by cytokines. J. Biol. Chem. 1996, 271, 20458–20464. [Google Scholar] [CrossRef]
- Rennel, E.; Mellberg, S.; Dimberg, A.; Petersson, L.; Botling, J.; Ameur, A.; Westholm, J.O.; Komorowski, J.; Lassalle, P.; Cross, M.J.; et al. Endocan is a VEGF-A and PI3K regulated gene with increased expression in human renal cancer. Exp. Cell Res. 2007, 313, 1285–1294. [Google Scholar] [CrossRef]
- Shin, J.W.; Huggenberger, R.; Detmar, M. Transcriptional profiling of VEGF-A and VEGF-C target genes in lymphatic endothelium reveals endothelial-specific molecule-1 as a novel mediator of lymphangiogenesis. Blood 2008, 112, 2318–2326. [Google Scholar] [CrossRef]
- Scherpereel, A.; Depontieu, F.; Grigoriu, B.; Cavestri, B.; Tsicopoulos, A.; Gentina, T.; Jourdain, M.; Pugin, J.; Tonnel, A.B.; Lassalle, P. Endocan, a new endothelial marker in human sepsis. Crit. Care Med. 2006, 34, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Grigoriu, B.D.; Depontieu, F.; Scherpereel, A.; Gourcerol, D.; Devos, P.; Ouatas, T.; Lafitte, J.J.; Copin, M.C.; Tonnel, A.B.; Lassalle, P. Endocan expression and relationship with survival in human non-small cell lung cancer. Clin. Cancer Res. 2006, 12, 4575–4582. [Google Scholar] [CrossRef] [PubMed]
- Leroy, X.; Aubert, S.; Zini, L.; Franquet, H.; Kervoaze, G.; Villers, A.; Delehedde, M.; Copin, M.C.; Lassalle, P. Vascular endocan (ESM-1) is markedly overexpressed in clear cell renal cell carcinoma. Histopathology 2010, 56, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Abid, M.R.; Yi, X.; Yano, K.; Shih, S.C.; Aird, W.C. Vascular endocan is preferentially expressed in tumor endothelium. Microvasc. Res. 2006, 72, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.H.; Ji, N.Y.; Han, S.R.; Lee, C.I.; Kim, J.W.; Yeom, Y.I.; Kim, Y.H.; Chun, H.K.; Kim, J.W.; Chung, J.W.; et al. ESM-1 regulates cell growth and metastatic process through activation of NF-κB in colorectal cancer. Cell. Signal. 2012, 24, 1940–1949. [Google Scholar] [CrossRef]
- Hatfield, K.J.; Lassalle, P.; Leiva, R.A.; Lindås, R.; Wendelboe, Ø.; Bruserud, Ø. Serum levels of endothelium-derived endocan are increased in patients with untreated acute myeloid leukemia. Hematology 2011, 16, 351–356. [Google Scholar] [CrossRef]
- Sagara, A.; Igarashi, K.; Otsuka, M.; Kodama, A.; Yamashita, M.; Sugiura, R.; Karasawa, T.; Arakawa, K.; Narita, M.; Kuzumaki, N.; et al. Endocan as a prognostic biomarker of triple-negative breast cancer. Breast Cancer Res. Treat. 2017, 161, 269–278. [Google Scholar] [CrossRef]
- Scherpereel, A.; Gentina, T.; Grigoriu, B.; Sénéchal, S.; Janin, A.; Tsicopoulos, A.; Plénat, F.; Béchard, D.; Tonnel, A.B.; Lassalle, P. Overexpression of endocan induces tumor formation. Cancer Res. 2003, 63, 6084–6089. [Google Scholar]
- Intlekofer, A.M.; Finley, L.W.S. Metabolic signatures of cancer cells and stem cells. Nat. Metab. 2019, 1, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Benedicto, A.; Romayor, I.; Arteta, B. Role of liver ICAM-1 in metastasis. Oncol. Lett. 2017, 14, 3883–3892. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.H.; Kim, Y.K.; Kim, M.R.; Jang, J.H.; Lee, S. Emerging Roles of Vascular Cell Adhesion Molecule-1 (VCAM-1) in Immunological Disorders and Cancer. Int. J. Mol. Sci. 2018, 19, 1057. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Shen, S.; Verma, I.M. NF-κB, an active player in human cancers. Cancer Immunol. Res. 2014, 2, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Multhoff, G.; Molls, M.; Radons, J. Chronic inflammation in cancer development. Front. Immunol. 2012, 2, 98. [Google Scholar] [CrossRef]
- Karagiota, A.; Kourti, M.; Simos, G.; Mylonis, I. HIF-1α-derived cell-penetrating peptides inhibit ERK-dependent activation of HIF-1 and trigger apoptosis of cancer cells under hypoxia. Cell. Mol. Life Sci. 2019, 76, 809–825. [Google Scholar] [CrossRef]
- Wang, X.; Chen, Q.; Xing, D. Focal adhesion kinase activates NF-κB via the ERK1/2 and p38MAPK pathways in amyloid-β25-35-Induced apoptosis in PC12 cells. J. Alzheimer’s Dis. 2012, 32, 77–94. [Google Scholar] [CrossRef]
- Chen, B.; Liu, J.; Ho, T.T.; Ding, X.; Mo, Y.Y. ERK-mediated NF-κB activation through ASIC1 in response to acidosis. Oncogenesis 2016, 5, e279. [Google Scholar] [CrossRef]
- Taniguchi, K.; Karin, M. NF-κB, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol. 2018, 18, 309–324. [Google Scholar] [CrossRef]
- Robinson, R.L.; Sharma, A.; Bai, S.; Heneidi, S.; Lee, T.J.; Kodeboyina, S.K.; Patel, N.; Sharma, S. Comparative STAT3-regulated gene expression profile in renal cell carcinoma subtypes. Front. Oncol. 2019, 9, 72. [Google Scholar] [CrossRef]
- Ding, D.; Han, S.; Zhang, H.; He, Y.; Li, Y. Predictive biomarkers of colorectal cancer. Comput. Biol. Chem. 2019, 83, 107106. [Google Scholar] [CrossRef] [PubMed]
- Ji, N.Y.; Kim, Y.H.; Jang, Y.J.; Kang, Y.H.; Lee, C.I.; Kim, J.W.; Yeom, Y.I.; Chun, H.K.; Choi, Y.H.; Kim, J.H.; et al. Identification of endothelial cell-specific molecule-1 as a potential serum marker for colorectal cancer. Cancer Sci. 2010, 101, 2248–2253. [Google Scholar] [CrossRef]
- Jin, H.; Eun, S.Y.; Lee, J.S.; Park, S.W.; Lee, J.H.; Chang, K.C.; Kim, H.J. P2Y2 receptor activation by nucleotides released from highly metastatic breast cancer cells increases tumor growth and invasion via crosstalk with endothelial cells. Breast Cancer Res. 2014, 16, R77. [Google Scholar] [CrossRef] [PubMed]
- Arsenic, R. Immunohistochemical analysis of PDK1 expression in breast cancer. Diagn. Pathol. 2014, 9, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, C.; Falasca, M. Targeting PDK1 in Cancer. Curr. Med. Chem. 2011, 18, 2763–2769. [Google Scholar] [CrossRef]
- Du, J.; Yang, M.; Chen, S.; Li, D.; Chang, Z.; Dong, Z. PDK1 promotes tumor growth and metastasis in a spontaneous breast cancer model. Oncogene 2016, 35, 3314–3323. [Google Scholar] [CrossRef]
- Islam, S.M.A.; Patel, R.; Acevedo-Duncan, M. Protein Kinase C-ζ stimulates colorectal cancer cell carcinogenesis via PKC-ζ/Rac1/Pak1/β-Catenin signaling cascade. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 650–664. [Google Scholar] [CrossRef]
- Cooke, M.; Magimaidas, A.; Casado-Medrano, V.; Kazanietz, M.G. Protein kinase C in cancer: The top five unanswered questions. Mol. Carcinog. 2017, 56, 1531–1542. [Google Scholar] [CrossRef]
- Hwang, Y.P.; Jeong, H.G. Metformin blocks migration and invasion of tumour cells by inhibition of matrix metalloproteinase-9 activation through a calcium and protein kinase Cα-dependent pathway: Phorbol-12-myristate-13-acetate-induced/ extracellular signal-regulated kinase/activa. Br. J. Pharmacol. 2010, 160, 1195–1211. [Google Scholar] [CrossRef]
- Bartholomeusz, C.; Gonzalez-Angulo, A.M.; Liu, P.; Hayashi, N.; Lluch, A.; Ferrer-Lozano, J.; Hortobágyi, G.N. High ERK protein expression levels correlate with shorter survival in triple-negative breast cancer patients. Oncologist 2012, 17, 766–774. [Google Scholar] [CrossRef]
| Gene Symbol | Fold Change (RT-R-MDA-MB-231 vs. MDA-MB-231) |
|---|---|
| ESM1 | 318.75 |
| MMP1 | 203.77 |
| SEL1L3 | 131.56 |
| MAGEB2 | 118.04 |
| ALPK2 | 116.37 |
| LOC100134317 | 100.11 |
| PHGDH | 98.82 |
| LOC284412 | 97.10 |
| PLCB4 | 89.00 |
| PLCH2 | 82.37 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, H.; Rugira, T.; Ko, Y.S.; Park, S.W.; Yun, S.P.; Kim, H.J. ESM-1 Overexpression is Involved in Increased Tumorigenesis of Radiotherapy-Resistant Breast Cancer Cells. Cancers 2020, 12, 1363. https://doi.org/10.3390/cancers12061363
Jin H, Rugira T, Ko YS, Park SW, Yun SP, Kim HJ. ESM-1 Overexpression is Involved in Increased Tumorigenesis of Radiotherapy-Resistant Breast Cancer Cells. Cancers. 2020; 12(6):1363. https://doi.org/10.3390/cancers12061363
Chicago/Turabian StyleJin, Hana, Trojan Rugira, Young Shin Ko, Sang Won Park, Seung Pil Yun, and Hye Jung Kim. 2020. "ESM-1 Overexpression is Involved in Increased Tumorigenesis of Radiotherapy-Resistant Breast Cancer Cells" Cancers 12, no. 6: 1363. https://doi.org/10.3390/cancers12061363
APA StyleJin, H., Rugira, T., Ko, Y. S., Park, S. W., Yun, S. P., & Kim, H. J. (2020). ESM-1 Overexpression is Involved in Increased Tumorigenesis of Radiotherapy-Resistant Breast Cancer Cells. Cancers, 12(6), 1363. https://doi.org/10.3390/cancers12061363






