Stress Management: Death Receptor Signalling and Cross-Talks with the Unfolded Protein Response in Cancer
Abstract
1. Introduction
2. Cellular Roles and Molecular Determinants of DR Signalling and Current Directions for Its Targeting in Oncology
2.1. TNFR1 Signalling, Cellular Roles and Main Directions for Its Targeting in Oncology
2.2. CD95 Signalling, Cellular Roles and Main Directions for Its Targeting in Oncology
2.3. TRAIL-R1/2 Signalling, Cellular Roles and the Main Directions for Its Targeting in Oncology
3. Cross-Talks between the UPR and DR Signalling in Cancer and Potential Vulnerability Points
3.1. Main Actors of the Unfolded Protein Response
3.2. Cross-Talk between the UPR and DR Signalling: Molecular Mechanisms and Impacts on Cellular Outcomes
4. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Itoh, N.; Nagata, S. A novel protein domain required for apoptosis. Mutational analysis of human Fas antigen. J. Biol. Chem. 1993, 268, 10932–10937. [Google Scholar] [PubMed]
- Tartaglia, L.A.; Ayres, T.M.; Wong, G.H.; Goeddel, D.V. A novel domain within the 55 kd TNF receptor signals cell death. Cell 1993, 74, 845–853. [Google Scholar] [CrossRef]
- Lowin, B.; Beermann, F.; Schmidt, A.; Tschopp, J. A null mutation in the perforin gene impairs cytolytic T lymphocyte- and natural killer cell-mediated cytotoxicity. Proc. Natl. Acad. Sci. USA 1994, 91, 11571–11575. [Google Scholar] [CrossRef] [PubMed]
- Shanker, A.; Brooks, A.D.; Jacobsen, K.M.; Wine, J.W.; Wiltrout, R.H.; Yagita, H.; Sayers, T.J. Antigen presented by tumors in vivo determines the nature of CD8+ T-cell cytotoxicity. Cancer Res. 2009, 69, 6615–6623. [Google Scholar] [CrossRef] [PubMed]
- Prager, I.; Liesche, C.; van Ooijen, H.; Urlaub, D.; Verron, Q.; Sandstrom, N.; Fasbender, F.; Claus, M.; Eils, R.; Beaudouin, J.; et al. NK cells switch from granzyme B to death receptor-mediated cytotoxicity during serial killing. J. Exp. Med. 2019, 216, 2113–2127. [Google Scholar] [CrossRef] [PubMed]
- Ratner, A.; Clark, W.R. Role of TNF-alpha in CD8+ cytotoxic T lymphocyte-mediated lysis. J. Immunol. 1993, 150, 4303–4314. [Google Scholar] [PubMed]
- Kagi, D.; Vignaux, F.; Ledermann, B.; Burki, K.; Depraetere, V.; Nagata, S.; Hengartner, H.; Golstein, P. Fas and perforin pathways as major mechanisms of T cell-mediated cytotoxicity. Science 1994, 265, 528–530. [Google Scholar] [CrossRef]
- Lowin, B.; Hahne, M.; Mattmann, C.; Tschopp, J. Cytolytic T-cell cytotoxicity is mediated through perforin and Fas lytic pathways. Nature 1994, 370, 650–652. [Google Scholar] [CrossRef]
- Oshimi, Y.; Oda, S.; Honda, Y.; Nagata, S.; Miyazaki, S. Involvement of Fas ligand and Fas-mediated pathway in the cytotoxicity of human natural killer cells. J. Immunol. 1996, 157, 2909–2915. [Google Scholar]
- Takeda, K.; Hayakawa, Y.; Smyth, M.J.; Kayagaki, N.; Yamaguchi, N.; Kakuta, S.; Iwakura, Y.; Yagita, H.; Okumura, K. Involvement of tumor necrosis factor-related apoptosis-inducing ligand in surveillance of tumor metastasis by liver natural killer cells. Nat. Med. 2001, 7, 94–100. [Google Scholar] [CrossRef]
- Kearney, C.J.; Vervoort, S.J.; Hogg, S.J.; Ramsbottom, K.M.; Freeman, A.J.; Lalaoui, N.; Pijpers, L.; Michie, J.; Brown, K.K.; Knight, D.A.; et al. Tumor immune evasion arises through loss of TNF sensitivity. Sci. Immunol. 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Cubillos-Ruiz, J.R.; Bettigole, S.E.; Glimcher, L.H. Tumorigenic and Immunosuppressive Effects of Endoplasmic Reticulum Stress in Cancer. Cell 2017, 168, 692–706. [Google Scholar] [CrossRef] [PubMed]
- Urra, H.; Dufey, E.; Avril, T.; Chevet, E.; Hetz, C. Endoplasmic Reticulum Stress and the Hallmarks of Cancer. Trends Cancer 2016, 2, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Limia, C.M.; Sauzay, C.; Urra, H.; Hetz, C.; Chevet, E.; Avril, T. Emerging Roles of the Endoplasmic Reticulum Associated Unfolded Protein Response in Cancer Cell Migration and Invasion. Cancers 2019, 11, 631. [Google Scholar] [CrossRef] [PubMed]
- Madden, E.; Logue, S.E.; Healy, S.J.; Manie, S.; Samali, A. The role of the unfolded protein response in cancer progression: From oncogenesis to chemoresistance. Biol. Cell 2019, 111, 1–17. [Google Scholar] [CrossRef]
- Von Karstedt, S.; Conti, A.; Nobis, M.; Montinaro, A.; Hartwig, T.; Lemke, J.; Legler, K.; Annewanter, F.; Campbell, A.D.; Taraborrelli, L.; et al. Cancer cell-autonomous TRAIL-R signaling promotes KRAS-driven cancer progression, invasion, and metastasis. Cancer Cell 2015, 27, 561–573. [Google Scholar] [CrossRef]
- Seyrek, K.; Richter, M.; Lavrik, I.N. Decoding the sweet regulation of apoptosis: The role of glycosylation and galectins in apoptotic signaling pathways. Cell Death Differ. 2019, 26, 981–993. [Google Scholar] [CrossRef]
- Chakrabandhu, K.; Herincs, Z.; Huault, S.; Dost, B.; Peng, L.; Conchonaud, F.; Marguet, D.; He, H.T.; Hueber, A.O. Palmitoylation is required for efficient Fas cell death signaling. EMBO J. 2007, 26, 209–220. [Google Scholar] [CrossRef]
- Feig, C.; Tchikov, V.; Schutze, S.; Peter, M.E. Palmitoylation of CD95 facilitates formation of SDS-stable receptor aggregates that initiate apoptosis signaling. EMBO J. 2007, 26, 221–231. [Google Scholar] [CrossRef]
- Cruz, A.C.; Ramaswamy, M.; Ouyang, C.; Klebanoff, C.A.; Sengupta, P.; Yamamoto, T.N.; Meylan, F.; Thomas, S.K.; Richoz, N.; Eil, R.; et al. Fas/CD95 prevents autoimmunity independently of lipid raft localization and efficient apoptosis induction. Nat. Commun. 2016, 7, 13895. [Google Scholar] [CrossRef]
- Plenchette, S.; Romagny, S.; Laurens, V.; Bettaieb, A. S-Nitrosylation in TNF superfamily signaling pathway: Implication in cancer. Redox Biol. 2015, 6, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Herrero, R.; Kajikawa, O.; Matute-Bello, G.; Wang, Y.; Hagimoto, N.; Mongovin, S.; Wong, V.; Park, D.R.; Brot, N.; Heinecke, J.W.; et al. The biological activity of FasL in human and mouse lungs is determined by the structure of its stalk region. J. Clin. Investig. 2011, 121, 1174–1190. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Delanghe, T.; Dondelinger, Y.; Bertrand, M.J.M. RIPK1 Kinase-Dependent Death: A Symphony of Phosphorylation Events. Trends Cell Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Lafont, E.; Hartwig, T.; Walczak, H. Paving TRAIL’s Path with Ubiquitin. Trends Biochem. Sci. 2018, 43, 44–60. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, L.A.; Tai, L.; Lee, L.; Kruse, E.A.; Grabow, S.; Fairlie, W.D.; Haynes, N.M.; Tarlinton, D.M.; Zhang, J.G.; Belz, G.T.; et al. Membrane-bound Fas ligand only is essential for Fas-induced apoptosis. Nature 2009, 461, 659–663. [Google Scholar] [CrossRef]
- Newton, K.; Wickliffe, K.E.; Dugger, D.L.; Maltzman, A.; Roose-Girma, M.; Dohse, M.; Komuves, L.; Webster, J.D.; Dixit, V.M. Cleavage of RIPK1 by caspase-8 is crucial for limiting apoptosis and necroptosis. Nature 2019, 574, 428–431. [Google Scholar] [CrossRef]
- Tao, P.; Sun, J.; Wu, Z.; Wang, S.; Wang, J.; Li, W.; Pan, H.; Bai, R.; Zhang, J.; Wang, Y.; et al. A dominant autoinflammatory disease caused by non-cleavable variants of RIPK1. Nature 2020, 577, 109–114. [Google Scholar] [CrossRef]
- Lalaoui, N.; Boyden, S.E.; Oda, H.; Wood, G.M.; Stone, D.L.; Chau, D.; Liu, L.; Stoffels, M.; Kratina, T.; Lawlor, K.E.; et al. Mutations that prevent caspase cleavage of RIPK1 cause autoinflammatory disease. Nature 2020, 577, 103–108. [Google Scholar] [CrossRef]
- Tracey, K.J.; Beutler, B.; Lowry, S.F.; Merryweather, J.; Wolpe, S.; Milsark, I.W.; Hariri, R.J.; Fahey, T.J., 3rd; Zentella, A.; Albert, J.D.; et al. Shock and tissue injury induced by recombinant human cachectin. Science 1986, 234, 470–474. [Google Scholar] [CrossRef]
- Micheau, O.; Tschopp, J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 2003, 114, 181–190. [Google Scholar] [CrossRef]
- Schlicher, L.; Brauns-Schubert, P.; Schubert, F.; Maurer, U. SPATA2: More than a missing link. Cell Death Differ. 2017, 24, 1142–1147. [Google Scholar] [CrossRef] [PubMed]
- Lafont, E.; Draber, P.; Rieser, E.; Reichert, M.; Kupka, S.; de Miguel, D.; Draberova, H.; von Massenhausen, A.; Bhamra, A.; Henderson, S.; et al. TBK1 and IKKepsilon prevent TNF-induced cell death by RIPK1 phosphorylation. Nat. Cell. Biol. 2018, 20, 1389–1399. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Jin, T.; Zhu, H.; Chen, H.; Ofengeim, D.; Zou, C.; Mifflin, L.; Pan, L.; Amin, P.; Li, W.; et al. TBK1 Suppresses RIPK1-Driven Apoptosis and Inflammation during Development and in Aging. Cell 2018, 174, 1477–1491.e19. [Google Scholar] [CrossRef]
- Fong, Y.; Tracey, K.J.; Moldawer, L.L.; Hesse, D.G.; Manogue, K.B.; Kenney, J.S.; Lee, A.T.; Kuo, G.C.; Allison, A.C.; Lowry, S.F. Antibodies to cachectin/tumor necrosis factor reduce interleukin 1 beta and interleukin 6 appearance during lethal bacteremia. J. Exp. Med. 1989, 170, 1627–1633. [Google Scholar] [CrossRef] [PubMed]
- Brennan, F.M.; Chantry, D.; Jackson, A.; Maini, R.; Feldmann, M. Inhibitory effect of TNF alpha antibodies on synovial cell interleukin-1 production in rheumatoid arthritis. Lancet 1989, 2, 244–247. [Google Scholar] [CrossRef]
- Kalliolias, G.D.; Ivashkiv, L.B. TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat. Rev. Rheumatol. 2016, 12, 49–62. [Google Scholar] [CrossRef]
- Kalden, J.R.; Schulze-Koops, H. Immunogenicity and loss of response to TNF inhibitors: Implications for rheumatoid arthritis treatment. Nat. Rev. Rheumatol. 2017, 13, 707–718. [Google Scholar] [CrossRef]
- Annibaldi, A.; Walczak, H. Death Receptors and Their Ligands in Inflammatory Disease and Cancer. Cold Spring Harb. Perspect. Biol. 2020. [Google Scholar] [CrossRef]
- Gerlach, B.; Cordier, S.M.; Schmukle, A.C.; Emmerich, C.H.; Rieser, E.; Haas, T.L.; Webb, A.I.; Rickard, J.A.; Anderton, H.; Wong, W.W.; et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 2011, 471, 591–596. [Google Scholar] [CrossRef]
- Duprez, L.; Takahashi, N.; Van Hauwermeiren, F.; Vandendriessche, B.; Goossens, V.; Vanden Berghe, T.; Declercq, W.; Libert, C.; Cauwels, A.; Vandenabeele, P. RIP kinase-dependent necrosis drives lethal systemic inflammatory response syndrome. Immunity 2011, 35, 908–918. [Google Scholar] [CrossRef]
- Van Hauwermeiren, F.; Armaka, M.; Karagianni, N.; Kranidioti, K.; Vandenbroucke, R.E.; Loges, S.; Van Roy, M.; Staelens, J.; Puimege, L.; Palagani, A.; et al. Safe TNF-based antitumor therapy following p55TNFR reduction in intestinal epithelium. J. Clin. Investig. 2013, 123, 2590–2603. [Google Scholar] [CrossRef] [PubMed]
- Rickard, J.A.; Anderton, H.; Etemadi, N.; Nachbur, U.; Darding, M.; Peltzer, N.; Lalaoui, N.; Lawlor, K.E.; Vanyai, H.; Hall, C.; et al. TNFR1-dependent cell death drives inflammation in Sharpin-deficient mice. Elife 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Redouane, Y.; Lopez-Mosqueda, J.; Shiraishi, R.; Romanowska, M.; Lutzmayer, S.; Kuiper, J.; Martinez, C.; Dikic, I.; Pasparakis, M.; et al. Sharpin prevents skin inflammation by inhibiting TNFR1-induced keratinocyte apoptosis. Elife 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Newton, K.; Dugger, D.L.; Maltzman, A.; Greve, J.M.; Hedehus, M.; Martin-McNulty, B.; Carano, R.A.; Cao, T.C.; van Bruggen, N.; Bernstein, L.; et al. RIPK3 deficiency or catalytically inactive RIPK1 provides greater benefit than MLKL deficiency in mouse models of inflammation and tissue injury. Cell Death Differ. 2016, 23, 1565–1576. [Google Scholar] [CrossRef] [PubMed]
- Peltzer, N.; Darding, M.; Montinaro, A.; Draber, P.; Draberova, H.; Kupka, S.; Rieser, E.; Fisher, A.; Hutchinson, C.; Taraborrelli, L.; et al. LUBAC is essential for embryogenesis by preventing cell death and enabling haematopoiesis. Nature 2018, 557, 112–117. [Google Scholar] [CrossRef]
- Taraborrelli, L.; Peltzer, N.; Montinaro, A.; Kupka, S.; Rieser, E.; Hartwig, T.; Sarr, A.; Darding, M.; Draber, P.; Haas, T.L.; et al. LUBAC prevents lethal dermatitis by inhibiting cell death induced by TNF, TRAIL and CD95L. Nat. Commun. 2018, 9, 3910. [Google Scholar] [CrossRef]
- Eggermont, A.M.; Schraffordt Koops, H.; Klausner, J.M.; Kroon, B.B.; Schlag, P.M.; Lienard, D.; van Geel, A.N.; Hoekstra, H.J.; Meller, I.; Nieweg, O.E.; et al. Isolated limb perfusion with tumor necrosis factor and melphalan for limb salvage in 186 patients with locally advanced soft tissue extremity sarcomas. The cumulative multicenter European experience. Ann. Surg. 1996, 224, 756–764. [Google Scholar] [CrossRef]
- Fraker, D.L.; Alexander, H.R.; Andrich, M.; Rosenberg, S.A. Treatment of patients with melanoma of the extremity using hyperthermic isolated limb perfusion with melphalan, tumor necrosis factor, and interferon gamma: Results of a tumor necrosis factor dose-escalation study. J. Clin. Oncol. 1996, 14, 479–489. [Google Scholar] [CrossRef]
- Eggermont, A.M.; de Wilt, J.H.; ten Hagen, T.L. Current uses of isolated limb perfusion in the clinic and a model system for new strategies. Lancet Oncol. 2003, 4, 429–437. [Google Scholar] [CrossRef]
- Elia, A.R.; Grioni, M.; Basso, V.; Curnis, F.; Freschi, M.; Corti, A.; Mondino, A.; Bellone, M. Targeting Tumor Vasculature with TNF Leads Effector T Cells to the Tumor and Enhances Therapeutic Efficacy of Immune Checkpoint Blockers in Combination with Adoptive Cell Therapy. Clin. Cancer Res. 2018, 24, 2171–2181. [Google Scholar] [CrossRef]
- Huyghe, L.; Van Parys, A.; Cauwels, A.; Van Lint, S.; De Munter, S.; Bultinck, J.; Zabeau, L.; Hostens, J.; Goethals, A.; Vanderroost, N.; et al. Safe eradication of large established tumors using neovasculature-targeted tumor necrosis factor-based therapies. EMBO Mol. Med. 2020, 12, e11223. [Google Scholar] [CrossRef] [PubMed]
- Annibaldi, A.; Wicky John, S.; Vanden Berghe, T.; Swatek, K.N.; Ruan, J.; Liccardi, G.; Bianchi, K.; Elliott, P.R.; Choi, S.M.; Van Coillie, S.; et al. Ubiquitin-Mediated Regulation of RIPK1 Kinase Activity Independent of IKK and MK2. Mol. Cell 2018, 69, 566–580.e5. [Google Scholar] [CrossRef] [PubMed]
- Dondelinger, Y.; Jouan-Lanhouet, S.; Divert, T.; Theatre, E.; Bertin, J.; Gough, P.J.; Giansanti, P.; Heck, A.J.; Dejardin, E.; Vandenabeele, P.; et al. NF-κB-Independent Role of IKKα/IKKβ in Preventing RIPK1 Kinase-Dependent Apoptotic and Necroptotic Cell Death during TNF Signaling. Mol. Cell 2015, 60, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Dondelinger, Y.; Delanghe, T.; Priem, D.; Wynosky-Dolfi, M.A.; Sorobetea, D.; Rojas-Rivera, D.; Giansanti, P.; Roelandt, R.; Gropengiesser, J.; Ruckdeschel, K.; et al. Serine 25 phosphorylation inhibits RIPK1 kinase-dependent cell death in models of infection and inflammation. Nat. Commun. 2019, 10, 1729. [Google Scholar] [CrossRef]
- Dondelinger, Y.; Delanghe, T.; Rojas-Rivera, D.; Priem, D.; Delvaeye, T.; Bruggeman, I.; Van Herreweghe, F.; Vandenabeele, P.; Bertrand, M.J.M. MK2 phosphorylation of RIPK1 regulates TNF-mediated cell death. Nat. Cell. Biol. 2017, 19, 1237–1247. [Google Scholar] [CrossRef]
- Jaco, I.; Annibaldi, A.; Lalaoui, N.; Wilson, R.; Tenev, T.; Laurien, L.; Kim, C.; Jamal, K.; Wicky John, S.; Liccardi, G.; et al. MK2 Phosphorylates RIPK1 to Prevent TNF-Induced Cell Death. Mol. Cell 2017. [Google Scholar] [CrossRef]
- Menon, M.B.; Gropengiesser, J.; Fischer, J.; Novikova, L.; Deuretzbacher, A.; Lafera, J.; Schimmeck, H.; Czymmeck, N.; Ronkina, N.; Kotlyarov, A.; et al. p38MAPK/MK2-dependent phosphorylation controls cytotoxic RIPK1 signalling in inflammation and infection. Nat. Cell. Biol. 2017, 19, 1248–1259. [Google Scholar] [CrossRef]
- Van Antwerp, D.J.; Martin, S.J.; Kafri, T.; Green, D.R.; Verma, I.M. Suppression of TNF-alpha-induced apoptosis by NF-kappaB. Science 1996, 274, 787–789. [Google Scholar] [CrossRef]
- Yatim, N.; Cullen, S.; Albert, M.L. Dying cells actively regulate adaptive immune responses. Nat. Rev. Immunol. 2017, 17, 262–275. [Google Scholar] [CrossRef]
- Legrand, A.J.; Konstantinou, M.; Goode, E.F.; Meier, P. The Diversification of Cell Death and Immunity: Memento Mori. Mol. Cell 2019, 76, 232–242. [Google Scholar] [CrossRef]
- Holler, N.; Zaru, R.; Micheau, O.; Thome, M.; Attinger, A.; Valitutti, S.; Bodmer, J.L.; Schneider, P.; Seed, B.; Tschopp, J. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat. Immunol. 2000, 1, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Meurette, O.; Rebillard, A.; Huc, L.; Le Moigne, G.; Merino, D.; Micheau, O.; Lagadic-Gossmann, D.; Dimanche-Boitrel, M.T. TRAIL induces receptor-interacting protein 1-dependent and caspase-dependent necrosis-like cell death under acidic extracellular conditions. Cancer Res. 2007, 67, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Geserick, P.; Hupe, M.; Moulin, M.; Wong, W.W.; Feoktistova, M.; Kellert, B.; Gollnick, H.; Silke, J.; Leverkus, M. Cellular IAPs inhibit a cryptic CD95-induced cell death by limiting RIP1 kinase recruitment. J. Cell Biol. 2009, 187, 1037–1054. [Google Scholar] [CrossRef] [PubMed]
- Karl, I.; Jossberger-Werner, M.; Schmidt, N.; Horn, S.; Goebeler, M.; Leverkus, M.; Wajant, H.; Giner, T. TRAF2 inhibits TRAIL- and CD95L-induced apoptosis and necroptosis. Cell Death Dis. 2014, 5, e1444. [Google Scholar] [CrossRef]
- Degterev, A.; Huang, Z.; Boyce, M.; Li, Y.; Jagtap, P.; Mizushima, N.; Cuny, G.D.; Mitchison, T.J.; Moskowitz, M.A.; Yuan, J. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat. Chem. Biol. 2005, 1, 112–119. [Google Scholar] [CrossRef]
- Petrie, E.J.; Czabotar, P.E.; Murphy, J.M. The Structural Basis of Necroptotic Cell Death Signaling. Trends Biochem. Sci. 2019, 44, 53–63. [Google Scholar] [CrossRef]
- Koo, G.-B.; Morgan, M.J.; Lee, D.-G.; Kim, W.-J.; Yoon, J.-H.; Koo, J.S.; Kim, S.I.; Kim, S.J.; Son, M.K.; Hong, S.S.; et al. Methylation-dependent loss of RIP3 expression in cancer represses programmed necrosis in response to chemotherapeutics. Cell Res. 2015, 25, 707–725. [Google Scholar] [CrossRef]
- Aaes, T.L.; Kaczmarek, A.; Delvaeye, T.; De Craene, B.; De Koker, S.; Heyndrickx, L.; Delrue, I.; Taminau, J.; Wiernicki, B.; De Groote, P.; et al. Vaccination with Necroptotic Cancer Cells Induces Efficient Anti-tumor Immunity. Cell Rep. 2016, 15, 274–287. [Google Scholar] [CrossRef]
- Zhu, K.; Liang, W.; Ma, Z.; Xu, D.; Cao, S.; Lu, X.; Liu, N.; Shan, B.; Qian, L.; Yuan, J. Necroptosis promotes cell-autonomous activation of proinflammatory cytokine gene expression. Cell Death Dis. 2018, 9, 500. [Google Scholar] [CrossRef]
- Kearney, C.J.; Cullen, S.P.; Tynan, G.A.; Henry, C.M.; Clancy, D.; Lavelle, E.C.; Martin, S.J. Necroptosis suppresses inflammation via termination of TNF- or LPS-induced cytokine and chemokine production. Cell Death Differ. 2015, 22, 1313–1327. [Google Scholar] [CrossRef]
- Douanne, T.; Andre-Gregoire, G.; Trillet, K.; Thys, A.; Papin, A.; Feyeux, M.; Hulin, P.; Chiron, D.; Gavard, J.; Bidere, N. Pannexin-1 limits the production of proinflammatory cytokines during necroptosis. EMBO Rep. 2019, 20, e47840. [Google Scholar] [CrossRef] [PubMed]
- Tanzer, M.C.; Frauenstein, A.; Stafford, C.A.; Phulphagar, K.; Mann, M.; Meissner, F. Quantitative and Dynamic Catalogs of Proteins Released during Apoptotic and Necroptotic Cell Death. Cell Rep. 2020, 30, 1260–1270.e5. [Google Scholar] [CrossRef] [PubMed]
- Orozco, S.L.; Daniels, B.P.; Yatim, N.; Messmer, M.N.; Quarato, G.; Chen-Harris, H.; Cullen, S.P.; Snyder, A.G.; Ralli-Jain, P.; Frase, S.; et al. RIPK3 Activation Leads to Cytokine Synthesis that Continues after Loss of Cell Membrane Integrity. Cell Rep. 2019, 28, 2275–2287.e5. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Zhang, A.; Choksi, S.; Li, W.; Li, T.; Zhang, X.M.; Liu, Z.G. Activation of cell-surface proteases promotes necroptosis, inflammation and cell migration. Cell Res. 2016, 26, 886–900. [Google Scholar] [CrossRef] [PubMed]
- Medina, C.B.; Mehrotra, P.; Arandjelovic, S.; Perry, J.S.A.; Guo, Y.; Morioka, S.; Barron, B.; Walk, S.F.; Ghesquière, B.; Krupnick, A.S.; et al. Metabolites released from apoptotic cells act as tissue messengers. Nature 2020. [Google Scholar] [CrossRef] [PubMed]
- Yatim, N.; Jusforgues-Saklani, H.; Orozco, S.; Schulz, O.; Barreira da Silva, R.; Reis e Sousa, C.; Green, D.R.; Oberst, A.; Albert, M.L. RIPK1 and NF-kappaB signaling in dying cells determines cross-priming of CD8(+) T cells. Science 2015, 350, 328–334. [Google Scholar] [CrossRef]
- Li, L.; Thomas, R.M.; Suzuki, H.; De Brabander, J.K.; Wang, X.; Harran, P.G. A small molecule Smac mimic potentiates TRAIL- and TNFalpha-mediated cell death. Science 2004, 305, 1471–1474. [Google Scholar] [CrossRef]
- Bertrand, M.J.; Milutinovic, S.; Dickson, K.M.; Ho, W.C.; Boudreault, A.; Durkin, J.; Gillard, J.W.; Jaquith, J.B.; Morris, S.J.; Barker, P.A. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol. Cell 2008, 30, 689–700. [Google Scholar] [CrossRef]
- Oost, T.K.; Sun, C.; Armstrong, R.C.; Al-Assaad, A.S.; Betz, S.F.; Deckwerth, T.L.; Ding, H.; Elmore, S.W.; Meadows, R.P.; Olejniczak, E.T.; et al. Discovery of potent antagonists of the antiapoptotic protein XIAP for the treatment of cancer. J. Med. Chem. 2004, 47, 4417–4426. [Google Scholar] [CrossRef]
- Varfolomeev, E.; Blankenship, J.W.; Wayson, S.M.; Fedorova, A.V.; Kayagaki, N.; Garg, P.; Zobel, K.; Dynek, J.N.; Elliott, L.O.; Wallweber, H.J.; et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell 2007, 131, 669–681. [Google Scholar] [CrossRef]
- Vince, J.E.; Wong, W.W.; Khan, N.; Feltham, R.; Chau, D.; Ahmed, A.U.; Benetatos, C.A.; Chunduru, S.K.; Condon, S.M.; McKinlay, M.; et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell 2007, 131, 682–693. [Google Scholar] [CrossRef] [PubMed]
- Vince, J.E.; Pantaki, D.; Feltham, R.; Mace, P.D.; Cordier, S.M.; Schmukle, A.C.; Davidson, A.J.; Callus, B.A.; Wong, W.W.; Gentle, I.E.; et al. TRAF2 must bind to cellular inhibitors of apoptosis for tumor necrosis factor (tnf) to efficiently activate nf-{kappa}b and to prevent tnf-induced apoptosis. J. Biol. Chem. 2009, 284, 35906–35915. [Google Scholar] [CrossRef] [PubMed]
- Gonzalvez, F.; Lawrence, D.; Yang, B.; Yee, S.; Pitti, R.; Marsters, S.; Pham, V.C.; Stephan, J.-P.; Lill, J.; Ashkenazi, A. TRAF2 Sets a threshold for extrinsic apoptosis by tagging caspase-8 with a ubiquitin shutoff timer. Mol. Cell 2012, 48, 888–899. [Google Scholar] [CrossRef]
- Etemadi, N.; Chopin, M.; Anderton, H.; Tanzer, M.C.; Rickard, J.A.; Abeysekera, W.; Hall, C.; Spall, S.K.; Wang, B.; Xiong, Y.; et al. TRAF2 regulates TNF and NF-kappaB signalling to suppress apoptosis and skin inflammation independently of Sphingosine kinase 1. Elife 2015, 4. [Google Scholar] [CrossRef]
- Kearney, C.J.; Lalaoui, N.; Freeman, A.J.; Ramsbottom, K.M.; Silke, J.; Oliaro, J. PD-L1 and IAPs co-operate to protect tumors from cytotoxic lymphocyte-derived TNF. Cell Death Differ. 2017, 24, 1705–1716. [Google Scholar] [CrossRef]
- Vredevoogd, D.W.; Kuilman, T.; Ligtenberg, M.A.; Boshuizen, J.; Stecker, K.E.; de Bruijn, B.; Krijgsman, O.; Huang, X.; Kenski, J.C.N.; Lacroix, R.; et al. Augmenting Immunotherapy Impact by Lowering Tumor TNF Cytotoxicity Threshold. Cell 2019, 178, 585–599.e15. [Google Scholar] [CrossRef]
- Moore, R.J.; Owens, D.M.; Stamp, G.; Arnott, C.; Burke, F.; East, N.; Holdsworth, H.; Turner, L.; Rollins, B.; Pasparakis, M.; et al. Mice deficient in tumor necrosis factor-alpha are resistant to skin carcinogenesis. Nat. Med. 1999, 5, 828–831. [Google Scholar] [CrossRef]
- Suganuma, M.; Okabe, S.; Marino, M.W.; Sakai, A.; Sueoka, E.; Fujiki, H. Essential role of tumor necrosis factor alpha (TNF-alpha) in tumor promotion as revealed by TNF-alpha-deficient mice. Cancer Res. 1999, 59, 4516–4518. [Google Scholar]
- Pikarsky, E.; Porat, R.M.; Stein, I.; Abramovitch, R.; Amit, S.; Kasem, S.; Gutkovich-Pyest, E.; Urieli-Shoval, S.; Galun, E.; Ben-Neriah, Y. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature 2004, 431, 461–466. [Google Scholar] [CrossRef]
- Balkwill, F. Tumour necrosis factor and cancer. Nat. Rev. Cancer 2009, 9, 361–371. [Google Scholar] [CrossRef]
- Cordero, J.B.; Macagno, J.P.; Stefanatos, R.K.; Strathdee, K.E.; Cagan, R.L.; Vidal, M. Oncogenic Ras diverts a host TNF tumor suppressor activity into tumor promoter. Dev. Cell 2010, 18, 999–1011. [Google Scholar] [CrossRef] [PubMed]
- Park, E.J.; Lee, J.H.; Yu, G.Y.; He, G.; Ali, S.R.; Holzer, R.G.; Osterreicher, C.H.; Takahashi, H.; Karin, M. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell 2010, 140, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, F.; Rochotte, J.; Colacios, C.; Montfort, A.; Tilkin-Mariame, A.F.; Touriol, C.; Rochaix, P.; Lajoie-Mazenc, I.; Andrieu-Abadie, N.; Levade, T.; et al. Blocking Tumor Necrosis Factor alpha Enhances CD8 T-cell-Dependent Immunity in Experimental Melanoma. Cancer Res. 2015, 75, 2619–2628. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, F.; Montfort, A.; Marcheteau, E.; Imbert, C.; Gilhodes, J.; Filleron, T.; Rochaix, P.; Andrieu-Abadie, N.; Levade, T.; Meyer, N.; et al. TNFalpha blockade overcomes resistance to anti-PD-1 in experimental melanoma. Nat. Commun. 2017, 8, 2256. [Google Scholar] [CrossRef]
- Montfort, A.; Colacios, C.; Levade, T.; Andrieu-Abadie, N.; Meyer, N.; Segui, B. The TNF Paradox in Cancer Progression and Immunotherapy. Front. Immunol. 2019, 10, 1818. [Google Scholar] [CrossRef]
- Perez-Ruiz, E.; Minute, L.; Otano, I.; Alvarez, M.; Ochoa, M.C.; Belsue, V.; de Andrea, C.; Rodriguez-Ruiz, M.E.; Perez-Gracia, J.L.; Marquez-Rodas, I.; et al. Prophylactic TNF blockade uncouples efficacy and toxicity in dual CTLA-4 and PD-1 immunotherapy. Nature 2019, 569, 428–432. [Google Scholar] [CrossRef]
- Ogasawara, J.; Watanabe-Fukunaga, R.; Adachi, M.; Matsuzawa, A.; Kasugai, T.; Kitamura, Y.; Itoh, N.; Suda, T.; Nagata, S. Lethal effect of the anti-Fas antibody in mice. Nature 1993, 364, 806–809. [Google Scholar] [CrossRef]
- Rieux-Laucat, F. What’s up in the ALPS. Curr. Opin. Immunol. 2017, 49, 79–86. [Google Scholar] [CrossRef]
- Fisher, G.H.; Rosenberg, F.J.; Straus, S.E.; Dale, J.K.; Middleton, L.A.; Lin, A.Y.; Strober, W.; Lenardo, M.J.; Puck, J.M. Dominant interfering Fas gene mutations impair apoptosis in a human autoimmune lymphoproliferative syndrome. Cell 1995, 81, 935–946. [Google Scholar] [CrossRef]
- Rieux-Laucat, F.; Le Deist, F.; Hivroz, C.; Roberts, I.A.; Debatin, K.M.; Fischer, A.; de Villartay, J.P. Mutations in Fas associated with human lymphoproliferative syndrome and autoimmunity. Science 1995, 268, 1347–1349. [Google Scholar] [CrossRef]
- Desbarats, J.; Birge, R.B.; Mimouni-Rongy, M.; Weinstein, D.E.; Palerme, J.S.; Newell, M.K. Fas engagement induces neurite growth through ERK activation and p35 upregulation. Nat. Cell. Biol. 2003, 5, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Senju, S.; Negishi, I.; Motoyama, N.; Wang, F.; Nakayama, K.; Nakayama, K.; Lucas, P.J.; Hatakeyama, S.; Zhang, Q.; Yonehara, S.; et al. Functional significance of the Fas molecule in naive lymphocytes. Int. Immunol. 1996, 8, 423–431. [Google Scholar] [CrossRef]
- Karray, S.; Kress, C.; Cuvellier, S.; Hue-Beauvais, C.; Damotte, D.; Babinet, C.; Levi-Strauss, M. Complete loss of Fas ligand gene causes massive lymphoproliferation and early death, indicating a residual activity of gld allele. J. Immunol. 2004, 172, 2118–2125. [Google Scholar] [CrossRef]
- Rossin, A.; Miloro, G.; Hueber, A.O. TRAIL and FasL Functions in Cancer and Autoimmune Diseases: Towards an Increasing Complexity. Cancers 2019, 11, 639. [Google Scholar] [CrossRef] [PubMed]
- Bosque, A.; Aguilo, J.I.; del Rey, M.; Paz-Artal, E.; Allende, L.M.; Naval, J.; Anel, A. Cell cycle regulation by FasL and Apo2L/TRAIL in human T-cell blasts. Implications for autoimmune lymphoproliferative syndromes. J. Leukoc. Biol. 2008, 84, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Klebanoff, C.A.; Scott, C.D.; Leonardi, A.J.; Yamamoto, T.N.; Cruz, A.C.; Ouyang, C.; Ramaswamy, M.; Roychoudhuri, R.; Ji, Y.; Eil, R.L.; et al. Memory T cell-driven differentiation of naive cells impairs adoptive immunotherapy. J. Clin. Investig. 2016, 126, 318–334. [Google Scholar] [CrossRef]
- Straus, S.E.; Jaffe, E.S.; Puck, J.M.; Dale, J.K.; Elkon, K.B.; Rosen-Wolff, A.; Peters, A.M.; Sneller, M.C.; Hallahan, C.W.; Wang, J.; et al. The development of lymphomas in families with autoimmune lymphoproliferative syndrome with germline Fas mutations and defective lymphocyte apoptosis. Blood 2001, 98, 194–200. [Google Scholar] [CrossRef]
- Zornig, M.; Grzeschiczek, A.; Kowalski, M.B.; Hartmann, K.U.; Moroy, T. Loss of Fas/Apo-1 receptor accelerates lymphomagenesis in E mu L-MYC transgenic mice but not in animals infected with MoMuLV. Oncogene 1995, 10, 2397–2401. [Google Scholar]
- Afshar-Sterle, S.; Zotos, D.; Bernard, N.J.; Scherger, A.K.; Rodling, L.; Alsop, A.E.; Walker, J.; Masson, F.; Belz, G.T.; Corcoran, L.M.; et al. Fas ligand-mediated immune surveillance by T cells is essential for the control of spontaneous B cell lymphomas. Nat. Med. 2014, 20, 283–290. [Google Scholar] [CrossRef]
- Xu, Y.; Szalai, A.J.; Zhou, T.; Zinn, K.R.; Chaudhuri, T.R.; Li, X.; Koopman, W.J.; Kimberly, R.P. Fc gamma Rs modulate cytotoxicity of anti-Fas antibodies: Implications for agonistic antibody-based therapeutics. J. Immunol. 2003, 171, 562–568. [Google Scholar] [CrossRef]
- Holler, N.; Tardivel, A.; Kovacsovics-Bankowski, M.; Hertig, S.; Gaide, O.; Martinon, F.; Tinel, A.; Deperthes, D.; Calderara, S.; Schulthess, T.; et al. Two adjacent trimeric Fas ligands are required for Fas signaling and formation of a death-inducing signaling complex. Mol. Cell. Biol. 2003, 23, 1428–1440. [Google Scholar] [CrossRef] [PubMed]
- Verbrugge, I.; Wissink, E.H.; Rooswinkel, R.W.; Jongsma, J.; Beltraminelli, N.; Dupuis, M.; Borst, J.; Verheij, M. Combining radiotherapy with APO010 in cancer treatment. Clin. Cancer Res. 2009, 15, 2031–2038. [Google Scholar] [CrossRef] [PubMed]
- Eisele, G.; Roth, P.; Hasenbach, K.; Aulwurm, S.; Wolpert, F.; Tabatabai, G.; Wick, W.; Weller, M. APO010, a synthetic hexameric CD95 ligand, induces human glioma cell death in vitro and in vivo. Neuro-Oncology 2011, 13, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Park, S.M.; Tumanov, A.V.; Hau, A.; Sawada, K.; Feig, C.; Turner, J.R.; Fu, Y.X.; Romero, I.L.; Lengyel, E.; et al. CD95 promotes tumour growth. Nature 2010, 465, 492–496. [Google Scholar] [CrossRef]
- Qadir, A.S.; Stults, A.M.; Murmann, A.E.; Peter, M.E. The mechanism of how CD95/Fas activates the Type I IFN/STAT1 axis, driving cancer stemness in breast cancer. Sci. Rep. 2020, 10, 1310. [Google Scholar] [CrossRef]
- Qadir, A.S.; Ceppi, P.; Brockway, S.; Law, C.; Mu, L.; Khodarev, N.N.; Kim, J.; Zhao, J.C.; Putzbach, W.; Murmann, A.E.; et al. CD95/Fas Increases Stemness in Cancer Cells by Inducing a STAT1-Dependent Type I Interferon Response. Cell Rep. 2017, 18, 2373–2386. [Google Scholar] [CrossRef]
- Teodorczyk, M.; Kleber, S.; Wollny, D.; Sefrin, J.P.; Aykut, B.; Mateos, A.; Herhaus, P.; Sancho-Martinez, I.; Hill, O.; Gieffers, C.; et al. CD95 promotes metastatic spread via Sck in pancreatic ductal adenocarcinoma. Cell Death Differ. 2015, 22, 1192–1202. [Google Scholar] [CrossRef]
- Letellier, E.; Kumar, S.; Sancho-Martinez, I.; Krauth, S.; Funke-Kaiser, A.; Laudenklos, S.; Konecki, K.; Klussmann, S.; Corsini, N.S.; Kleber, S.; et al. CD95-ligand on peripheral myeloid cells activates Syk kinase to trigger their recruitment to the inflammatory site. Immunity 2010, 32, 240–252. [Google Scholar] [CrossRef]
- Gao, L.; Gulculer, G.S.; Golbach, L.; Block, H.; Zarbock, A.; Martin-Villalba, A. Endothelial cell-derived CD95 ligand serves as a chemokine in induction of neutrophil slow rolling and adhesion. Elife 2016, 5. [Google Scholar] [CrossRef]
- Cullen, S.P.; Henry, C.M.; Kearney, C.J.; Logue, S.E.; Feoktistova, M.; Tynan, G.A.; Lavelle, E.C.; Leverkus, M.; Martin, S.J. Fas/CD95-induced chemokines can serve as “find-me” signals for apoptotic cells. Mol. Cell 2013, 49, 1034–1048. [Google Scholar] [CrossRef]
- Motz, G.T.; Santoro, S.P.; Wang, L.P.; Garrabrant, T.; Lastra, R.R.; Hagemann, I.S.; Lal, P.; Feldman, M.D.; Benencia, F.; Coukos, G. Tumor endothelium FasL establishes a selective immune barrier promoting tolerance in tumors. Nat. Med. 2014, 20, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Lakins, M.A.; Ghorani, E.; Munir, H.; Martins, C.P.; Shields, J.D. Cancer-associated fibroblasts induce antigen-specific deletion of CD8 (+) T Cells to protect tumour cells. Nat. Commun. 2018, 9, 948. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Powis de Tenbossche, C.G.; Cane, S.; Colau, D.; van Baren, N.; Lurquin, C.; Schmitt-Verhulst, A.M.; Liljestrom, P.; Uyttenhove, C.; Van den Eynde, B.J. Resistance to cancer immunotherapy mediated by apoptosis of tumor-infiltrating lymphocytes. Nat. Commun. 2017, 8, 1404. [Google Scholar] [CrossRef] [PubMed]
- Hoogwater, F.J.; Nijkamp, M.W.; Smakman, N.; Steller, E.J.; Emmink, B.L.; Westendorp, B.F.; Raats, D.A.; Sprick, M.R.; Schaefer, U.; Van Houdt, W.J.; et al. Oncogenic K-Ras turns death receptors into metastasis-promoting receptors in human and mouse colorectal cancer cells. Gastroenterology 2010, 138, 2357–2367. [Google Scholar] [CrossRef]
- Steller, E.J.; Ritsma, L.; Raats, D.A.; Hoogwater, F.J.; Emmink, B.L.; Govaert, K.M.; Laoukili, J.; Rinkes, I.H.; van Rheenen, J.; Kranenburg, O. The death receptor CD95 activates the cofilin pathway to stimulate tumour cell invasion. EMBO Rep. 2011, 12, 931–937. [Google Scholar] [CrossRef]
- Raats, D.A.; Frenkel, N.; van Schelven, S.J.; Rinkes, I.H.; Laoukili, J.; Kranenburg, O. CD95 ligand induces senescence in mismatch repair-deficient human colon cancer via chronic caspase-mediated induction of DNA damage. Cell Death Dis. 2017, 8, e2669. [Google Scholar] [CrossRef]
- Kleber, S.; Sancho-Martinez, I.; Wiestler, B.; Beisel, A.; Gieffers, C.; Hill, O.; Thiemann, M.; Mueller, W.; Sykora, J.; Kuhn, A.; et al. Yes and PI3K bind CD95 to signal invasion of glioblastoma. Cancer Cell 2008, 13, 235–248. [Google Scholar] [CrossRef]
- Wick, W.; Fricke, H.; Junge, K.; Kobyakov, G.; Martens, T.; Heese, O.; Wiestler, B.; Schliesser, M.G.; von Deimling, A.; Pichler, J.; et al. A phase II, randomized, study of weekly APG101+reirradiation versus reirradiation in progressive glioblastoma. Clin. Cancer Res. 2014, 20, 6304–6313. [Google Scholar] [CrossRef]
- Blaes, J.; Thome, C.M.; Pfenning, P.N.; Rubmann, P.; Sahm, F.; Wick, A.; Bunse, T.; Schmenger, T.; Sykora, J.; von Deimling, A.; et al. Inhibition of CD95/CD95L (FAS/FASLG) Signaling with APG101 Prevents Invasion and Enhances Radiation Therapy for Glioblastoma. Mol. Cancer Res. 2018, 16, 767–776. [Google Scholar] [CrossRef]
- Boch, T.; Luft, T.; Metzgeroth, G.; Mossner, M.; Jann, J.C.; Nowak, D.; Meir, F.; Schumann, C.; Klemmer, J.; Brendel, S.; et al. Safety and efficacy of the CD95-ligand inhibitor asunercept in transfusion-dependent patients with low and intermediate risk MDS. Leuk. Res. 2018, 68, 62–69. [Google Scholar] [CrossRef]
- Leon-Bollotte, L.; Subramaniam, S.; Cauvard, O.; Plenchette-Colas, S.; Paul, C.; Godard, C.; Martinez-Ruiz, A.; Legembre, P.; Jeannin, J.F.; Bettaieb, A. S-nitrosylation of the death receptor fas promotes fas ligand-mediated apoptosis in cancer cells. Gastroenterology 2011, 140, 2009–2018.e4. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.M.; Frederiksen, J.K.; Zacharias, D.A.; Chan, F.K.; Johnson, M.; Lynch, D.; Tsien, R.Y.; Lenardo, M.J. Fas preassociation required for apoptosis signaling and dominant inhibition by pathogenic mutations. Science 2000, 288, 2354–2357. [Google Scholar] [CrossRef] [PubMed]
- Papoff, G.; Hausler, P.; Eramo, A.; Pagano, M.G.; Di Leve, G.; Signore, A.; Ruberti, G. Identification and characterization of a ligand-independent oligomerization domain in the extracellular region of the CD95 death receptor. J. Biol. Chem. 1999, 274, 38241–38250. [Google Scholar] [CrossRef] [PubMed]
- Chakrabandhu, K.; Huault, S.; Durivault, J.; Lang, K.; Ta Ngoc, L.; Bole, A.; Doma, E.; Derijard, B.; Gerard, J.P.; Pierres, M.; et al. An Evolution-Guided Analysis Reveals a Multi-Signaling Regulation of Fas by Tyrosine Phosphorylation and its Implication in Human Cancers. PLoS Biol. 2016, 14, e1002401. [Google Scholar] [CrossRef]
- Gulculer Balta, G.S.; Monzel, C.; Kleber, S.; Beaudouin, J.; Balta, E.; Kaindl, T.; Chen, S.; Gao, L.; Thiemann, M.; Wirtz, C.R.; et al. 3D Cellular Architecture Modulates Tyrosine Kinase Activity, Thereby Switching CD95-Mediated Apoptosis to Survival. Cell Rep. 2019, 29, 2295–2306.e6. [Google Scholar] [CrossRef]
- Tanaka, M.; Suda, T.; Haze, K.; Nakamura, N.; Sato, K.; Kimura, F.; Motoyoshi, K.; Mizuki, M.; Tagawa, S.; Ohga, S.; et al. Fas ligand in human serum. Nat. Med. 1996, 2, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Itai, T.; Adachi, M.; Nagata, S. Downregulation of Fas ligand by shedding. Nat. Med. 1998, 4, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Hohlbaum, A.M.; Moe, S.; Marshak-Rothstein, A. Opposing effects of transmembrane and soluble Fas ligand expression on inflammation and tumor cell survival. J. Exp. Med. 2000, 191, 1209–1220. [Google Scholar] [CrossRef] [PubMed]
- Matsuno, H.; Yudoh, K.; Watanabe, Y.; Nakazawa, F.; Aono, H.; Kimura, T. Stromelysin-1 (MMP-3) in synovial fluid of patients with rheumatoid arthritis has potential to cleave membrane bound Fas ligand. J. Rheumatol. 2001, 28, 22–28. [Google Scholar]
- Vargo-Gogola, T.; Crawford, H.C.; Fingleton, B.; Matrisian, L.M. Identification of novel matrix metalloproteinase-7 (matrilysin) cleavage sites in murine and human Fas ligand. Arch. Biochem. Biophys. 2002, 408, 155–161. [Google Scholar] [CrossRef]
- Schulte, M.; Reiss, K.; Lettau, M.; Maretzky, T.; Ludwig, A.; Hartmann, D.; de Strooper, B.; Janssen, O.; Saftig, P. ADAM10 regulates FasL cell surface expression and modulates FasL-induced cytotoxicity and activation-induced cell death. Cell Death Differ. 2007, 14, 1040–1049. [Google Scholar] [CrossRef] [PubMed]
- Kirkin, V.; Cahuzac, N.; Guardiola-Serrano, F.; Huault, S.; Lückerath, K.; Friedmann, E.; Novac, N.; Wels, W.S.; Martoglio, B.; Hueber, A.O.; et al. The Fas ligand intracellular domain is released by ADAM10 and SPPL2a cleavage in T-cells. Cell Death Differ. 2007, 14, 1678–1687. [Google Scholar] [CrossRef] [PubMed]
- Kiaei, M.; Kipiani, K.; Calingasan, N.Y.; Wille, E.; Chen, J.; Heissig, B.; Rafii, S.; Lorenzl, S.; Beal, M.F. Matrix metalloproteinase-9 regulates TNF-alpha and FasL expression in neuronal, glial cells and its absence extends life in a transgenic mouse model of amyotrophic lateral sclerosis. Exp. Neurol. 2007, 205, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, H.; Tanaka, M.; Suda, T.; Tomita, T.; Hayashida, K.; Takeuchi, E.; Kaneko, M.; Takano, H.; Nagata, S.; Ochi, T. Soluble Fas ligand in the joints of patients with rheumatoid arthritis and osteoarthritis. Arthritis Rheum. 1998, 41, 657–662. [Google Scholar] [CrossRef]
- Oliveira, J.B.; Bleesing, J.J.; Dianzani, U.; Fleisher, T.A.; Jaffe, E.S.; Lenardo, M.J.; Rieux-Laucat, F.; Siegel, R.M.; Su, H.C.; Teachey, D.T.; et al. Revised diagnostic criteria and classification for the autoimmune lymphoproliferative syndrome (ALPS): Report from the 2009 NIH International Workshop. Blood 2010, 116, e35–e40. [Google Scholar] [CrossRef]
- Poissonnier, A.; Sanséau, D.; Le Gallo, M.; Malleter, M.; Levoin, N.; Viel, R.; Morere, L.; Penna, A.; Blanco, P.; Dupuy, A.; et al. CD95-Mediated Calcium Signaling Promotes T Helper 17 Trafficking to Inflamed Organs in Lupus-Prone Mice. Immunity 2016, 45, 209–223. [Google Scholar] [CrossRef]
- Schneider, P.; Holler, N.; Bodmer, J.L.; Hahne, M.; Frei, K.; Fontana, A.; Tschopp, J. Conversion of membrane-bound Fas(CD95) ligand to its soluble form is associated with downregulation of its proapoptotic activity and loss of liver toxicity. J. Exp. Med. 1998, 187, 1205–1213. [Google Scholar] [CrossRef]
- Suda, T.; Hashimoto, H.; Tanaka, M.; Ochi, T.; Nagata, S. Membrane Fas ligand kills human peripheral blood T lymphocytes, and soluble Fas ligand blocks the killing. J. Exp. Med. 1997, 186, 2045–2050. [Google Scholar] [CrossRef]
- Bajou, K.; Peng, H.; Laug, W.E.; Maillard, C.; Noel, A.; Foidart, J.M.; Martial, J.A.; DeClerck, Y.A. Plasminogen activator inhibitor-1 protects endothelial cells from FasL-mediated apoptosis. Cancer Cell 2008, 14, 324–334. [Google Scholar] [CrossRef]
- Neumann, S.; Hasenauer, J.; Pollak, N.; Scheurich, P. Dominant negative effects of tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) receptor 4 on TRAIL receptor 1 signaling by formation of heteromeric complexes. J. Biol. Chem. 2014, 289, 16576–16587. [Google Scholar] [CrossRef]
- Merino, D.; Lalaoui, N.; Morizot, A.; Schneider, P.; Solary, E.; Micheau, O. Differential inhibition of TRAIL-mediated DR5-DISC formation by decoy receptors 1 and 2. Mol. Cell. Biol. 2006, 26, 7046–7055. [Google Scholar] [CrossRef] [PubMed]
- Muhlenbeck, F.; Schneider, P.; Bodmer, J.L.; Schwenzer, R.; Hauser, A.; Schubert, G.; Scheurich, P.; Moosmayer, D.; Tschopp, J.; Wajant, H. The tumor necrosis factor-related apoptosis-inducing ligand receptors TRAIL-R1 and TRAIL-R2 have distinct cross-linking requirements for initiation of apoptosis and are non-redundant in JNK activation. J. Biol. Chem. 2000, 275, 32208–32213. [Google Scholar] [CrossRef] [PubMed]
- Dufour, F.; Rattier, T.; Constantinescu, A.A.; Zischler, L.; Morle, A.; Ben Mabrouk, H.; Humblin, E.; Jacquemin, G.; Szegezdi, E.; Delacote, F.; et al. TRAIL receptor gene editing unveils TRAIL-R1 as a master player of apoptosis induced by TRAIL and ER stress. Oncotarget 2017, 8, 9974–9985. [Google Scholar] [CrossRef] [PubMed]
- Sedger, L.M.; Katewa, A.; Pettersen, A.K.; Osvath, S.R.; Farrell, G.C.; Stewart, G.J.; Bendall, L.J.; Alexander, S.I. Extreme lymphoproliferative disease and fatal autoimmune thrombocytopenia in FasL and TRAIL double-deficient mice. Blood 2010, 115, 3258–3268. [Google Scholar] [CrossRef] [PubMed]
- Zerafa, N.; Westwood, J.A.; Cretney, E.; Mitchell, S.; Waring, P.; Iezzi, M.; Smyth, M.J. Cutting edge: TRAIL deficiency accelerates hematological malignancies. J. Immunol. 2005, 175, 5586–5590. [Google Scholar] [CrossRef]
- Finnberg, N.; Klein-Szanto, A.J.; El-Deiry, W.S. TRAIL-R deficiency in mice promotes susceptibility to chronic inflammation and tumorigenesis. J. Clin. Investig. 2008, 118, 111–123. [Google Scholar] [CrossRef]
- Smyth, M.J.; Cretney, E.; Takeda, K.; Wiltrout, R.H.; Sedger, L.M.; Kayagaki, N.; Yagita, H.; Okumura, K. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) contributes to interferon gamma-dependent natural killer cell protection from tumor metastasis. J. Exp. Med. 2001, 193, 661–670. [Google Scholar] [CrossRef]
- Takeda, K.; Smyth, M.J.; Cretney, E.; Hayakawa, Y.; Yamaguchi, N.; Yagita, H.; Okumura, K. Involvement of tumor necrosis factor-related apoptosis-inducing ligand in NK cell-mediated and IFN-gamma-dependent suppression of subcutaneous tumor growth. Cell. Immunol. 2001, 214, 194–200. [Google Scholar] [CrossRef]
- Walczak, H.; Miller, R.E.; Ariail, K.; Gliniak, B.; Griffith, T.S.; Kubin, M.; Chin, W.; Jones, J.; Woodward, A.; Le, T.; et al. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat. Med. 1999, 5, 157–163. [Google Scholar] [CrossRef]
- Ashkenazi, A.; Pai, R.C.; Fong, S.; Leung, S.; Lawrence, D.A.; Marsters, S.A.; Blackie, C.; Chang, L.; McMurtrey, A.E.; Hebert, A.; et al. Safety and antitumor activity of recombinant soluble Apo2 ligand. J. Clin. Investig. 1999, 104, 155–162. [Google Scholar] [CrossRef]
- Von Karstedt, S.; Montinaro, A.; Walczak, H. Exploring the TRAILs less travelled: TRAIL in cancer biology and therapy. Nat. Rev. Cancer 2017. [Google Scholar] [CrossRef] [PubMed]
- Wajant, H. Molecular Mode of Action of TRAIL Receptor Agonists-Common Principles and Their Translational Exploitation. Cancers 2019, 11, 954. [Google Scholar] [CrossRef] [PubMed]
- Naval, J.; de Miguel, D.; Gallego-Lleyda, A.; Anel, A.; Martinez-Lostao, L. Importance of TRAIL Molecular Anatomy in Receptor Oligomerization and Signaling. Implications for Cancer Therapy. Cancers 2019, 11, 444. [Google Scholar] [CrossRef] [PubMed]
- Kretz, A.L.; Trauzold, A.; Hillenbrand, A.; Knippschild, U.; Henne-Bruns, D.; von Karstedt, S.; Lemke, J. TRAILblazing Strategies for Cancer Treatment. Cancers 2019, 11, 456. [Google Scholar] [CrossRef]
- Leverkus, M.; Sprick, M.R.; Wachter, T.; Denk, A.; Brocker, E.B.; Walczak, H.; Neumann, M. TRAIL-induced apoptosis and gene induction in HaCaT keratinocytes: Differential contribution of TRAIL receptors 1 and 2. J. Investig. Derm. 2003, 121, 149–155. [Google Scholar] [CrossRef]
- MacFarlane, M.; Kohlhaas, S.L.; Sutcliffe, M.J.; Dyer, M.J.; Cohen, G.M. TRAIL receptor-selective mutants signal to apoptosis via TRAIL-R1 in primary lymphoid malignancies. Cancer Res. 2005, 65, 11265–11270. [Google Scholar] [CrossRef]
- Lemke, J.; Noack, A.; Adam, D.; Tchikov, V.; Bertsch, U.; Roder, C.; Schutze, S.; Wajant, H.; Kalthoff, H.; Trauzold, A. TRAIL signaling is mediated by DR4 in pancreatic tumor cells despite the expression of functional DR5. J. Mol. Med. 2010, 88, 729–740. [Google Scholar] [CrossRef]
- Stadel, D.; Mohr, A.; Ref, C.; MacFarlane, M.; Zhou, S.; Humphreys, R.; Bachem, M.; Cohen, G.; Moller, P.; Zwacka, R.M.; et al. TRAIL-induced apoptosis is preferentially mediated via TRAIL receptor 1 in pancreatic carcinoma cells and profoundly enhanced by XIAP inhibitors. Clin. Cancer Res. 2010, 16, 5734–5749. [Google Scholar] [CrossRef]
- Graves, J.D.; Kordich, J.J.; Huang, T.H.; Piasecki, J.; Bush, T.L.; Sullivan, T.; Foltz, I.N.; Chang, W.; Douangpanya, H.; Dang, T.; et al. Apo2L/TRAIL and the death receptor 5 agonist antibody AMG 655 cooperate to promote receptor clustering and antitumor activity. Cancer Cell 2014, 26, 177–189. [Google Scholar] [CrossRef]
- Tuthill, M.H.; Montinaro, A.; Zinngrebe, J.; Prieske, K.; Draber, P.; Prieske, S.; Newsom-Davis, T.; von Karstedt, S.; Graves, J.; Walczak, H. TRAIL-R2-specific antibodies and recombinant TRAIL can synergise to kill cancer cells. Oncogene 2015, 34, 2138–2144. [Google Scholar] [CrossRef]
- Dominguez, G.A.; Condamine, T.; Mony, S.; Hashimoto, A.; Wang, F.; Liu, Q.; Forero, A.; Bendell, J.; Witt, R.; Hockstein, N.; et al. Selective Targeting of Myeloid-Derived Suppressor Cells in Cancer Patients Using DS-8273a, an Agonistic TRAIL-R2 Antibody. Clin. Cancer Res. 2017, 23, 2942–2950. [Google Scholar] [CrossRef] [PubMed]
- Wilson, N.S.; Yang, A.; Yang, B.; Couto, S.; Stern, H.; Gogineni, A.; Pitti, R.; Marsters, S.; Weimer, R.M.; Singh, M.; et al. Proapoptotic activation of death receptor 5 on tumor endothelial cells disrupts the vasculature and reduces tumor growth. Cancer Cell 2012, 22, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Ehrhardt, H.; Fulda, S.; Schmid, I.; Hiscott, J.; Debatin, K.-M.; Jeremias, I. TRAIL induced survival and proliferation in cancer cells resistant towards TRAIL-induced apoptosis mediated by NF-kappaB. Oncogene 2003, 22, 3842–3852. [Google Scholar] [CrossRef] [PubMed]
- Somasekharan, S.P.; Koc, M.; Morizot, A.; Micheau, O.; Sorensen, P.H.; Gaide, O.; Andera, L.; Martinou, J.C. TRAIL promotes membrane blebbing, detachment and migration of cells displaying a dysfunctional intrinsic pathway of apoptosis. Apoptosis Int. J. Program. Cell Death 2013, 18, 324–336. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Wang, W.; Zhang, Y.; Liu, S.; Liu, Y.; Zheng, D. TRAIL receptor mediates inflammatory cytokine release in an NF-kappaB-dependent manner. Cell Res. 2009, 19, 758–767. [Google Scholar] [CrossRef]
- Hartwig, T.; Montinaro, A.; von Karstedt, S.; Sevko, A.; Surinova, S.; Chakravarthy, A.; Taraborrelli, L.; Draber, P.; Lafont, E.; Arce Vargas, F.; et al. The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2. Mol. Cell 2017, 65, 730–742.e5. [Google Scholar] [CrossRef]
- Geismann, C.; Erhart, W.; Grohmann, F.; Schreiber, S.; Schneider, G.; Schafer, H.; Arlt, A. TRAIL/NF-kappaB/CX3CL1 Mediated Onco-Immuno Crosstalk Leading to TRAIL Resistance of Pancreatic Cancer Cell Lines. Int. J. Mol. Sci. 2018, 19, 1661. [Google Scholar] [CrossRef]
- Lafont, E.; Kantari-Mimoun, C.; Draber, P.; De Miguel, D.; Hartwig, T.; Reichert, M.; Kupka, S.; Shimizu, Y.; Taraborrelli, L.; Spit, M.; et al. The linear ubiquitin chain assembly complex regulates TRAIL-induced gene activation and cell death. EMBO J. 2017. [Google Scholar] [CrossRef]
- Henry, C.M.; Martin, S.J. Caspase-8 Acts in a Non-enzymatic Role as a Scaffold for Assembly of a Pro-inflammatory “FADDosome” Complex upon TRAIL Stimulation. Mol. Cell 2017, 65, 715–729.e5. [Google Scholar] [CrossRef]
- Varfolomeev, E.; Maecker, H.; Sharp, D.; Lawrence, D.; Renz, M.; Vucic, D.; Ashkenazi, A. Molecular determinants of kinase pathway activation by Apo2 ligand/tumor necrosis factor-related apoptosis-inducing ligand. J. Biol. Chem. 2005, 280, 40599–40608. [Google Scholar] [CrossRef]
- Lin, Y.; Devin, A.; Cook, A.; Keane, M.M.; Kelliher, M.; Lipkowitz, S.; Liu, Z.G. The death domain kinase RIP is essential for TRAIL (Apo2L)-induced activation of IkappaB kinase and c-Jun N-terminal kinase. Mol. Cell. Biol. 2000, 20, 6638–6645. [Google Scholar] [CrossRef] [PubMed]
- Fullsack, S.; Rosenthal, A.; Wajant, H.; Siegmund, D. Redundant and receptor-specific activities of TRADD, RIPK1 and FADD in death receptor signaling. Cell Death Dis. 2019, 10, 122. [Google Scholar] [CrossRef] [PubMed]
- Azijli, K.; Yuvaraj, S.; Peppelenbosch, M.P.; Wurdinger, T.; Dekker, H.; Joore, J.; van Dijk, E.; Quax, W.J.; Peters, G.J.; de Jong, S.; et al. Kinome profiling of non-canonical TRAIL signaling reveals RIP1-Src-STAT3-dependent invasion in resistant non-small cell lung cancer cells. J. Cell Sci. 2012, 125, 4651–4661. [Google Scholar] [CrossRef] [PubMed]
- Haselmann, V.; Kurz, A.; Bertsch, U.; Hubner, S.; Olempska-Muller, M.; Fritsch, J.; Hasler, R.; Pickl, A.; Fritsche, H.; Annewanter, F.; et al. Nuclear death receptor TRAIL-R2 inhibits maturation of let-7 and promotes proliferation of pancreatic and other tumor cells. Gastroenterology 2014, 146, 278–290. [Google Scholar] [CrossRef]
- Kischkel, F.C.; Hellbardt, S.; Behrmann, I.; Germer, M.; Pawlita, M.; Krammer, P.H.; Peter, M.E. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 1995, 14, 5579–5588. [Google Scholar] [CrossRef]
- Sprick, M.R.; Weigand, M.A.; Rieser, E.; Rauch, C.T.; Juo, P.; Blenis, J.; Krammer, P.H.; Walczak, H. FADD/MORT1 and caspase-8 are recruited to TRAIL receptors 1 and 2 and are essential for apoptosis mediated by TRAIL receptor 2. Immunity 2000, 12, 599–609. [Google Scholar] [CrossRef]
- Schleich, K.; Warnken, U.; Fricker, N.; Ozturk, S.; Richter, P.; Kammerer, K.; Schnolzer, M.; Krammer, P.H.; Lavrik, I.N. Stoichiometry of the CD95 death-inducing signaling complex: Experimental and modeling evidence for a death effector domain chain model. Mol. Cell 2012, 47, 306–319. [Google Scholar] [CrossRef]
- Dickens, L.S.; Boyd, R.S.; Jukes-Jones, R.; Hughes, M.A.; Robinson, G.L.; Fairall, L.; Schwabe, J.W.; Cain, K.; Macfarlane, M. A death effector domain chain DISC model reveals a crucial role for caspase-8 chain assembly in mediating apoptotic cell death. Mol. Cell 2012, 47, 291–305. [Google Scholar] [CrossRef]
- Fu, T.M.; Li, Y.; Lu, A.; Li, Z.; Vajjhala, P.R.; Cruz, A.C.; Srivastava, D.B.; DiMaio, F.; Penczek, P.A.; Siegel, R.M.; et al. Cryo-EM Structure of Caspase-8 Tandem DED Filament Reveals Assembly and Regulation Mechanisms of the Death-Inducing Signaling Complex. Mol. Cell 2016, 64, 236–250. [Google Scholar] [CrossRef]
- Jost, P.J.; Grabow, S.; Gray, D.; McKenzie, M.D.; Nachbur, U.; Huang, D.C.; Bouillet, P.; Thomas, H.E.; Borner, C.; Silke, J.; et al. XIAP discriminates between type I and type II FAS-induced apoptosis. Nature 2009, 460, 1035–1039. [Google Scholar] [CrossRef]
- Hughes, M.A.; Powley, I.R.; Jukes-Jones, R.; Horn, S.; Feoktistova, M.; Fairall, L.; Schwabe, J.W.; Leverkus, M.; Cain, K.; MacFarlane, M. Co-operative and Hierarchical Binding of c-FLIP and Caspase-8: A Unified Model Defines How c-FLIP Isoforms Differentially Control Cell Fate. Mol. Cell 2016, 61, 834–849. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, L.M.; Fox, J.P.; Higgins, C.A.; Majkut, J.; Sessler, T.; McLaughlin, K.; McCann, C.; Roberts, J.Z.; Crawford, N.T.; McDade, S.S.; et al. A revised model of TRAIL-R2 DISC assembly explains how FLIP(L) can inhibit or promote apoptosis. EMBO Rep. 2020, 21, e49254. [Google Scholar] [CrossRef] [PubMed]
- Hillert, L.K.; Ivanisenko, N.V.; Espe, J.; Konig, C.; Ivanisenko, V.A.; Kahne, T.; Lavrik, I.N. Long and short isoforms of c-FLIP act as control checkpoints of DED filament assembly. Oncogene 2020, 39, 1756–1772. [Google Scholar] [CrossRef]
- Jin, Z.; Li, Y.; Pitti, R.; Lawrence, D.; Pham, V.C.; Lill, J.R.; Ashkenazi, A. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell 2009, 137, 721–735. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, L.; Yao, Q.; Li, L.; Dong, N.; Rong, J.; Gao, W.; Ding, X.; Sun, L.; Chen, X.; et al. Pathogen blocks host death receptor signalling by arginine GlcNAcylation of death domains. Nature 2013, 501, 242–246. [Google Scholar] [CrossRef] [PubMed]
- Pearson, J.S.; Giogha, C.; Ong, S.Y.; Kennedy, C.L.; Kelly, M.; Robinson, K.S.; Lung, T.W.; Mansell, A.; Riedmaier, P.; Oates, C.V.; et al. A type III effector antagonizes death receptor signalling during bacterial gut infection. Nature 2013, 501, 247–251. [Google Scholar] [CrossRef]
- Scott, N.E.; Giogha, C.; Pollock, G.L.; Kennedy, C.L.; Webb, A.I.; Williamson, N.A.; Pearson, J.S.; Hartland, E.L. The bacterial arginine glycosyltransferase effector NleB preferentially modifies Fas-associated death domain protein (FADD). J. Biol. Chem. 2017, 292, 17337–17350. [Google Scholar] [CrossRef]
- Ehrenschwender, M.; Siegmund, D.; Wicovsky, A.; Kracht, M.; Dittrich-Breiholz, O.; Spindler, V.; Waschke, J.; Kalthoff, H.; Trauzold, A.; Wajant, H. Mutant PIK3CA licenses TRAIL and CD95L to induce non-apoptotic caspase-8-mediated ROCK activation. Cell Death Differ. 2010, 17, 1435–1447. [Google Scholar] [CrossRef]
- Chen, X.; Iliopoulos, D.; Zhang, Q.; Tang, Q.; Greenblatt, M.B.; Hatziapostolou, M.; Lim, E.; Tam, W.L.; Ni, M.; Chen, Y.; et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1alpha pathway. Nature 2014, 508, 103–107. [Google Scholar] [CrossRef]
- Bertolotti, A.; Zhang, Y.; Hendershot, L.M.; Harding, H.P.; Ron, D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell. Biol. 2000, 2, 326–332. [Google Scholar] [CrossRef]
- Shen, J.; Chen, X.; Hendershot, L.; Prywes, R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 2002, 3, 99–111. [Google Scholar] [CrossRef]
- Gardner, B.M.; Walter, P. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science 2011, 333, 1891–1894. [Google Scholar] [CrossRef] [PubMed]
- Karagoz, G.E.; Acosta-Alvear, D.; Nguyen, H.T.; Lee, C.P.; Chu, F.; Walter, P. An unfolded protein-induced conformational switch activates mammalian IRE1. Elife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Li, J.; Tao, J.; Sha, B. The luminal domain of the ER stress sensor protein PERK binds misfolded proteins and thereby triggers PERK oligomerization. J. Biol. Chem. 2018, 293, 4110–4121. [Google Scholar] [CrossRef]
- Calfon, M.; Zeng, H.; Urano, F.; Till, J.H.; Hubbard, S.R.; Harding, H.P.; Clark, S.G.; Ron, D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 2002, 415, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Sato, T.; Matsui, T.; Sato, M.; Okada, T.; Yoshida, H.; Harada, A.; Mori, K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev. Cell 2007, 13, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Hollien, J.; Weissman, J.S. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 2006, 313, 104–107. [Google Scholar] [CrossRef]
- Maurel, M.; Chevet, E.; Tavernier, J.; Gerlo, S. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem. Sci. 2014, 39, 245–254. [Google Scholar] [CrossRef]
- Lerner, A.G.; Upton, J.P.; Praveen, P.V.; Ghosh, R.; Nakagawa, Y.; Igbaria, A.; Shen, S.; Nguyen, V.; Backes, B.J.; Heiman, M.; et al. IRE1alpha induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metab. 2012, 16, 250–264. [Google Scholar] [CrossRef]
- Logue, S.E.; McGrath, E.P.; Cleary, P.; Greene, S.; Mnich, K.; Almanza, A.; Chevet, E.; Dwyer, R.M.; Oommen, A.; Legembre, P.; et al. Inhibition of IRE1 RNase activity modulates the tumor cell secretome and enhances response to chemotherapy. Nat. Commun. 2018, 9, 3267. [Google Scholar] [CrossRef]
- Han, D.; Lerner, A.G.; Vande Walle, L.; Upton, J.P.; Xu, W.; Hagen, A.; Backes, B.J.; Oakes, S.A.; Papa, F.R. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell 2009, 138, 562–575. [Google Scholar] [CrossRef] [PubMed]
- Korennykh, A.V.; Egea, P.F.; Korostelev, A.A.; Finer-Moore, J.; Zhang, C.; Shokat, K.M.; Stroud, R.M.; Walter, P. The unfolded protein response signals through high-order assembly of Ire1. Nature 2009, 457, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Bouchecareilh, M.; Higa, A.; Fribourg, S.; Moenner, M.; Chevet, E. Peptides derived from the bifunctional kinase/RNase enzyme IRE1alpha modulate IRE1alpha activity and protect cells from endoplasmic reticulum stress. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2011, 25, 3115–3129. [Google Scholar] [CrossRef]
- Ghosh, R.; Wang, L.; Wang, E.S.; Perera, B.G.; Igbaria, A.; Morita, S.; Prado, K.; Thamsen, M.; Caswell, D.; Macias, H.; et al. Allosteric inhibition of the IRE1alpha RNase preserves cell viability and function during endoplasmic reticulum stress. Cell 2014, 158, 534–548. [Google Scholar] [CrossRef] [PubMed]
- Tam, A.B.; Koong, A.C.; Niwa, M. Ire1 has distinct catalytic mechanisms for XBP1/HAC1 splicing and RIDD. Cell Rep. 2014, 9, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, Y.; Song, L.; Zeng, L.; Yi, W.; Liu, T.; Chen, H.; Wang, M.; Ju, Z.; Cong, Y.S. A critical role of DDRGK1 in endoplasmic reticulum homoeostasis via regulation of IRE1alpha stability. Nat. Commun. 2017, 8, 14186. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C.; Papa, F.R. The Unfolded Protein Response and Cell Fate Control. Mol. Cell 2018, 69, 169–181. [Google Scholar] [CrossRef]
- Papaioannou, A.; Metais, A.; Maurel, M.; Negroni, L.; González-Quiroz, M.; Golchesmeh, E.Z.; Blondel, A.; Koong, A.C.; Hetz, C.; Pedeux, R.; et al. Stress-induced tyrosine phosphorylation of RtcB modulates IRE1 activity and signaling outputs. bioRxiv 2020. [Google Scholar] [CrossRef]
- Urano, F.; Wang, X.; Bertolotti, A.; Zhang, Y.; Chung, P.; Harding, H.P.; Ron, D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 2000, 287, 664–666. [Google Scholar] [CrossRef]
- Hu, P.; Han, Z.; Couvillon, A.D.; Kaufman, R.J.; Exton, J.H. Autocrine tumor necrosis factor alpha links endoplasmic reticulum stress to the membrane death receptor pathway through IRE1alpha-mediated NF-kappaB activation and down-regulation of TRAF2 expression. Mol. Cell. Biol. 2006, 26, 3071–3084. [Google Scholar] [CrossRef]
- Yamamoto, K.; Ichijo, H.; Korsmeyer, S.J. BCL-2 is phosphorylated and inactivated by an ASK1/Jun N-terminal protein kinase pathway normally activated at G(2)/M. Mol. Cell. Biol. 1999, 19, 8469–8478. [Google Scholar] [CrossRef] [PubMed]
- Obacz, J.; Avril, T.; Rubio-Patino, C.; Bossowski, J.P.; Igbaria, A.; Ricci, J.E.; Chevet, E. Regulation of tumor-stroma interactions by the unfolded protein response. FEBS J. 2019, 286, 279–296. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Patino, C.; Bossowski, J.P.; Chevet, E.; Ricci, J.E. Reshaping the Immune Tumor Microenvironment Through IRE1 Signaling. Trends Mol. Med. 2018, 24, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Patino, C.; Bossowski, J.P.; De Donatis, G.M.; Mondragon, L.; Villa, E.; Aira, L.E.; Chiche, J.; Mhaidly, R.; Lebeaupin, C.; Marchetti, S.; et al. Low-Protein Diet Induces IRE1alpha-Dependent Anticancer Immunosurveillance. Cell Metab. 2018, 27, 828–842.e7. [Google Scholar] [CrossRef] [PubMed]
- Obacz, J.; Archambeau, J.; Sicari, D.; Le Reste, P.J.; Pineau, R.; Martin, S.; Barroso, K.; Vlachavas, E.; Voutetakis, K.; Fainsod-Levi, T.; et al. Novel IRE1-dependent proinflammatory signaling controls tumor infiltration by myeloid cells. bioRxiv 2020. [Google Scholar] [CrossRef]
- Urra, H.; Henriquez, D.R.; Canovas, J.; Villarroel-Campos, D.; Carreras-Sureda, A.; Pulgar, E.; Molina, E.; Hazari, Y.M.; Limia, C.M.; Alvarez-Rojas, S.; et al. IRE1alpha governs cytoskeleton remodelling and cell migration through a direct interaction with filamin A. Nat. Cell. Biol. 2018, 20, 942–953. [Google Scholar] [CrossRef]
- Carreras-Sureda, A.; Jana, F.; Urra, H.; Durand, S.; Mortenson, D.E.; Sagredo, A.; Bustos, G.; Hazari, Y.; Ramos-Fernandez, E.; Sassano, M.L.; et al. Non-canonical function of IRE1alpha determines mitochondria-associated endoplasmic reticulum composition to control calcium transfer and bioenergetics. Nat. Cell. Biol. 2019, 21, 755–767. [Google Scholar] [CrossRef]
- Walter, F.; Schmid, J.; Dussmann, H.; Concannon, C.G.; Prehn, J.H. Imaging of single cell responses to ER stress indicates that the relative dynamics of IRE1/XBP1 and PERK/ATF4 signalling rather than a switch between signalling branches determine cell survival. Cell Death Differ. 2015, 22, 1502–1516. [Google Scholar] [CrossRef]
- Liu, C.Y.; Schroder, M.; Kaufman, R.J. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J. Biol. Chem. 2000, 275, 24881–24885. [Google Scholar] [CrossRef]
- Sarcinelli, C.; Dragic, H.; Piecyk, M.; Barbet, V.; Duret, C.; Barthelaix, A.; Ferraro-Peyret, C.; Fauvre, J.; Renno, T.; Chaveroux, C.; et al. ATF4-Dependent NRF2 Transcriptional Regulation Promotes Antioxidant Protection during Endoplasmic Reticulum Stress. Cancers 2020, 12, 569. [Google Scholar] [CrossRef]
- Cullinan, S.B.; Zhang, D.; Hannink, M.; Arvisais, E.; Kaufman, R.J.; Diehl, J.A. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol. Cell. Biol. 2003, 23, 7198–7209. [Google Scholar] [CrossRef] [PubMed]
- Cullinan, S.B.; Diehl, J.A. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. J. Biol. Chem. 2004, 279, 20108–20117. [Google Scholar] [CrossRef]
- Tam, A.B.; Mercado, E.L.; Hoffmann, A.; Niwa, M. ER stress activates NF-kappaB by integrating functions of basal IKK activity, IRE1 and PERK. PLoS ONE 2012, 7, e45078. [Google Scholar] [CrossRef]
- Puthalakath, H.; O’Reilly, L.A.; Gunn, P.; Lee, L.; Kelly, P.N.; Huntington, N.D.; Hughes, P.D.; Michalak, E.M.; McKimm-Breschkin, J.; Motoyama, N.; et al. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell 2007, 129, 1337–1349. [Google Scholar] [CrossRef] [PubMed]
- McCullough, K.D.; Martindale, J.L.; Klotz, L.O.; Aw, T.Y.; Holbrook, N.J. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol. Cell. Biol. 2001, 21, 1249–1259. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Aviles, G.; Liu, Y.; Tian, R.; Unger, B.A.; Lin, Y.T.; Wiita, A.P.; Xu, K.; Correia, M.A.; Kampmann, M. Mitochondrial stress is relayed to the cytosol by an OMA1-DELE1-HRI pathway. Nature 2020. [Google Scholar] [CrossRef]
- Fessler, E.; Eckl, E.M.; Schmitt, S.; Mancilla, I.A.; Meyer-Bender, M.F.; Hanf, M.; Philippou-Massier, J.; Krebs, S.; Zischka, H.; Jae, L.T. A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol. Nature 2020. [Google Scholar] [CrossRef]
- Taniuchi, S.; Miyake, M.; Tsugawa, K.; Oyadomari, M.; Oyadomari, S. Integrated stress response of vertebrates is regulated by four eIF2alpha kinases. Sci. Rep. 2016, 6, 32886. [Google Scholar] [CrossRef]
- Connor, J.H.; Weiser, D.C.; Li, S.; Hallenbeck, J.M.; Shenolikar, S. Growth arrest and DNA damage-inducible protein GADD34 assembles a novel signaling complex containing protein phosphatase 1 and inhibitor 1. Mol. Cell. Biol. 2001, 21, 6841–6850. [Google Scholar] [CrossRef]
- Novoa, I.; Zeng, H.; Harding, H.P.; Ron, D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J. Cell Biol. 2001, 153, 1011–1022. [Google Scholar] [CrossRef]
- Marciniak, S.J.; Yun, C.Y.; Oyadomari, S.; Novoa, I.; Zhang, Y.; Jungreis, R.; Nagata, K.; Harding, H.P.; Ron, D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004, 18, 3066–3077. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.X.; Sokol, E.S.; Del Vecchio, C.A.; Sanduja, S.; Claessen, J.H.; Proia, T.A.; Jin, D.X.; Reinhardt, F.; Ploegh, H.L.; Wang, Q.; et al. Epithelial-to-mesenchymal transition activates PERK-eIF2alpha and sensitizes cells to endoplasmic reticulum stress. Cancer Discov. 2014, 4, 702–715. [Google Scholar] [CrossRef] [PubMed]
- Pommier, A.; Anaparthy, N.; Memos, N.; Kelley, Z.L.; Gouronnec, A.; Yan, R.; Auffray, C.; Albrengues, J.; Egeblad, M.; Iacobuzio-Donahue, C.A.; et al. Unresolved endoplasmic reticulum stress engenders immune-resistant, latent pancreatic cancer metastases. Science 2018, 360. [Google Scholar] [CrossRef] [PubMed]
- Haze, K.; Yoshida, H.; Yanagi, H.; Yura, T.; Mori, K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 1999, 10, 3787–3799. [Google Scholar] [CrossRef]
- Ye, J.; Rawson, R.B.; Komuro, R.; Chen, X.; Dave, U.P.; Prywes, R.; Brown, M.S.; Goldstein, J.L. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 2000, 6, 1355–1364. [Google Scholar] [CrossRef]
- Yoshida, H.; Matsui, T.; Yamamoto, A.; Okada, T.; Mori, K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 2001, 107, 881–891. [Google Scholar] [CrossRef]
- Lee, K.; Tirasophon, W.; Shen, X.; Michalak, M.; Prywes, R.; Okada, T.; Yoshida, H.; Mori, K.; Kaufman, R.J. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002, 16, 452–466. [Google Scholar] [CrossRef]
- Iurlaro, R.; Munoz-Pinedo, C. Cell death induced by endoplasmic reticulum stress. FEBS J. 2016, 283, 2640–2652. [Google Scholar] [CrossRef]
- Li, T.; Su, L.; Lei, Y.; Liu, X.; Zhang, Y.; Liu, X. DDIT3 and KAT2A Proteins Regulate TNFRSF10A and TNFRSF10B Expression in Endoplasmic Reticulum Stress-mediated Apoptosis in Human Lung Cancer Cells. J. Biol. Chem. 2015, 290, 11108–11118. [Google Scholar] [CrossRef]
- Sasaki, K.; Yoshida, H. Golgi stress response and organelle zones. FEBS Lett. 2019, 593, 2330–2340. [Google Scholar] [CrossRef]
- He, Q.; Lee, D.I.; Rong, R.; Yu, M.; Luo, X.; Klein, M.; El-Deiry, W.S.; Huang, Y.; Hussain, A.; Sheikh, M.S. Endoplasmic reticulum calcium pool depletion-induced apoptosis is coupled with activation of the death receptor 5 pathway. Oncogene 2002, 21, 2623–2633. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, H.; Wang, H.G. CHOP is involved in endoplasmic reticulum stress-induced apoptosis by enhancing DR5 expression in human carcinoma cells. J. Biol. Chem. 2004, 279, 45495–45502. [Google Scholar] [CrossRef] [PubMed]
- Cazanave, S.C.; Mott, J.L.; Bronk, S.F.; Werneburg, N.W.; Fingas, C.D.; Meng, X.W.; Finnberg, N.; El-Deiry, W.S.; Kaufmann, S.H.; Gores, G.J. Death receptor 5 signaling promotes hepatocyte lipoapoptosis. J. Biol. Chem. 2011, 286, 39336–39348. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.; Lawrence, D.A.; Ashkenazi, A.; Walter, P. Confirming a critical role for death receptor 5 and caspase-8 in apoptosis induction by endoplasmic reticulum stress. Cell Death Differ. 2018, 25, 1530–1531. [Google Scholar] [CrossRef]
- van Raam, B.J.; Lacina, T.; Lindemann, R.K.; Reiling, J.H. Secretory stressors induce intracellular death receptor accumulation to control apoptosis. Cell Death Dis. 2017, 8, e3069. [Google Scholar] [CrossRef]
- Jiang, C.C.; Chen, L.H.; Gillespie, S.; Kiejda, K.A.; Mhaidat, N.; Wang, Y.F.; Thorne, R.; Zhang, X.D.; Hersey, P. Tunicamycin sensitizes human melanoma cells to tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by up-regulation of TRAIL-R2 via the unfolded protein response. Cancer Res. 2007, 67, 5880–5888. [Google Scholar] [CrossRef]
- Iurlaro, R.; Puschel, F.; Leon-Annicchiarico, C.L.; O’Connor, H.; Martin, S.J.; Palou-Gramon, D.; Lucendo, E.; Munoz-Pinedo, C. Glucose Deprivation Induces ATF4-Mediated Apoptosis through TRAIL Death Receptors. Mol. Cell. Biol. 2017, 37. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Bhalla, K.; Wang, H.G. Bax plays a pivotal role in thapsigargin-induced apoptosis of human colon cancer HCT116 cells by controlling Smac/Diablo and Omi/HtrA2 release from mitochondria. Cancer Res. 2003, 63, 1483–1489. [Google Scholar]
- Cano-Gonzalez, A.; Mauro-Lizcano, M.; Iglesias-Serret, D.; Gil, J.; Lopez-Rivas, A. Involvement of both caspase-8 and Noxa-activated pathways in endoplasmic reticulum stress-induced apoptosis in triple-negative breast tumor cells. Cell Death Dis. 2018, 9, 134. [Google Scholar] [CrossRef]
- Pan, L.; Fu, T.M.; Zhao, W.; Zhao, L.; Chen, W.; Qiu, C.; Liu, W.; Liu, Z.; Piai, A.; Fu, Q.; et al. Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling. Cell 2019, 176, 1477–1489.e14. [Google Scholar] [CrossRef]
- Wang, M.; Law, M.E.; Davis, B.J.; Yaaghubi, E.; Ghilardi, A.F.; Ferreira, R.B.; Chiang, C.W.; Guryanova, O.A.; Kopinke, D.; Heldermon, C.D.; et al. Disulfide bond-disrupting agents activate the tumor necrosis family-related apoptosis-inducing ligand/death receptor 5 pathway. Cell Death Discov. 2019, 5, 153. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.; Marsters, S.A.; Ashkenazi, A.; Walter, P. Misfolded proteins bind and activate death receptor 5 to induce apoptosis during unresolved endoplasmic reticulum stress. Elife 2020, 9. [Google Scholar] [CrossRef]
- Clancy, L.; Mruk, K.; Archer, K.; Woelfel, M.; Mongkolsapaya, J.; Screaton, G.; Lenardo, M.J.; Chan, F.K. Preligand assembly domain-mediated ligand-independent association between TRAIL receptor 4 (TR4) and TR2 regulates TRAIL-induced apoptosis. Proc. Natl. Acad. Sci. USA 2005, 102, 18099–18104. [Google Scholar] [CrossRef] [PubMed]
- Smulski, C.R.; Decossas, M.; Chekkat, N.; Beyrath, J.; Willen, L.; Guichard, G.; Lorenzetti, R.; Rizzi, M.; Eibel, H.; Schneider, P.; et al. Hetero-oligomerization between the TNF receptor superfamily members CD40, Fas and TRAILR2 modulate CD40 signalling. Cell Death Dis. 2017, 8, e2601. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Lawrence, D.A.; Marsters, S.; Acosta-Alvear, D.; Kimmig, P.; Mendez, A.S.; Paton, A.W.; Paton, J.C.; Walter, P.; Ashkenazi, A. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science 2014, 345, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.H.; Li, H.; Yasumura, D.; Cohen, H.R.; Zhang, C.; Panning, B.; Shokat, K.M.; Lavail, M.M.; Walter, P. IRE1 signaling affects cell fate during the unfolded protein response. Science 2007, 318, 944–949. [Google Scholar] [CrossRef]
- Chang, T.K.; Lawrence, D.A.; Lu, M.; Tan, J.; Harnoss, J.M.; Marsters, S.A.; Liu, P.; Sandoval, W.; Martin, S.E.; Ashkenazi, A. Coordination between Two Branches of the Unfolded Protein Response Determines Apoptotic Cell Fate. Mol. Cell 2018, 71, 629–636.e5. [Google Scholar] [CrossRef]
- Sullivan, G.P.; O’Connor, H.; Henry, C.M.; Davidovich, P.; Clancy, D.M.; Albert, M.L.; Cullen, S.P.; Martin, S.J. TRAIL Receptors Serve as Stress-Associated Molecular Patterns to Promote ER-Stress-Induced Inflammation. Dev. Cell 2020, 52, 714–730.e5. [Google Scholar] [CrossRef]
- Püschel, F.; Favaro, F.; Redondo-Pedraza, J.; Lucendo, E.; Iurlaro, R.; Marchetti, S.; Majem, B.; Eldering, E.; Nadal, E.; Ricci, J.-E.; et al. Starvation and antimetabolic therapy promote cytokine release and recruitment of immune cells. Proc. Natl. Acad. Sci. USA 2020. [Google Scholar] [CrossRef]
- Teng, Y.; Gao, M.; Wang, J.; Kong, Q.; Hua, H.; Luo, T.; Jiang, Y. Inhibition of eIF2alpha dephosphorylation enhances TRAIL-induced apoptosis in hepatoma cells. Cell Death Dis. 2014, 5, e1060. [Google Scholar] [CrossRef]
- Chen, L.H.; Jiang, C.C.; Kiejda, K.A.; Wang, Y.F.; Thorne, R.F.; Zhang, X.D.; Hersey, P. Thapsigargin sensitizes human melanoma cells to TRAIL-induced apoptosis by up-regulation of TRAIL-R2 through the unfolded protein response. Carcinogenesis 2007, 28, 2328–2336. [Google Scholar] [CrossRef]
- Shiraishi, T.; Yoshida, T.; Nakata, S.; Horinaka, M.; Wakada, M.; Mizutani, Y.; Miki, T.; Sakai, T. Tunicamycin enhances tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis in human prostate cancer cells. Cancer Res. 2005, 65, 6364–6370. [Google Scholar] [CrossRef] [PubMed]
- Estornes, Y.; Dondelinger, Y.; Weber, K.; Bruggeman, I.; Peall, A.; MacFarlane, M.; Lebecque, S.; Vandenabeele, P.; Bertrand, M.J.M. N-glycosylation of mouse TRAIL-R restrains TRAIL-induced apoptosis. Cell Death Dis. 2018, 9, 494. [Google Scholar] [CrossRef] [PubMed]
- Dufour, F.; Rattier, T.; Shirley, S.; Picarda, G.; Constantinescu, A.A.; Morle, A.; Zakaria, A.B.; Marcion, G.; Causse, S.; Szegezdi, E.; et al. N-glycosylation of mouse TRAIL-R and human TRAIL-R1 enhances TRAIL-induced death. Cell Death Differ. 2017, 24, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Martin-Perez, R.; Niwa, M.; Lopez-Rivas, A. ER stress sensitizes cells to TRAIL through down-regulation of FLIP and Mcl-1 and PERK-dependent up-regulation of TRAIL-R2. Apoptosis Int. J. Program. Cell Death 2012, 17, 349–363. [Google Scholar] [CrossRef]
- Koltai, T. Nelfinavir and other protease inhibitors in cancer: Mechanisms involved in anticancer activity. F1000 Res. 2015, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- De Gassart, A.; Bujisic, B.; Zaffalon, L.; Decosterd, L.A.; Di Micco, A.; Frera, G.; Tallant, R.; Martinon, F. An inhibitor of HIV-1 protease modulates constitutive eIF2alpha dephosphorylation to trigger a specific integrated stress response. Proc. Natl. Acad. Sci. USA 2016, 113, E117–E126. [Google Scholar] [CrossRef]
- Tian, X.; Ye, J.; Alonso-Basanta, M.; Hahn, S.M.; Koumenis, C.; Dorsey, J.F. Modulation of CCAAT/enhancer binding protein homologous protein (CHOP)-dependent DR5 expression by nelfinavir sensitizes glioblastoma multiforme cells to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). J. Biol. Chem. 2011, 286, 29408–29416. [Google Scholar] [CrossRef]
- Song, M.; Sandoval, T.A.; Chae, C.S.; Chopra, S.; Tan, C.; Rutkowski, M.R.; Raundhal, M.; Chaurio, R.A.; Payne, K.K.; Konrad, C.; et al. IRE1alpha-XBP1 controls T cell function in ovarian cancer by regulating mitochondrial activity. Nature 2018, 562, 423–428. [Google Scholar] [CrossRef]
- Dong, H.; Adams, N.M.; Xu, Y.; Cao, J.; Allan, D.S.J.; Carlyle, J.R.; Chen, X.; Sun, J.C.; Glimcher, L.H. The IRE1 endoplasmic reticulum stress sensor activates natural killer cell immunity in part by regulating c-Myc. Nat. Immunol. 2019, 20, 865–878. [Google Scholar] [CrossRef]
- Condamine, T.; Kumar, V.; Ramachandran, I.R.; Youn, J.I.; Celis, E.; Finnberg, N.; El-Deiry, W.S.; Winograd, R.; Vonderheide, R.H.; English, N.R.; et al. ER stress regulates myeloid-derived suppressor cell fate through TRAIL-R-mediated apoptosis. J. Clin. Investig. 2014, 124, 2626–2639. [Google Scholar] [CrossRef] [PubMed]
- Timmins, J.M.; Ozcan, L.; Seimon, T.A.; Li, G.; Malagelada, C.; Backs, J.; Backs, T.; Bassel-Duby, R.; Olson, E.N.; Anderson, M.E.; et al. Calcium/calmodulin-dependent protein kinase II links ER stress with Fas and mitochondrial apoptosis pathways. J. Clin. Investig. 2009, 119, 2925–2941. [Google Scholar] [CrossRef] [PubMed]
- Martinon, F.; Chen, X.; Lee, A.H.; Glimcher, L.H. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat. Immunol. 2010, 11, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Kim, Y.S.; Lin, Y.; Lewis, J.; Neckers, L.; Liu, Z.G. Tumour necrosis factor receptor 1 mediates endoplasmic reticulum stress-induced activation of the MAP kinase JNK. EMBO Rep. 2006, 7, 622–627. [Google Scholar] [CrossRef] [PubMed]
- Estornes, Y.; Aguileta, M.A.; Dubuisson, C.; De Keyser, J.; Goossens, V.; Kersse, K.; Samali, A.; Vandenabeele, P.; Bertrand, M.J. RIPK1 promotes death receptor-independent caspase-8-mediated apoptosis under unresolved ER stress conditions. Cell Death Dis. 2014, 5, e1555. [Google Scholar] [CrossRef] [PubMed]
- Saveljeva, S.; Mc Laughlin, S.L.; Vandenabeele, P.; Samali, A.; Bertrand, M.J. Endoplasmic reticulum stress induces ligand-independent TNFR1-mediated necroptosis in L929 cells. Cell Death Dis. 2015, 6, e1587. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ibi, D.; Taniguchi, K.; Lee, J.; Herrema, H.; Akosman, B.; Mucka, P.; Salazar Hernandez, M.A.; Uyar, M.F.; Park, S.W.; et al. Inflammation Improves Glucose Homeostasis through IKKbeta-XBP1s Interaction. Cell 2016, 167, 1052–1066.e18. [Google Scholar] [CrossRef]
- Yousaf, N.; Gould, D.J.; Aganna, E.; Hammond, L.; Mirakian, R.M.; Turner, M.D.; Hitman, G.A.; McDermott, M.F.; Chernajovsky, Y. Tumor necrosis factor receptor I from patients with tumor necrosis factor receptor-associated periodic syndrome interacts with wild-type tumor necrosis factor receptor I and induces ligand-independent NF-kappaB activation. Arthritis Rheum. 2005, 52, 2906–2916. [Google Scholar] [CrossRef]
- Lobito, A.A.; Kimberley, F.C.; Muppidi, J.R.; Komarow, H.; Jackson, A.J.; Hull, K.M.; Kastner, D.L.; Screaton, G.R.; Siegel, R.M. Abnormal disulfide-linked oligomerization results in ER retention and altered signaling by TNFR1 mutants in TNFR1-associated periodic fever syndrome (TRAPS). Blood 2006, 108, 1320–1327. [Google Scholar] [CrossRef]
- Greco, E.; Aita, A.; Galozzi, P.; Gava, A.; Sfriso, P.; Negm, O.H.; Tighe, P.; Caso, F.; Navaglia, F.; Dazzo, E.; et al. The novel S59P mutation in the TNFRSF1A gene identified in an adult onset TNF receptor associated periodic syndrome (TRAPS) constitutively activates NF-κB pathway. Arthritis Res. Ther. 2015, 17, 93. [Google Scholar] [CrossRef]
- Nozaki, S.; Sledge, G.W., Jr.; Nakshatri, H. Repression of GADD153/CHOP by NF-kappaB: A possible cellular defense against endoplasmic reticulum stress-induced cell death. Oncogene 2001, 20, 2178–2185. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Warnakulasuriyarachchi, D.; Cerquozzi, S.; Cheung, H.H.; Holcik, M. Translational induction of the inhibitor of apoptosis protein HIAP2 during endoplasmic reticulum stress attenuates cell death and is mediated via an inducible internal ribosome entry site element. J. Biol. Chem. 2004, 279, 17148–17157. [Google Scholar] [CrossRef] [PubMed]
- Hamanaka, R.B.; Bobrovnikova-Marjon, E.; Ji, X.; Liebhaber, S.A.; Diehl, J.A. PERK-dependent regulation of IAP translation during ER stress. Oncogene 2009, 28, 910–920. [Google Scholar] [CrossRef]
- Hu, P.; Han, Z.; Couvillon, A.D.; Exton, J.H. Critical role of endogenous Akt/IAPs and MEK1/ERK pathways in counteracting endoplasmic reticulum stress-induced cell death. J. Biol. Chem. 2004, 279, 49420–49429. [Google Scholar] [CrossRef] [PubMed]
- Abhari, B.A.; McCarthy, N.; Le Berre, M.; Kilcoyne, M.; Joshi, L.; Agostinis, P.; Fulda, S. Smac mimetic suppresses tunicamycin-induced apoptosis via resolution of ER stress. Cell Death Dis. 2019, 10, 155. [Google Scholar] [CrossRef]
- Hetz, C.; Bernasconi, P.; Fisher, J.; Lee, A.H.; Bassik, M.C.; Antonsson, B.; Brandt, G.S.; Iwakoshi, N.N.; Schinzel, A.; Glimcher, L.H.; et al. Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science 2006, 312, 572–576. [Google Scholar] [CrossRef]
- Shemorry, A.; Harnoss, J.M.; Guttman, O.; Marsters, S.A.; Komuves, L.G.; Lawrence, D.A.; Ashkenazi, A. Caspase-mediated cleavage of IRE1 controls apoptotic cell commitment during endoplasmic reticulum stress. Elife 2019, 8. [Google Scholar] [CrossRef]
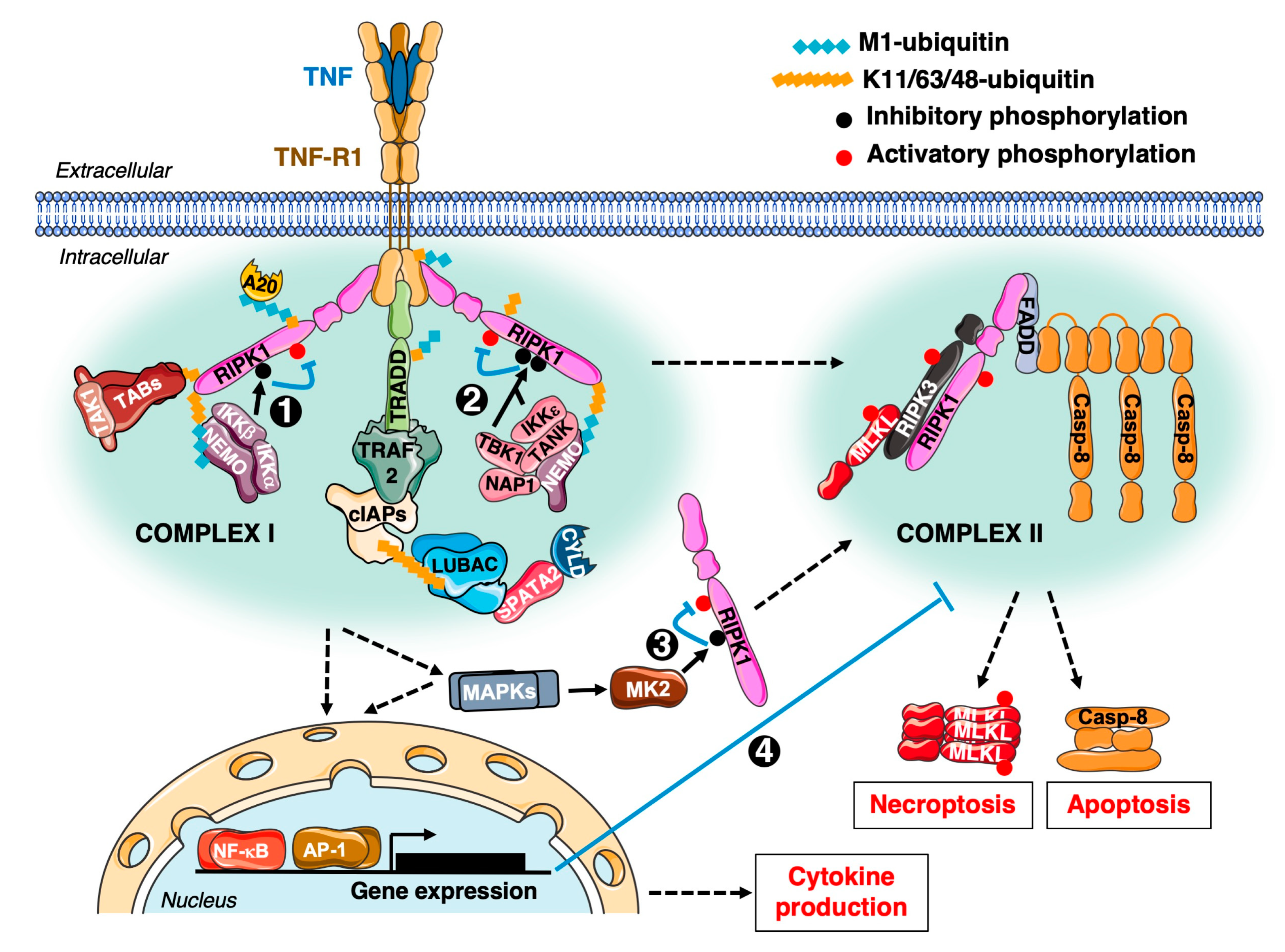
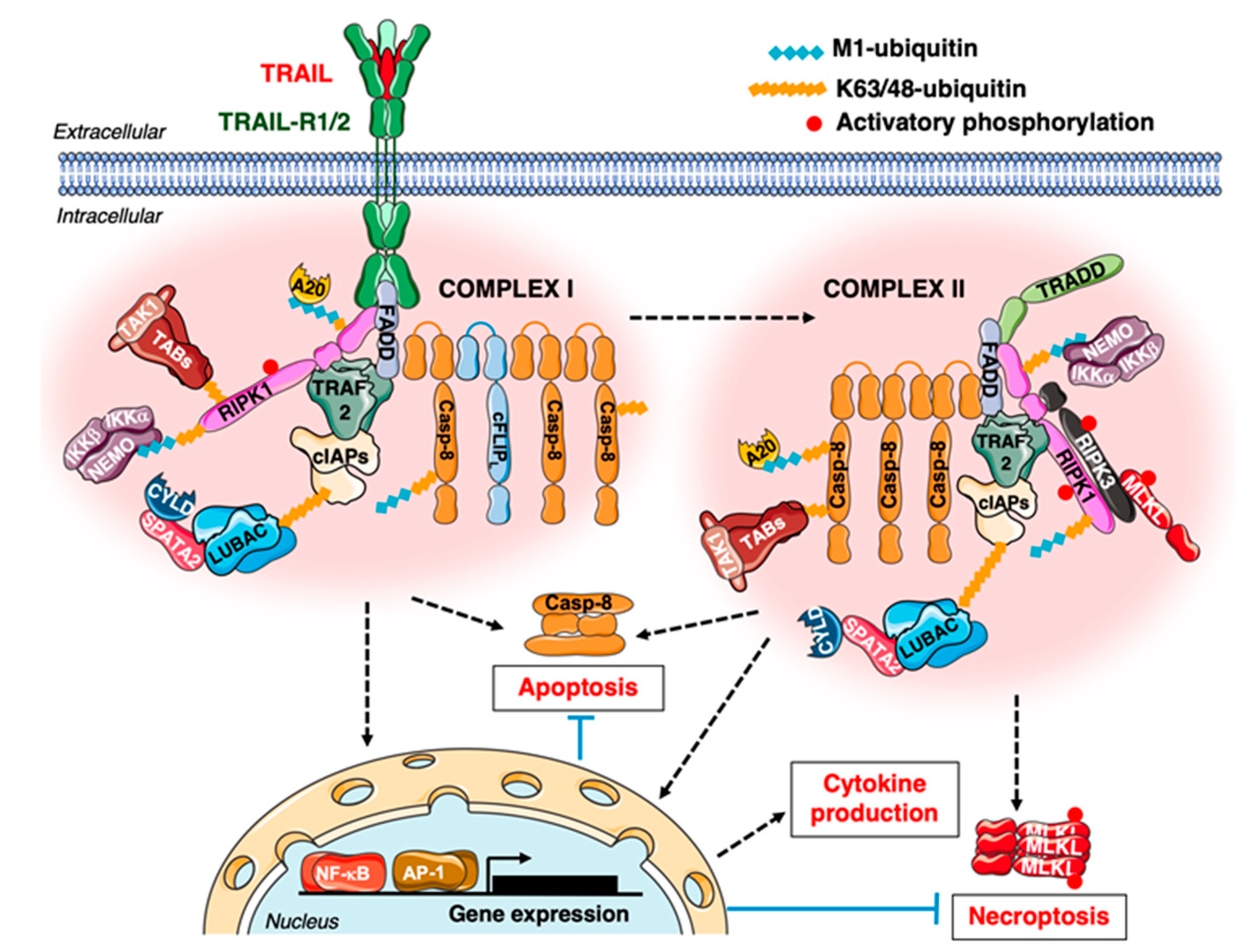
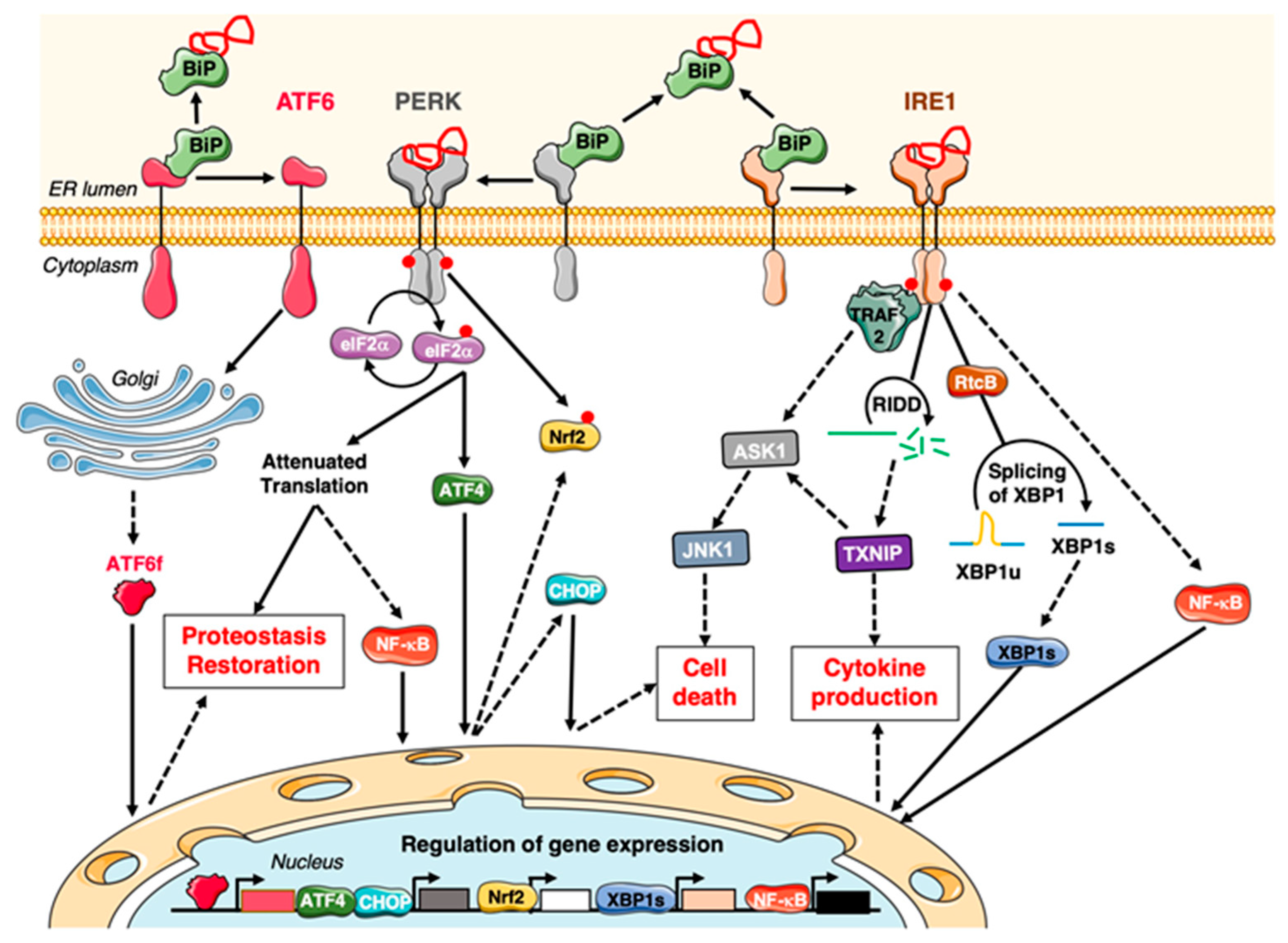
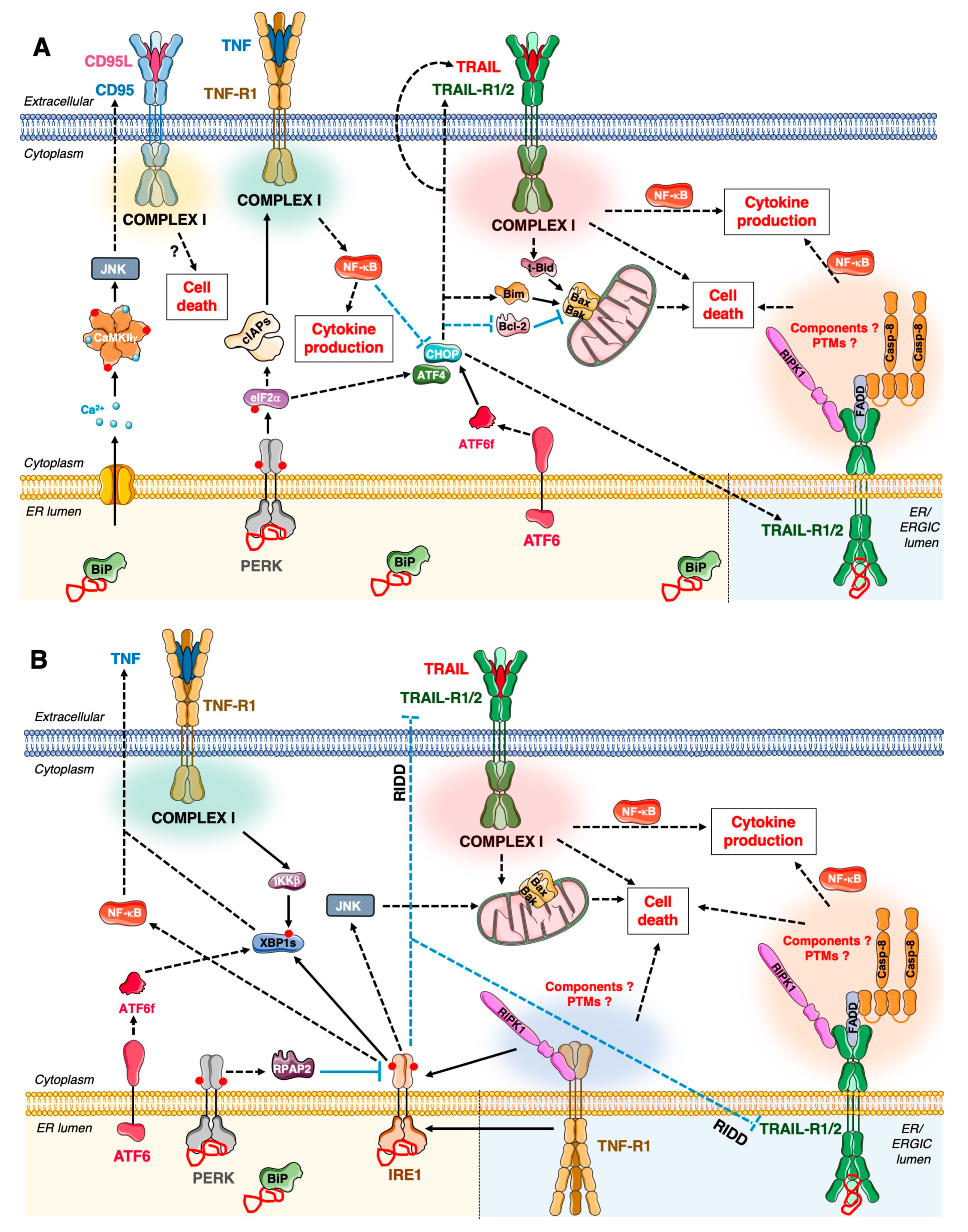
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lafont, E. Stress Management: Death Receptor Signalling and Cross-Talks with the Unfolded Protein Response in Cancer. Cancers 2020, 12, 1113. https://doi.org/10.3390/cancers12051113
Lafont E. Stress Management: Death Receptor Signalling and Cross-Talks with the Unfolded Protein Response in Cancer. Cancers. 2020; 12(5):1113. https://doi.org/10.3390/cancers12051113
Chicago/Turabian StyleLafont, Elodie. 2020. "Stress Management: Death Receptor Signalling and Cross-Talks with the Unfolded Protein Response in Cancer" Cancers 12, no. 5: 1113. https://doi.org/10.3390/cancers12051113
APA StyleLafont, E. (2020). Stress Management: Death Receptor Signalling and Cross-Talks with the Unfolded Protein Response in Cancer. Cancers, 12(5), 1113. https://doi.org/10.3390/cancers12051113




