Role of CYP3A5 in Modulating Androgen Receptor Signaling and Its Relevance to African American Men with Prostate Cancer
Abstract
1. Introduction
2. Results
2.1. Differential Expression of CYP3A5 between African American and Non-Hispanic White Americans Origin AR Positive Prostate Cancer Cell Lines
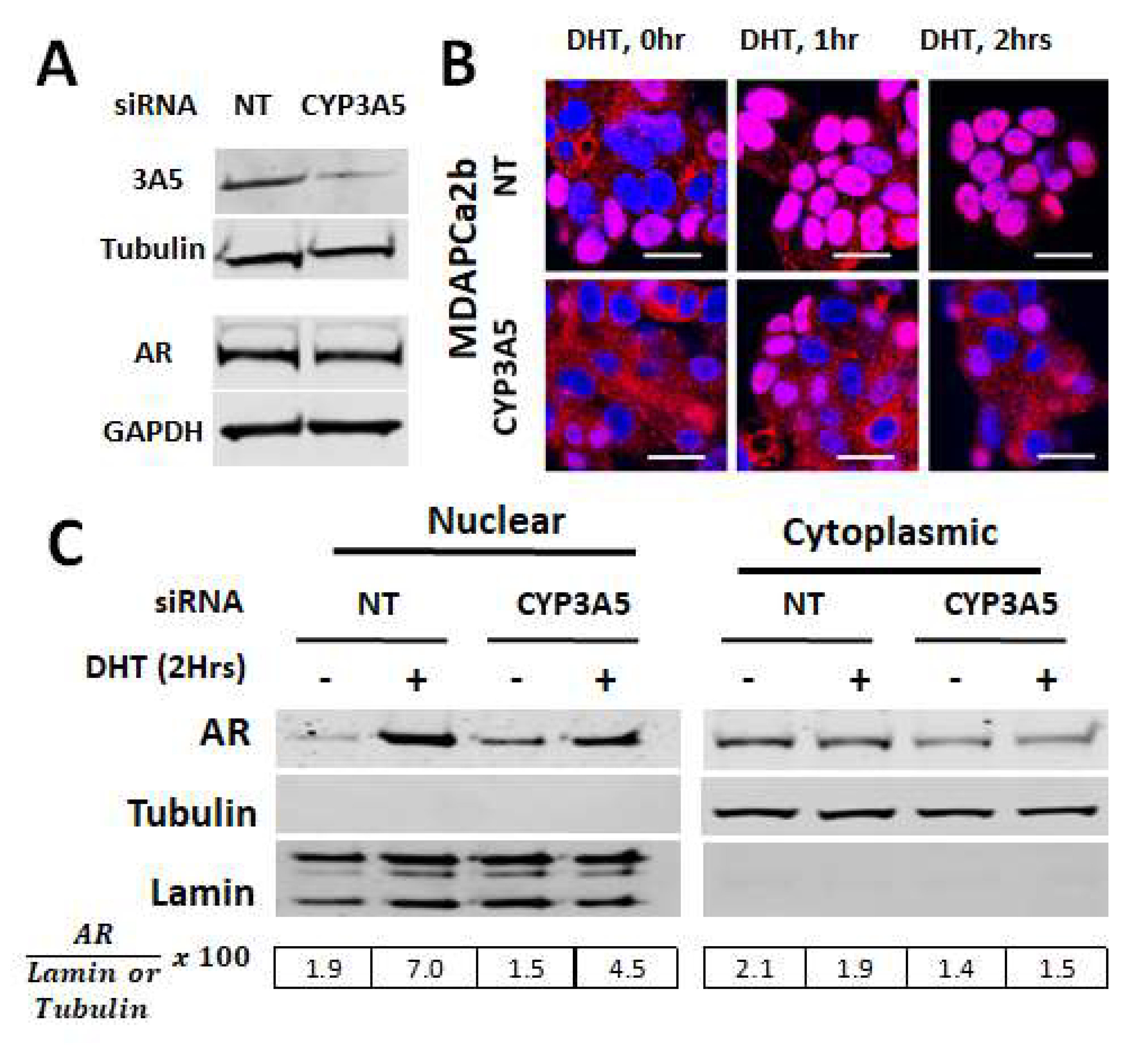
2.2. CYP3A5siRNA Downregulates AR Nuclear Translocation in MDAPCa2b Cells Expressing Wild Type CYP3A5 (*1/*3)
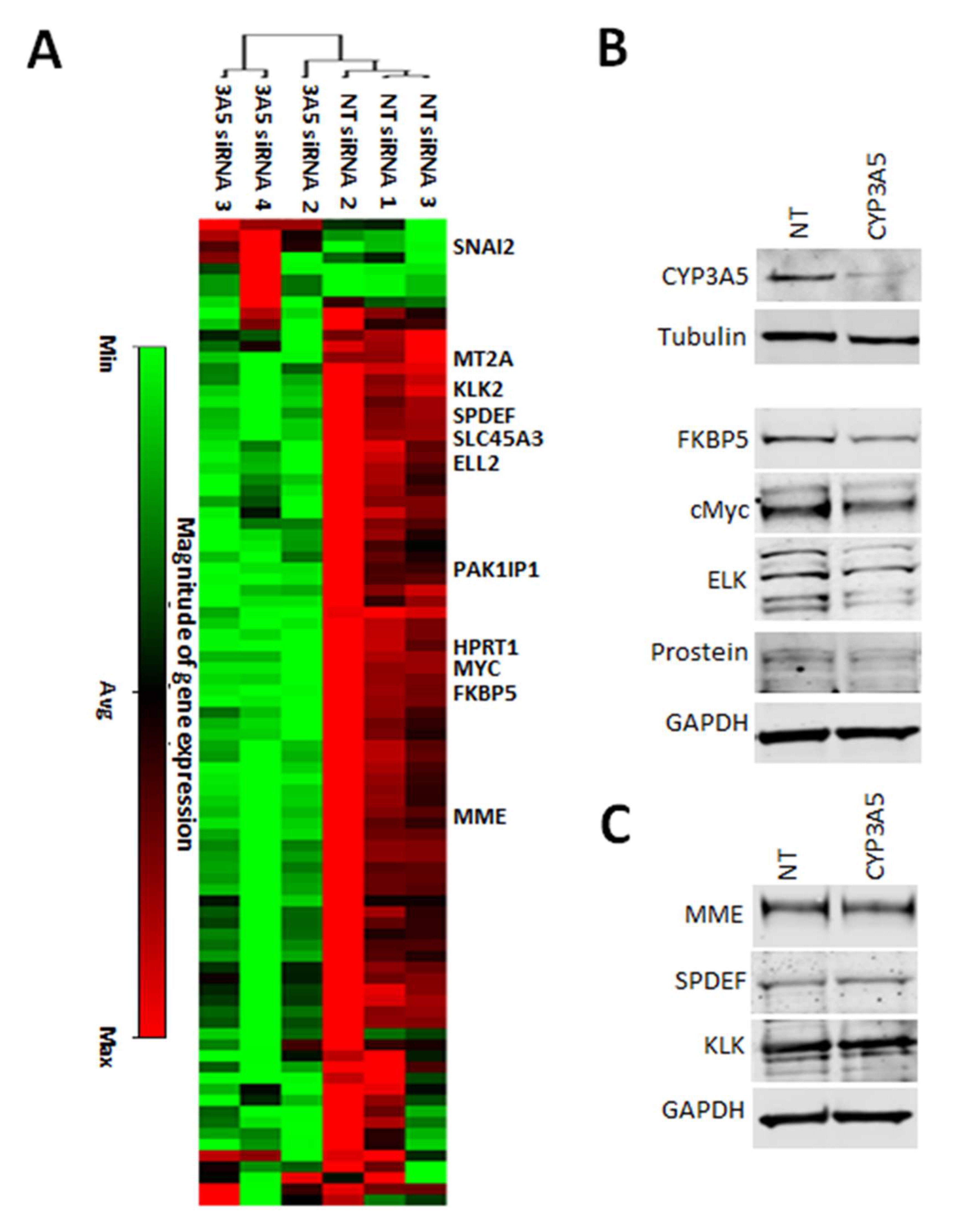
2.3. CYP3A5 siRNA Downregulates Expression of AR Regulated Genes in MDAPCA2b Cells
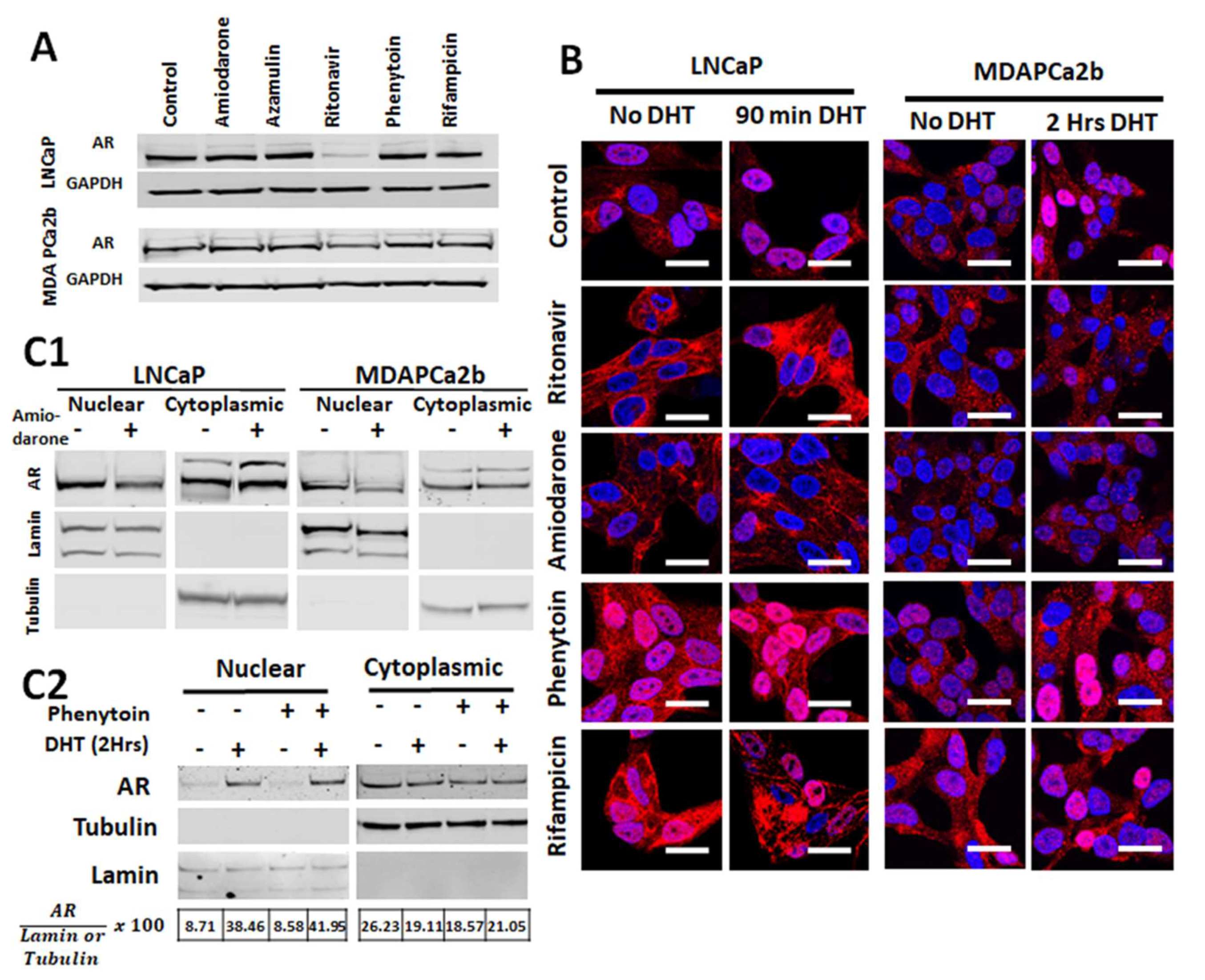
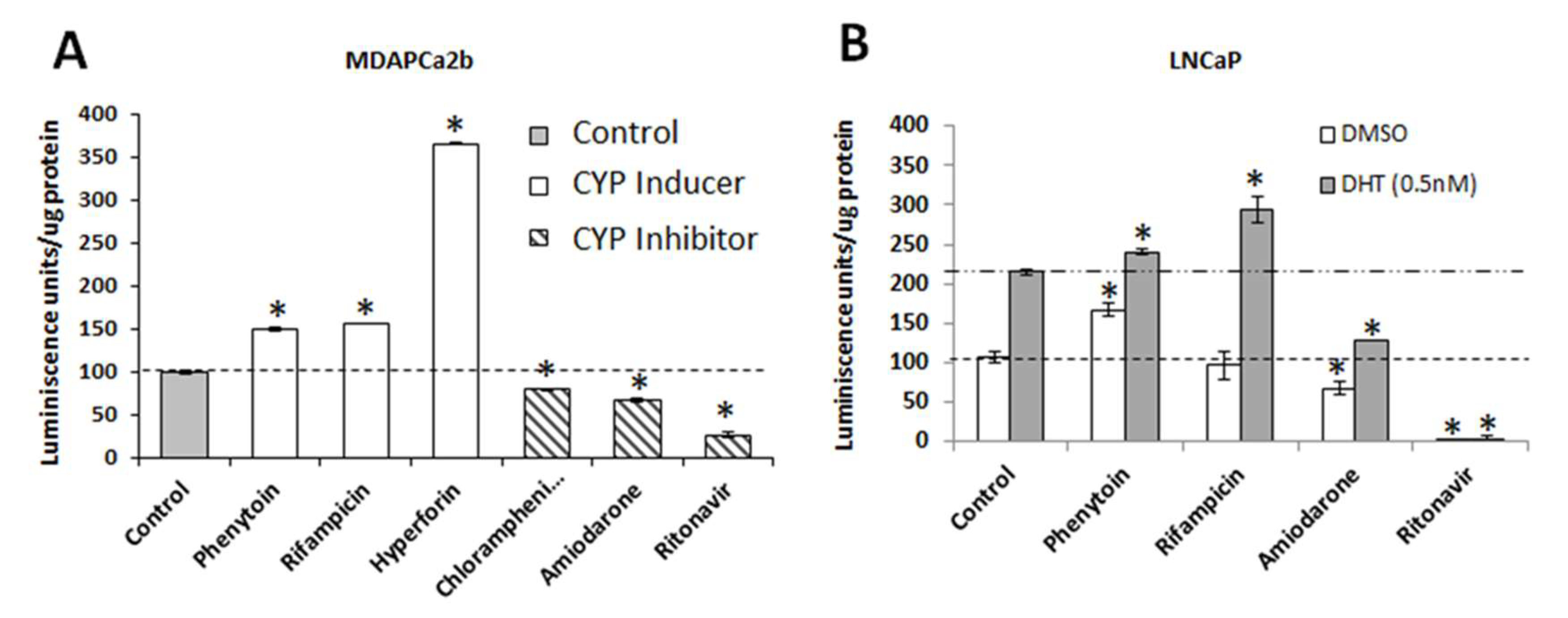
2.4. Commonly Co-Prescribed CYP3A5 Inducers/Inhibitors can Alter AR Nuclear Translocation
2.5. Changes in AR Activation by CYP3A Inducers are due to Their Effect on CYP3A5 Activity
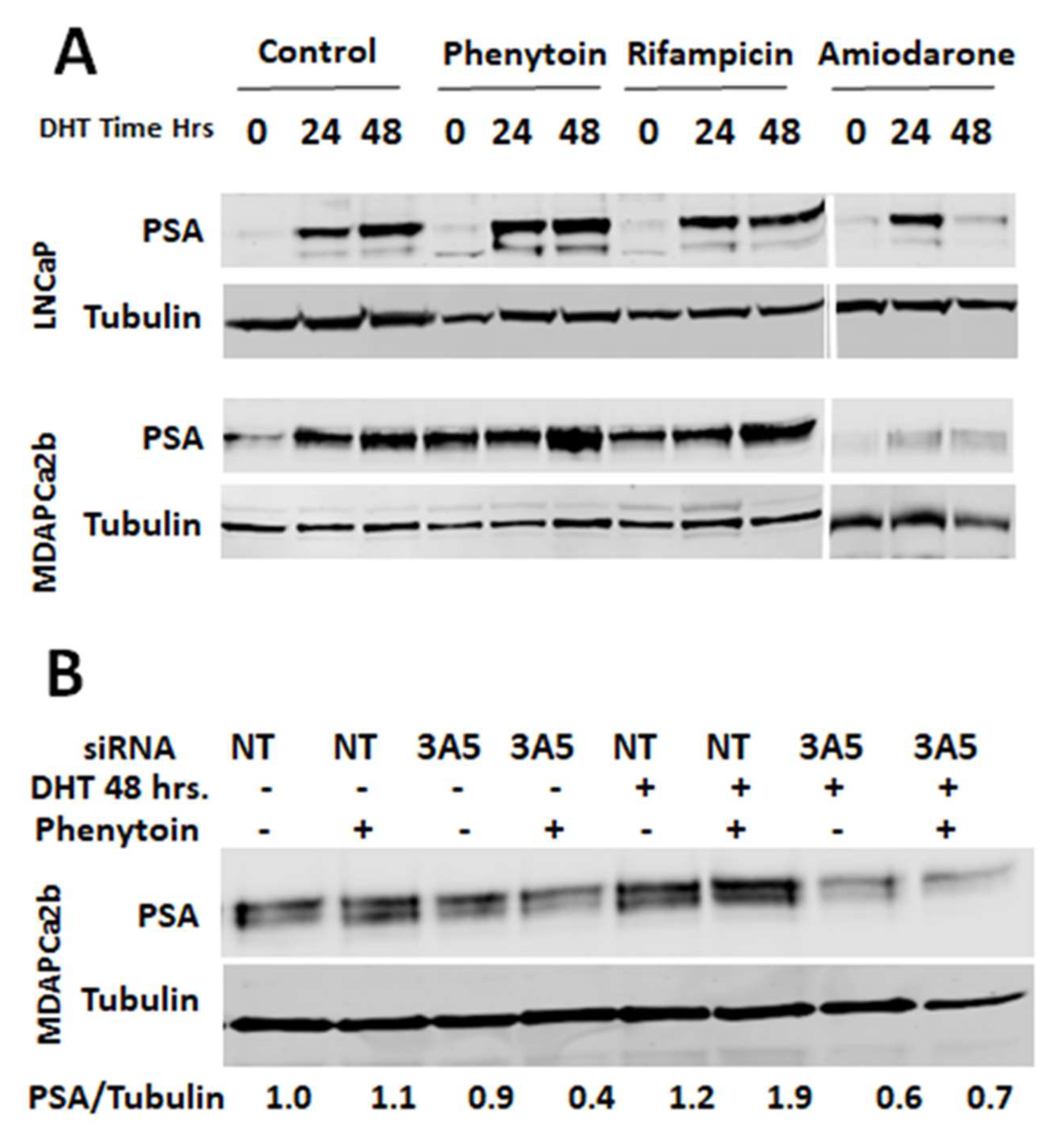
2.6. CYP3A5 Inhibitors and Inducers Alter PSA Levels
2.7. CYP3A5 Modulating Drugs Affect AR Downstream Signaling
2.8. CYP3A can Regulate PCa Cell Growth by Modifying AR Activation
3. Discussion
4. Materials and Methods
4.1. Cell Lines, Drugs, and Antibodies
4.2. Western Blotting
4.3. Cell Fractionation
4.4. siRNA Inhibition
4.5. Confocal Microscopy
4.6. qPCR and RT2 Profiler Assays
4.7. Luciferase Assay
4.8. Genotyping Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Huggins, C.; Hodges, C.V. Studies on prostatic cancer. I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. J. Urol. 2002, 167, 948–951. [Google Scholar] [CrossRef]
- Sadi, M.V.; Walsh, P.C.; Barrack, E.R. Immunohistochemical study of androgen receptors in metastatic prostate cancer. Comparison of receptor content and response to hormonal therapy. Cancer 1991, 67, 3057–3064. [Google Scholar] [CrossRef]
- van der Kwast, T.H.; Schalken, J.; Ruizeveld de Winter, J.A.; van Vroonhoven, C.C.; Mulder, E.; Boersma, W.; Trapman, J. Androgen receptors in endocrine-therapy-resistant human prostate cancer. Int. J. Cancer 1991, 48, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Chodak, G.W.; Kranc, D.M.; Puy, L.A.; Takeda, H.; Johnson, K.; Chang, C. Nuclear localization of androgen receptor in heterogeneous samples of normal, hyperplastic and neoplastic human prostate. J. Urol. 1992, 147, 798–803. [Google Scholar] [CrossRef]
- Heinlein, C.A.; Chang, C. Androgen Receptor in Prostate Cancer. Endocr. Rev. 2004, 25, 276–308. [Google Scholar] [CrossRef] [PubMed]
- de Bono, J.S.; Logothetis, C.J.; Molina, A.; Fizazi, K.; North, S.; Chu, L.; Chi, K.N.; Jones, R.J.; Goodman, O.B., Jr.; Saad, F.; et al. Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 2011, 364, 1995–2005. [Google Scholar] [CrossRef] [PubMed]
- Bianchini, D.; Lorente, D.; Rodriguez-Vida, A.; Omlin, A.; Pezaro, C.; Ferraldeschi, R.; Zivi, A.; Attard, G.; Chowdhury, S.; de Bono, J.S. Antitumour activity of enzalutamide (MDV3100) in patients with metastatic castration-resistant prostate cancer (CRPC) pre-treated with docetaxel and abiraterone. Eur. J. Cancer 2014, 50, 78–84. [Google Scholar] [CrossRef]
- Darshan, M.S.; Loftus, M.S.; Thadani-Mulero, M.; Levy, B.P.; Escuin, D.; Zhou, X.K.; Gjyrezi, A.; Chanel-Vos, C.; Shen, R.; Tagawa, S.T.; et al. Taxane-induced blockade to nuclear accumulation of the androgen receptor predicts clinical responses in metastatic prostate cancer. Cancer Res. 2011, 71, 6019–6029. [Google Scholar] [CrossRef]
- Mezynski, J.; Pezaro, C.; Bianchini, D.; Zivi, A.; Sandhu, S.; Thompson, E.; Hunt, J.; Sheridan, E.; Baikady, B.; Sarvadikar, A.; et al. Antitumour activity of docetaxel following treatment with the CYP17A1 inhibitor abiraterone: Clinical evidence for cross-resistance? Ann. Oncol. 2012, 23, 2943–2947. [Google Scholar] [CrossRef]
- Mitra, R.; Goodman, O.B. CYP3A5 regulates prostate cancer cell growth by facilitating nuclear translocation of AR. Prostate 2015, 75, 527–538. [Google Scholar] [CrossRef]
- Zanger, U.M.; Schwab, M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef]
- Yamakoshi, Y.; Kishimoto, T.; Sugimura, K.; Kawashima, H. Human prostate CYP3A5: Identification of a unique 5′-untranslated sequence and characterization of purified recombinant protein. Biochem. Biophys. Res. Commun. 1999, 260, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Hou, G.; Zheng, Y.; Wei, D.; Li, X.; Wang, F.; Tian, J.; Zhang, G.; Yan, F.; Zhu, Z.; Meng, P.; et al. Development and validation of a SEER-based prognostic nomogram for patients with bone metastatic prostate cancer. Medicine (Baltimore) 2019, 98, e17197. [Google Scholar] [CrossRef]
- Tompkins, L.M.; Wallace, A.D. Mechanisms of cytochrome P450 induction. J. Biochem. Mol. Toxicol. 2007, 21, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Lolodi, O.; Wang, Y.M.; Wright, W.C.; Chen, T. Differential regulation of CYP3A4 and CYP3A5 and its implication in drug discovery. Curr. Drug Metab. 2017, 18, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Nem, D.; Baranyai, D.; Qiu, H.; Godtel-Armbrust, U.; Nestler, S.; Wojnowski, L. Pregnane X receptor and yin yang 1 contribute to the differential tissue expression and induction of CYP3A5 and CYP3A4. PLoS ONE 2012, 7, e30895. [Google Scholar] [CrossRef] [PubMed]
- Finnstrom, N.; Bjelfman, C.; Soderstrom, T.G.; Smith, G.; Egevad, L.; Norlen, B.J.; Wolf, C.R.; Rane, A. Detection of cytochrome P450 mRNA transcripts in prostate samples by RT-PCR. Eur. J. Clin. Investig. 2001, 31, 880–886. [Google Scholar] [CrossRef] [PubMed]
- Leskela, S.; Honrado, E.; Montero-Conde, C.; Landa, I.; Cascon, A.; Leton, R.; Talavera, P.; Cozar, J.M.; Concha, A.; Robledo, M.; et al. Cytochrome P450 3A5 is highly expressed in normal prostate cells but absent in prostate cancer. Endocr. Relat. Cancer 2007, 14, 645–654. [Google Scholar] [CrossRef]
- Moilanen, A.M.; Hakkola, J.; Vaarala, M.H.; Kauppila, S.; Hirvikoski, P.; Vuoristo, J.T.; Edwards, R.J.; Paavonen, T.K. Characterization of androgen-regulated expression of CYP3A5 in human prostate. Carcinogenesis 2007, 28, 916–921. [Google Scholar] [CrossRef]
- Kuehl, P.; Zhang, J.; Lin, Y.; Lamba, J.; Assem, M.; Schuetz, J.; Watkins, P.B.; Daly, A.; Wrighton, S.A.; Hall, S.D.; et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat. Genet. 2001, 27, 383–391. [Google Scholar] [CrossRef]
- Hooker, S.E.; Woods-Burnham, L.; Bathina, M.; Lloyd, S.M.; Gorjala, P.; Mitra, R.; Nonn, L.; Kimbro, K.S.; Kittles, R. Genetic ancestry analysis reveals misclassification of commonly used cancer cell lines. Cancer Epidemiol. Biomark. Prev. 2019. [Google Scholar] [CrossRef] [PubMed]
- Lynch, T.; Price, A. The effect of cytochrome P450 metabolism on drug response, interactions, and adverse effects. Am. Fam. Physician 2007, 76, 391–396. [Google Scholar] [PubMed]
- Schuetz, E.G.; Relling, M.V.; Kishi, S.; Yang, W.; Das, S.; Chen, P.; Cook, E.H.; Rosner, G.L.; Pui, C.H.; Blanco, J.G.; et al. PharmGKB update: II. CYP3A5, cytochrome P450, family 3, subfamily A, polypeptide 5. Pharmacol. Rev. 2004, 56, 159. [Google Scholar] [CrossRef] [PubMed]
- Lamba, J.K.; Lin, Y.S.; Schuetz, E.G.; Thummel, K.E. Genetic contribution to variable human CYP3A-mediated metabolism. Adv. Drug Deliv. Rev. 2002, 54, 1271–1294. [Google Scholar] [CrossRef]
- Lamba, J.; Hebert, J.M.; Schuetz, E.G.; Klein, T.E.; Altman, R.B. PharmGKB summary: Very important pharmacogene information for CYP3A5. Pharm. Genome 2012, 22, 555–558. [Google Scholar] [CrossRef]
- Xu, J.; Kalos, M.; Stolk, J.A.; Zasloff, E.J.; Zhang, X.; Houghton, R.L.; Filho, A.M.; Nolasco, M.; Badaro, R.; Reed, S.G. Identification and characterization of prostein, a novel prostate-specific protein. Cancer Res. 2001, 61, 1563–1568. [Google Scholar]
- Kalos, M.; Askaa, J.; Hylander, B.L.; Repasky, E.A.; Cai, F.; Vedvick, T.; Reed, S.G.; Wright, G.L., Jr.; Fanger, G.R. Prostein expression is highly restricted to normal and malignant prostate tissues. Prostate 2004, 60, 246–256. [Google Scholar] [CrossRef]
- Avellino, R.; Romano, S.; Parasole, R.; Bisogni, R.; Lamberti, A.; Poggi, V.; Venuta, S.; Romano, M.F. Rapamycin stimulates apoptosis of childhood acute lymphoblastic leukemia cells. Blood 2005, 106, 1400–1406. [Google Scholar] [CrossRef]
- Pei, H.; Li, L.; Fridley, B.L.; Jenkins, G.D.; Kalari, K.R.; Lingle, W.; Petersen, G.; Lou, Z.; Wang, L. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell 2009, 16, 259–266. [Google Scholar] [CrossRef]
- Bolton, E.C.; So, A.Y.; Chaivorapol, C.; Haqq, C.M.; Li, H.; Yamamoto, K.R. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev. 2007, 21, 2005–2017. [Google Scholar] [CrossRef]
- Ni, L.; Yang, C.S.; Gioeli, D.; Frierson, H.; Toft, D.O.; Paschal, B.M. FKBP51 promotes assembly of the Hsp90 chaperone complex and regulates androgen receptor signaling in prostate cancer cells. Mol. Cell. Biol. 2010, 30, 1243–1253. [Google Scholar] [CrossRef] [PubMed]
- Grad, J.M.; Dai, J.L.; Wu, S.; Burnstein, K.L. Multiple androgen response elements and a Myc consensus site in the androgen receptor (AR) coding region are involved in androgen-mediated up-regulation of AR messenger RNA. Mol. Endocrinol. 1999, 13, 1896–1911. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.M.; Bieberich, C.J.; Dang, C.V.; Nelson, W.G.; Yegnasubramanian, S.; De Marzo, A.M. MYC and prostate cancer. Genes Cancer 2010, 1, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Tomlins, S.A.; Mehra, R.; Rhodes, D.R.; Cao, X.; Wang, L.; Dhanasekaran, S.M.; Kalyana-Sundaram, S.; Wei, J.T.; Rubin, M.A.; Pienta, K.J.; et al. Integrative molecular concept modeling of prostate cancer progression. Nat. Genet. 2007, 39, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Cao, S.; Jin, L.; Kobelski, M.; Schouest, B.; Wang, X.; Ungerleider, N.; Baddoo, M.; Zhang, W.; Corey, E.; et al. A positive role of c-Myc in regulating androgen receptor and its splice variants in prostate cancer. Oncogene 2019. [Google Scholar] [CrossRef]
- Gao, L.; Schwartzman, J.; Gibbs, A.; Lisac, R.; Kleinschmidt, R.; Wilmot, B.; Bottomly, D.; Coleman, I.; Nelson, P.; McWeeney, S.; et al. Androgen receptor promotes ligand-independent prostate cancer progression through c-Myc upregulation. PLoS ONE 2013, 8, e63563. [Google Scholar] [CrossRef]
- Qiu, X.; Pascal, L.E.; Song, Q.; Zang, Y.; Ai, J.; O’Malley, K.J.; Nelson, J.B.; Wang, Z. Physical and functional Interactions between ELL2 and RB in the suppression of prostate cancer cell proliferation, migration, and invasion. Neoplasia 2017, 19, 207–215. [Google Scholar] [CrossRef]
- Sims, R.J., III; Belotserkovskaya, R.; Reinberg, D. Elongation by RNA polymerase II: The short and long of it. Genes Dev. 2004, 18, 2437–2468. [Google Scholar] [CrossRef]
- Hieronymus, H.; Lamb, J.; Ross, K.N.; Peng, X.P.; Clement, C.; Rodina, A.; Nieto, M.; Du, J.; Stegmaier, K.; Raj, S.M.; et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell 2006, 10, 321–330. [Google Scholar] [CrossRef]
- Pascal, L.E.; Masoodi, K.Z.; Liu, J.; Qiu, X.; Song, Q.; Wang, Y.; Zang, Y.; Yang, T.; Wang, Y.; Rigatti, L.H.; et al. Conditional deletion of ELL2 induces murine prostate intraepithelial neoplasia. J. Endocrinol. 2017, 235, 123–136. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, B.; Wu, W.; Li, L.; Broom, B.M.; Basourakos, S.P.; Korentzelos, D.; Luan, Y.; Wang, J.; Yang, G.; et al. Targeting the MYCN-PARP-DNA damage response pathway in neuroendocrine prostate cancer. Clin. Cancer Res. 2018, 24, 696–707. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Mikolajczyk, S.D.; Goel, A.S.; Millar, L.S.; Saedi, M.S. Expression of pro form of prostate-specific antigen by mammalian cells and its conversion to mature, active form by human kallikrein 2. Cancer Res. 1997, 57, 3111–3114. [Google Scholar] [PubMed]
- Haese, A.; Graefen, M.; Becker, C.; Noldus, J.; Katz, J.; Cagiannos, I.; Kattan, M.; Scardino, P.T.; Huland, E.; Huland, H.; et al. The role of human glandular kallikrein 2 for prediction of pathologically organ confined prostate cancer. Prostate 2003, 54, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Stephan, C.; Jung, K.; Nakamura, T.; Yousef, G.M.; Kristiansen, G.; Diamandis, E.P. Serum human glandular kallikrein 2 (hK2) for distinguishing stage and grade of prostate cancer. Int. J. Urol. 2006, 13, 238–243. [Google Scholar] [CrossRef]
- Steuber, T.; Vickers, A.J.; Serio, A.M.; Vaisanen, V.; Haese, A.; Pettersson, K.; Eastham, J.A.; Scardino, P.T.; Huland, H.; Lilja, H. Comparison of free and total forms of serum human kallikrein 2 and prostate-specific antigen for prediction of locally advanced and recurrent prostate cancer. Clin. Chem. 2007, 53, 233–240. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shang, Z.; Niu, Y.; Cai, Q.; Chen, J.; Tian, J.; Yeh, S.; Lai, K.P.; Chang, C. Human kallikrein 2 (KLK2) promotes prostate cancer cell growth via function as a modulator to promote the ARA70-enhanced androgen receptor transactivation. Tumor Biol. 2014, 35, 1881–1890. [Google Scholar] [CrossRef] [PubMed]
- Theodore, S.; Sharp, S.; Zhou, J.; Turner, T.; Li, H.; Miki, J.; Ji, Y.; Patel, V.; Yates, C.; Rhim, J.S. Establishment and characterization of a pair of non-malignant and malignant tumor derived cell lines from an African American prostate cancer patient. Int. J. Oncol. 2010, 37, 1477–1482. [Google Scholar] [CrossRef][Green Version]


| Cell Line | Genotype | Origin |
|---|---|---|
| LNCaP | *3/*3 | NHWA |
| 22RV1 | *3/*3 | NHWA |
| C4-2 | *3/*3 | NHWA |
| E006AAhT | *3/*3 | NHWA |
| MDAPCa2b | *1/*3 | AA |
| RC77 T/E Tumor | *1/*3 | AA |
| RC77 N/E Normal | *1/*3 | AA |
| Gene Symbol | Fold Regulation | p Value |
|---|---|---|
| SLC45A3 | −4.56 | 0.002 |
| FKBP5 | −4.43 | 0.002 |
| MYC | −3.68 | 0.001 |
| MME | −3.34 | 0.016 |
| PAK1IP1 | −3.25 | 0.016 |
| ELL2 | −3.25 | 0.004 |
| KLK2 | −2.82 | 0.009 |
| HPRT1 | −2.65 | 0.005 |
| SPDEF | −2.58 | 0.012 |
| MT2A | −2.45 | 0.001 |
| SNAI2 | 3.32 | 0.005 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gorjala, P.; Kittles, R.A.; Goodman Jr., O.B.; Mitra, R. Role of CYP3A5 in Modulating Androgen Receptor Signaling and Its Relevance to African American Men with Prostate Cancer. Cancers 2020, 12, 989. https://doi.org/10.3390/cancers12040989
Gorjala P, Kittles RA, Goodman Jr. OB, Mitra R. Role of CYP3A5 in Modulating Androgen Receptor Signaling and Its Relevance to African American Men with Prostate Cancer. Cancers. 2020; 12(4):989. https://doi.org/10.3390/cancers12040989
Chicago/Turabian StyleGorjala, Priyatham, Rick A. Kittles, Oscar B. Goodman Jr., and Ranjana Mitra. 2020. "Role of CYP3A5 in Modulating Androgen Receptor Signaling and Its Relevance to African American Men with Prostate Cancer" Cancers 12, no. 4: 989. https://doi.org/10.3390/cancers12040989
APA StyleGorjala, P., Kittles, R. A., Goodman Jr., O. B., & Mitra, R. (2020). Role of CYP3A5 in Modulating Androgen Receptor Signaling and Its Relevance to African American Men with Prostate Cancer. Cancers, 12(4), 989. https://doi.org/10.3390/cancers12040989




