An Updated Perspective on Current Prognostic and Predictive Biomarkers in Chronic Lymphocytic Leukemia in the Context of Chemoimmunotherapy and Novel Targeted Therapy
Abstract
1. Introduction
2. Today’s Most Important Prognostic Biomarkers
3. Discussing the Role of These Prognosticators as Predictive Biomarkers
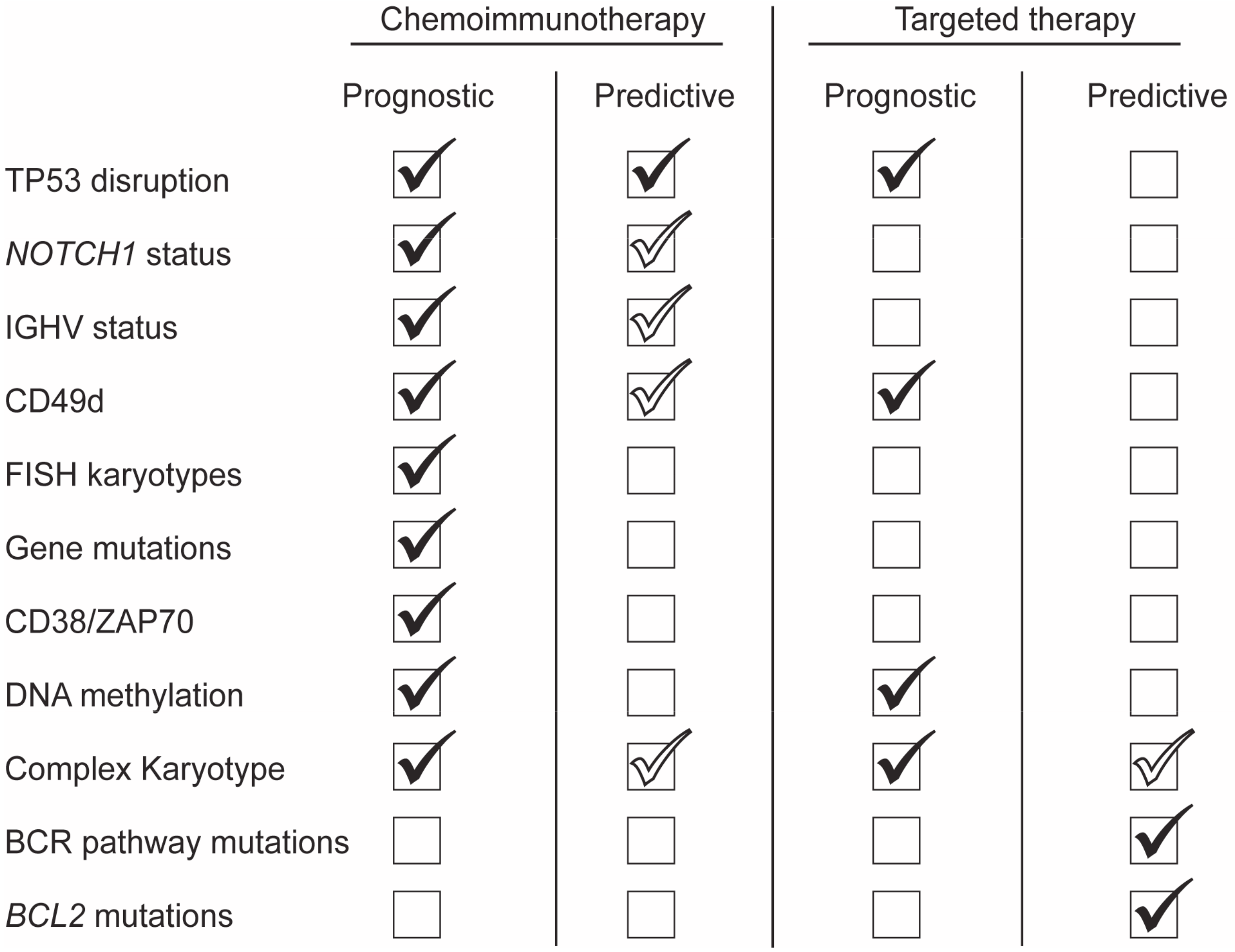
3.1. TP53 Disruption
3.2. Immunoglobulin Mutational Status
3.3. NOTCH1 Gene Mutation
3.4. CD49d
3.5. DNA Methylation
3.6. Complex Karyotype
4. Other Predictive Biomarkers in the Chemo-Free Era
4.1. BCR Pathway Mutations
4.2. BCL2 Mutations
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- An international prognostic index for patients with chronic lymphocytic leukaemia (CLL-IPI): A meta-analysis of individual patient data. Lancet Oncol. 2016, 17, 779–790. [CrossRef]
- Bulian, P.; Rossi, D.; Forconi, F.; Del, P.G.; Bertoni, F.; Zucca, E.; Montillo, M.; Pozzato, G.; D’Arena, G.; Efremov, D.G.; et al. IGHV gene mutational status and 17p deletion are independent molecular predictors in a comprehensive clinical-biological prognostic model for overall survival prediction in chronic lymphocytic leukemia. J. Transl. Med. 2012, 10, e18. [Google Scholar] [CrossRef]
- Pflug, N.; Bahlo, J.; Shanafelt, T.D.; Eichhorst, B.F.; Bergmann, M.A.; Elter, T.; Bauer, K.; Malchau, G.; Rabe, K.G.; Stilgenbauer, S.; et al. Development of a comprehensive prognostic index for patients with chronic lymphocytic leukemia. Blood 2014, 124, 49–62. [Google Scholar] [CrossRef]
- Wierda, W.G.; O’Brien, S.; Wang, X.; Faderl, S.; Ferrajoli, A.; Do, K.A.; Cortes, J.; Thomas, D.; Garcia-Manero, G.; Koller, C.; et al. Prognostic nomogram and index for overall survival in previously untreated patients with chronic lymphocytic leukemia. Blood 2007, 109, 4679–4685. [Google Scholar] [CrossRef]
- Network, N.C.C. Chronic Lymphocytic Leukemia/Small Lymphocytic Lymphoma (Version 4.2020). Available online: https://www.nccn.org/professionals/physician_gls/pdf/cll.pdf (accessed on 20 December 2019).
- Chevallier, P.; Penther, D.; Avet-Loiseau, H.; Robillard, N.; Ifrah, N.; Mahe, B.; Hamidou, M.; Maisonneuve, H.; Moreau, P.; Jardel, H.; et al. CD38 expression and secondary 17p deletion are important prognostic factors in chronic lymphocytic leukaemia. Br. J. Haematol. 2002, 116, 142–150. [Google Scholar] [CrossRef] [PubMed]
- D’Arena, G.; Musto, P.; Cascavilla, N.; Dell’Olio, M.; Di Renzo, N.; Perla, G.; Savino, L.; Carotenuto, M. CD38 expression correlates with adverse biological features and predicts poor clinical outcome in B-cell chronic lymphocytic leukemia. Leuk. Lymphoma 2001, 42, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Ghia, P.; Guida, G.; Stella, S.; Gottardi, D.; Geuna, M.; Strola, G.; Scielzo, C.; Caligaris-Cappio, F. The pattern of CD38 expression defines a distinct subset of chronic lymphocytic leukemia (CLL) patients at risk of disease progression. Blood 2003, 101, 1262–1269. [Google Scholar] [CrossRef]
- Ibrahim, S.; Keating, M.; Do, K.A.; O’Brien, S.; Huh, Y.O.; Jilani, I.; Lerner, S.; Kantarjian, H.M.; Albitar, M. CD38 expression as an important prognostic factor in B-cell chronic lymphocytic leukemia. Blood 2001, 98, 181–186. [Google Scholar] [CrossRef]
- Rassenti, L.Z.; Jain, S.; Keating, M.J.; Wierda, W.G.; Grever, M.R.; Byrd, J.C.; Kay, N.E.; Brown, J.R.; Gribben, J.G.; Neuberg, D.S.; et al. Relative value of ZAP-70, CD38, and immunoglobulin mutation status in predicting aggressive disease in chronic lymphocytic leukemia. Blood 2008, 112, 1923–1930. [Google Scholar] [CrossRef]
- Cro, L.; Ferrario, A.; Lionetti, M.; Bertoni, F.; Zucal, N.N.; Nobili, L.; Fabris, S.; Todoerti, K.; Cortelezzi, A.; Guffanti, A.; et al. The clinical and biological features of a series of immunophenotypic variant of B-CLL. Eur. J. Haematol. 2010, 85, 120–129. [Google Scholar] [CrossRef]
- Gattei, V.; Bulian, P.; Del Principe, M.I.; Zucchetto, A.; Maurillo, L.; Buccisano, F.; Bomben, R.; Dal-Bo, M.; Luciano, F.; Rossi, F.M.; et al. Relevance of CD49d protein expression as overall survival and progressive disease prognosticator in chronic lymphocytic leukemia. Blood 2008, 111, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Kurtova, A.; Sivina, M.; Quiroga, M.P.; Wierda, W.G.; Keating, M.J.; Burger, J.A. The Immunophenotype Signature CD49d+CD38+ Identifies Chronic Lymphocytic Leukemia Cases with a Higher Potential for Migration Beneath Marrow Stromal Cells. Blood 2009, 114, 356. [Google Scholar] [CrossRef]
- Majid, A.; Lin, T.T.; Best, G.; Fishlock, K.; Hewamana, S.; Pratt, G.; Yallop, D.; Buggins, A.G.; Wagner, S.; Kennedy, B.J.; et al. CD49d is an independent prognostic marker that is associated with CXCR4 expression in CLL. Leuk. Res. 2011, 35, 750–756. [Google Scholar] [CrossRef]
- Nuckel, H.; Switala, M.; Collins, C.H.; Sellmann, L.; Grosse-Wilde, H.; Duhrsen, U.; Rebmann, V. High CD49d protein and mRNA expression predicts poor outcome in chronic lymphocytic leukemia. Clin. Immunol. 2009, 131, 472–480. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Zucchetto, A.; Rossi, F.M.; Capello, D.; Cerri, M.; Deambrogi, C.; Cresta, S.; Rasi, S.; De Paoli, L.; Bodoni, C.L.; et al. CD49d expression is an independent risk factor of progressive disease in early stage chronic lymphocytic leukemia. Haematologica 2008, 93, 1575–1579. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shanafelt, T.D.; Drake, M.T.; Maurer, M.J.; Allmer, C.; Rabe, K.G.; Slager, S.L.; Weiner, G.J.; Call, T.G.; Link, B.K.; Zent, C.S.; et al. Vitamin D insufficiency and prognosis in chronic lymphocytic leukemia. Blood 2011, 117, 1492–1498. [Google Scholar] [CrossRef] [PubMed]
- Shanafelt, T.D.; Geyer, S.M.; Bone, N.D.; Tschumper, R.C.; Witzig, T.E.; Nowakowski, G.S.; Zent, C.S.; Call, T.G.; LaPlant, B.; Dewald, G.W.; et al. CD49d expression is an independent predictor of overall survival in patients with chronic lymphocytic leukaemia: A prognostic parameter with therapeutic potential. Br. J. Haematol. 2008, 140, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Bulian, P.; Shanafelt, T.D.; Fegan, C.; Zucchetto, A.; Cro, L.; Nuckel, H.; Baldini, L.; Kurtova, A.V.; Ferrajoli, A.; Burger, J.A.; et al. CD49d is the strongest flow cytometry-based predictor of overall survival in chronic lymphocytic leukemia. J. Clin. Oncol. 2014, 32, 897–904. [Google Scholar] [CrossRef]
- Fabbri, G.; Rasi, S.; Rossi, D.; Trifonov, V.; Khiabanian, H.; Ma, J.; Grunn, A.; Fangazio, M.; Capello, D.; Monti, S.; et al. Analysis of the chronic lymphocytic leukemia coding genome: Role of NOTCH1 mutational activation. J. Exp. Med. 2011, 208, 1389–1401. [Google Scholar] [CrossRef]
- Landau, D.A.; Tausch, E.; Taylor-Weiner, A.N.; Stewart, C.; Reiter, J.G.; Bahlo, J.; Kluth, S.; Bozic, I.; Lawrence, M.; Bottcher, S.; et al. Mutations driving CLL and their evolution in progression and relapse. Nature 2015, 526, 525–530. [Google Scholar] [CrossRef]
- Oscier, D.G.; Rose-Zerilli, M.J.; Winkelmann, N.; Gonzalez de Castro, D.; Gomez, B.; Forster, J.; Parker, H.; Parker, A.; Gardiner, A.; Collins, A.; et al. The clinical significance of NOTCH1 and SF3B1 mutations in the UK LRF CLL4 trial. Blood 2013, 121, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Puente, X.S.; Bea, S.; Valdes-Mas, R.; Villamor, N.; Gutierrez-Abril, J.; Martin-Subero, J.I.; Munar, M.; Rubio-Perez, C.; Jares, P.; Aymerich, M.; et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature 2015, 526, 519–524. [Google Scholar] [CrossRef] [PubMed]
- Puente, X.S.; Pinyol, M.; Quesada, V.; Conde, L.; Ordonez, G.R.; Villamor, N.; Escaramis, G.; Jares, P.; Bea, S.; Gonzalez-Diaz, M.; et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011, 475, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Bruscaggin, A.; Spina, V.; Rasi, S.; Khiabanian, H.; Messina, M.; Fangazio, M.; Vaisitti, T.; Monti, S.; Chiaretti, S.; et al. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: Association with progression and fludarabine-refractoriness. Blood 2011, 118, 6904–6908. [Google Scholar] [CrossRef]
- Rossi, D.; Fangazio, M.; Rasi, S.; Vaisitti, T.; Monti, S.; Cresta, S.; Chiaretti, S.; Del Giudice, I.; Fabbri, G.; Bruscaggin, A.; et al. Disruption of BIRC3 associates with fludarabine chemorefractoriness in TP53 wild-type chronic lymphocytic leukemia. Blood 2012, 119, 2854–2862. [Google Scholar] [CrossRef]
- Wang, L.; Lawrence, M.S.; Wan, Y.; Stojanov, P.; Sougnez, C.; Stevenson, K.; Werner, L.; Sivachenko, A.; DeLuca, D.S.; Zhang, L.; et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med. 2011, 365, 2497–2506. [Google Scholar] [CrossRef]
- Dal Bo, M.; Bulian, P.; Bomben, R.; Zucchetto, A.; Rossi, F.M.; Pozzo, F.; Tissino, E.; Benedetti, D.; Bittolo, T.; Nanni, P.; et al. CD49d prevails over the novel recurrent mutations as independent prognosticator of overall survival in chronic lymphocytic leukemia. Leukemia 2016, 30, 2011–2018. [Google Scholar] [CrossRef]
- Rossi, D.; Rasi, S.; Spina, V.; Bruscaggin, A.; Monti, S.; Ciardullo, C.; Deambrogi, C.; Khiabanian, H.; Serra, R.; Bertoni, F.; et al. Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood 2013, 121, 1403–1412. [Google Scholar] [CrossRef]
- Toledo, F.; Wahl, G.M. Regulating the p53 pathway: In vitro hypotheses, in vivo veritas. Nat. Rev. Cancer 2006, 6, 909–923. [Google Scholar] [CrossRef]
- Muller, P.A.; Vousden, K.H. p53 mutations in cancer. Nat. Cell Biol. 2013, 15, 2–8. [Google Scholar] [CrossRef]
- Xu-Monette, Z.Y.; Medeiros, L.J.; Li, Y.; Orlowski, R.Z.; Andreeff, M.; Bueso-Ramos, C.E.; Greiner, T.C.; McDonnell, T.J.; Young, K.H. Dysfunction of the TP53 tumor suppressor gene in lymphoid malignancies. Blood 2012, 119, 3668–3683. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Cerri, M.; Deambrogi, C.; Sozzi, E.; Cresta, S.; Rasi, S.; De Paoli, L.; Spina, V.; Gattei, V.; Capello, D.; et al. The prognostic value of TP53 mutations in chronic lymphocytic leukemia is independent of Del17p13: Implications for overall survival and chemorefractoriness. Clin. Cancer Res. 2009, 15, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- Bomben, R.; Rossi, F.M.; D’Agaro, T.; Bittolo, T.; Vit, F.; Zucchetto, A.; Tissino, E.; Pozzo, F.; Ballerini, M.; Bulian, P.; et al. Clinical Impact of Clonal and Subclonal TP53 Mutations and Deletions in Chronic Lymphocytic Leukemia: An Italian Multicenter Experience. Blood Annu. Meet. Abstr. 2019, 134, 480. [Google Scholar] [CrossRef]
- Rossi, D.; Khiabanian, H.; Spina, V.; Ciardullo, C.; Bruscaggin, A.; Fama, R.; Rasi, S.; Monti, S.; Deambrogi, C.; De Paoli, L.; et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood 2014, 123, 2139–2147. [Google Scholar] [CrossRef] [PubMed]
- da Silva-Coelho, P.; Kroeze, L.I.; Yoshida, K.; Koorenhof-Scheele, T.N.; Knops, R.; van de Locht, L.T.; de Graaf, A.O.; Massop, M.; Sandmann, S.; Dugas, M.; et al. Clonal evolution in myelodysplastic syndromes. Nat. Commun. 2017, 8, e15099. [Google Scholar] [CrossRef]
- Wong, T.N.; Miller, C.A.; Klco, J.M.; Petti, A.; Demeter, R.; Helton, N.M.; Li, T.; Fulton, R.S.; Heath, S.E.; Mardis, E.R.; et al. Rapid expansion of preexisting nonleukemic hematopoietic clones frequently follows induction therapy for de novo AML. Blood 2016, 127, 893–897. [Google Scholar] [CrossRef]
- Eichhorst, B.; Robak, T.; Montserrat, E.; Ghia, P.; Hillmen, P.; Hallek, M.; Buske, C. Chronic lymphocytic leukaemia: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015, 26 (Suppl. 5), v78–v84. [Google Scholar] [CrossRef]
- Hallek, M.; Cheson, B.D.; Catovsky, D.; Caligaris-Cappio, F.; Dighiero, G.; Dohner, H.; Hillmen, P.; Keating, M.; Montserrat, E.; Chiorazzi, N.; et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood 2018, 131, 2745–2760. [Google Scholar] [CrossRef]
- Schuh, A.H.; Parry-Jones, N.; Appleby, N.; Bloor, A.; Dearden, C.E.; Fegan, C.; Follows, G.; Fox, C.P.; Iyengar, S.; Kennedy, B.; et al. Guideline for the treatment of chronic lymphocytic leukaemia: A British Society for Haematology Guideline. Br. J. Haematol. 2018, 182, 344–359. [Google Scholar] [CrossRef]
- Wierda, W.G.; Byrd, J.C.; Abramson, J.S.; Bilgrami, S.F.; Bociek, G.; Brander, D.; Brown, J.; Chanan-Khan, A.A.; Chavez, J.C.; Coutre, S.E.; et al. NCCN Guidelines Insights: Chronic Lymphocytic Leukemia/Small Lymphocytic Lymphoma, Version 2.2019. J. Natl. Compr. Cancer Netw. 2019, 17, 12–20. [Google Scholar] [CrossRef]
- Hallek, M. Chronic lymphocytic leukemia: 2020 update on diagnosis, risk stratification and treatment. Am. J. Hematol. 2019, 94, 1266–1287. [Google Scholar] [CrossRef] [PubMed]
- Hallek, M.; Fischer, K.; Fingerle-Rowson, G.; Fink, A.M.; Busch, R.; Mayer, J.; Hensel, M.; Hopfinger, G.; Hess, G.; von Grunhagen, U.; et al. Addition of rituximab to fludarabine and cyclophosphamide in patients with chronic lymphocytic leukaemia: A randomised, open-label, phase 3 trial. Lancet 2010, 376, 1164–1174. [Google Scholar] [CrossRef]
- Stilgenbauer, S.; Schnaiter, A.; Paschka, P.; Zenz, T.; Rossi, M.; Dohner, K.; Buhler, A.; Bottcher, S.; Ritgen, M.; Kneba, M.; et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: Results from the CLL8 trial. Blood 2014, 123, 3247–3254. [Google Scholar] [CrossRef]
- Gonzalez, D.; Martinez, P.; Wade, R.; Hockley, S.; Oscier, D.; Matutes, E.; Dearden, C.E.; Richards, S.M.; Catovsky, D.; Morgan, G.J. Mutational status of the TP53 gene as a predictor of response and survival in patients with chronic lymphocytic leukemia: Results from the LRF CLL4 trial. J. Clin. Oncol. 2011, 29, 2223–2229. [Google Scholar] [CrossRef] [PubMed]
- Zenz, T.; Eichhorst, B.; Busch, R.; Denzel, T.; Habe, S.; Winkler, D.; Buhler, A.; Edelmann, J.; Bergmann, M.; Hopfinger, G.; et al. TP53 mutation and survival in chronic lymphocytic leukemia. J. Clin. Oncol. 2010, 28, 4473–4479. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A.; O’Brien, S. Evolution of CLL treatment-from chemoimmunotherapy to targeted and individualized therapy. Nat. Rev. Clin. Oncol. 2018, 15, 510–527. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.R.; Hallek, M.J.; Pagel, J.M. Chemoimmunotherapy Versus Targeted Treatment in Chronic Lymphocytic Leukemia: When, How Long, How Much, and in Which Combination? Am. Soc. Clin. Oncol. Educ. Book 2016, 35, e387–e398. [Google Scholar] [CrossRef]
- Badoux, X.C.; Keating, M.J.; Wang, X.; O’Brien, S.M.; Ferrajoli, A.; Faderl, S.; Burger, J.; Koller, C.; Lerner, S.; Kantarjian, H.; et al. Fludarabine, cyclophosphamide, and rituximab chemoimmunotherapy is highly effective treatment for relapsed patients with CLL. Blood 2011, 117, 3016–3024. [Google Scholar] [CrossRef]
- Fischer, K.; Cramer, P.; Busch, R.; Stilgenbauer, S.; Bahlo, J.; Schweighofer, C.D.; Bottcher, S.; Staib, P.; Kiehl, M.; Eckart, M.J.; et al. Bendamustine combined with rituximab in patients with relapsed and/or refractory chronic lymphocytic leukemia: A multicenter phase II trial of the German Chronic Lymphocytic Leukemia Study Group. J. Clin. Oncol. 2011, 29, 3559–3566. [Google Scholar] [CrossRef]
- Furman, R.R.; Sharman, J.P.; Coutre, S.E.; Cheson, B.D.; Pagel, J.M.; Hillmen, P.; Barrientos, J.C.; Zelenetz, A.D.; Kipps, T.J.; Flinn, I.; et al. Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N. Engl. J. Med. 2014, 370, 997–1007. [Google Scholar] [CrossRef]
- O’Brien, S.; Furman, R.R.; Coutre, S.; Flinn, I.W.; Burger, J.A.; Blum, K.; Sharman, J.; Wierda, W.; Jones, J.; Zhao, W.; et al. Single-agent ibrutinib in treatment-naive and relapsed/refractory chronic lymphocytic leukemia: A 5-year experience. Blood 2018, 131, 1910–1919. [Google Scholar] [CrossRef] [PubMed]
- Stilgenbauer, S.; Eichhorst, B.; Schetelig, J.; Coutre, S.; Seymour, J.F.; Munir, T.; Puvvada, S.D.; Wendtner, C.M.; Roberts, A.W.; Jurczak, W.; et al. Venetoclax in relapsed or refractory chronic lymphocytic leukaemia with 17p deletion: A multicentre, open-label, phase 2 study. Lancet Oncol. 2016, 17, 768–778. [Google Scholar] [CrossRef]
- Ahn, I.E.; Farooqui, M.Z.H.; Tian, X.; Valdez, J.; Sun, C.; Soto, S.; Lotter, J.; Housel, S.; Stetler-Stevenson, M.; Yuan, C.M.; et al. Depth and durability of response to ibrutinib in CLL: 5-year follow-up of a phase 2 study. Blood 2018, 131, 2357–2366. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A.; Sivina, M.; Jain, N.; Kim, E.; Kadia, T.; Estrov, Z.; Nogueras-Gonzalez, G.M.; Huang, X.; Jorgensen, J.; Li, J.; et al. Randomized trial of ibrutinib vs ibrutinib plus rituximab in patients with chronic lymphocytic leukemia. Blood 2019, 133, 1011–1019. [Google Scholar] [CrossRef]
- Farooqui, M.Z.; Valdez, J.; Martyr, S.; Aue, G.; Saba, N.; Niemann, C.U.; Herman, S.E.; Tian, X.; Marti, G.; Soto, S.; et al. Ibrutinib for previously untreated and relapsed or refractory chronic lymphocytic leukaemia with TP53 aberrations: A phase 2, single-arm trial. Lancet Oncol. 2015, 16, 169–176. [Google Scholar] [CrossRef]
- Fischer, K.; Al-Sawaf, O.; Bahlo, J.; Fink, A.M.; Tandon, M.; Dixon, M.; Robrecht, S.; Warburton, S.; Humphrey, K.; Samoylova, O.; et al. Venetoclax and Obinutuzumab in Patients with CLL and Coexisting Conditions. N. Engl. J. Med. 2019, 380, 2225–2236. [Google Scholar] [CrossRef]
- Moreno, C.; Greil, R.; Demirkan, F.; Tedeschi, A.; Anz, B.; Larratt, L.; Simkovic, M.; Samoilova, O.; Novak, J.; Ben-Yehuda, D.; et al. Ibrutinib plus obinutuzumab versus chlorambucil plus obinutuzumab in first-line treatment of chronic lymphocytic leukaemia (iLLUMINATE): A multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2019, 20, 43–56. [Google Scholar] [CrossRef]
- Byrd, J.C.; Furman, R.R.; Coutre, S.E.; Burger, J.A.; Blum, K.A.; Coleman, M.; Wierda, W.G.; Jones, J.A.; Zhao, W.; Heerema, N.A.; et al. Three-year follow-up of treatment-naive and previously treated patients with CLL and SLL receiving single-agent ibrutinib. Blood 2015, 125, 2497–2506. [Google Scholar] [CrossRef]
- Thompson, P.A.; O’Brien, S.M.; Wierda, W.G.; Ferrajoli, A.; Stingo, F.; Smith, S.C.; Burger, J.A.; Estrov, Z.; Jain, N.; Kantarjian, H.M.; et al. Complex karyotype is a stronger predictor than del(17p) for an inferior outcome in relapsed or refractory chronic lymphocytic leukemia patients treated with ibrutinib-based regimens. Cancer 2015, 121, 3612–3621. [Google Scholar] [CrossRef]
- Winqvist, M.; Asklid, A.; Andersson, P.O.; Karlsson, K.; Karlsson, C.; Lauri, B.; Lundin, J.; Mattsson, M.; Norin, S.; Sandstedt, A.; et al. Real-world results of ibrutinib in patients with relapsed or refractory chronic lymphocytic leukemia: Data from 95 consecutive patients treated in a compassionate use program. A study from the Swedish Chronic Lymphocytic Leukemia Group. Haematologica 2016, 101, 1573–1580. [Google Scholar] [CrossRef]
- Barrientos, J.C.; Ghia, P.; Pagel, J.; Salles, G.A.; Sharman, J.P.; Stilgenbauer, S.; Gurtovaya, O.; Kim, Y.; Philip, B.; Zelenetz, A.D. Outcomes of anticoagulant (AC) or antiplatelet (AP) use in patients (pts) with chronic lymphocytic leukemia (CLL) or indolent non-Hodgkin’s lymphoma (iNHL) in idelalisib (IDELA) trials. J. Clin. Oncol. 2015, 33, 8563. [Google Scholar] [CrossRef]
- Roberts, A.W.; Davids, M.S.; Pagel, J.M.; Kahl, B.S.; Puvvada, S.D.; Gerecitano, J.F.; Kipps, T.J.; Anderson, M.A.; Brown, J.R.; Gressick, L.; et al. Targeting BCL2 with Venetoclax in Relapsed Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2016, 374, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Damle, R.N.; Wasil, T.; Fais, F.; Ghiotto, F.; Valetto, A.; Allen, S.L.; Buchbinder, A.; Budman, D.; Dittmar, K.; Kolitz, J.; et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood 1999, 94, 1840–1847. [Google Scholar] [CrossRef] [PubMed]
- Hamblin, T.J.; Davis, Z.; Gardiner, A.; Oscier, D.G.; Stevenson, F.K. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood 1999, 94, 1848–1854. [Google Scholar] [CrossRef]
- Jain, P.; Nogueras Gonzalez, G.M.; Kanagal-Shamanna, R.; Rozovski, U.; Sarwari, N.; Tam, C.; Wierda, W.G.; Thompson, P.A.; Jain, N.; Luthra, R.; et al. The absolute percent deviation of IGHV mutation rather than a 98% cut-off predicts survival of chronic lymphocytic leukaemia patients treated with fludarabine, cyclophosphamide and rituximab. Br. J. Haematol. 2018, 180, 33–40. [Google Scholar] [CrossRef]
- Morabito, F.; Shanafelt, T.D.; Gentile, M.; Reda, G.; Mauro, F.R.; Rossi, D.; Di Renzo, N.; Molica, S.; Angrilli, F.; Chiarenza, A.; et al. Immunoglobulin heavy chain variable region gene and prediction of time to first treatment in patients with chronic lymphocytic leukemia: Mutational load or mutational status? Analysis of 1003 cases. Am. J. Hematol. 2018, 93, 216–219. [Google Scholar] [CrossRef]
- Fischer, K.; Bahlo, J.; Fink, A.M.; Goede, V.; Herling, C.D.; Cramer, P.; Langerbeins, P.; von Tresckow, J.; Engelke, A.; Maurer, C.; et al. Long-term remissions after FCR chemoimmunotherapy in previously untreated patients with CLL: Updated results of the CLL8 trial. Blood 2016, 127, 208–215. [Google Scholar] [CrossRef]
- Shanafelt, T.D.; Wang, X.V.; Kay, N.E.; Hanson, C.A.; O’Brien, S.; Barrientos, J.; Jelinek, D.F.; Braggio, E.; Leis, J.F.; Zhang, C.C.; et al. Ibrutinib-Rituximab or Chemoimmunotherapy for Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2019, 381, 432–443. [Google Scholar] [CrossRef]
- Thompson, P.A.; Tam, C.S.; O’Brien, S.M.; Wierda, W.G.; Stingo, F.; Plunkett, W.; Smith, S.C.; Kantarjian, H.M.; Freireich, E.J.; Keating, M.J. Fludarabine, cyclophosphamide, and rituximab treatment achieves long-term disease-free survival in IGHV-mutated chronic lymphocytic leukemia. Blood 2016, 127, 303–309. [Google Scholar] [CrossRef]
- Shanafelt, T.D.; Wang, V.; Kay, N.E.; Hanson, C.A.; O’Brien, S.M.; Barrientos, J.C.; Jelinek, D.F.; Braggio, E.; Leis, J.F.; Zhang, C.C.; et al. Ibrutinib and Rituximab Provides Superior Clinical Outcome Compared to FCR in Younger Patients with Chronic Lymphocytic Leukemia (CLL): Extended Follow-up from the E1912 Trial. Blood 2019, 134, 33. [Google Scholar] [CrossRef]
- Woyach, J.A.; Ruppert, A.S.; Heerema, N.A.; Zhao, W.; Booth, A.M.; Ding, W.; Bartlett, N.L.; Brander, D.M.; Barr, P.M.; Rogers, K.A.; et al. Ibrutinib Regimens versus Chemoimmunotherapy in Older Patients with Untreated CLL. N. Engl. J. Med. 2018, 379, 2517–2528. [Google Scholar] [CrossRef]
- Del Giudice, I.; Rossi, D.; Chiaretti, S.; Marinelli, M.; Tavolaro, S.; Gabrielli, S.; Laurenti, L.; Marasca, R.; Rasi, S.; Fangazio, M.; et al. NOTCH1 mutations in +12 chronic lymphocytic leukemia (CLL) confer an unfavorable prognosis, induce a distinctive transcriptional profiling and refine the intermediate prognosis of +12 CLL. Haematologica 2012, 97, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Rosati, E.; Sabatini, R.; Rampino, G.; Tabilio, A.; Di Ianni, M.; Fettucciari, K.; Bartoli, A.; Coaccioli, S.; Screpanti, I.; Marconi, P. Constitutively activated Notch signaling is involved in survival and apoptosis resistance of B-CLL cells. Blood 2009, 113, 856–865. [Google Scholar] [CrossRef] [PubMed]
- Bray, S.J. Notch signalling: A simple pathway becomes complex. Nat. Rev. Mol Cell Biol 2006, 7, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Bray, S.J. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016, 17, 722–735. [Google Scholar] [CrossRef] [PubMed]
- Castel, D.; Mourikis, P.; Bartels, S.J.; Brinkman, A.B.; Tajbakhsh, S.; Stunnenberg, H.G. Dynamic binding of RBPJ is determined by Notch signaling status. Genes Dev. 2013, 27, 1059–1071. [Google Scholar] [CrossRef]
- Herranz, D.; Ambesi-Impiombato, A.; Palomero, T.; Schnell, S.A.; Belver, L.; Wendorff, A.A.; Xu, L.; Castillo-Martin, M.; Llobet-Navas, D.; Cordon-Cardo, C.; et al. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat. Med. 2014, 20, 1130–1137. [Google Scholar] [CrossRef]
- Palomero, T.; Lim, W.K.; Odom, D.T.; Sulis, M.L.; Real, P.J.; Margolin, A.; Barnes, K.C.; O’Neil, J.; Neuberg, D.; Weng, A.P.; et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl. Acad. Sci. USA 2006, 103, 18261–18266. [Google Scholar] [CrossRef]
- Yashiro-Ohtani, Y.; Wang, H.; Zang, C.; Arnett, K.L.; Bailis, W.; Ho, Y.; Knoechel, B.; Lanauze, C.; Louis, L.; Forsyth, K.S.; et al. Long-range enhancer activity determines Myc sensitivity to Notch inhibitors in T cell leukemia. Proc. Natl. Acad. Sci. USA 2014, 111, E4946–E4953. [Google Scholar] [CrossRef]
- Pozzo, F.; Bittolo, T.; Arruga, F.; Bulian, P.; Macor, P.; Tissino, E.; Gizdic, B.; Rossi, F.M.; Bomben, R.; Zucchetto, A.; et al. NOTCH1 mutations associate with low CD20 level in chronic lymphocytic leukemia: Evidence for a NOTCH1 mutation-driven epigenetic dysregulation. Leukemia 2016, 30, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Tedder, T.F.; Streuli, M.; Schlossman, S.F.; Saito, H. Isolation and structure of a cDNA encoding the B1 (CD20) cell-surface antigen of human B lymphocytes. Proc. Natl. Acad. Sci. USA 1988, 85, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A. Targeting the microenvironment in chronic lymphocytic leukemia is changing the therapeutic landscape. Curr. Opin. Oncol. 2012, 24, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Zucchetto, A.; Bomben, R.; Dal Bo, M.; Bulian, P.; Benedetti, D.; Nanni, P.; Del Poeta, G.; Degan, M.; Gattei, V. CD49d in B-cell chronic lymphocytic leukemia: Correlated expression with CD38 and prognostic relevance. Leukemia 2006, 20, 523–525. [Google Scholar] [CrossRef] [PubMed]
- Zucchetto, A.; Vaisitti, T.; Benedetti, D.; Tissino, E.; Bertagnolo, V.; Rossi, D.; Bomben, R.; Dal Bo, M.; Del Principe, M.I.; Gorgone, A.; et al. The CD49d/CD29 complex is physically and functionally associated with CD38 in B-cell chronic lymphocytic leukemia cells. Leukemia 2012, 26, 1301–1312. [Google Scholar] [CrossRef] [PubMed]
- Brachtl, G.; Pinon Hofbauer, J.; Greil, R.; Hartmann, T.N. The pathogenic relevance of the prognostic markers CD38 and CD49d in chronic lymphocytic leukemia. Ann. Hematol. 2014, 93, 361–374. [Google Scholar] [CrossRef]
- Tissino, E.; Benedetti, D.; Herman, S.E.M.; Ten Hacken, E.; Ahn, I.E.; Chaffee, K.G.; Rossi, F.M.; Dal Bo, M.; Bulian, P.; Bomben, R.; et al. Functional and clinical relevance of VLA-4 (CD49d/CD29) in ibrutinib-treated chronic lymphocytic leukemia. J. Exp. Med. 2018, 215, 681–697. [Google Scholar] [CrossRef]
- Tissino, E.; Pozzo, F.; Benedetti, D.; Caldana, C.; Bittolo, T.; Rossi, F.M.; Bomben, R.; Nanni, P.; Chivilo, H.; Cattarossi, I.; et al. CD49d promotes disease progression in chronic lymphocytic leukemia: New insights from CD49d bimodal expression. Blood 2020. [Google Scholar] [CrossRef]
- de Rooij, M.F.; Kuil, A.; Geest, C.R.; Eldering, E.; Chang, B.Y.; Buggy, J.J.; Pals, S.T.; Spaargaren, M. The clinically active BTK inhibitor PCI-32765 targets B-cell receptor- and chemokine-controlled adhesion and migration in chronic lymphocytic leukemia. Blood 2012, 119, 2590–2594. [Google Scholar] [CrossRef]
- Herman, S.E.; Mustafa, R.Z.; Jones, J.; Wong, D.H.; Farooqui, M.; Wiestner, A. Treatment with Ibrutinib Inhibits BTK- and VLA-4-Dependent Adhesion of Chronic Lymphocytic Leukemia Cells In Vivo. Clin. Cancer Res. 2015, 21, 4642–4651. [Google Scholar] [CrossRef]
- Herman, S.E.; Niemann, C.U.; Farooqui, M.; Jones, J.; Mustafa, R.Z.; Lipsky, A.; Saba, N.; Martyr, S.; Soto, S.; Valdez, J.; et al. Ibrutinib-induced lymphocytosis in patients with chronic lymphocytic leukemia: Correlative analyses from a phase II study. Leukemia 2014, 28, 2188–2196. [Google Scholar] [CrossRef]
- Thompson, P.A.; Ferrajoli, A.; O’Brien, S.; Wierda, W.G.; Keating, M.J.; Burger, J.A. Trisomy 12 is associated with an abbreviated redistribution lymphocytosis during treatment with the BTK inhibitor ibrutinib in patients with chronic lymphocytic leukaemia. Br. J. Haematol. 2015, 170, 125–128. [Google Scholar] [CrossRef] [PubMed]
- Heyn, H.; Esteller, M. DNA methylation profiling in the clinic: Applications and challenges. Nat. Rev. Genet. 2012, 13, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Kulis, M.; Heath, S.; Bibikova, M.; Queiros, A.C.; Navarro, A.; Clot, G.; Martinez-Trillos, A.; Castellano, G.; Brun-Heath, I.; Pinyol, M.; et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat. Genet. 2012, 44, 1236–1242. [Google Scholar] [CrossRef] [PubMed]
- Queiros, A.C.; Villamor, N.; Clot, G.; Martinez-Trillos, A.; Kulis, M.; Navarro, A.; Penas, E.M.; Jayne, S.; Majid, A.; Richter, J.; et al. A B-cell epigenetic signature defines three biologic subgroups of chronic lymphocytic leukemia with clinical impact. Leukemia 2015, 29, 598–605. [Google Scholar] [CrossRef]
- Catovsky, D.; Richards, S.; Matutes, E.; Oscier, D.; Dyer, M.; Bezares, R.F.; Pettitt, A.R.; Hamblin, T.; Milligan, D.W.; Child, J.A.; et al. Assessment of fludarabine plus cyclophosphamide for patients with chronic lymphocytic leukaemia (the LRF CLL4 Trial): A randomised controlled trial. Lancet 2007, 370, 230–239. [Google Scholar] [CrossRef]
- Munir, T.; Howard, D.R.; McParland, L.; Pocock, C.; Rawstron, A.C.; Hockaday, A.; Varghese, A.; Hamblin, M.; Bloor, A.; Pettitt, A.; et al. Results of the randomized phase IIB ADMIRE trial of FCR with or without mitoxantrone in previously untreated CLL. Leukemia 2017, 31, 2085–2093. [Google Scholar] [CrossRef]
- Howard, D.R.; Munir, T.; McParland, L.; Rawstron, A.C.; Milligan, D.; Schuh, A.; Hockaday, A.; Allsup, D.J.; Marshall, S.; Duncombe, A.S.; et al. Results of the randomized phase IIB ARCTIC trial of low-dose rituximab in previously untreated CLL. Leukemia 2017, 31, 2416–2425. [Google Scholar] [CrossRef]
- Wojdacz, T.K.; Amarasinghe, H.E.; Kadalayil, L.; Beattie, A.; Forster, J.; Blakemore, S.J.; Parker, H.; Bryant, D.; Larrayoz, M.; Clifford, R.; et al. Clinical significance of DNA methylation in chronic lymphocytic leukemia patients: Results from 3 UK clinical trials. Blood Adv. 2019, 3, 2474–2481. [Google Scholar] [CrossRef]
- Dohner, H.; Stilgenbauer, S.; Benner, A.; Leupolt, E.; Krober, A.; Bullinger, L.; Dohner, K.; Bentz, M.; Lichter, P. Genomic aberrations and survival in chronic lymphocytic leukemia. N. Engl J. Med. 2000, 343, 1910–1916. [Google Scholar] [CrossRef]
- Juliusson, G.; Oscier, D.G.; Fitchett, M.; Ross, F.M.; Stockdill, G.; Mackie, M.J.; Parker, A.C.; Castoldi, G.L.; Guneo, A.; Knuutila, S.; et al. Prognostic subgroups in B-cell chronic lymphocytic leukemia defined by specific chromosomal abnormalities. N. Engl. J. Med. 1990, 323, 720–724. [Google Scholar] [CrossRef]
- Baliakas, P.; Iskas, M.; Gardiner, A.; Davis, Z.; Plevova, K.; Nguyen-Khac, F.; Malcikova, J.; Anagnostopoulos, A.; Glide, S.; Mould, S.; et al. Chromosomal translocations and karyotype complexity in chronic lymphocytic leukemia: A systematic reappraisal of classic cytogenetic data. Am. J. Hematol. 2014, 89, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Herling, C.D.; Klaumunzer, M.; Rocha, C.K.; Altmuller, J.; Thiele, H.; Bahlo, J.; Kluth, S.; Crispatzu, G.; Herling, M.; Schiller, J.; et al. Complex karyotypes and KRAS and POT1 mutations impact outcome in CLL after chlorambucil-based chemotherapy or chemoimmunotherapy. Blood 2016, 128, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Le Bris, Y.; Struski, S.; Guieze, R.; Rouvellat, C.; Prade, N.; Troussard, X.; Tournilhac, O.; Bene, M.C.; Delabesse, E.; Ysebaert, L. Major prognostic value of complex karyotype in addition to TP53 and IGHV mutational status in first-line chronic lymphocytic leukemia. Hematol. Oncol. 2017, 35, 664–670. [Google Scholar] [CrossRef]
- Rigolin, G.M.; Cavallari, M.; Quaglia, F.M.; Formigaro, L.; Lista, E.; Urso, A.; Guardalben, E.; Liberatore, C.; Faraci, D.; Saccenti, E.; et al. In CLL, comorbidities and the complex karyotype are associated with an inferior outcome independently of CLL-IPI. Blood J. Am. Soc. Hematol. 2017, 129, 3495–3498. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.A.; Tam, C.; Lew, T.E.; Juneja, S.; Juneja, M.; Westerman, D.; Wall, M.; Lade, S.; Gorelik, A.; Huang, D.C.S.; et al. Clinicopathological features and outcomes of progression of CLL on the BCL2 inhibitor venetoclax. Blood 2017, 129, 3362–3370. [Google Scholar] [CrossRef]
- Visentin, A.; Bonaldi, L.; Rigolin, G.M.; Mauro, F.R.; Martines, A.; Frezzato, F.; Imbergamo, S.; Scomazzon, E.; Pravato, S.; Bardi, M.A.; et al. The combination of complex karyotype subtypes and IGHV mutational status identifies new prognostic and predictive groups in chronic lymphocytic leukaemia. Br. J. Cancer 2019, 121, 150–156. [Google Scholar] [CrossRef]
- Pozzo, F.; Bittolo, T.; Vendramini, E.; Bomben, R.; Bulian, P.; Rossi, F.M.; Zucchetto, A.; Tissino, E.; Degan, M.; D’Arena, G.; et al. NOTCH1-mutated chronic lymphocytic leukemia cells are characterized by a MYC-related overexpression of nucleophosmin 1 and ribosome-associated components. Leukemia 2017, 31, 2407–2415. [Google Scholar] [CrossRef]
- Pozzo, F.; Dal Bo, M.; Peragine, N.; Bomben, R.; Zucchetto, A.; Rossi, F.; Degan, M.; Rossi, D.; Chiarenza, A.; Grossi, A.; et al. Detection of TP53 dysfunction in chronic lymphocytic leukemia by an in vitro functional assay based on TP53 activation by the non-genotoxic drug Nutlin-3: A proposal for clinical application. J. Hematol. Oncol. 2013, 6, e83. [Google Scholar] [CrossRef][Green Version]
- Zucchetto, A.; Benedetti, D.; Tripodo, C.; Bomben, R.; Dal Bo, M.; Marconi, D.; Bossi, F.; Lorenzon, D.; Degan, M.; Rossi, F.M.; et al. CD38/CD31, the CCL3 and CCL4 chemokines, and CD49d/vascular cell adhesion molecule-1 are interchained by sequential events sustaining chronic lymphocytic leukemia cell survival. Cancer Res. 2009, 69, 4001–4009. [Google Scholar] [CrossRef]
- Arruga, F.; Bracciama, V.; Vitale, N.; Vaisitti, T.; Gizzi, K.; Yeomans, A.; Coscia, M.; D’Arena, G.; Gaidano, G.; Allan, J.N.; et al. Bidirectional linkage between the B-cell receptor and NOTCH1 in chronic lymphocytic leukemia and in Richter’s syndrome: Therapeutic implications. Leukemia 2020, 34, 462–477. [Google Scholar] [CrossRef]
- Arruga, F.; Gizdic, B.; Bologna, C.; Cignetto, S.; Buonincontri, R.; Serra, S.; Vaisitti, T.; Gizzi, K.; Vitale, N.; Garaffo, G.; et al. Mutations in NOTCH1 PEST domain orchestrate CCL19-driven homing of chronic lymphocytic leukemia cells by modulating the tumor suppressor gene DUSP22. Leukemia 2017, 31, 1882–1893. [Google Scholar] [CrossRef]
- Benedetti, D.; Tissino, E.; Pozzo, F.; Bittolo, T.; Caldana, C.; Perini, C.; Martorelli, D.; Bravin, V.; D’Agaro, T.; Rossi, F.M.; et al. NOTCH1 mutations are associated with high CD49d expression in chronic lymphocytic leukemia: Link between the NOTCH1 and the NF-kappaB pathways. Leukemia 2018, 32, 654–662. [Google Scholar] [CrossRef]
- Stamatopoulos, K.; Agathangelidis, A.; Rosenquist, R.; Ghia, P. Antigen receptor stereotypy in chronic lymphocytic leukemia. Leukemia 2017, 31, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Stamatopoulos, K.; Belessi, C.; Moreno, C.; Boudjograh, M.; Guida, G.; Smilevska, T.; Belhoul, L.; Stella, S.; Stavroyianni, N.; Crespo, M.; et al. Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: Pathogenetic implications and clinical correlations. Blood 2007, 109, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Woyach, J.A.; Johnson, A.J.; Byrd, J.C. The B-cell receptor signaling pathway as a therapeutic target in CLL. Blood 2012, 120, 1175–1184. [Google Scholar] [CrossRef]
- Maddocks, K.J.; Ruppert, A.S.; Lozanski, G.; Heerema, N.A.; Zhao, W.; Abruzzo, L.; Lozanski, A.; Davis, M.; Gordon, A.; Smith, L.L.; et al. Etiology of Ibrutinib Therapy Discontinuation and Outcomes in Patients With Chronic Lymphocytic Leukemia. JAMA Oncol. 2015, 1, 80–87. [Google Scholar] [CrossRef]
- Woyach, J.A.; Furman, R.R.; Liu, T.M.; Ozer, H.G.; Zapatka, M.; Ruppert, A.S.; Xue, L.; Li, D.H.; Steggerda, S.M.; Versele, M.; et al. Resistance mechanisms for the Bruton’s tyrosine kinase inhibitor ibrutinib. N. Engl. J. Med. 2014, 370, 2286–2294. [Google Scholar] [CrossRef] [PubMed]
- Woyach, J.A.; Ruppert, A.S.; Guinn, D.; Lehman, A.; Blachly, J.S.; Lozanski, A.; Heerema, N.A.; Zhao, W.; Coleman, J.; Jones, D.; et al. BTK(C481S)-Mediated Resistance to Ibrutinib in Chronic Lymphocytic Leukemia. J. Clin. Oncol. 2017, 35, 1437–1443. [Google Scholar] [CrossRef] [PubMed]
- Hockenbery, D.; Nunez, G.; Milliman, C.; Schreiber, R.D.; Korsmeyer, S.J. Bcl-2 is an inner mitochondrial membrane protein that blocks programmed cell death. Nature 1990, 348, 334–336. [Google Scholar] [CrossRef]
- Hanada, M.; Delia, D.; Aiello, A.; Stadtmauer, E.; Reed, J.C. bcl-2 gene hypomethylation and high-level expression in B-cell chronic lymphocytic leukemia. Blood 1993, 82, 1820–1828. [Google Scholar] [CrossRef]
- Coutre, S.; Choi, M.; Furman, R.R.; Eradat, H.; Heffner, L.; Jones, J.A.; Chyla, B.; Zhou, L.; Agarwal, S.; Waskiewicz, T.; et al. Venetoclax for patients with chronic lymphocytic leukemia who progressed during or after idelalisib therapy. Blood 2018, 131, 1704–1711. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.A.; Mato, A.R.; Wierda, W.G.; Davids, M.S.; Choi, M.; Cheson, B.D.; Furman, R.R.; Lamanna, N.; Barr, P.M.; Zhou, L.; et al. Venetoclax for chronic lymphocytic leukaemia progressing after ibrutinib: An interim analysis of a multicentre, open-label, phase 2 trial. Lancet Oncol. 2018, 19, 65–75. [Google Scholar] [CrossRef]
- Seymour, J.F.; Kipps, T.J.; Eichhorst, B.; Hillmen, P.; D’Rozario, J.; Assouline, S.; Owen, C.; Gerecitano, J.; Robak, T.; De la Serna, J.; et al. Venetoclax-Rituximab in Relapsed or Refractory Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2018, 378, 1107–1120. [Google Scholar] [CrossRef] [PubMed]
- Seymour, J.F.; Ma, S.; Brander, D.M.; Choi, M.Y.; Barrientos, J.; Davids, M.S.; Anderson, M.A.; Beaven, A.W.; Rosen, S.T.; Tam, C.S.; et al. Venetoclax plus rituximab in relapsed or refractory chronic lymphocytic leukaemia: A phase 1b study. Lancet Oncol. 2017, 18, 230–240. [Google Scholar] [CrossRef]
- Davids, M.S.; Hallek, M.; Wierda, W.; Roberts, A.W.; Stilgenbauer, S.; Jones, J.A.; Gerecitano, J.F.; Kim, S.Y.; Potluri, J.; Busman, T.; et al. Comprehensive Safety Analysis of Venetoclax Monotherapy for Patients with Relapsed/Refractory Chronic Lymphocytic Leukemia. Clin. Cancer Res. 2018, 24, 4371–4379. [Google Scholar] [CrossRef]
- Blombery, P.; Anderson, M.A.; Gong, J.N.; Thijssen, R.; Birkinshaw, R.W.; Thompson, E.R.; Teh, C.E.; Nguyen, T.; Xu, Z.; Flensburg, C.; et al. Acquisition of the Recurrent Gly101Val Mutation in BCL2 Confers Resistance to Venetoclax in Patients with Progressive Chronic Lymphocytic Leukemia. Cancer Discov. 2019, 9, 342–353. [Google Scholar] [CrossRef]
- Blombery, P.; Thompson, E.R.; Nguyen, T.; Birkinshaw, R.W.; Gong, J.-n.; Chen, X.; McBean, M.; Thijssen, R.; Conway, T.; Anderson, M.A.; et al. Multiple BCL2 mutations cooccurring with Gly101Val emerge in chronic lymphocytic leukemia progression on venetoclax. Blood 2020, 135, 773–777. [Google Scholar] [CrossRef]
- Tausch, E.; Close, W.; Dolnik, A.; Bloehdorn, J.; Chyla, B.; Bullinger, L.; Dohner, H.; Mertens, D.; Stilgenbauer, S. Venetoclax resistance and acquired BCL2 mutations in chronic lymphocytic leukemia. Haematologica 2019, 104, e434. [Google Scholar] [CrossRef]
| Category | Biomarkers |
|---|---|
| Host Factors | Age, Gender, Ethnicity |
| Disease Markers | Lymph node involvement (size, site(s) of involvement), Hepatomegaly, Splenomegaly, LDT, WBC count, ALC, Anemia, Thrombocytopenia, |
| Antigen Expression | CD38, ZAP70, CD49d/VLA-4 |
| Serology | β2M, TK, LDH, IL-8 |
| Genetics | del17p, TP53 mutation, del11q, del13q, trisomy 12, NOTCH1 mutation, DNA methylation, complex karyotype, SF3B1 mutation, BIRC3 mutation, BRAF mutation, miR-223, miR-29c, miR-155 |
| Immunogenetics | IGHV sequence, BCR structure |
| Society | Recommendation | Timing |
|---|---|---|
| iwCLL | ||
| TP53 disruption | Always | Prior to treatment |
| IGHV gene mutational status | Always | Prior to treatment |
| BCSH | ||
| TP53 disruption | Always | Prior to treatment |
| IGHV gene mutational status | “Should be considered” | Prior to treatment |
| NCCN | ||
| TP53 disruption | Always | At diagnosis or prior to treatment 1 |
| IGHV gene mutational status | Always | At diagnosis or prior to treatment |
| ESMO | ||
| TP53 disruption | Always | Prior to treatment |
| IGHV gene mutational status | “Desirable” | Prior to treatment |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cohen, J.A.; Bomben, R.; Pozzo, F.; Tissino, E.; Härzschel, A.; Hartmann, T.N.; Zucchetto, A.; Gattei, V. An Updated Perspective on Current Prognostic and Predictive Biomarkers in Chronic Lymphocytic Leukemia in the Context of Chemoimmunotherapy and Novel Targeted Therapy. Cancers 2020, 12, 894. https://doi.org/10.3390/cancers12040894
Cohen JA, Bomben R, Pozzo F, Tissino E, Härzschel A, Hartmann TN, Zucchetto A, Gattei V. An Updated Perspective on Current Prognostic and Predictive Biomarkers in Chronic Lymphocytic Leukemia in the Context of Chemoimmunotherapy and Novel Targeted Therapy. Cancers. 2020; 12(4):894. https://doi.org/10.3390/cancers12040894
Chicago/Turabian StyleCohen, Jared A., Riccardo Bomben, Federico Pozzo, Erika Tissino, Andrea Härzschel, Tanja Nicole Hartmann, Antonella Zucchetto, and Valter Gattei. 2020. "An Updated Perspective on Current Prognostic and Predictive Biomarkers in Chronic Lymphocytic Leukemia in the Context of Chemoimmunotherapy and Novel Targeted Therapy" Cancers 12, no. 4: 894. https://doi.org/10.3390/cancers12040894
APA StyleCohen, J. A., Bomben, R., Pozzo, F., Tissino, E., Härzschel, A., Hartmann, T. N., Zucchetto, A., & Gattei, V. (2020). An Updated Perspective on Current Prognostic and Predictive Biomarkers in Chronic Lymphocytic Leukemia in the Context of Chemoimmunotherapy and Novel Targeted Therapy. Cancers, 12(4), 894. https://doi.org/10.3390/cancers12040894






