Biological and Clinical Significance of the CCR5/CCL5 Axis in Hepatocellular Carcinoma
Abstract
1. Introduction
2. Results
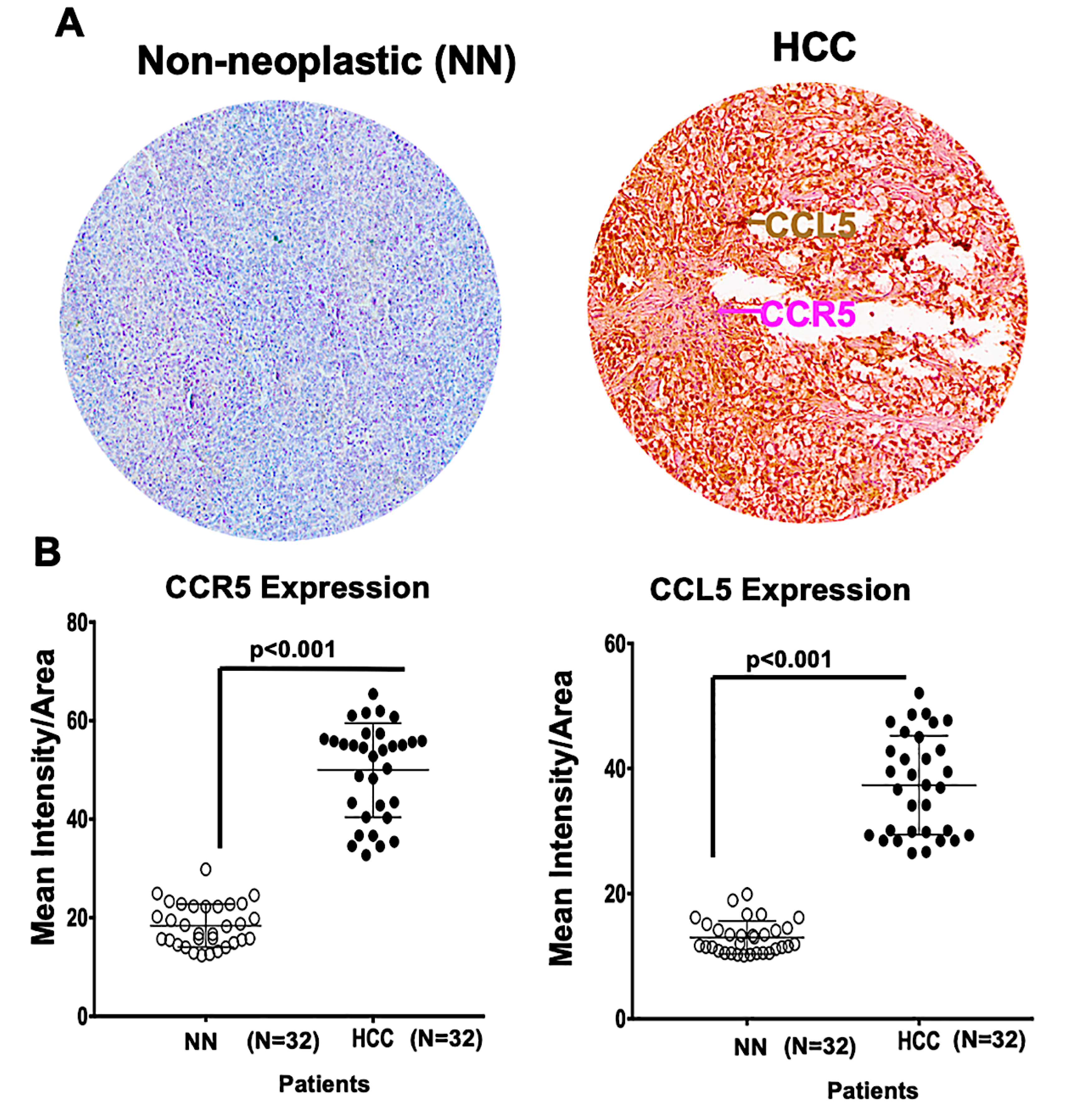
2.1. A Microarray of Human HCC Tissues Shows Upregulated CCR5 and CCL5 Levels
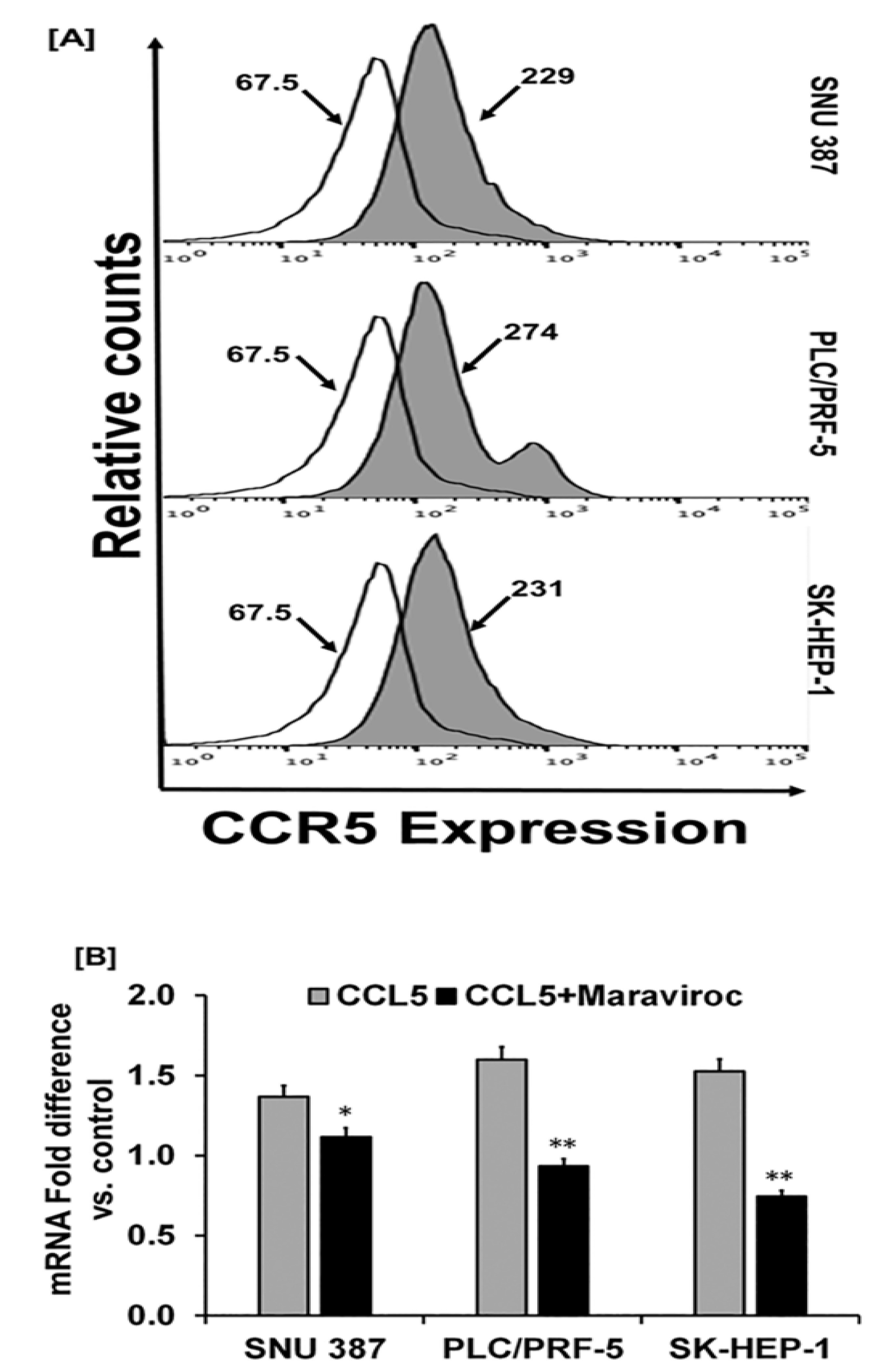
2.2. Human-Derived HCC Cells Display Higher Levels of CCR5 Expression
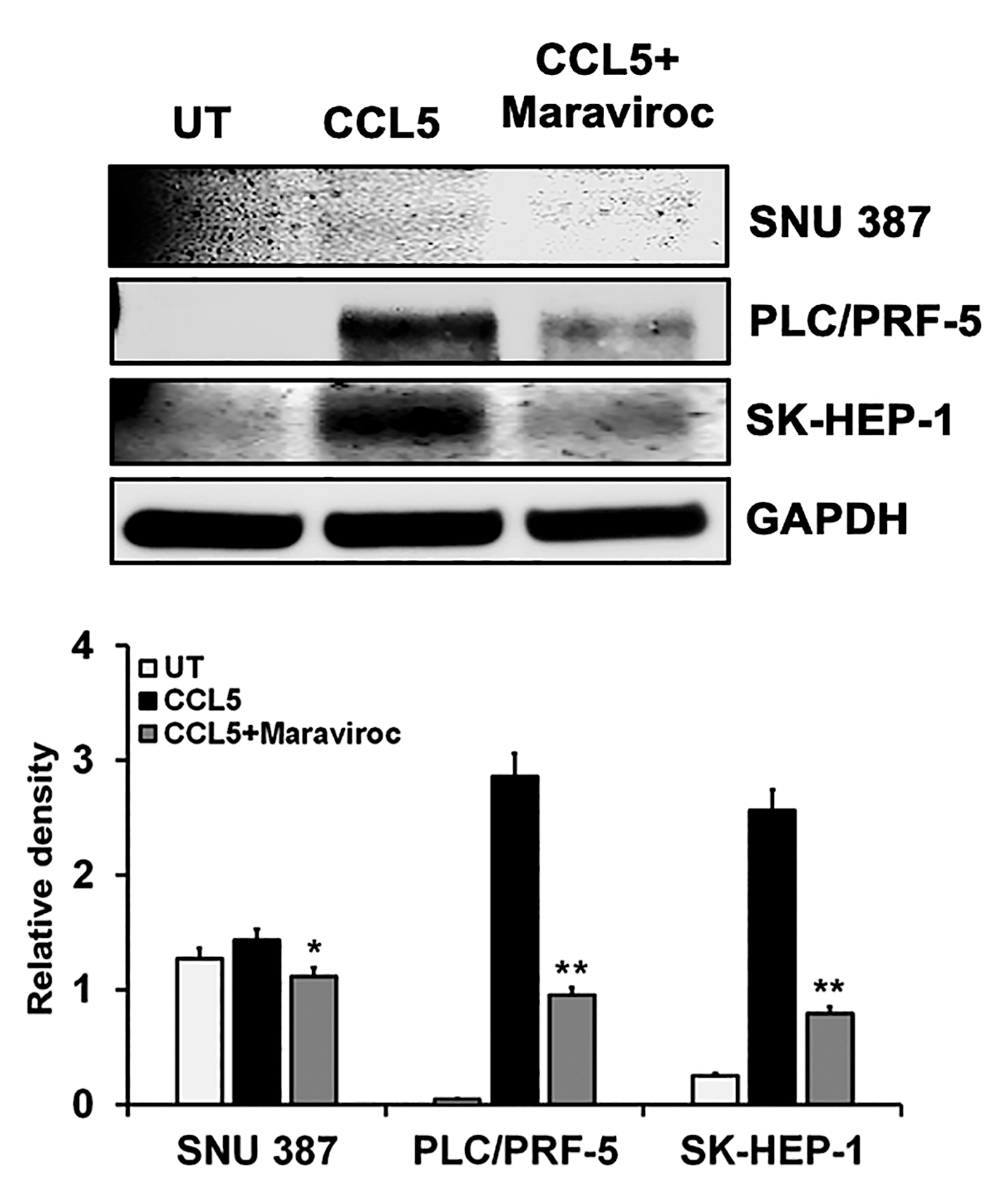
2.3. CCL5 Enhances the CCR5 Expression in HCC Cell Lines
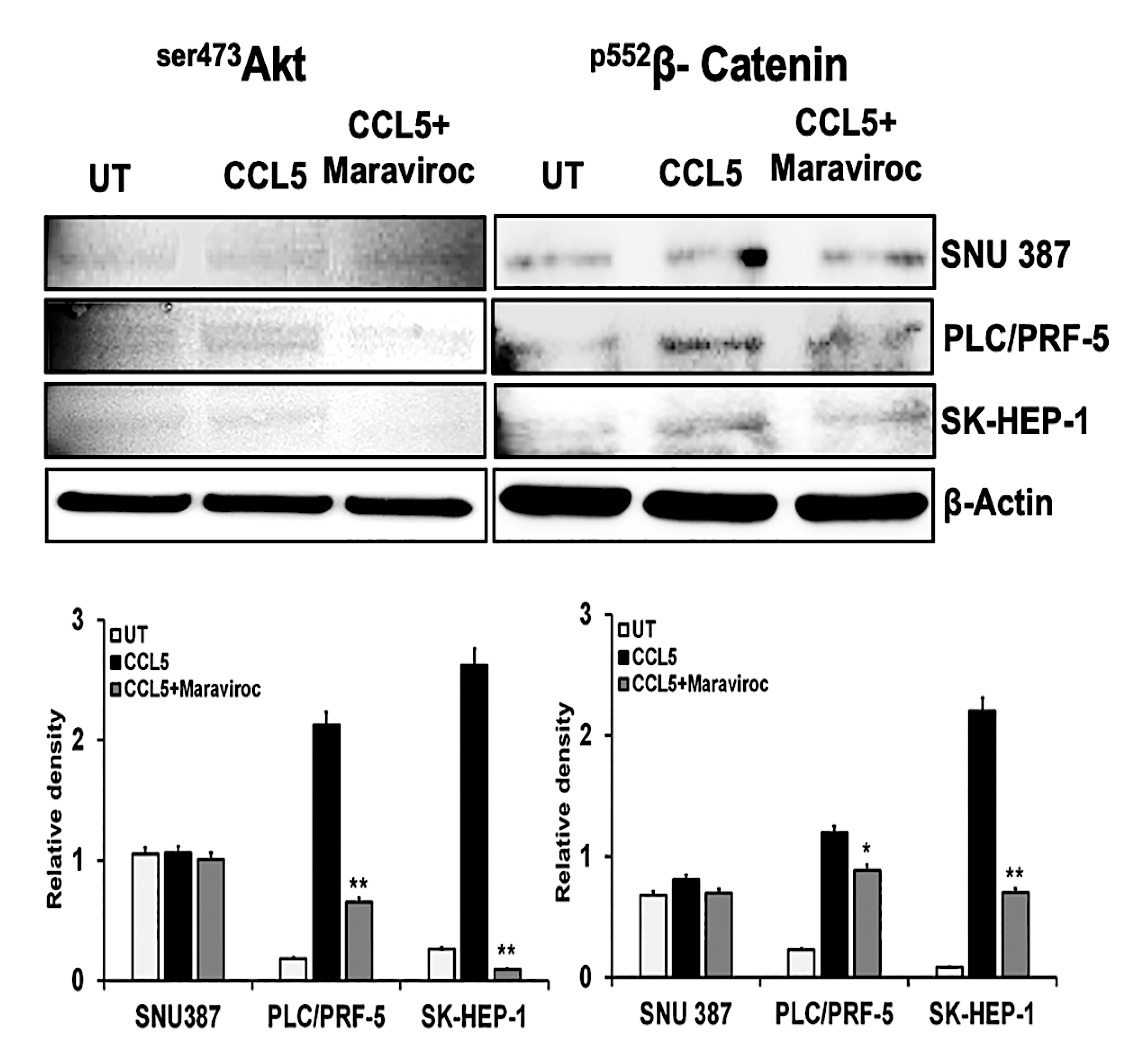
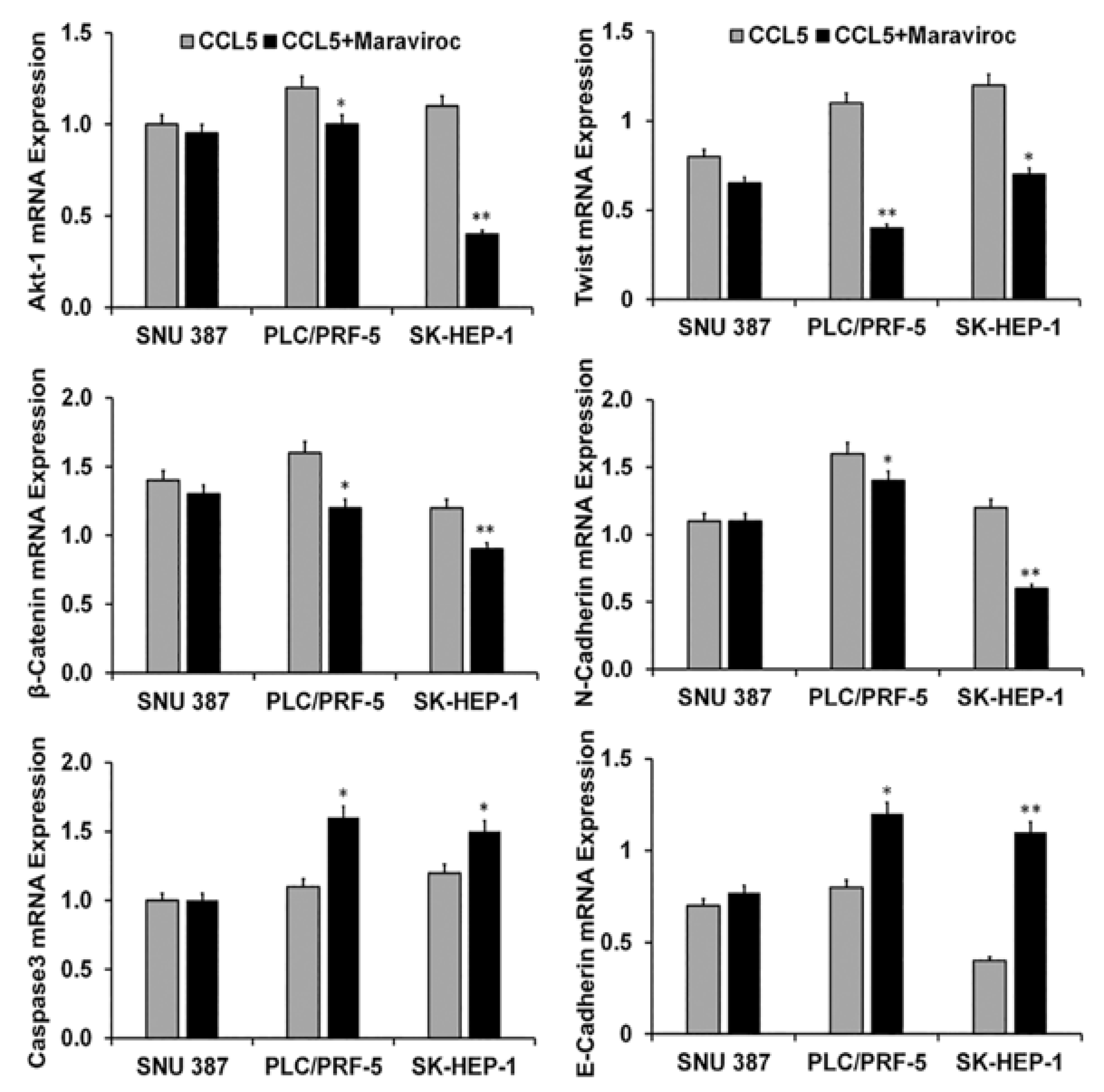
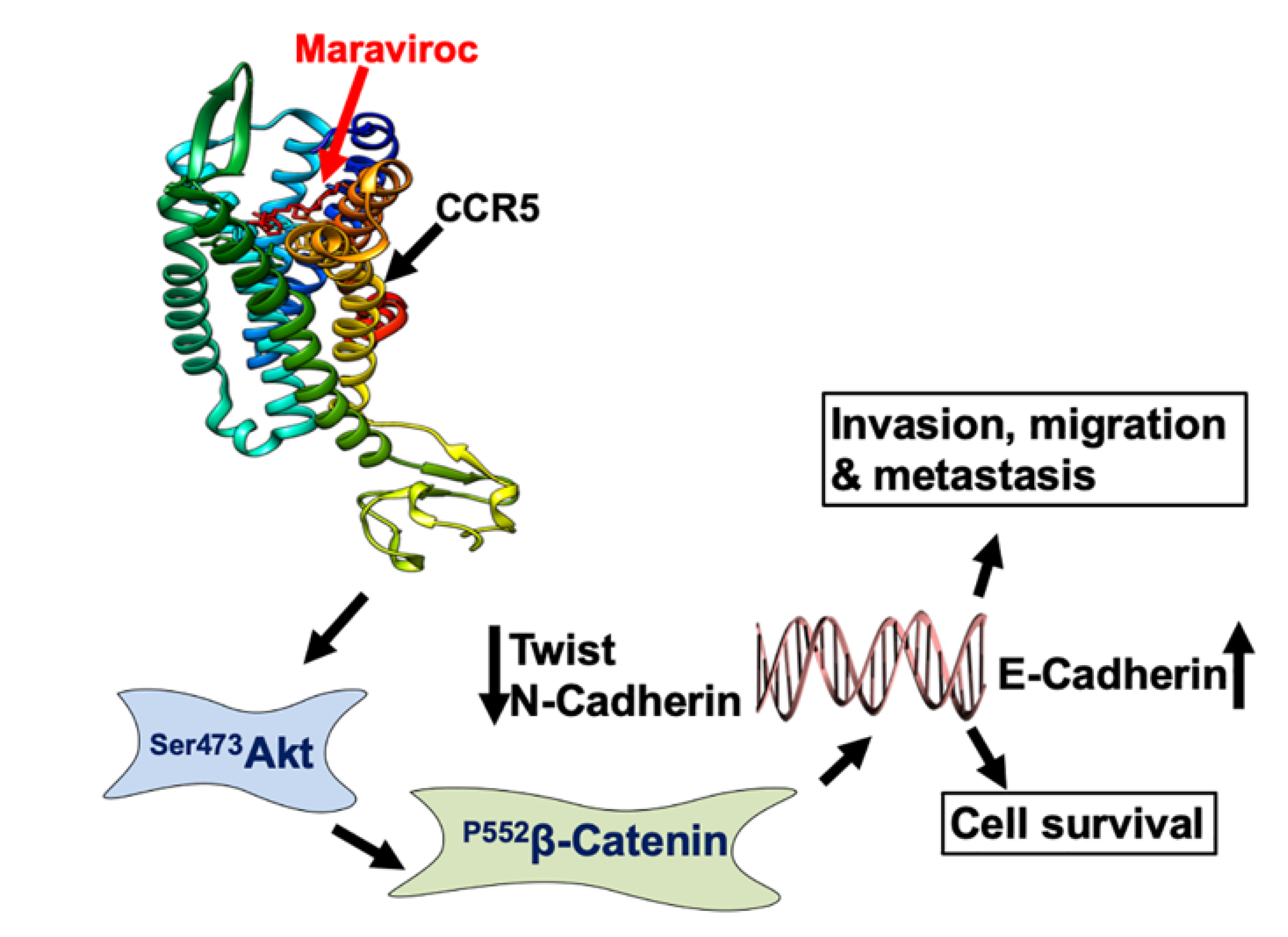
2.4. CCR5/CCL5 Interaction Indicates the Migration Potential of HCC Cells by Enhancing the Expression of Akt and Epithelial to Mesenchymal Transition (EMT) Markers
2.5. Inhibition of the CCR5/CCL5 Interaction Leads to Apoptosis of HCC Cells
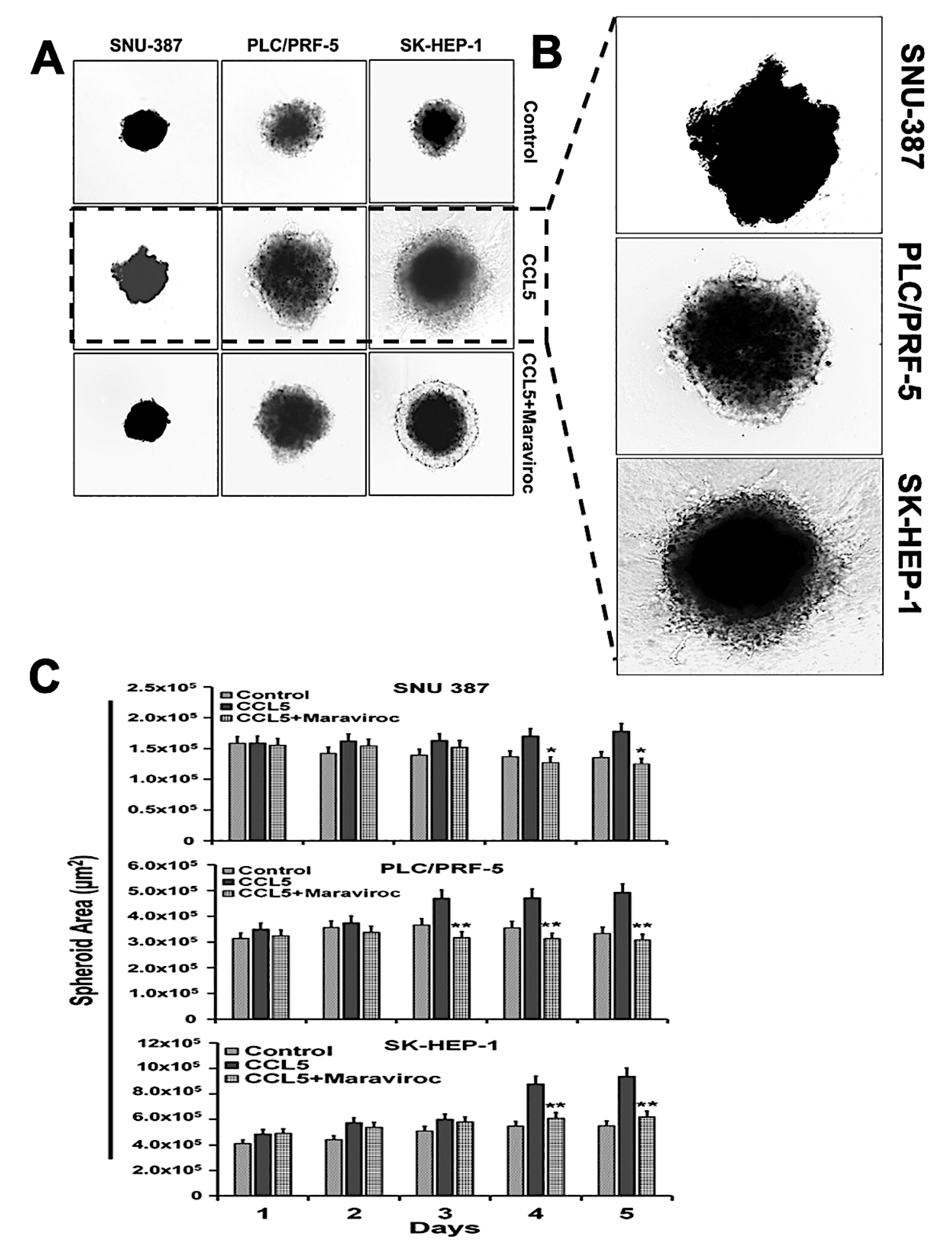
2.6. The CCR5/ CCL5 Interaction Induces Metastatic Behavior in HCC Cells
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cultures
4.2. Immunohistochemistry (IHC) of Clinical Samples
4.3. Flow Cytometry
4.4. Immunofluorescence
4.5. Western Blot Analysis
4.6. Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
4.7. 3 D Cell Migration and Invasion Assay
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Suk, F.M.; Liu, C.L.; Hsu, M.H.; Chuang, Y.T.; Wang, J.P.; Liao, Y.J. Treatment with a new benzimidazole derivative bearing a pyrrolidine side chain overcomes sorafenib resistance in hepatocellular carcinoma. Sci. Rep. 2019, 9, 17259. [Google Scholar] [CrossRef]
- Kummar, S.; Shafi, N.Q. Metastatic hepatocellular carcinoma. Clin. Oncol. (R. Coll. Radiol.) 2003, 15, 288–294. [Google Scholar] [CrossRef]
- Anwanwan, D.; Singh, S.K.; Singh, S.; Saikam, V.; Singh, R. Challenges in liver cancer and possible treatment approaches. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188314. [Google Scholar] [CrossRef]
- Kaemmerer, D.; Schindler, R.; Mussbach, F.; Dahmen, U.; Altendorf-Hofmann, A.; Dirsch, O.; Sanger, J.; Schulz, S.; Lupp, A. Somatostatin and CXCR4 chemokine receptor expression in hepatocellular and cholangiocellular carcinomas: Tumor capillaries as promising targets. BMC Cancer 2017, 17, 896. [Google Scholar] [CrossRef]
- Schlageter, M.; Terracciano, L.M.; D’Angelo, S.; Sorrentino, P. Histopathology of hepatocellular carcinoma. World J. Gastroenterol. 2014, 20, 15955–15964. [Google Scholar] [CrossRef]
- Greten, T.F.; Papendorf, F.; Bleck, J.S.; Kirchhoff, T.; Wohlberedt, T.; Kubicka, S.; Klempnauer, J.; Galanski, M.; Manns, M.P. Survival rate in patients with hepatocellular carcinoma: A retrospective analysis of 389 patients. Br. J. Cancer 2005, 92, 1862–1868. [Google Scholar] [CrossRef]
- Altekruse, S.F.; McGlynn, K.A.; Reichman, M.E. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J. Clin. Oncol. 2009, 27, 1485–1491. [Google Scholar] [CrossRef]
- Gomez, D.; Lobo, D.N. Malignant liver tumours. Surgery 2011, 29, 632–639. [Google Scholar] [CrossRef]
- Bromley, S.K.; Mempel, T.R.; Luster, A.D. Orchestrating the orchestrators: Chemokines in control of T cell traffic. Nat. Immunol. 2008, 9, 970–980. [Google Scholar] [CrossRef]
- Murphy, P.M. International Union of Pharmacology. XXX. Update on chemokine receptor nomenclature. Pharmacol. Rev. 2002, 54, 227–229. [Google Scholar] [CrossRef]
- Muehlhoefer, A.; Saubermann, L.J.; Gu, X.; Luedtke-Heckenkamp, K.; Xavier, R.; Blumberg, R.S.; Podolsky, D.K.; MacDermott, R.P.; Reinecker, H.C. Fractalkine is an epithelial and endothelial cell-derived chemoattractant for intraepithelial lymphocytes in the small intestinal mucosa. J. Immunol. 2000, 164, 3368–3376. [Google Scholar] [CrossRef]
- Hughes, C.E.; Nibbs, R.J.B. A guide to chemokines and their receptors. FEBS J. 2018, 285, 2944–2971. [Google Scholar] [CrossRef]
- Singh, S.K.; Mishra, M.K.; Singh, R. Hypoxia-inducible factor-1alpha induces CX3CR1 expression and promotes the epithelial to mesenchymal transition (EMT) in ovarian cancer cells. J. Ovarian Res. 2019, 12, 42. [Google Scholar] [CrossRef]
- Vaday, G.G.; Peehl, D.M.; Kadam, P.A.; Lawrence, D.M. Expression of CCL5 (RANTES) and CCR5 in prostate cancer. Prostate 2006, 66, 124–134. [Google Scholar] [CrossRef]
- Li, W.; Gomez, E.; Zhang, Z. Immunohistochemical expression of stromal cell-derived factor-1 (SDF-1) and CXCR4 ligand receptor system in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2007, 26, 527–533. [Google Scholar]
- Fujii, H.; Itoh, Y.; Yamaguchi, K.; Yamauchi, N.; Harano, Y.; Nakajima, T.; Minami, M.; Okanoue, T. Chemokine CCL20 enhances the growth of HuH7 cells via phosphorylation of p44/42 MAPK in vitro. Biochem. Biophys. Res. Commun. 2004, 322, 1052–1058. [Google Scholar] [CrossRef]
- Sutton, A.; Friand, V.; Papy-Garcia, D.; Dagouassat, M.; Martin, L.; Vassy, R.; Haddad, O.; Sainte-Catherine, O.; Kraemer, M.; Saffar, L.; et al. Glycosaminoglycans and their synthetic mimetics inhibit RANTES-induced migration and invasion of human hepatoma cells. Mol. Cancer Ther. 2007, 6, 2948–2958. [Google Scholar] [CrossRef]
- Singh, S.K.; Mishra, M.K.; Eltoum, I.A.; Bae, S.; Lillard, J.W., Jr.; Singh, R. CCR5/CCL5 axis interaction promotes migratory and invasiveness of pancreatic cancer cells. Sci. Rep. 2018, 8, 1323. [Google Scholar] [CrossRef]
- Appay, V.; Rowland-Jones, S.L. RANTES: A versatile and controversial chemokine. Trends Immunol. 2001, 22, 83–87. [Google Scholar] [CrossRef]
- Aldinucci, D.; Colombatti, A. The inflammatory chemokine CCL5 and cancer progression. Med. Inflamm. 2014, 2014, 292376. [Google Scholar] [CrossRef]
- Soria, G.; Ben-Baruch, A. The inflammatory chemokines CCL2 and CCL5 in breast cancer. Cancer Lett. 2008, 267, 271–285. [Google Scholar] [CrossRef]
- Cambien, B.; Richard-Fiardo, P.; Karimdjee, B.F.; Martini, V.; Ferrua, B.; Pitard, B.; Schmid-Antomarchi, H.; Schmid-Alliana, A. CCL5 neutralization restricts cancer growth and potentiates the targeting of PDGFRbeta in colorectal carcinoma. PLoS ONE 2011, 6, e28842. [Google Scholar] [CrossRef]
- Halama, N.; Zoernig, I.; Berthel, A.; Kahlert, C.; Klupp, F.; Suarez-Carmona, M.; Suetterlin, T.; Brand, K.; Krauss, J.; Lasitschka, F.; et al. Tumoral Immune Cell Exploitation in Colorectal Cancer Metastases Can Be Targeted Effectively by Anti-CCR5 Therapy in Cancer Patients. Cancer Cell 2016, 29, 587–601. [Google Scholar] [CrossRef]
- Wang, S.W.; Wu, H.H.; Liu, S.C.; Wang, P.C.; Ou, W.C.; Chou, W.Y.; Shen, Y.S.; Tang, C.H. CCL5 and CCR5 interaction promotes cell motility in human osteosarcoma. PLoS ONE 2012, 7, e35101. [Google Scholar] [CrossRef]
- Aldinucci, D.; Casagrande, N. Inhibition of the CCL5/CCR5 Axis against the Progression of Gastric Cancer. Int. J. Mol. Sci. 2018, 19, 1477. [Google Scholar] [CrossRef]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef]
- Zhou, Q.; Lui, V.W.; Yeo, W. Targeting the PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Future Oncol. 2011, 7, 1149–1167. [Google Scholar] [CrossRef]
- Jiao, X.; Nawab, O.; Patel, T.; Kossenkov, A.V.; Halama, N.; Jaeger, D.; Pestell, R.G. Recent Advances Targeting CCR5 for Cancer and Its Role in Immuno-Oncology. Cancer Res. 2019, 79, 4801–4807. [Google Scholar] [CrossRef]
- Karnoub, A.E.; Dash, A.B.; Vo, A.P.; Sullivan, A.; Brooks, M.W.; Bell, G.W.; Richardson, A.L.; Polyak, K.; Tubo, R.; Weinberg, R.A. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature 2007, 449, 557–563. [Google Scholar] [CrossRef]
- Lin, Y.L.; Liu, C.C.; Chuang, J.I.; Lei, H.Y.; Yeh, T.M.; Lin, Y.S.; Huang, Y.H.; Liu, H.S. Involvement of oxidative stress, NF-IL-6, and RANTES expression in dengue-2-virus-infected human liver cells. Virology 2000, 276, 114–126. [Google Scholar] [CrossRef]
- Nahon, P.; Sutton, A.; Rufat, P.; Simon, C.; Trinchet, J.C.; Gattegno, L.; Beaugrand, M.; Charnaux, N. Chemokine system polymorphisms, survival and hepatocellular carcinoma occurrence in patients with hepatitis C virus-related cirrhosis. World J. Gastroenterol. 2008, 14, 713–719. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Faried, L.S.; Faried, A.; Kanuma, T.; Nakazato, T.; Tamura, T.; Kuwano, H.; Minegishi, T. Inhibition of the mammalian target of rapamycin (mTOR) by rapamycin increases chemosensitivity of CaSki cells to paclitaxel. Eur. J. Cancer 2006, 42, 934–947. [Google Scholar] [CrossRef] [PubMed]
- VanderWeele, D.J.; Zhou, R.; Rudin, C.M. Akt up-regulation increases resistance to microtubule-directed chemotherapeutic agents through mammalian target of rapamycin. Mol. Cancer Ther. 2004, 3, 1605–1613. [Google Scholar] [PubMed]
- Yap, T.A.; Garrett, M.D.; Walton, M.I.; Raynaud, F.; de Bono, J.S.; Workman, P. Targeting the PI3K-AKT-mTOR pathway: Progress, pitfalls, and promises. Curr. Opin. Pharmacol. 2008, 8, 393–412. [Google Scholar] [CrossRef]
- Dunlop, E.A.; Tee, A.R. Mammalian target of rapamycin complex 1: Signalling inputs, substrates and feedback mechanisms. Cell Signal. 2009, 21, 827–835. [Google Scholar] [CrossRef]
- Yoshida, G.J. Emerging role of epithelial-mesenchymal transition in hepatic cancer. J. Exp. Clin. Cancer Res. 2016, 35, 141. [Google Scholar] [CrossRef]
- Yasuhara, R.; Irie, T.; Suzuki, K.; Sawada, T.; Miwa, N.; Sasaki, A.; Tsunoda, Y.; Nakamura, S.; Mishima, K. The beta-catenin signaling pathway induces aggressive potential in breast cancer by up-regulating the chemokine CCL5. Exp. Cell Res. 2015, 338, 22–31. [Google Scholar] [CrossRef]
- Singh, S.K.; Singh, R. Liver cancer incidence and mortality: Disparities based on age, ethnicity, health and nutrition, molecular factors, and geography. Cancer Health Disparities 2019. [Google Scholar] [CrossRef]
- Abdolmohammadi, R.; Shahbazi Azar, S.; Khosravi, A.; Shahbazi, M. CCR5 Polymorphism as a Protective Factor for Hepatocellular Carcinoma in Hepatitis B Virus-Infected Iranian Patients. Asian Pac. J. Cancer Prev. 2016, 17, 4643–4646. [Google Scholar] [CrossRef]
- Suneetha, P.V.; Sarin, S.K.; Goyal, A.; Kumar, G.T.; Shukla, D.K.; Hissar, S. Association between vitamin D receptor, CCR5, TNF-alpha and TNF-beta gene polymorphisms and HBV infection and severity of liver disease. J. Hepatol. 2006, 44, 856–863. [Google Scholar] [CrossRef]
- Gonzalez-Martin, A.; Mira, E.; Manes, S. CCR5 in cancer immunotherapy: More than an “attractive” receptor for T cells. Oncoimmunology 2012, 1, 106–108. [Google Scholar] [CrossRef] [PubMed]
- Velasco-Velazquez, M.; Xolalpa, W.; Pestell, R.G. The potential to target CCL5/CCR5 in breast cancer. Expert Opin. Ther. Targets 2014, 18, 1265–1275. [Google Scholar] [CrossRef] [PubMed]
- Hawila, E.; Razon, H.; Wildbaum, G.; Blattner, C.; Sapir, Y.; Shaked, Y.; Umansky, V.; Karin, N. CCR5 Directs the Mobilization of CD11b(+)Gr1(+)Ly6C(low) Polymorphonuclear Myeloid Cells from the Bone Marrow to the Blood to Support Tumor Development. Cell Rep. 2017, 21, 2212–2222. [Google Scholar] [CrossRef] [PubMed]
- Vinci, M.; Gowan, S.; Boxall, F.; Patterson, L.; Zimmermann, M.; Court, W.; Lomas, C.; Mendiola, M.; Hardisson, D.; Eccles, S.A. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Robertson, G.; Pedersen, L.; Lim, E.; Hernandez-Herrera, A.; Rowat, A.C.; Patil, S.L.; Chan, C.K.; Wen, Y.; Zhang, X.; et al. Correction: MiR-509-3p is clinically significant and strongly attenuates cellular migration and multi-cellular spheroids in ovarian cancer. Oncotarget 2017, 8, 17406. [Google Scholar] [CrossRef]

| Gene | Sense | Antisense |
|---|---|---|
| 18S | 5′-GGCCCTGTAATTGGAATGAGTC-3′ | 5′-CCAAGATCCAACTACGAGCTT-3′ |
| CCR5 | 5′-GCAAGGAGACCACCAACAG-3X | 5′-CCCTCACTTCCAACCCAAATC-3′ |
| Akt-1 | 5′-ATGGACAGGGAGAGCAAACG-3′ | 5′-CTGGCCACAGCCTCTGATG-3′ |
| β-catenin | 5′-TCCTCAGATGGTGTCTGCTA-3′ | 5′-GATGATGGGAAAGGTTATGC-3′ |
| Caspase-3 | 5′-CTCTGGTTTTCGGTGGGTGT-3′ | 5′-CGCTTCCATGTATGATCTTTGGTT-3′ |
| Twist | 5′-AGCTGAGCAAGATTCAGACC-3′ | 5′-CAGCTTGCCATCTTGGAGT-3′ |
| N-cadherin | 5′-TACAGACATGGAAGGAATCCCC-3X | 5′-ATGGCAGTAAACTCTGGAGGA-3′ |
| E-cadherin | 5′-CGTCCTGGGCAGAGTGAAT-3′ | 5′-TTTGAATCGGGTGTCGAGGG-3′ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, S.K.; Mishra, M.K.; Rivers, B.M.; Gordetsky, J.B.; Bae, S.; Singh, R. Biological and Clinical Significance of the CCR5/CCL5 Axis in Hepatocellular Carcinoma. Cancers 2020, 12, 883. https://doi.org/10.3390/cancers12040883
Singh SK, Mishra MK, Rivers BM, Gordetsky JB, Bae S, Singh R. Biological and Clinical Significance of the CCR5/CCL5 Axis in Hepatocellular Carcinoma. Cancers. 2020; 12(4):883. https://doi.org/10.3390/cancers12040883
Chicago/Turabian StyleSingh, Santosh K., Manoj K. Mishra, Brian M. Rivers, Jennifer B. Gordetsky, Sejong Bae, and Rajesh Singh. 2020. "Biological and Clinical Significance of the CCR5/CCL5 Axis in Hepatocellular Carcinoma" Cancers 12, no. 4: 883. https://doi.org/10.3390/cancers12040883
APA StyleSingh, S. K., Mishra, M. K., Rivers, B. M., Gordetsky, J. B., Bae, S., & Singh, R. (2020). Biological and Clinical Significance of the CCR5/CCL5 Axis in Hepatocellular Carcinoma. Cancers, 12(4), 883. https://doi.org/10.3390/cancers12040883








