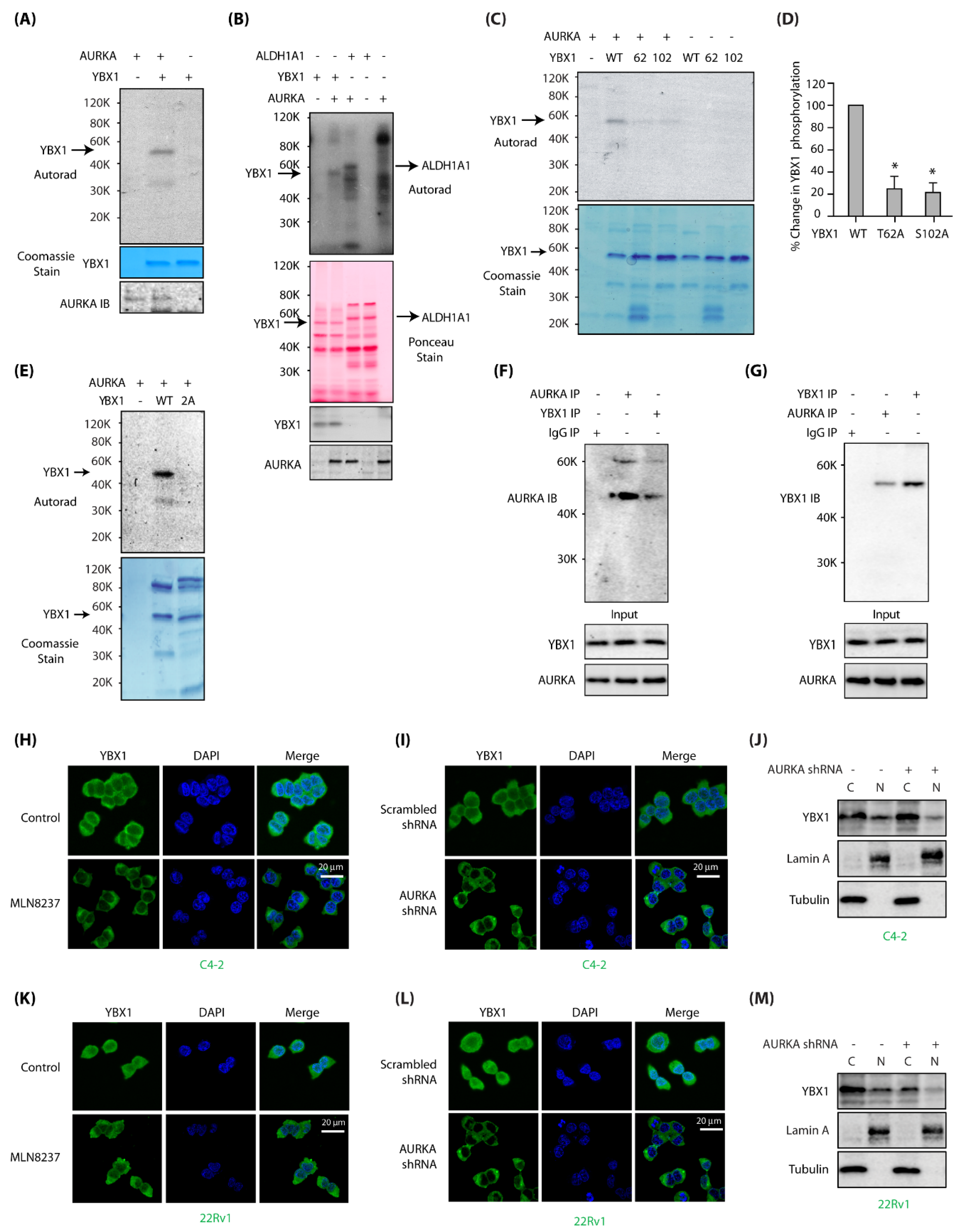
Figure 1.
Aurora Kinase-A (AURKA) associates and directly phosphorylates Y-box binding protein-1 (YBX1) at two sites and regulates its localization in C4-2 and 22Rv1 cells. (A) YBX1 is a substrate of AURKA in vitro. AURKA-TPX2 was mixed with either [32P]ATP (lane 1), or 6x-His–YBX1 and [32P]ATP (lane 2) in a kinase assay for 30 min. Lane 3 shows YBX1 mixed with [32P]ATP. (B) YBX1 shows comparable phosphorylation levels as ALDH1A1, a known AURKA substrate. YBX1 and ALDH1A1 were subjected to kinase assay with AURKA and [32P]ATP (lanes 2 and 3, respectively). Lanes 1 and 4 are controls with YBX1 and ALDH1A1, respectively, with [32P]ATP. Lane 5 shows AURKA with [32P]ATP. (C) AURKA phosphorylates YBX1 at T62 and S102. Phospho-resistant YBX1 single mutants were exposed to an in vitro kinase assay using AURKA-TPX2 and [32P] ATP (lanes 3 and 4, respectively). WT YBX1 with [32P] ATP was used as a positive control (lane 2). Lanes 5, 6 and 7 show WT, T62 and S102 mutants exposed to [32P] ATP, respectively in the absence of AURKA. (D) Histogram shows % change in phosphorylation of WT and phospho-dead single mutants of YBX1 from three independent experiments. (E) T62 and S102 are the only AURKA sites on YBX1, as 2A-YBX1 mutant is not phosphorylated by AURKA (lane 3). Lane 2 shows WT YBX1 with AURKA and [32P] ATP. (F) YBX1 associates with AURKA in cells. YBX1 IP was conducted in C4-2 cells, and AURKA levels analyzed (lane 3). IgG (lane 1) and AURKA (lane 2) were used as negative and positive controls, respectively. (G) AURKA binds YBX1 in C4-2 cells. AURKA IP was conducted in C4-2 cells, and YBX1 levels analyzed (lane 2). IgG (lane 1) and YBX1 (lane 3) were used as negative and positive controls, respectively. (H) AURKA inhibition using MLN8237 inhibits nuclear translocation of YBX1 in C4-2 cells. C4-2 cells were treated with 1 μM MLN8237 for 12 h, and AURKA subcellular localization analyzed using AURKA-specific antibody (green). DAPI is shown in blue. (I) AURKA depletion inhibits nuclear translocation of YBX1. C4-2 cells were exposed to control or AURKA shRNA for 30h, fixed and stained with YBX1 antibody (green) or DAPI (blue). >100 cells were analyzed from multiple random frames. (J) Subcellular fractionation of scrambled shRNA-treated and AURKA shRNA-treated C4-2 cells confirms reduced nuclear translocation of YBX1 upon AURKA depletion. C4-2 cells were exposed to control or AURKA shRNA for 30 h prior to fractionation. (K) AURKA inhibition using MLN8237 prevents nuclear translocation of YBX1 in 22Rv1 cells. 22Rv1 cells were treated with 1 μM MLN8237 for 12 h, and AURKA subcellular localization analyzed using AURKA-specific antibody (green). (L) AURKA depletion inhibits nuclear translocation of YBX1 in 22Rv1 cells. 22Rv1 cells were exposed to control or AURKA shRNA for 30 h, fixed and stained with YBX1 antibody (green) or DAPI (blue). (M) Subcellular fractionation confirms reduced nuclear translocation of YBX1 upon AURKA depletion in 22Rv1 cells. 22Rv1 cells were exposed to control or AURKA shRNA for 30 h prior to fractionation.
Figure 1.
Aurora Kinase-A (AURKA) associates and directly phosphorylates Y-box binding protein-1 (YBX1) at two sites and regulates its localization in C4-2 and 22Rv1 cells. (A) YBX1 is a substrate of AURKA in vitro. AURKA-TPX2 was mixed with either [32P]ATP (lane 1), or 6x-His–YBX1 and [32P]ATP (lane 2) in a kinase assay for 30 min. Lane 3 shows YBX1 mixed with [32P]ATP. (B) YBX1 shows comparable phosphorylation levels as ALDH1A1, a known AURKA substrate. YBX1 and ALDH1A1 were subjected to kinase assay with AURKA and [32P]ATP (lanes 2 and 3, respectively). Lanes 1 and 4 are controls with YBX1 and ALDH1A1, respectively, with [32P]ATP. Lane 5 shows AURKA with [32P]ATP. (C) AURKA phosphorylates YBX1 at T62 and S102. Phospho-resistant YBX1 single mutants were exposed to an in vitro kinase assay using AURKA-TPX2 and [32P] ATP (lanes 3 and 4, respectively). WT YBX1 with [32P] ATP was used as a positive control (lane 2). Lanes 5, 6 and 7 show WT, T62 and S102 mutants exposed to [32P] ATP, respectively in the absence of AURKA. (D) Histogram shows % change in phosphorylation of WT and phospho-dead single mutants of YBX1 from three independent experiments. (E) T62 and S102 are the only AURKA sites on YBX1, as 2A-YBX1 mutant is not phosphorylated by AURKA (lane 3). Lane 2 shows WT YBX1 with AURKA and [32P] ATP. (F) YBX1 associates with AURKA in cells. YBX1 IP was conducted in C4-2 cells, and AURKA levels analyzed (lane 3). IgG (lane 1) and AURKA (lane 2) were used as negative and positive controls, respectively. (G) AURKA binds YBX1 in C4-2 cells. AURKA IP was conducted in C4-2 cells, and YBX1 levels analyzed (lane 2). IgG (lane 1) and YBX1 (lane 3) were used as negative and positive controls, respectively. (H) AURKA inhibition using MLN8237 inhibits nuclear translocation of YBX1 in C4-2 cells. C4-2 cells were treated with 1 μM MLN8237 for 12 h, and AURKA subcellular localization analyzed using AURKA-specific antibody (green). DAPI is shown in blue. (I) AURKA depletion inhibits nuclear translocation of YBX1. C4-2 cells were exposed to control or AURKA shRNA for 30h, fixed and stained with YBX1 antibody (green) or DAPI (blue). >100 cells were analyzed from multiple random frames. (J) Subcellular fractionation of scrambled shRNA-treated and AURKA shRNA-treated C4-2 cells confirms reduced nuclear translocation of YBX1 upon AURKA depletion. C4-2 cells were exposed to control or AURKA shRNA for 30 h prior to fractionation. (K) AURKA inhibition using MLN8237 prevents nuclear translocation of YBX1 in 22Rv1 cells. 22Rv1 cells were treated with 1 μM MLN8237 for 12 h, and AURKA subcellular localization analyzed using AURKA-specific antibody (green). (L) AURKA depletion inhibits nuclear translocation of YBX1 in 22Rv1 cells. 22Rv1 cells were exposed to control or AURKA shRNA for 30 h, fixed and stained with YBX1 antibody (green) or DAPI (blue). (M) Subcellular fractionation confirms reduced nuclear translocation of YBX1 upon AURKA depletion in 22Rv1 cells. 22Rv1 cells were exposed to control or AURKA shRNA for 30 h prior to fractionation.
![Cancers 12 00660 g001 Cancers 12 00660 g001]()
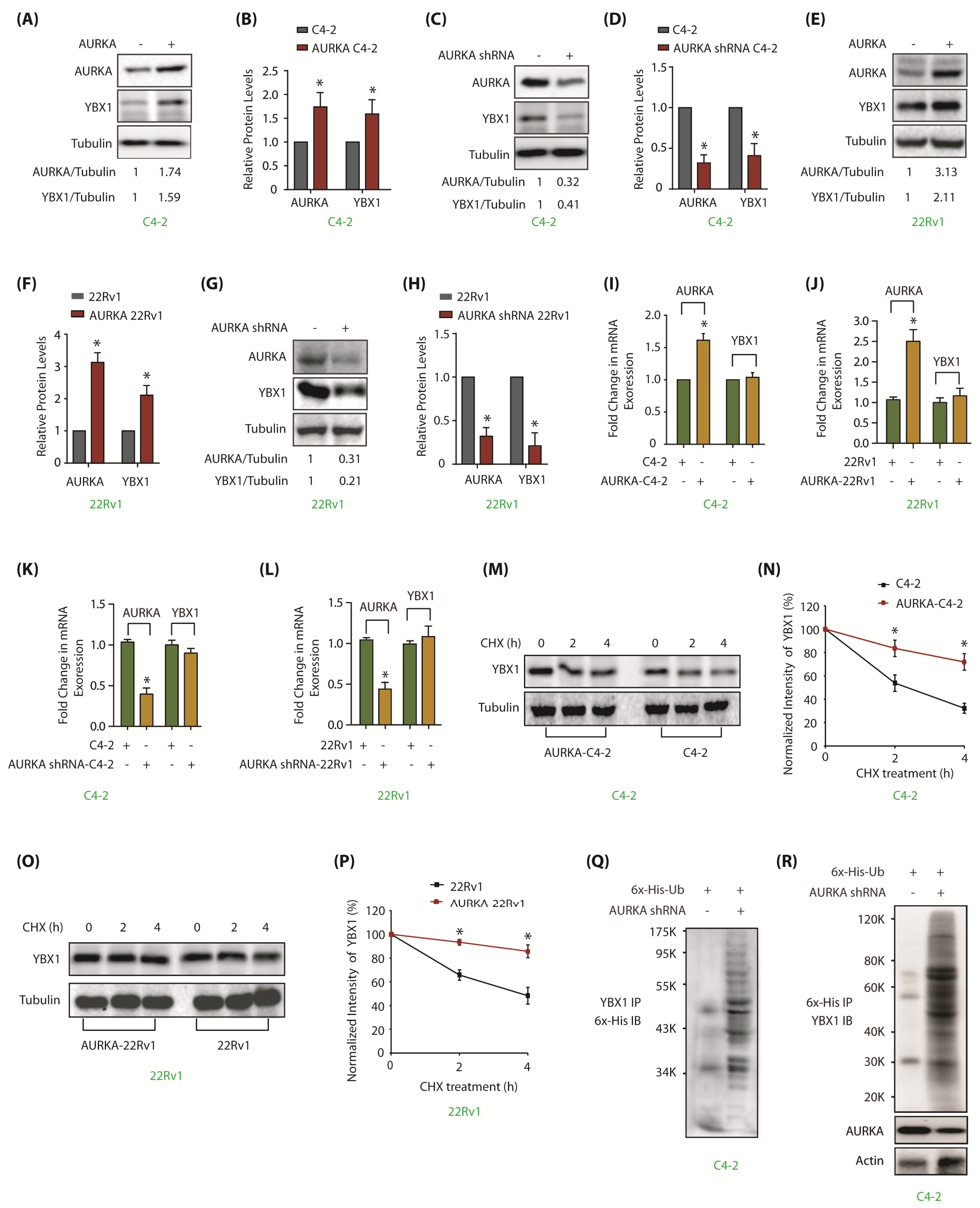
Figure 2.
AURKA upregulates YBX1 levels by inhibiting its ubiquitylation in C4-2 and 22Rv1 cells. (A) Overexpression of AURKA increases YBX1 levels in C4-2 cells. (B) The data represents mean ± SEM of three independent experiments in C4-2 cells. * signifies significance at p < 0.05 compared to control. (C) AURKA ablation using AURKA shRNA depletes YBX1. C4-2 cells were exposed to control or AURKA shRNA for 30 h. (D) Histogram shows relative band intensities of AURKA and YBX1 normalized to the corresponding tubulin level from three independent experiments. (E) Overexpression of AURKA increases YBX1 levels in 22Rv1 cells. (F) Histogram shows relative band intensities of AURKA and YBX1 normalized to the corresponding tubulin level from three independent experiments. (G) AURKA ablation using AURKA shRNA (treated for 30 h) inhibits YBX1 protein levels in 22Rv1 cells. (H) Histogram shows relative band intensities of AURKA and YBX1 normalized to the corresponding tubulin level from three independent experiments. (I,J) AURKA does not regulate the mRNA levels of YBX1 in C4-2 and 22Rv1 cells, respectively. AURKA was stably overexpressed and YBX1 levels analyzed using real time qPCR. (K,L) AURKA does not regulate the mRNA levels of YBX1 in C4-2 and 22Rv1 cells, respectively. AURKA was knocked-down and YBX1 mRNA levels analyzed. C4-2 and 22Rv1 cells were exposed to control or AURKA shRNA for 30 h. (M) AURKA prevents YBX1 degradation. AURKA-C4-2 and C4-2 cells were treated with cycloheximide for 2 and 4 h, and YBX1 levels analyzed. (N) Graphical representation of YBX1 degradation rate. The YBX1 band intensity was normalized to tubulin and then normalized to the t=0 controls. Data are shown as mean± SEM from three different experiments (n = 3) * p < 0.05. (O) AURKA prevents YBX1 degradation in 22Rv1 cells. AURKA-22Rv1 and 22Rv1 cells were treated with cycloheximide for 2 and 4 h, and YBX1 levels analyzed. (P) Graphical representation of YBX1 degradation rate in 22Rv1 cells. The YBX1 band intensity was normalized to tubulin and then normalized to the t=0 controls. Data are shown as mean± SEM from three different experiments (n = 3) * p < 0.05. (Q) AURKA stabilizes YBX1 by inhibiting its ubiquitylation. 6x-His-Ubiquitin-expressing C4-2 cells were infected with either scrambled or AURKA shRNA lentivirus for 30 h and then treated with MG132 for 12 h. YBX1 was isolated and ubiquitylation analyzed using 6x-His antibody. (R) AURKA stabilizes YBX1 by inhibiting its ubiquitylation. Ubiquitylation was performed as described above, except ubiqitylated proteins were isolated using Ni-NTA beads followed by YBX1 IB. Each experiment was done at least three independent times and representative data are shown.
Figure 2.
AURKA upregulates YBX1 levels by inhibiting its ubiquitylation in C4-2 and 22Rv1 cells. (A) Overexpression of AURKA increases YBX1 levels in C4-2 cells. (B) The data represents mean ± SEM of three independent experiments in C4-2 cells. * signifies significance at p < 0.05 compared to control. (C) AURKA ablation using AURKA shRNA depletes YBX1. C4-2 cells were exposed to control or AURKA shRNA for 30 h. (D) Histogram shows relative band intensities of AURKA and YBX1 normalized to the corresponding tubulin level from three independent experiments. (E) Overexpression of AURKA increases YBX1 levels in 22Rv1 cells. (F) Histogram shows relative band intensities of AURKA and YBX1 normalized to the corresponding tubulin level from three independent experiments. (G) AURKA ablation using AURKA shRNA (treated for 30 h) inhibits YBX1 protein levels in 22Rv1 cells. (H) Histogram shows relative band intensities of AURKA and YBX1 normalized to the corresponding tubulin level from three independent experiments. (I,J) AURKA does not regulate the mRNA levels of YBX1 in C4-2 and 22Rv1 cells, respectively. AURKA was stably overexpressed and YBX1 levels analyzed using real time qPCR. (K,L) AURKA does not regulate the mRNA levels of YBX1 in C4-2 and 22Rv1 cells, respectively. AURKA was knocked-down and YBX1 mRNA levels analyzed. C4-2 and 22Rv1 cells were exposed to control or AURKA shRNA for 30 h. (M) AURKA prevents YBX1 degradation. AURKA-C4-2 and C4-2 cells were treated with cycloheximide for 2 and 4 h, and YBX1 levels analyzed. (N) Graphical representation of YBX1 degradation rate. The YBX1 band intensity was normalized to tubulin and then normalized to the t=0 controls. Data are shown as mean± SEM from three different experiments (n = 3) * p < 0.05. (O) AURKA prevents YBX1 degradation in 22Rv1 cells. AURKA-22Rv1 and 22Rv1 cells were treated with cycloheximide for 2 and 4 h, and YBX1 levels analyzed. (P) Graphical representation of YBX1 degradation rate in 22Rv1 cells. The YBX1 band intensity was normalized to tubulin and then normalized to the t=0 controls. Data are shown as mean± SEM from three different experiments (n = 3) * p < 0.05. (Q) AURKA stabilizes YBX1 by inhibiting its ubiquitylation. 6x-His-Ubiquitin-expressing C4-2 cells were infected with either scrambled or AURKA shRNA lentivirus for 30 h and then treated with MG132 for 12 h. YBX1 was isolated and ubiquitylation analyzed using 6x-His antibody. (R) AURKA stabilizes YBX1 by inhibiting its ubiquitylation. Ubiquitylation was performed as described above, except ubiqitylated proteins were isolated using Ni-NTA beads followed by YBX1 IB. Each experiment was done at least three independent times and representative data are shown.
![Cancers 12 00660 g002 Cancers 12 00660 g002]()
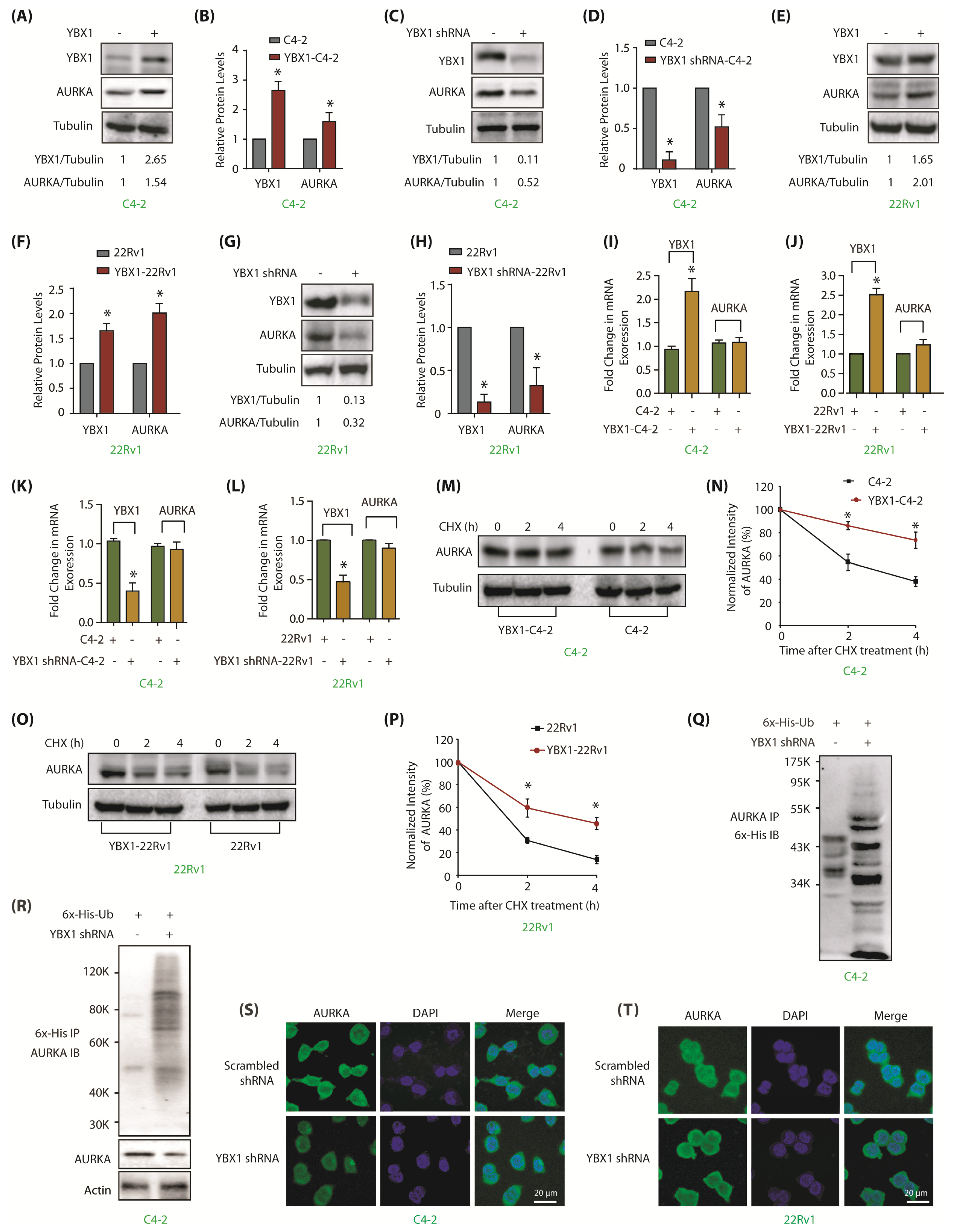
Figure 3.
YBX1 stabilizes AURKA protein in C4-2 and 22Rv1 cells. (A) Overexpression of YBX1 enhances AURKA levels. (B) Histogram shows relative band intensities of YBX1 and AURKA normalized to the corresponding tubulin level from three independent experiments. * signifies significance at p < 0.05 compared to control. (C) YBX1 knock-down using YBX1 shRNA decreases AURKA in C4-2 cells. Cells were infected with control shRNA lentivirus (lane 1), or YBX1-shRNA lentivirus (lane 2), and AURKA and YBX1 levels were analyzed after 30 h. (D) Histogram shows relative band intensities of YBX1 and AURKA normalized to the corresponding tubulin level from three independent experiments. (E) YBX1 overexpression enhances AURKA levels in 22Rv1 cells. (F) Histogram shows relative band intensities of YBX1 and AURKA normalized to the corresponding tubulin level from three independent experiments. (G) YBX1 knockdown depletes AURKA protein levels in 22Rv1 cells. Cells were infected with control shRNA lentivirus (lane 1), or YBX1-shRNA lentivirus (lane 2), and AURKA and YBX1 levels were analyzed after 30 h. (H) Histogram showing relative band intensities of YBX1 and AURKA normalized to the corresponding tubulin level from three independent experiments. (I,J) YBX1 does not regulate the mRNA levels of AURKA in C4-2 and 22Rv1 cells, respectively. YBX1 was overexpressed and AURKA mRNA levels analyzed. (K,L) YBX1 does not regulate the mRNA levels of AURKA in C4-2 and 22Rv1 cells, respectively. YBX1 was knocked-down and AURKA mRNA levels analyzed. (M) YBX1 inhibits AURKA degradation. YBX1-C4-2 and C4-2 cells were treated with cycloheximide and AURKA levels evaluated. (N) Graphical representation of AURKA half-life in C4-2 cells. The results of densitometric scanning are shown with AURKA levels normalized to tubulin levels. (O) YBX1 prevents AURKA degradation in 22Rv1 cells. YBX1-22Rv1 and 22Rv1 cells were treated with cycloheximide and AURKA levels evaluated. (P) Graphical representation of YBX1 degradation rate in 22Rv1 cells. * p < 0.05. (Q) YBX1 stabilizes AURKA by inhibiting its ubiquitylation. 6x-His-Ubiquitin-expressing C4-2 cells were infected with either scrambled or YBX1 shRNA lentivirus for 30 h and then treated with MG132 for 12 h. AURKA was isolated and ubiquitylation analyzed using 6x-His antibody. (R) YBX1 stabilizes AURKA by inhibiting its ubiquitylation. Ubiquitylation was performed as described above, except that ubiqitylated proteins were isolated using Ni-NTA beads followed by AURKA IB. (S,T) YBX1 depletion does not impact the subcellular localization of AURKA in C4-2 and 22Rv1 cells, respectively. Cells were exposed to control or YBX1 shRNA for 30 h, fixed and stained with AURKA antibody (green) or DAPI (blue). >100 cells were analyzed from multiple random frames. Each experiment was done at least three independent times. Representative data are shown.
Figure 3.
YBX1 stabilizes AURKA protein in C4-2 and 22Rv1 cells. (A) Overexpression of YBX1 enhances AURKA levels. (B) Histogram shows relative band intensities of YBX1 and AURKA normalized to the corresponding tubulin level from three independent experiments. * signifies significance at p < 0.05 compared to control. (C) YBX1 knock-down using YBX1 shRNA decreases AURKA in C4-2 cells. Cells were infected with control shRNA lentivirus (lane 1), or YBX1-shRNA lentivirus (lane 2), and AURKA and YBX1 levels were analyzed after 30 h. (D) Histogram shows relative band intensities of YBX1 and AURKA normalized to the corresponding tubulin level from three independent experiments. (E) YBX1 overexpression enhances AURKA levels in 22Rv1 cells. (F) Histogram shows relative band intensities of YBX1 and AURKA normalized to the corresponding tubulin level from three independent experiments. (G) YBX1 knockdown depletes AURKA protein levels in 22Rv1 cells. Cells were infected with control shRNA lentivirus (lane 1), or YBX1-shRNA lentivirus (lane 2), and AURKA and YBX1 levels were analyzed after 30 h. (H) Histogram showing relative band intensities of YBX1 and AURKA normalized to the corresponding tubulin level from three independent experiments. (I,J) YBX1 does not regulate the mRNA levels of AURKA in C4-2 and 22Rv1 cells, respectively. YBX1 was overexpressed and AURKA mRNA levels analyzed. (K,L) YBX1 does not regulate the mRNA levels of AURKA in C4-2 and 22Rv1 cells, respectively. YBX1 was knocked-down and AURKA mRNA levels analyzed. (M) YBX1 inhibits AURKA degradation. YBX1-C4-2 and C4-2 cells were treated with cycloheximide and AURKA levels evaluated. (N) Graphical representation of AURKA half-life in C4-2 cells. The results of densitometric scanning are shown with AURKA levels normalized to tubulin levels. (O) YBX1 prevents AURKA degradation in 22Rv1 cells. YBX1-22Rv1 and 22Rv1 cells were treated with cycloheximide and AURKA levels evaluated. (P) Graphical representation of YBX1 degradation rate in 22Rv1 cells. * p < 0.05. (Q) YBX1 stabilizes AURKA by inhibiting its ubiquitylation. 6x-His-Ubiquitin-expressing C4-2 cells were infected with either scrambled or YBX1 shRNA lentivirus for 30 h and then treated with MG132 for 12 h. AURKA was isolated and ubiquitylation analyzed using 6x-His antibody. (R) YBX1 stabilizes AURKA by inhibiting its ubiquitylation. Ubiquitylation was performed as described above, except that ubiqitylated proteins were isolated using Ni-NTA beads followed by AURKA IB. (S,T) YBX1 depletion does not impact the subcellular localization of AURKA in C4-2 and 22Rv1 cells, respectively. Cells were exposed to control or YBX1 shRNA for 30 h, fixed and stained with AURKA antibody (green) or DAPI (blue). >100 cells were analyzed from multiple random frames. Each experiment was done at least three independent times. Representative data are shown.
![Cancers 12 00660 g003 Cancers 12 00660 g003]()
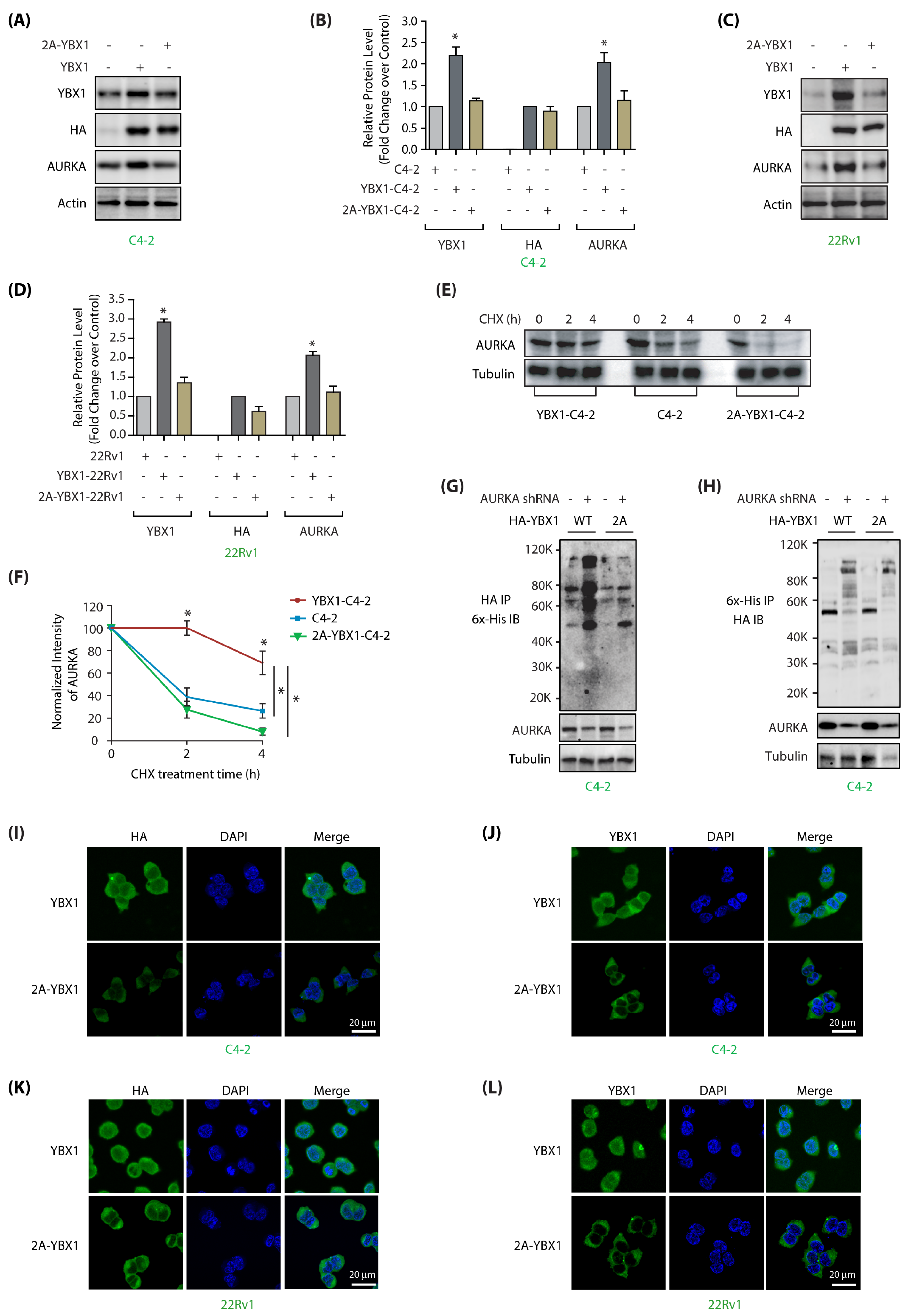
Figure 4.
AURKA regulates stability and subcellular localization of YBX1 by phosphorylation in C4-2 and 22Rv1 cells. (A) AURKA increases YBX1 levels by phosphorylation. C4-2 cells were infected with HA-tagged wild-type YBX1 or 2A-YBX1 retrovirus. After 30 h, protein levels were analyzed using YBX1, AURKA, HA and actin antibodies. (B) Graphical representation of YBX1 and AURKA levels in C4-2 cells expressing either wild-type or mutant YBX1. Average values of wild-type and mutant YBX1 levels from three independent experiments are illustrated in the graph. The statistical significance was analyzed using one way ANOVA, * p < 0.05, compared to control. (C) AURKA increases YBX1 levels by phosphorylation in 22Rv1 cells, as 2A-YBX1 showed much less steady state levels. (D) Graphical representation of YBX1 and AURKA levels in 22Rv1 cells expressing either wild type or mutant YBX1 from three independent experiments. (E) AURKA increases the stability of YBX1 via phosphorylation. AURKA levels were analyzed in C4-2, YBX1-C4-2 and 2A-YBX1-C4-2 cells treated with cycloheximide. (F) Graphical representation of AURKA half-life in C4-2, YBX1-C4-2 and 2A-YBX1-C4-2 cells. (G) AURKA depletion increased the ubiquitylation of WT YBX1, but not 2A-YBX1 in C4-2 cells. 6x-His-Ubiquitin expressing WT YBX1-C4-2 and 2A-YBX1-C4-2 cells were treated with either scrambled or AURKA shRNA lentivirus for 30 h, followed by MG132 treatment for 12 h. YBX1 was isolated using HA antibody and ubiquitylation analyzed using 6x-His antibody. (H) AURKA depletion increased the ubiquitylation of WT YBX1, but not 2A-YBX1 in C4-2 cells. Ubiquitylation was performed as described above, except ubiqitylated proteins were isolated using Ni-NTA beads followed by HA IB. (I,J) AURKA regulates the subcellular localization of YBX1 via phosphorylation. HA-tagged wild-type and 2A-YBX1 expressing cells were stained using DAPI (blue), HA antibody (green in 4I) or YBX1 antibody (green in 4J). More than 100 cells were analyzed from multiple frames. Representative data are shown (>95%). (K,L) AURKA controls YBX1 subcellular localization via phosphorylation in 22Rv1 cells (analyzed using HA antibody in 4K and YBX1 in 4L).
Figure 4.
AURKA regulates stability and subcellular localization of YBX1 by phosphorylation in C4-2 and 22Rv1 cells. (A) AURKA increases YBX1 levels by phosphorylation. C4-2 cells were infected with HA-tagged wild-type YBX1 or 2A-YBX1 retrovirus. After 30 h, protein levels were analyzed using YBX1, AURKA, HA and actin antibodies. (B) Graphical representation of YBX1 and AURKA levels in C4-2 cells expressing either wild-type or mutant YBX1. Average values of wild-type and mutant YBX1 levels from three independent experiments are illustrated in the graph. The statistical significance was analyzed using one way ANOVA, * p < 0.05, compared to control. (C) AURKA increases YBX1 levels by phosphorylation in 22Rv1 cells, as 2A-YBX1 showed much less steady state levels. (D) Graphical representation of YBX1 and AURKA levels in 22Rv1 cells expressing either wild type or mutant YBX1 from three independent experiments. (E) AURKA increases the stability of YBX1 via phosphorylation. AURKA levels were analyzed in C4-2, YBX1-C4-2 and 2A-YBX1-C4-2 cells treated with cycloheximide. (F) Graphical representation of AURKA half-life in C4-2, YBX1-C4-2 and 2A-YBX1-C4-2 cells. (G) AURKA depletion increased the ubiquitylation of WT YBX1, but not 2A-YBX1 in C4-2 cells. 6x-His-Ubiquitin expressing WT YBX1-C4-2 and 2A-YBX1-C4-2 cells were treated with either scrambled or AURKA shRNA lentivirus for 30 h, followed by MG132 treatment for 12 h. YBX1 was isolated using HA antibody and ubiquitylation analyzed using 6x-His antibody. (H) AURKA depletion increased the ubiquitylation of WT YBX1, but not 2A-YBX1 in C4-2 cells. Ubiquitylation was performed as described above, except ubiqitylated proteins were isolated using Ni-NTA beads followed by HA IB. (I,J) AURKA regulates the subcellular localization of YBX1 via phosphorylation. HA-tagged wild-type and 2A-YBX1 expressing cells were stained using DAPI (blue), HA antibody (green in 4I) or YBX1 antibody (green in 4J). More than 100 cells were analyzed from multiple frames. Representative data are shown (>95%). (K,L) AURKA controls YBX1 subcellular localization via phosphorylation in 22Rv1 cells (analyzed using HA antibody in 4K and YBX1 in 4L).
![Cancers 12 00660 g004 Cancers 12 00660 g004]()
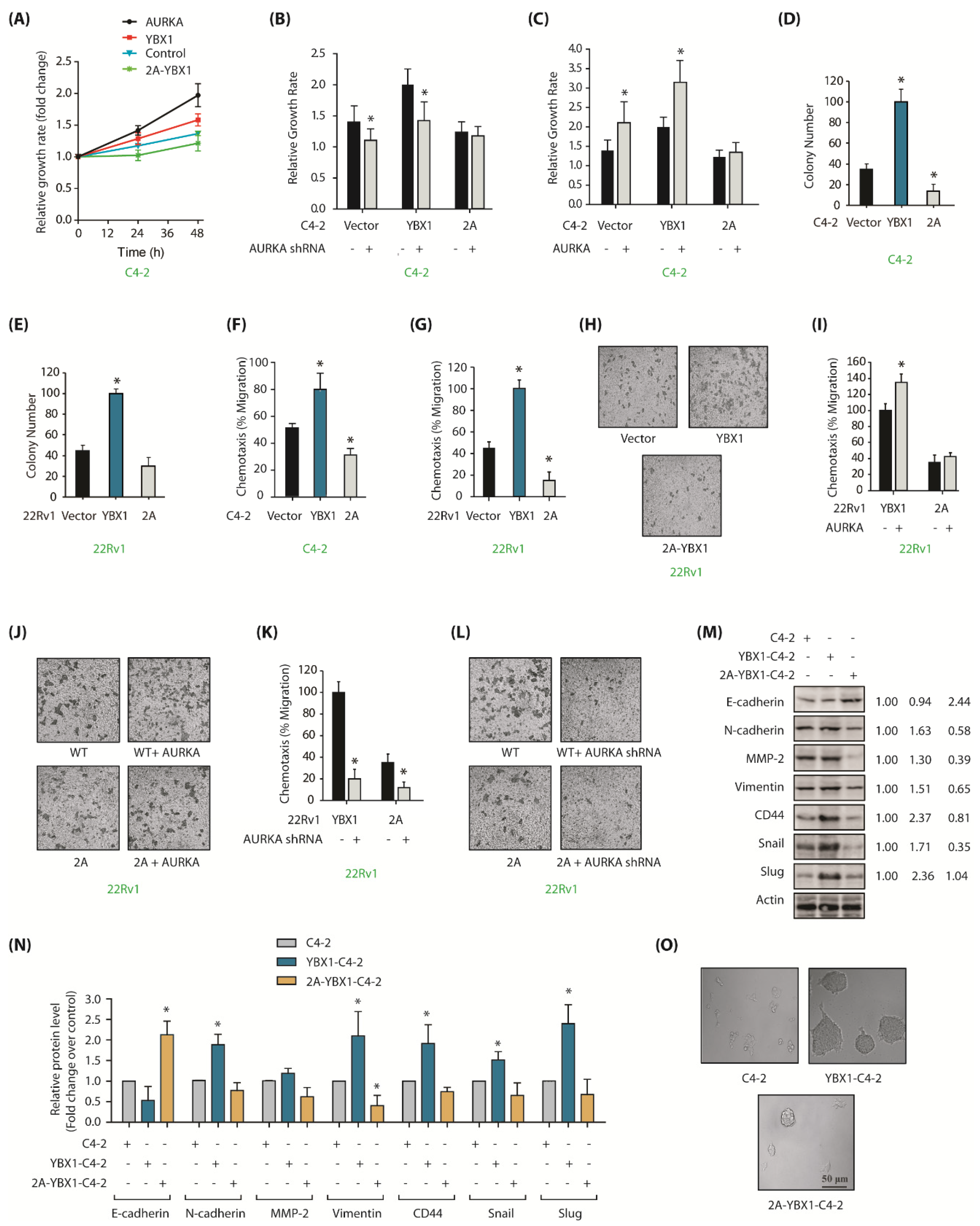
Figure 5.
YBX1 is an important cancer target of AURKA in CRPC cells. (A) YBX1 stimulates cell proliferation in C4-2 cells. C4-2, AURKA-C4-2, YBX1-C4-2 and 2A-YBX1-C4-2 cells were grown for 24 and 48 h, followed by an MTT assay. (B) AURKA knockdown decreases proliferation in C4-2 and YBX1-C4-2 cells, but not in 2A-C4-2 cells. C4-2, YBX1-C4-2 and 2A-YBX1-C4-2 cells were exposed to either scrambled shRNA or AURKA shRNA lentivirus for 48 h, followed by an MTT assay. (C) AURKA overexpression increases cell proliferation in C4-2 and YBX1-C4-2 cells, but not in 2A-C4-2 cells. C4-2, YBX1-C4-2 and 2A-C4-2 cells were exposed to either vector or AURKA retrovirus for 48 h, followed by an MTT assay. (D) Overexpression of WT-YBX1 shows higher colony formation compared to 2A-C4-2 cells using a soft agar assay. (E) Overexpression of WT-YBX1 shows higher colony formation in 22Rv1 cells compared to 2A-22Rv1 cells. (F) YBX1 promotes cell migration in C4-2, whereas 2A-YBX1 acts as dominant negative. Histogram shows mean ± SEM of three independent experiments. * p < 0.05 compared to vector-expressing control. (G) YBX1 promotes cell motility in 22Rv1 cells, whereas 2A-YBX1 acts as dominant negative. Histogram shows mean ± SEM of three independent experiments. * p < 0.05 compared to vector-expressing control. (H) YBX1 promotes cell motility in 22Rv1 cells. Representative pictures are shown. (I) AURKA overexpression accelerates proliferation in YBX1-22Rv1 cells, however not in 2A-YBX1-22Rv1 cells. Histogram shows mean ± SEM of three independent experiments. * p < 0.05 compared to vector-expressing control. (J) Representative pictures showing that AURKA overexpression accelerates proliferation in YBX1-22Rv1 cells, but not in 2A-YBX1-22Rv1 cells. These experiments were performed three independent times and representative pictures are shown. (K) AURKA knockdown inhibits cell motility in both YBX1-22Rv1 and 2A-YBX1-22Rv1 cells. C4-2, YBX1-C4-2 and 2A-C4-2 cells were exposed to either scrambled shRNA or AURKA shRNA lentivirus for 48 h, followed by chemotaxis assay. (L) Representative pictures showing that AURKA depletion decreases cell motility in both YBX1-22Rv1 and 2A-YBX1 22Rv1 cells. Magnification, 200×. (M) AURKA-mediated phosphorylation of YBX1 facilitates EMT. Ectopic expression of YBX1 increases EMT and CSC proteins, but decreases E-cadherin. 2A-YBX1 decreases EMT and CSC proteins but increases E-cadherin levels. (N) Histograms showing relative levels of EMT markers and E-cadherin from three independent experiments in C4-2, YBX1-C4-2 and 2A-YBX1-C4-2 cells. (O) YBX1 overexpression increases sphere-forming ability in C4-2 cells.
Figure 5.
YBX1 is an important cancer target of AURKA in CRPC cells. (A) YBX1 stimulates cell proliferation in C4-2 cells. C4-2, AURKA-C4-2, YBX1-C4-2 and 2A-YBX1-C4-2 cells were grown for 24 and 48 h, followed by an MTT assay. (B) AURKA knockdown decreases proliferation in C4-2 and YBX1-C4-2 cells, but not in 2A-C4-2 cells. C4-2, YBX1-C4-2 and 2A-YBX1-C4-2 cells were exposed to either scrambled shRNA or AURKA shRNA lentivirus for 48 h, followed by an MTT assay. (C) AURKA overexpression increases cell proliferation in C4-2 and YBX1-C4-2 cells, but not in 2A-C4-2 cells. C4-2, YBX1-C4-2 and 2A-C4-2 cells were exposed to either vector or AURKA retrovirus for 48 h, followed by an MTT assay. (D) Overexpression of WT-YBX1 shows higher colony formation compared to 2A-C4-2 cells using a soft agar assay. (E) Overexpression of WT-YBX1 shows higher colony formation in 22Rv1 cells compared to 2A-22Rv1 cells. (F) YBX1 promotes cell migration in C4-2, whereas 2A-YBX1 acts as dominant negative. Histogram shows mean ± SEM of three independent experiments. * p < 0.05 compared to vector-expressing control. (G) YBX1 promotes cell motility in 22Rv1 cells, whereas 2A-YBX1 acts as dominant negative. Histogram shows mean ± SEM of three independent experiments. * p < 0.05 compared to vector-expressing control. (H) YBX1 promotes cell motility in 22Rv1 cells. Representative pictures are shown. (I) AURKA overexpression accelerates proliferation in YBX1-22Rv1 cells, however not in 2A-YBX1-22Rv1 cells. Histogram shows mean ± SEM of three independent experiments. * p < 0.05 compared to vector-expressing control. (J) Representative pictures showing that AURKA overexpression accelerates proliferation in YBX1-22Rv1 cells, but not in 2A-YBX1-22Rv1 cells. These experiments were performed three independent times and representative pictures are shown. (K) AURKA knockdown inhibits cell motility in both YBX1-22Rv1 and 2A-YBX1-22Rv1 cells. C4-2, YBX1-C4-2 and 2A-C4-2 cells were exposed to either scrambled shRNA or AURKA shRNA lentivirus for 48 h, followed by chemotaxis assay. (L) Representative pictures showing that AURKA depletion decreases cell motility in both YBX1-22Rv1 and 2A-YBX1 22Rv1 cells. Magnification, 200×. (M) AURKA-mediated phosphorylation of YBX1 facilitates EMT. Ectopic expression of YBX1 increases EMT and CSC proteins, but decreases E-cadherin. 2A-YBX1 decreases EMT and CSC proteins but increases E-cadherin levels. (N) Histograms showing relative levels of EMT markers and E-cadherin from three independent experiments in C4-2, YBX1-C4-2 and 2A-YBX1-C4-2 cells. (O) YBX1 overexpression increases sphere-forming ability in C4-2 cells.
![Cancers 12 00660 g005 Cancers 12 00660 g005]()
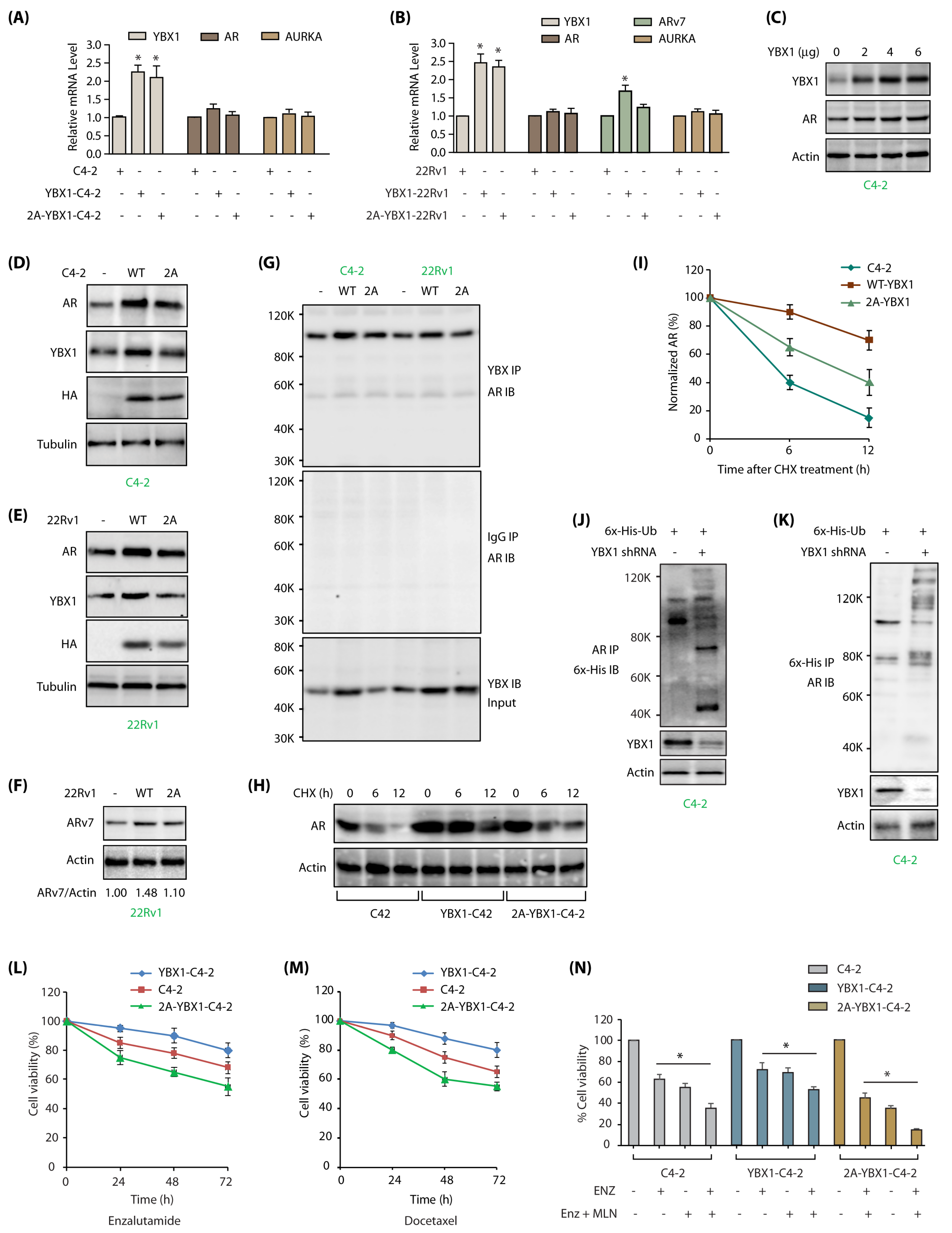
Figure 6.
YBX1-mediated regulation of AR and ARv7 in C4-2 cells. (A) YBX1 does not regulate AR or AURKA mRNA levels in C4-2 cells. YBX1, AR and AURKA mRNA levels were analyzed as described in Materials and Methods in C4-2, WT YBX1-C4-2 and 2A-YBX1-C4-2 cells. (B) YBX1 upregulation increases ARv7 mRNA levels in 22Rv1 cells. YBX1, AR, ARv7 and AURKA mRNA levels were analyzed in 22Rv1, WT YBX1-22Rv1 and 2A-YBX1-22Rv1 cells. (C) AR levels positively correlates with YBX1 levels in transfected C4-2 cells. Different amounts of YBX1 DNA was transfected in C4-2 cells. YBX1 and AR levels were analyzed after 30 h. (D) AR levels positively correlates with YBX1 levels in vector, YBX1 and 2A-YBX1-expressing C4-2 cells. (E) AR levels positively correlates with YBX1 levels in vector, YBX1 and 2A-YBX1-expressing 22Rv1 cells. (F) ARv7 levels positively correlates with YBX1 levels in vector, YBX1 and 2A-YBX1-expressing 22Rv1 cells. (G) AR binds YBX1 in vector, YBX1 and 2A-YBX1-expressing C4-2 and 22Rv1 cells. YBX1 was immunoprecipitated from vector, YBX1 and 2A-YBX1-expressing C4-2 and 22Rv1 cells, followed by AR immunoblotting (top panel). Middle panel shows IgG IP in the same six cell lines, followed by AR IB. Bottom panel shows YBX1 levels in the input. (H) YBX1 prevents AR degradation. AR levels were analyzed in C4-2, YBX1-C4-2 and 2A-YBX1-C4-2 cells following treatment with cycloheximide for 6 and 12 h. (I) Graphical representation of AR half-life. The results of densitometric scanning are shown graphically with YBX1 signal normalized to actin signal. * p < 0.05. (J) YBX1 stabilizes AR by inhibiting its ubiquitylation. 6x-His-Ubiquitin expressing C4-2 cells were treated with either scrambled or YBX1 shRNA lentivirus for 30 h, followed by MG132 treatment for 12 h. AR was isolated and ubiquitylation analyzed using 6x-His antibody. (K) YBX1 stabilizes AR by inhibiting its ubiquitylation. Ubiquitylation was performed as described above, except ubiqitylated proteins were isolated using Ni-NTA beads followed by AR IB. Each experiment was done at least three independent times. Representative data are shown. (L) AURKA-mediated phosphorylation of YBX1 promotes enzalutamide-resistance. C4-2, YBX1-C4-2 and 2A-YBX1-C4-2 cells were plated overnight, enzalutamide (1 µM) was added and cells grown for additional 24, 48 or 72 h, followed by MTT assay. (M) YBX1 overexpression increases docetaxel resistance in C4-2 cells. C4-2, YBX1-C4-2 and 2A-YBX1-C4-2 cells were plated overnight, followed by docetaxel (100 nM) treatment. The cells were grown for additional 24, 48 or 72 h, followed by MTT assay. (N) 2A-YBX1 overexpression sensitizes C4-2 cells to AURKA inhibitor MLN8237 and enzalutamide, whereas YBX1 overexpression confers resistance. C4-2, YBX1-C4-2 and 2A-YBX1-C4-2 cells were plated overnight, followed by enzalutamide (1 µM) or MLN8237 (1 µM) treatments either independently or in combination. The cells were grown for additional 48 h, followed by MTT assay.
Figure 6.
YBX1-mediated regulation of AR and ARv7 in C4-2 cells. (A) YBX1 does not regulate AR or AURKA mRNA levels in C4-2 cells. YBX1, AR and AURKA mRNA levels were analyzed as described in Materials and Methods in C4-2, WT YBX1-C4-2 and 2A-YBX1-C4-2 cells. (B) YBX1 upregulation increases ARv7 mRNA levels in 22Rv1 cells. YBX1, AR, ARv7 and AURKA mRNA levels were analyzed in 22Rv1, WT YBX1-22Rv1 and 2A-YBX1-22Rv1 cells. (C) AR levels positively correlates with YBX1 levels in transfected C4-2 cells. Different amounts of YBX1 DNA was transfected in C4-2 cells. YBX1 and AR levels were analyzed after 30 h. (D) AR levels positively correlates with YBX1 levels in vector, YBX1 and 2A-YBX1-expressing C4-2 cells. (E) AR levels positively correlates with YBX1 levels in vector, YBX1 and 2A-YBX1-expressing 22Rv1 cells. (F) ARv7 levels positively correlates with YBX1 levels in vector, YBX1 and 2A-YBX1-expressing 22Rv1 cells. (G) AR binds YBX1 in vector, YBX1 and 2A-YBX1-expressing C4-2 and 22Rv1 cells. YBX1 was immunoprecipitated from vector, YBX1 and 2A-YBX1-expressing C4-2 and 22Rv1 cells, followed by AR immunoblotting (top panel). Middle panel shows IgG IP in the same six cell lines, followed by AR IB. Bottom panel shows YBX1 levels in the input. (H) YBX1 prevents AR degradation. AR levels were analyzed in C4-2, YBX1-C4-2 and 2A-YBX1-C4-2 cells following treatment with cycloheximide for 6 and 12 h. (I) Graphical representation of AR half-life. The results of densitometric scanning are shown graphically with YBX1 signal normalized to actin signal. * p < 0.05. (J) YBX1 stabilizes AR by inhibiting its ubiquitylation. 6x-His-Ubiquitin expressing C4-2 cells were treated with either scrambled or YBX1 shRNA lentivirus for 30 h, followed by MG132 treatment for 12 h. AR was isolated and ubiquitylation analyzed using 6x-His antibody. (K) YBX1 stabilizes AR by inhibiting its ubiquitylation. Ubiquitylation was performed as described above, except ubiqitylated proteins were isolated using Ni-NTA beads followed by AR IB. Each experiment was done at least three independent times. Representative data are shown. (L) AURKA-mediated phosphorylation of YBX1 promotes enzalutamide-resistance. C4-2, YBX1-C4-2 and 2A-YBX1-C4-2 cells were plated overnight, enzalutamide (1 µM) was added and cells grown for additional 24, 48 or 72 h, followed by MTT assay. (M) YBX1 overexpression increases docetaxel resistance in C4-2 cells. C4-2, YBX1-C4-2 and 2A-YBX1-C4-2 cells were plated overnight, followed by docetaxel (100 nM) treatment. The cells were grown for additional 24, 48 or 72 h, followed by MTT assay. (N) 2A-YBX1 overexpression sensitizes C4-2 cells to AURKA inhibitor MLN8237 and enzalutamide, whereas YBX1 overexpression confers resistance. C4-2, YBX1-C4-2 and 2A-YBX1-C4-2 cells were plated overnight, followed by enzalutamide (1 µM) or MLN8237 (1 µM) treatments either independently or in combination. The cells were grown for additional 48 h, followed by MTT assay.
![Cancers 12 00660 g006 Cancers 12 00660 g006]()
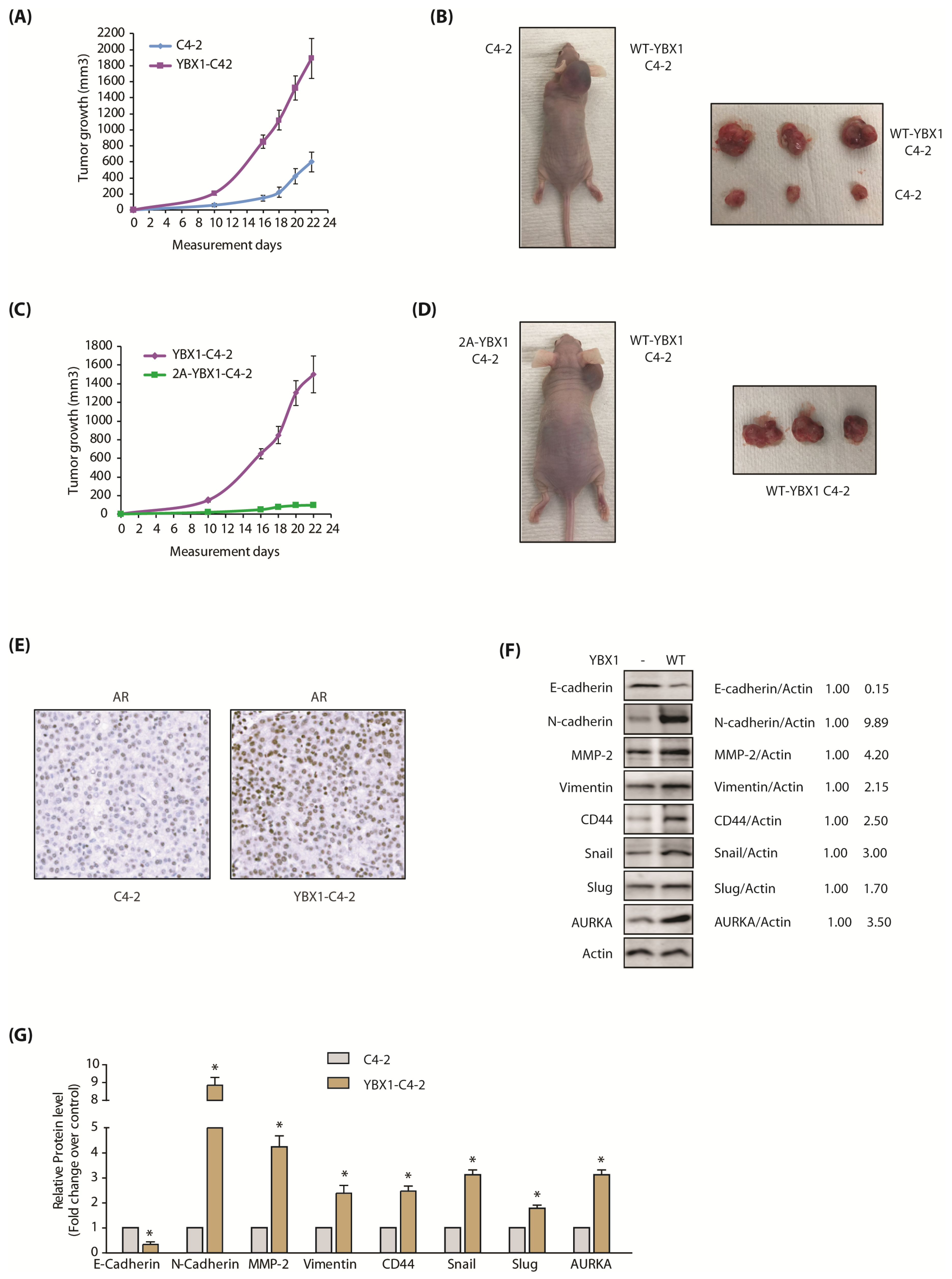
Figure 7.
The AURKA-YBX1 loop promotes tumorigenesis in vivo. (A) YBX1 overexpression enhances tumorigenesis in vivo. Three male nude athymic mice were injected with C4-2 cells and YBX1-C4-2 cells on left and right shoulders, respectively. (B) Nude athymic mice were inoculated with YBX1-C4-2 cells and C4-2 cells on the right and left shoulders, respectively. The pictures were taken 22 days following injection. Representative images are shown. (C) 2A-YBX1 expression inhibits tumor growth in vivo. Three nude mice were injected with 2A-YBX1-C4-2 and YBX1-C4-2 cells on the left and right shoulders, respectively. (D) Athymic nude mouse were inoculated with YBX1-C4-2 cells and 2A-YBX1-C4-2 cells on right and left shoulders, respectively. The pictures were taken 22 days following inoculation. Representative images are shown. (E) YBX1 upregulates AR levels in vivo. AR immunohistochemistry in C4-2 and YBX1-C4-2 xenografts. (F) Expression levels of AURKA, EMT proteins, E-cadherin and CSC proteins in control C4-2 and YBX1-C4-2 xenografts. Following euthanization, tumor tissues were isolated and flash-frozen in liquid nitrogen. Tumor tissues were homogenized and expression levels of various EMT proteins, E-Cadherin and AURKA analyzed. (G) Histogram shows relative band intensities normalized to the corresponding actin level. Data shown as mean ± SEM of three independent experiments. * signifies significance at p < 0.05 compared to control.
Figure 7.
The AURKA-YBX1 loop promotes tumorigenesis in vivo. (A) YBX1 overexpression enhances tumorigenesis in vivo. Three male nude athymic mice were injected with C4-2 cells and YBX1-C4-2 cells on left and right shoulders, respectively. (B) Nude athymic mice were inoculated with YBX1-C4-2 cells and C4-2 cells on the right and left shoulders, respectively. The pictures were taken 22 days following injection. Representative images are shown. (C) 2A-YBX1 expression inhibits tumor growth in vivo. Three nude mice were injected with 2A-YBX1-C4-2 and YBX1-C4-2 cells on the left and right shoulders, respectively. (D) Athymic nude mouse were inoculated with YBX1-C4-2 cells and 2A-YBX1-C4-2 cells on right and left shoulders, respectively. The pictures were taken 22 days following inoculation. Representative images are shown. (E) YBX1 upregulates AR levels in vivo. AR immunohistochemistry in C4-2 and YBX1-C4-2 xenografts. (F) Expression levels of AURKA, EMT proteins, E-cadherin and CSC proteins in control C4-2 and YBX1-C4-2 xenografts. Following euthanization, tumor tissues were isolated and flash-frozen in liquid nitrogen. Tumor tissues were homogenized and expression levels of various EMT proteins, E-Cadherin and AURKA analyzed. (G) Histogram shows relative band intensities normalized to the corresponding actin level. Data shown as mean ± SEM of three independent experiments. * signifies significance at p < 0.05 compared to control.
![Cancers 12 00660 g007 Cancers 12 00660 g007]()
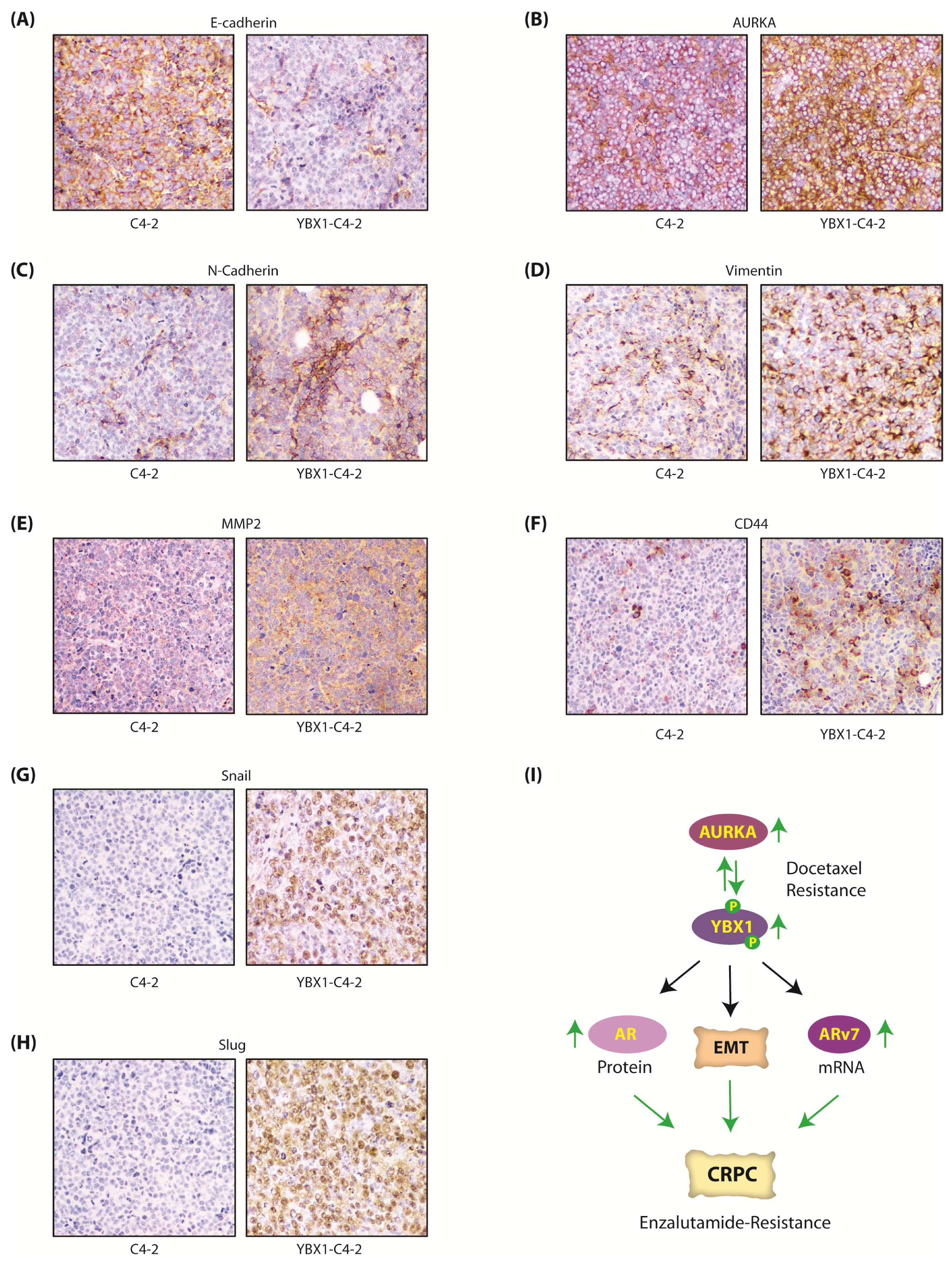
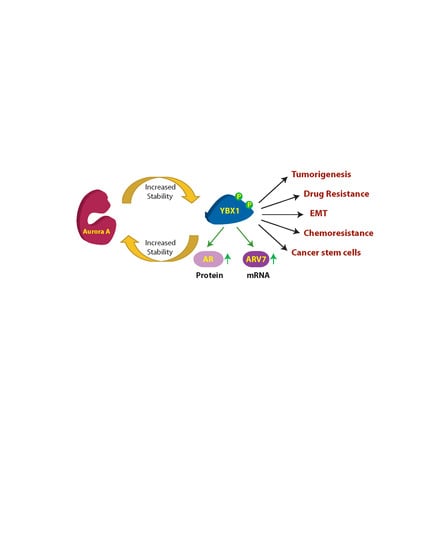
Figure 8.
The AURKA-YBX1 feedback loop facilitates EMT in vivo. (A) E-Cadherin (B) AURKA (C) N-Cadherin (D) Vimentin (E) MMP2 (F) CD44 (G) Snail and (H) Slug proteins in C4-2 and YBX1-C4-2 xenografts as determined by the immunohistochemistry (IHC) assay. (I) Proposed model showing the consequences of AURKA-YBX1 synergy in CRPC pathogenesis.
Figure 8.
The AURKA-YBX1 feedback loop facilitates EMT in vivo. (A) E-Cadherin (B) AURKA (C) N-Cadherin (D) Vimentin (E) MMP2 (F) CD44 (G) Snail and (H) Slug proteins in C4-2 and YBX1-C4-2 xenografts as determined by the immunohistochemistry (IHC) assay. (I) Proposed model showing the consequences of AURKA-YBX1 synergy in CRPC pathogenesis.