Sphere-Forming Culture for Expanding Genetically Distinct Patient-Derived Glioma Stem Cells by Cellular Growth Rate Screening
Abstract
1. Introduction
2. Results
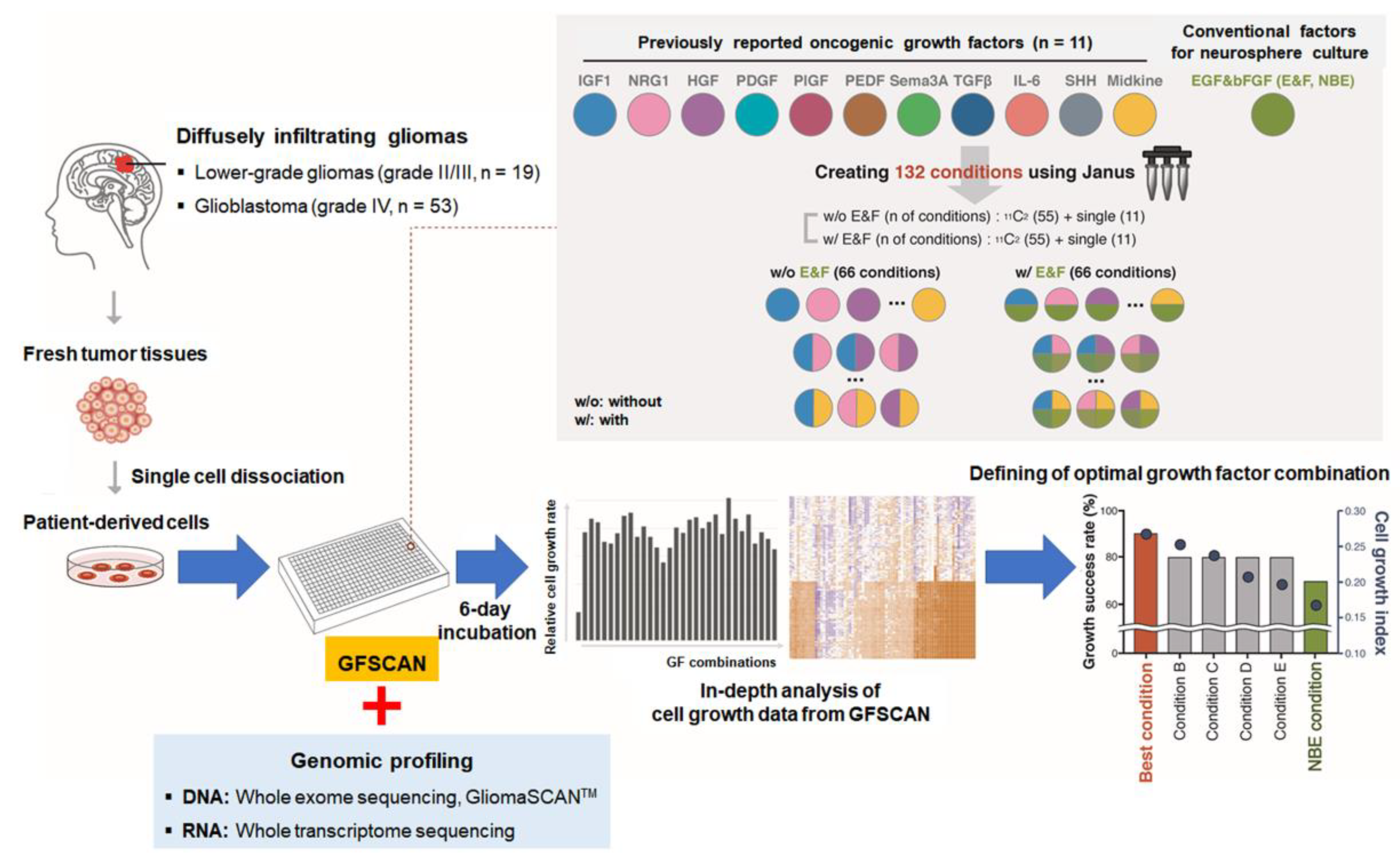
2.1. Reproducibility and Robustness of the GFSCAN Platform
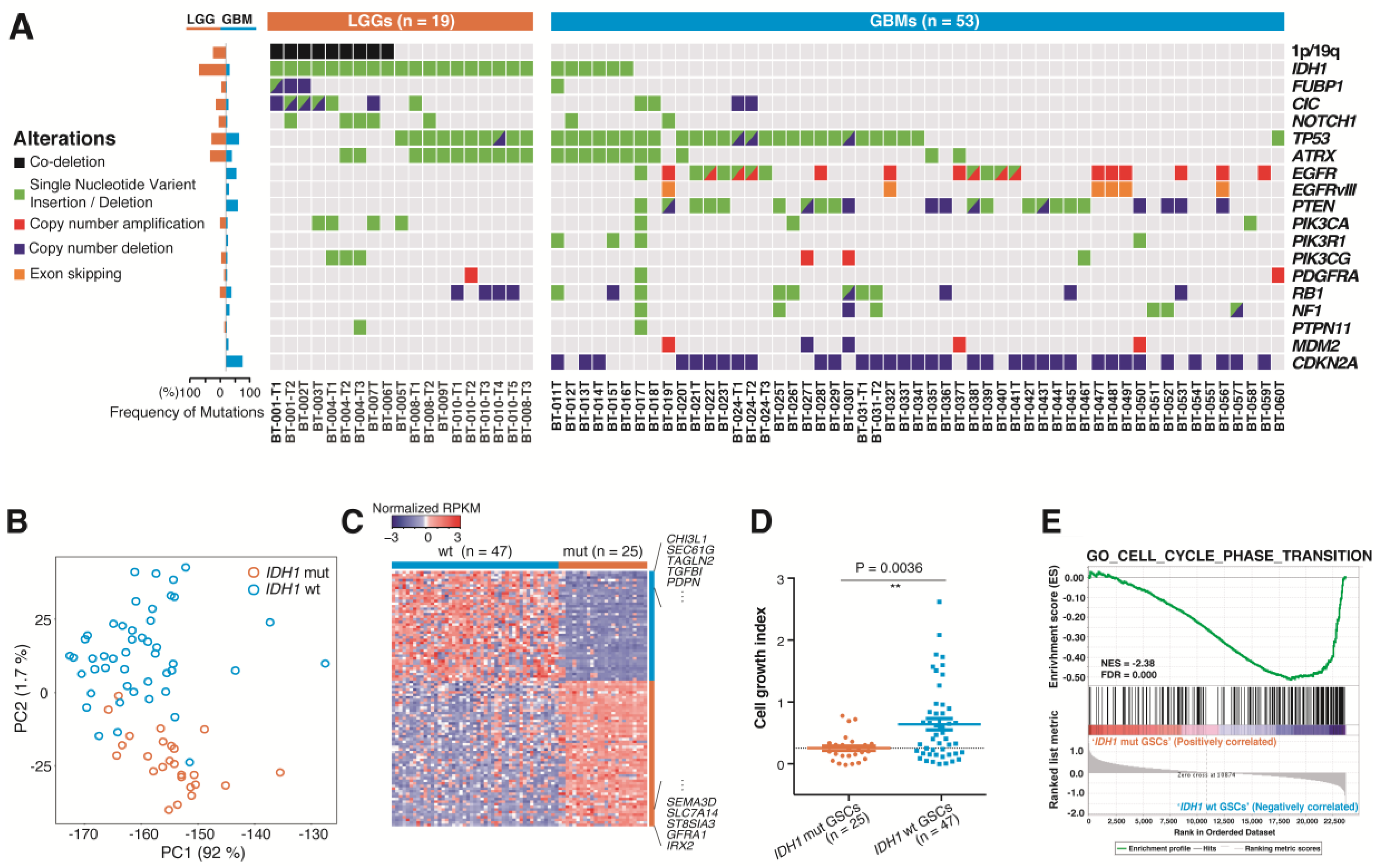
2.2. Genomic Profiles of 72 DIGs in the GFSCAN Cohort
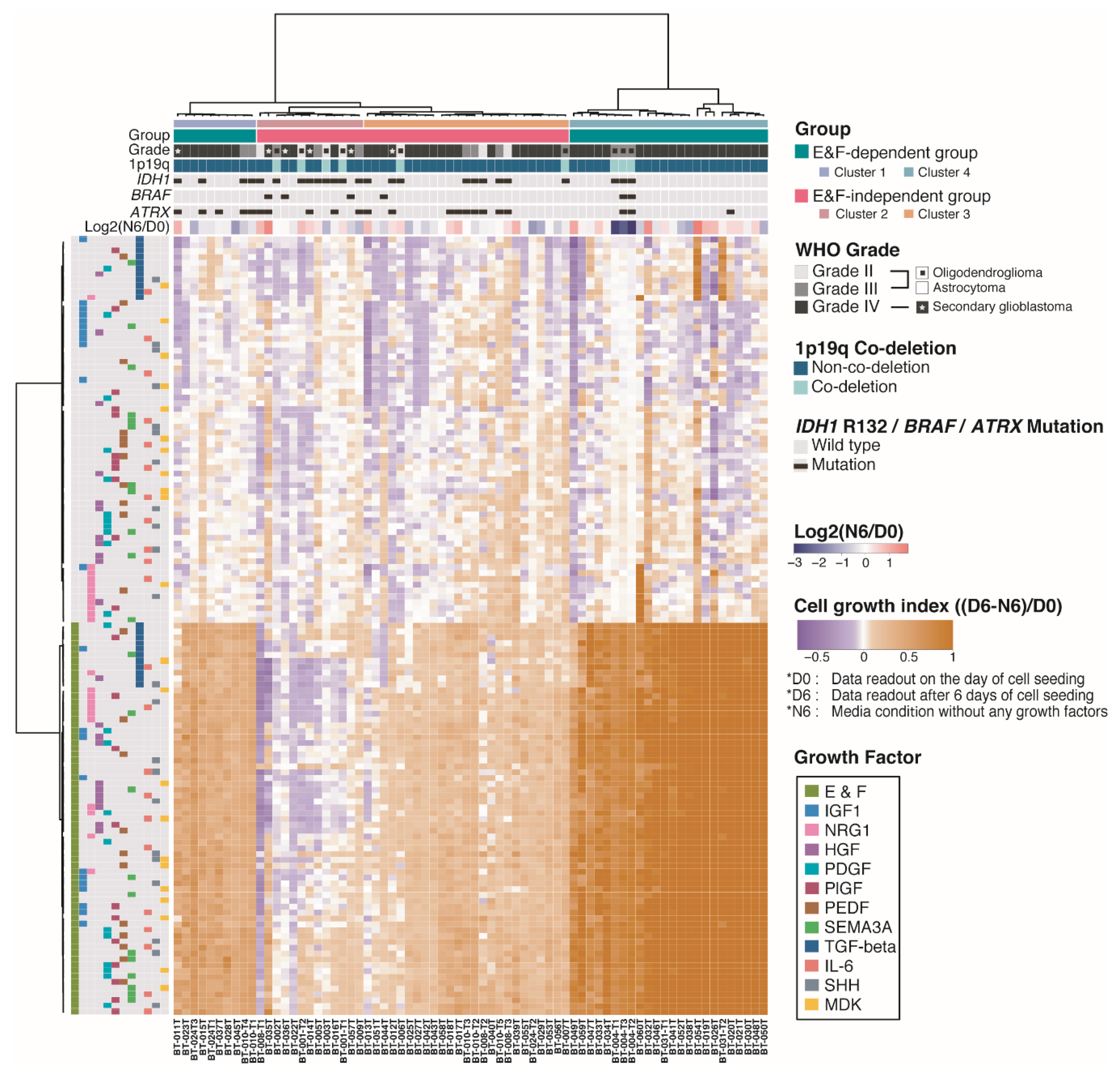
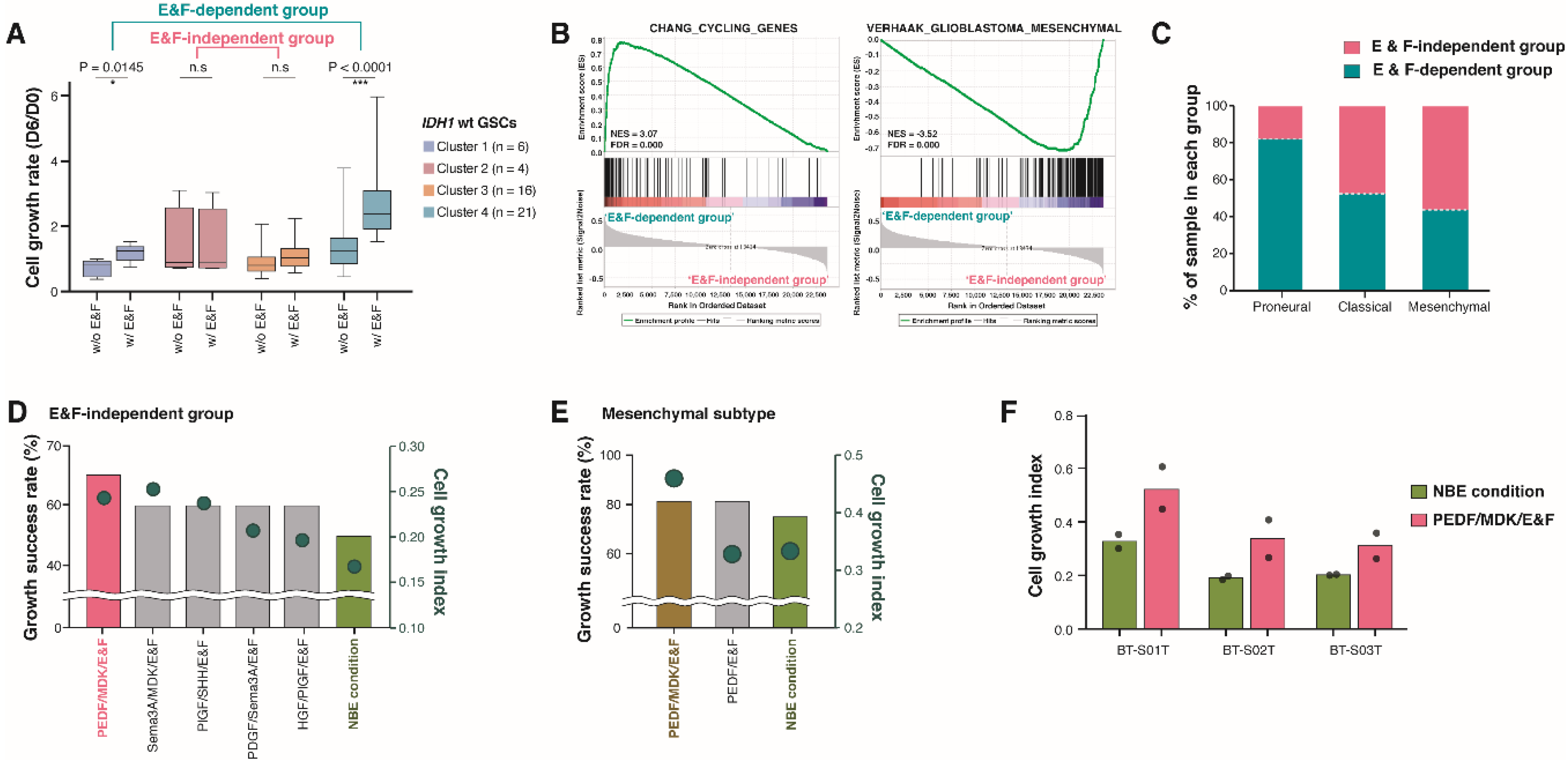
2.3. Molecular and Biological Characteristics of Patient-Derived GSCs with IDH1 Mutation
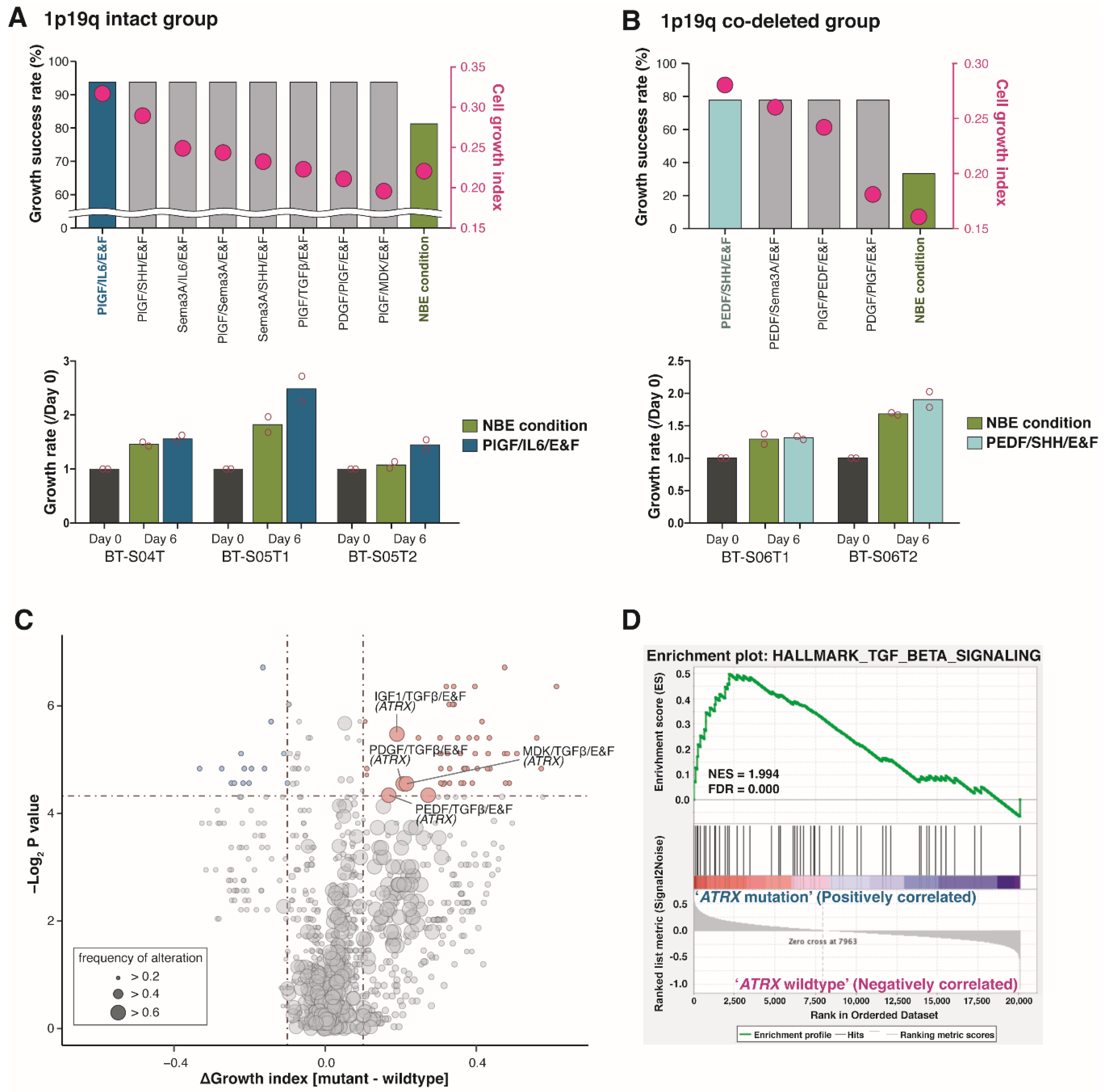
2.4. Pigment Epithelium-Derived Factor (PEDF)/Midkine (MDK)/E&F-Containing Media Promoted In Vitro GSC Propagation from IDH1-wt Glioblastomas
2.5. Newly Identified Conditions for the Cultivation of IDH1-mut GSCs
3. Discussion
4. Materials and Methods
4.1. Preparation of Patient-Derived GSCs from Human DIGs
4.2. Manufacturing GFSCAN Based on Patient-Derived GSCs
4.3. GFSCAN QC Criteria
4.4. Genomic and Transcriptomic Analysis
4.5. Unsupervised Hierarchical Clustering of Cell Growth Index from GFSCAN
4.6. Pathway Enrichment Analysis and GSEA
4.7. Glioblastoma Molecular Subtype Classification
4.8. External Gene Expression Data Sets
4.9. Statistical Analysis
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cancer Genome Atlas Research Network. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N. Engl. J. Med. 2015, 372, 2481–2498. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Pisapia, D.J. The Updated world health organization glioma classification: Cellular and molecular origins of adult infiltrating gliomas. Arch. Pathol. Lab. Med. 2017, 141, 1633–1645. [Google Scholar] [CrossRef]
- Ceccarelli, M.; Barthel, F.P.; Malta, T.M.; Sabedot, T.S.; Salama, S.R.; Murray, B.A.; Morozova, O.; Newton, Y.; Radenbaugh, A.; Pagnotta, S.M.; et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 2016, 164, 550–563. [Google Scholar] [CrossRef]
- Nomura, M.; Saito, K.; Aihara, K.; Nagae, G.; Yamamoto, S.; Tatsuno, K.; Ueda, H.; Fukuda, S.; Umeda, T.; Tanaka, S.; et al. DNA demethylation is associated with malignant progression of lower-grade gliomas. Sci. Rep. 2019, 9, 1903. [Google Scholar] [CrossRef] [PubMed]
- Hubert, C.G.; Rivera, M.; Spangler, L.C.; Wu, Q.; Mack, S.C.; Prager, B.C.; Couce, M.; McLendon, R.E.; Sloan, A.E.; Rich, J.N. A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 2016, 76, 2465–2477. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Salinas, R.D.; Zhang, D.Y.; Nguyen, P.T.T.; Schnoll, J.G.; Wong, S.Z.H.; Thokala, R.; Sheikh, S.; Saxena, D.; Prokop, S.; et al. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell 2019, 180, 188–204. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Eglen, R.M. Three-dimensional cell cultures in drug discovery and development. SLAS Discov. 2017, 22, 456–472. [Google Scholar] [PubMed]
- Quartararo, C.E.; Reznik, E.; deCarvalho, A.C.; Mikkelsen, T.; Stockwell, B.R. High-throughput screening of patient-derived cultures reveals potential for precision medicine in glioblastoma. ACS Med. Chem. Lett. 2015, 6, 948–952. [Google Scholar] [CrossRef] [PubMed]
- Patrizii, M.; Bartucci, M.; Pine, S.R.; Sabaawy, H.E. Utility of glioblastoma patient-derived orthotopic xenografts in drug discovery and personalized therapy. Front. Oncol. 2018, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Germano, I.M.; Binello, E. Stem cells and gliomas: Past, present, and future. J. Neurooncol. 2014, 119, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Meijer, T.G.; Naipal, K.A.; Jager, A.; van Gent, D.C. Ex vivo tumor culture systems for functional drug testing and therapy response prediction. Future Sci. OA 2017, 3, FSO190. [Google Scholar] [CrossRef] [PubMed]
- Cain, J. Exploratory implementation of a blended format escape room in a large enrollment pharmacy management class. Curr. Pharm. Teach. Learn. 2019, 11, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Liu, Z.; Sa, J.K.; Shin, S.; Wang, J.; Bordyuh, M.; Cho, H.J.; Elliott, O.; Chu, T.; Choi, S.W.; et al. Pharmacogenomic landscape of patient-derived tumor cells informs precision oncology therapy. Nat. Genet. 2018, 50, 1399–1411. [Google Scholar] [CrossRef] [PubMed]
- Caragher, S.; Chalmers, A.J.; Gomez-Roman, N. Glioblastoma’s next top model: Novel culture systems for brain cancer radiotherapy research. Cancers 2019, 11, 44. [Google Scholar] [CrossRef] [PubMed]
- Da Hora, C.C.; Schweiger, M.W.; Wurdinger, T.; Tannous, B.A. Patient-derived glioma models: From patients to dish to animals. Cells 2019, 8, 1177. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.V.; Stylli, S.S.; Kaye, A.H.; Mantamadiotis, T. Multilayered heterogeneity of glioblastoma stem cells: Biological and clinical significance. Adv. Exp. Med. Biol. 2019, 1139, 1–21. [Google Scholar]
- Lee, J.; Kotliarova, S.; Kotliarov, Y.; Li, A.; Su, Q.; Donin, N.M.; Pastorino, S.; Purow, B.W.; Christopher, N.; Zhang, W.; et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 2006, 9, 391–403. [Google Scholar] [CrossRef]
- Toda, M. Glioma stem cells and immunotherapy for the treatment of malignant gliomas. ISRN Oncol. 2013, 2013, 673793. [Google Scholar] [CrossRef]
- Galvao, R.P.; Kasina, A.; McNeill, R.S.; Harbin, J.E.; Foreman, O.; Verhaak, R.G.; Nishiyama, A.; Miller, C.R.; Zong, H. Transformation of quiescent adult oligodendrocyte precursor cells into malignant glioma through a multistep reactivation process. Proc. Natl. Acad. Sci. USA 2014, 111, E4214–E4223. [Google Scholar] [CrossRef]
- Phillips, J.J.; Aranda, D.; Ellison, D.W.; Judkins, A.R.; Croul, S.E.; Brat, D.J.; Ligon, K.L.; Horbinski, C.; Venneti, S.; Zadeh, G.; et al. PDGFRA amplification is common in pediatric and adult high-grade astrocytomas and identifies a poor prognostic group in IDH1 mutant glioblastoma. Brain. Pathol. 2013, 23, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Karsy, M.; Guan, J.; Cohen, A.L.; Jensen, R.L.; Colman, H. New molecular considerations for glioma: IDH, ATRX, BRAF, TERT, H3 K27M. Curr. Neurol. Neurosci. Rep. 2017, 17, 19. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C. Diffuse gliomas for nonneuropathologists: The new integrated molecular diagnostics. Arch. Pathol. Lab. Med. 2018, 142, 804–814. [Google Scholar] [CrossRef] [PubMed]
- Piaskowski, S.; Bienkowski, M.; Stoczynska-Fidelus, E.; Stawski, R.; Sieruta, M.; Szybka, M.; Papierz, W.; Wolanczyk, M.; Jaskolski, D.J.; Liberski, P.P.; et al. Glioma cells showing IDH1 mutation cannot be propagated in standard cell culture conditions. Br. J. Cancer 2011, 104, 968–970. [Google Scholar] [CrossRef] [PubMed]
- Mazor, T.; Chesnelong, C.; Pankov, A.; Jalbert, L.E.; Hong, C.; Hayes, J.; Smirnov, I.V.; Marshall, R.; Souza, C.F.; Shen, Y.; et al. Clonal expansion and epigenetic reprogramming following deletion or amplification of mutant IDH1. Proc. Natl. Acad. Sci. USA 2017, 114, 10743–10748. [Google Scholar] [CrossRef]
- Klink, B.; Miletic, H.; Stieber, D.; Huszthy, P.C.; Campos Valenzuela, J.A.; Balss, J.; Wang, J.; Schubert, M.; Sakariassen, P.O.; Sundstrom, T.; et al. A novel, diffusely infiltrative xenograft model of human anaplastic oligodendroglioma with mutations in FUBP1, CIC, and IDH1. PLoS ONE 2013, 8, e59773. [Google Scholar] [CrossRef]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef]
- Wang, Q.; Hu, B.; Hu, X.; Kim, H.; Squatrito, M.; Scarpace, L.; de Carvalho, A.C.; Lyu, S.; Li, P.; Li, Y.; et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 2017, 32, 42–56. [Google Scholar] [CrossRef]
- Han, S.; Shin, H.; Lee, J.K.; Liu, Z.; Rabadan, R.; Lee, J.; Shin, J.; Lee, C.; Yang, H.; Kim, D.; et al. Secretome analysis of patient-derived GBM tumor spheres identifies midkine as a potent therapeutic target. Exp. Mol. Med. 2019, 51, 147. [Google Scholar] [CrossRef]
- Bleeker, F.E.; Lamba, S.; Leenstra, S.; Troost, D.; Hulsebos, T.; Vandertop, W.P.; Frattini, M.; Molinari, F.; Knowles, M.; Cerrato, A.; et al. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum. Mutat. 2009, 30, 7–11. [Google Scholar] [CrossRef]
- Kloosterhof, N.K.; Bralten, L.B.; Dubbink, H.J.; French, P.J.; van den Bent, M.J. Isocitrate dehydrogenase-1 mutations: A fundamentally new understanding of diffuse glioma? Lancet Oncol. 2011, 12, 83–91. [Google Scholar] [CrossRef]
- Luchman, H.A.; Stechishin, O.D.; Dang, N.H.; Blough, M.D.; Chesnelong, C.; Kelly, J.J.; Nguyen, S.A.; Chan, J.A.; Weljie, A.M.; Cairncross, J.G.; et al. An in vivo patient-derived model of endogenous IDH1-mutant glioma. Neuro. Oncol. 2012, 14, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Killela, P.J.; Reitman, Z.J.; Rasheed, A.B.; Heaphy, C.M.; de Wilde, R.F.; Rodriguez, F.J.; Rosemberg, S.; Oba-Shinjo, S.M.; Nagahashi Marie, S.K.; et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget 2012, 3, 709–722. [Google Scholar] [CrossRef] [PubMed]
- Bhat, K.P.L.; Balasubramaniyan, V.; Vaillant, B.; Ezhilarasan, R.; Hummelink, K.; Hollingsworth, F.; Wani, K.; Heathcock, L.; James, J.D.; Goodman, L.D.; et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell 2013, 24, 331–346. [Google Scholar] [CrossRef]
- Ledur, P.F.; Onzi, G.R.; Zong, H.; Lenz, G. Culture conditions defining glioblastoma cells behavior: What is the impact for novel discoveries? Oncotarget 2017, 8, 69185–69197. [Google Scholar] [CrossRef]
- Cruickshanks, N.; Zhang, Y.; Yuan, F.; Pahuski, M.; Gibert, M.; Abounader, R. Role and therapeutic targeting of the HGF/MET pathway in glioblastoma. Cancers 2017, 9, 87. [Google Scholar] [CrossRef]
- Cheng, F.; Guo, D. MET in glioma: Signaling pathways and targeted therapies. J. Exp. Clin. Cancer Res. 2019, 38, 270. [Google Scholar] [CrossRef]
- Ritch, P.A.; Carroll, S.L.; Sontheimer, H. Neuregulin-1 enhances motility and migration of human astrocytic glioma cells. J. Biol. Chem. 2003, 278, 20971–20978. [Google Scholar] [CrossRef]
- Ritch, P.S.; Carroll, S.L.; Sontheimer, H. Neuregulin-1 enhances survival of human astrocytic glioma cells. Glia 2005, 51, 217–228. [Google Scholar] [CrossRef]
- Zhao, W.J.; Schachner, M. Neuregulin 1 enhances cell adhesion molecule l1 expression in human glioma cells and promotes their migration as a function of malignancy. J. Neuropathol. Exp. Neurol. 2013, 72, 244–255. [Google Scholar] [CrossRef]
- Lokker, N.A.; Sullivan, C.M.; Hollenbach, S.J.; Israel, M.A.; Giese, N.A. Platelet-derived growth factor (PDGF) autocrine signaling regulates survival and mitogenic pathways in glioblastoma cells: Evidence that the novel PDGF-C and PDGF-D ligands may play a role in the development of brain tumors. Cancer Res. 2002, 62, 3729–3735. [Google Scholar] [PubMed]
- Nazarenko, I.; Hede, S.M.; He, X.; Hedren, A.; Thompson, J.; Lindstrom, M.S.; Nister, M. PDGF and PDGF receptors in glioma. Ups. J. Med. Sci. 2012, 117, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Cantanhede, I.G.; de Oliveira, J.R.M. PDGF family expression in glioblastoma multiforme: Data compilation from ivy glioblastoma atlas project database. Sci. Rep. 2017, 7, 15271. [Google Scholar] [CrossRef] [PubMed]
- Bagci, T.; Wu, J.K.; Pfannl, R.; Ilag, L.L.; Jay, D.G. Autocrine semaphorin 3A signaling promotes glioblastoma dispersal. Oncogene 2009, 28, 3537–3550. [Google Scholar] [CrossRef] [PubMed]
- Law, J.W.; Lee, A.Y. The role of semaphorins and their receptors in gliomas. J. Signal Transduct. 2012, 2012, 902854. [Google Scholar] [CrossRef]
- Lee, J.; Shin, Y.J.; Lee, K.; Cho, H.J.; Sa, J.K.; Lee, S.Y.; Kim, S.H.; Lee, J.; Yoon, Y.; Nam, D.H. Anti-SEMA3A Antibody: A novel therapeutic agent to suppress glioblastoma tumor growth. Cancer Res. Treat. 2018, 50, 1009–1022. [Google Scholar] [CrossRef]
- Angelopoulou, E.; Piperi, C. Emerging role of plexins signaling in glioma progression and therapy. Cancer Lett. 2018, 414, 81–87. [Google Scholar] [CrossRef]
- Clement, V.; Sanchez, P.; de Tribolet, N.; Radovanovic, I.; Ruiz Altaba, A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr. Biol. 2007, 17, 165–172. [Google Scholar] [CrossRef]
- Uchida, H.; Arita, K.; Yunoue, S.; Yonezawa, H.; Shinsato, Y.; Kawano, H.; Hirano, H.; Hanaya, R.; Tokimura, H. Role of sonic hedgehog signaling in migration of cell lines established from CD133-positive malignant glioma cells. J. Neurooncol. 2011, 104, 697–704. [Google Scholar] [CrossRef]
- Takezaki, T.; Hide, T.; Takanaga, H.; Nakamura, H.; Kuratsu, J.; Kondo, T. Essential role of the Hedgehog signaling pathway in human glioma-initiating cells. Cancer Sci. 2011, 102, 1306–1312. [Google Scholar] [CrossRef]
- Tchirkov, A.; Khalil, T.; Chautard, E.; Mokhtari, K.; Veronese, L.; Irthum, B.; Vago, P.; Kemeny, J.L.; Verrelle, P. Interleukin-6 gene amplification and shortened survival in glioblastoma patients. Br. J. Cancer 2007, 96, 474–476. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lathia, J.D.; Wu, Q.; Wang, J.; Li, Z.; Heddleston, J.M.; Eyler, C.E.; Elderbroom, J.; Gallagher, J.; Schuschu, J.; et al. Targeting interleukin 6 signaling suppresses glioma stem cell survival and tumor growth. Stem Cells 2009, 27, 2393–2404. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.; He, X.; Song, W.; Han, D.; Niu, J.; Wang, J. Role of IL-6 in the invasiveness and prognosis of glioma. Int. J. Clin. Exp. Med. 2015, 8, 9114–9120. [Google Scholar] [PubMed]
- Jiang, Y.; Han, S.; Cheng, W.; Wang, Z.; Wu, A. NFAT1-regulated IL6 signalling contributes to aggressive phenotypes of glioma. Cell Commun. Signal 2017, 15, 54. [Google Scholar] [CrossRef] [PubMed]
- West, A.J.; Tsui, V.; Stylli, S.S.; Nguyen, H.P.T.; Morokoff, A.P.; Kaye, A.H.; Luwor, R.B. The role of interleukin-6-STAT3 signalling in glioblastoma. Oncol. Lett. 2018, 16, 4095–4104. [Google Scholar] [CrossRef] [PubMed]
- Penuelas, S.; Anido, J.; Prieto-Sanchez, R.M.; Folch, G.; Barba, I.; Cuartas, I.; Garcia-Dorado, D.; Poca, M.A.; Sahuquillo, J.; Baselga, J.; et al. TGF-beta increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell 2009, 15, 315–327. [Google Scholar] [CrossRef]
- Han, J.; Alvarez-Breckenridge, C.A.; Wang, Q.E.; Yu, J. TGF-beta signaling and its targeting for glioma treatment. Am. J. Cancer Res. 2015, 5, 945–955. [Google Scholar]
- Puputti, M.; Tynninen, O.; Sihto, H.; Blom, T.; Maenpaa, H.; Isola, J.; Paetau, A.; Joensuu, H.; Nupponen, N.N. Amplification of KIT, PDGFRA, VEGFR2, and EGFR in gliomas. Mol. Cancer Res. 2006, 4, 927–934. [Google Scholar] [CrossRef]
- Hitoshi, Y.; Harris, B.T.; Liu, H.; Popko, B.; Israel, M.A. Spinal glioma: Platelet-derived growth factor B-mediated oncogenesis in the spinal cord. Cancer Res. 2008, 68, 8507–8515. [Google Scholar] [CrossRef]
- Yin, J.; Park, G.; Kim, T.H.; Hong, J.H.; Kim, Y.J.; Jin, X.; Kang, S.; Jung, J.E.; Kim, J.Y.; Yun, H.; et al. Pigment epithelium-derived factor (PEDF) expression induced by EGFRvIII promotes self-renewal and tumor progression of glioma stem cells. PLoS Biol. 2015, 13, e1002152. [Google Scholar] [CrossRef]
- Cochrane, C.R.; Szczepny, A.; Watkins, D.N.; Cain, J.E. Hedgehog signaling in the maintenance of cancer stem cells. Cancers 2015, 7, 1554–1585. [Google Scholar] [CrossRef] [PubMed]
- Erdogan, S.; Doganlar, Z.B.; Doganlar, O.; Turkekul, K.; Serttas, R. Inhibition of midkine suppresses prostate cancer CD133(+) stem cell growth and migration. Am. J. Med. Sci. 2017, 354, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Castillejo, C.; Sanchez-Sanchez, F.; Andreu-Agullo, C.; Ferron, S.R.; Aroca-Aguilar, J.D.; Sanchez, P.; Mira, H.; Escribano, J.; Farinas, I. Pigment epithelium-derived factor is a niche signal for neural stem cell renewal. Nat. Neurosci. 2006, 9, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Wang, X.; Xia, Z.; Yang, L.; Ding, Z.; Chen, S.; Lai, B.; Zhang, N. Transcriptional factor specificity protein 1 (SP1) promotes the proliferation of glioma cells by up-regulating midkine (MDK). Mol. Biol. Cell 2015, 26, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.L.; Gao, Y.L.; Chen, X.B. Wnt/beta-catenin up-regulates Midkine expression in glioma cells. Int. J. Clin. Exp. Med. 2015, 8, 12644–12649. [Google Scholar]
- Sinha, S.; Koul, N.; Dixit, D.; Sharma, V.; Sen, E. IGF-1 induced HIF-1alpha-TLR9 cross talk regulates inflammatory responses in glioma. Cell Signal 2011, 23, 1869–1875. [Google Scholar] [CrossRef]
- Andreu-Agullo, C.; Morante-Redolat, J.M.; Delgado, A.C.; Farinas, I. Vascular niche factor PEDF modulates Notch-dependent stemness in the adult subependymal zone. Nat. Neurosci. 2009, 12, 1514–1523. [Google Scholar] [CrossRef]
- Pumiglia, K.; Temple, S. PEDF: Bridging neurovascular interactions in the stem cell niche. Nat. Neurosci. 2006, 9, 299–300. [Google Scholar] [CrossRef]
- Belenguer, G.; Domingo-Muelas, A.; Ferron, S.R.; Morante-Redolat, J.M.; Farinas, I. Isolation, culture and analysis of adult subependymal neural stem cells. Differentiation 2016, 91, 28–41. [Google Scholar] [CrossRef]
- Albonici, L.; Giganti, M.G.; Modesti, A.; Manzari, V.; Bei, R. Multifaceted role of the placental growth factor (PlGF) in the antitumor immune response and cancer progression. Int. J. Mol. Sci. 2019, 20, 2970. [Google Scholar] [CrossRef]
- Reardon, D.A.; Turner, S.; Peters, K.B.; Desjardins, A.; Gururangan, S.; Sampson, J.H.; McLendon, R.E.; Herndon, J.E.; Jones, L.W.; Kirkpatrick, J.P.; et al. A review of VEGF/VEGFR-targeted therapeutics for recurrent glioblastoma. J. Natl. Compr. Cancer Netw. 2011, 9, 414–427. [Google Scholar] [CrossRef] [PubMed]
- Atzori, M.G.; Tentori, L.; Ruffini, F.; Ceci, C.; Lisi, L.; Bonanno, E.; Scimeca, M.; Eskilsson, E.; Daubon, T.; Miletic, H.; et al. The anti-vascular endothelial growth factor receptor-1 monoclonal antibody D16F7 inhibits invasiveness of human glioblastoma and glioblastoma stem cells. J. Exp. Clin. Cancer Res. 2017, 36, 106. [Google Scholar] [CrossRef] [PubMed]
- Shahi, M.H.; Farheen, S.; Mariyath, M.P.; Castresana, J.S. Potential role of Shh-Gli1-BMI1 signaling pathway nexus in glioma chemoresistance. Tumour. Biol. 2016, 37, 15107–15114. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Yuan, X.; Liu, G.; Black, K.L.; Yu, J.S. Hedgehog signaling regulates brain tumor-initiating cell proliferation and portends shorter survival for patients with PTEN-coexpressing glioblastomas. Stem. Cells 2008, 26, 3018–3026. [Google Scholar] [CrossRef]
- Soeda, A.; Inagaki, A.; Oka, N.; Ikegame, Y.; Aoki, H.; Yoshimura, S.; Nakashima, S.; Kunisada, T.; Iwama, T. Epidermal growth factor plays a crucial role in mitogenic regulation of human brain tumor stem cells. J. Biol. Chem. 2008, 283, 10958–10966. [Google Scholar] [CrossRef]
- Nicolas, S.; Abdellatef, S.; Haddad, M.A.; Fakhoury, I.; El-Sibai, M. Hypoxia and EGF Stimulation regulate VEGF expression in human glioblastoma multiforme (GBM) cells by differential regulation of the PI3K/Rho-GTPase and MAPK Pathways. Cells 2019, 8, 1397. [Google Scholar] [CrossRef]
- An, Z.; Aksoy, O.; Zheng, T.; Fan, Q.W.; Weiss, W.A. Epidermal growth factor receptor and EGFRvIII in glioblastoma: Signaling pathways and targeted therapies. Oncogene 2018, 37, 1561–1575. [Google Scholar] [CrossRef]
- Jimenez-Pascual, A.; Siebzehnrubl, F.A. Fibroblast growth factor receptor functions in glioblastoma. Cells 2019, 8, 715. [Google Scholar] [CrossRef]
- Haley, E.M.; Kim, Y. The role of basic fibroblast growth factor in glioblastoma multiforme and glioblastoma stem cells and in their in vitro culture. Cancer Lett. 2014, 346, 1–5. [Google Scholar] [CrossRef]
- Joseph, J.V.; Balasubramaniyan, V.; Walenkamp, A.; Kruyt, F.A. TGF-beta as a therapeutic target in high grade gliomas—Promises and challenges. Biochem. Pharm. 2013, 85, 478–485. [Google Scholar] [CrossRef]
- Kaminska, B.; Kocyk, M.; Kijewska, M. TGF beta signaling and its role in glioma pathogenesis. Adv. Exp. Med. Biol. 2013, 986, 171–187. [Google Scholar] [PubMed]
- Wang, H.; Tang, C.; Na, M.; Ma, W.; Jiang, Z.; Gu, Y.; Ma, G.; Ge, H.; Shen, H.; Lin, Z. Mir-422a inhibits glioma proliferation and invasion by targeting IGF1 and IGF1R. Oncol. Res. 2017, 25, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Ho, K.H.; Chen, P.H.; Hsi, E.; Shih, C.M.; Chang, W.C.; Cheng, C.H.; Lin, C.W.; Chen, K.C. Identification of IGF-1-enhanced cytokine expressions targeted by miR-181d in glioblastomas via an integrative miRNA/mRNA regulatory network analysis. Sci. Rep. 2017, 7, 732. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Zhang, J.; Cui, Q.; Li, X.; Gao, G.; Wang, Y.; Xu, Y.; Gao, X. GSK1904529A, an insulin-like growth factor-1 receptor inhibitor, inhibits glioma tumor growth, induces apoptosis and inhibits migration. Mol. Med. Rep. 2015, 12, 3381–3385. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Mittal, S.; Berens, M.E. Targeting adaptive glioblastoma: An overview of proliferation and invasion. Neuro. Oncol. 2014, 16, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Hoque, M.O.; Wu, F.; Trink, B.; Sidransky, D.; Ratovitski, E.A. Midkine induces epithelial-mesenchymal transition through Notch2/Jak2-Stat3 signaling in human keratinocytes. Cell Cycle 2008, 7, 1613–1622. [Google Scholar] [CrossRef]
- Gungor, C.; Zander, H.; Effenberger, K.E.; Vashist, Y.K.; Kalinina, T.; Izbicki, J.R.; Yekebas, E.; Bockhorn, M. Notch signaling activated by replication stress-induced expression of midkine drives epithelial-mesenchymal transition and chemoresistance in pancreatic cancer. Cancer Res. 2011, 71, 5009–5019. [Google Scholar] [CrossRef]
- Kelly, J.J.; Blough, M.D.; Stechishin, O.D.; Chan, J.A.; Beauchamp, D.; Perizzolo, M.; Demetrick, D.J.; Steele, L.; Auer, R.N.; Hader, W.J.; et al. Oligodendroglioma cell lines containing t(1;19)(q10;p10). Neuro. Oncol. 2010, 12, 745–755. [Google Scholar] [CrossRef]
- Chen, Y.H.; McGowan, L.D.; Cimino, P.J.; Dahiya, S.; Leonard, J.R.; Lee, D.Y.; Gutmann, D.H. Mouse low-grade gliomas contain cancer stem cells with unique molecular and functional properties. Cell Rep. 2015, 10, 1899–1912. [Google Scholar] [CrossRef]
- Kim, J.; Lee, I.H.; Cho, H.J.; Park, C.K.; Jung, Y.S.; Kim, Y.; Nam, S.H.; Kim, B.S.; Johnson, M.D.; Kong, D.S.; et al. Spatiotemporal evolution of the primary glioblastoma genome. Cancer Cell 2015, 28, 318–328. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Hoshida, Y. Nearest template prediction: A single-sample-based flexible class prediction with confidence assessment. PLoS ONE 2010, 5, e15543. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Madhavan, S.; Zenklusen, J.C.; Kotliarov, Y.; Sahni, H.; Fine, H.A.; Buetow, K. Rembrandt: Helping personalized medicine become a reality through integrative translational research. Mol. Cancer Res. 2009, 7, 157–167. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, K.; Shin, H.; Cho, H.J.; Kang, H.; Lee, J.-K.; Seo, Y.J.; Shin, Y.J.; Kim, D.; Koo, H.; Kong, D.-S.; et al. Sphere-Forming Culture for Expanding Genetically Distinct Patient-Derived Glioma Stem Cells by Cellular Growth Rate Screening. Cancers 2020, 12, 549. https://doi.org/10.3390/cancers12030549
Shin K, Shin H, Cho HJ, Kang H, Lee J-K, Seo YJ, Shin YJ, Kim D, Koo H, Kong D-S, et al. Sphere-Forming Culture for Expanding Genetically Distinct Patient-Derived Glioma Stem Cells by Cellular Growth Rate Screening. Cancers. 2020; 12(3):549. https://doi.org/10.3390/cancers12030549
Chicago/Turabian StyleShin, Kayoung, Hyemi Shin, Hee Jin Cho, Hyunju Kang, Jin-Ku Lee, Yun Jee Seo, Yong Jae Shin, Donggeon Kim, Harim Koo, Doo-Sik Kong, and et al. 2020. "Sphere-Forming Culture for Expanding Genetically Distinct Patient-Derived Glioma Stem Cells by Cellular Growth Rate Screening" Cancers 12, no. 3: 549. https://doi.org/10.3390/cancers12030549
APA StyleShin, K., Shin, H., Cho, H. J., Kang, H., Lee, J.-K., Seo, Y. J., Shin, Y. J., Kim, D., Koo, H., Kong, D.-S., Seol, H. J., Lee, J.-I., Lee, H. W., & Nam, D.-H. (2020). Sphere-Forming Culture for Expanding Genetically Distinct Patient-Derived Glioma Stem Cells by Cellular Growth Rate Screening. Cancers, 12(3), 549. https://doi.org/10.3390/cancers12030549




