Melanoma-Derived Exosomal miR-125b-5p Educates Tumor Associated Macrophages (TAMs) by Targeting Lysosomal Acid Lipase A (LIPA)
Abstract
1. Introduction
2. Results
2.1. Isolation and Characterization of Exosomes Derived by Malignant Melanoma Cells
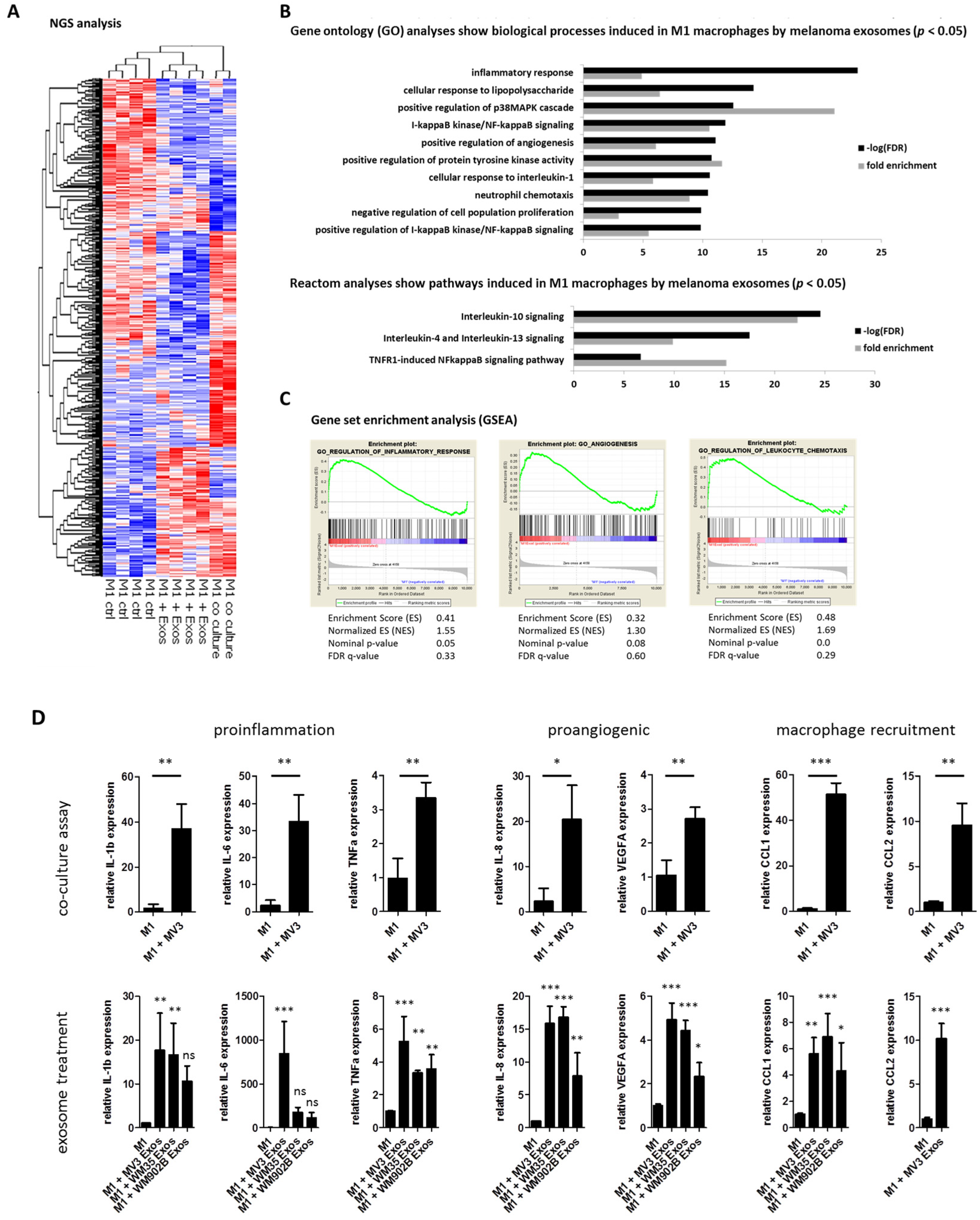
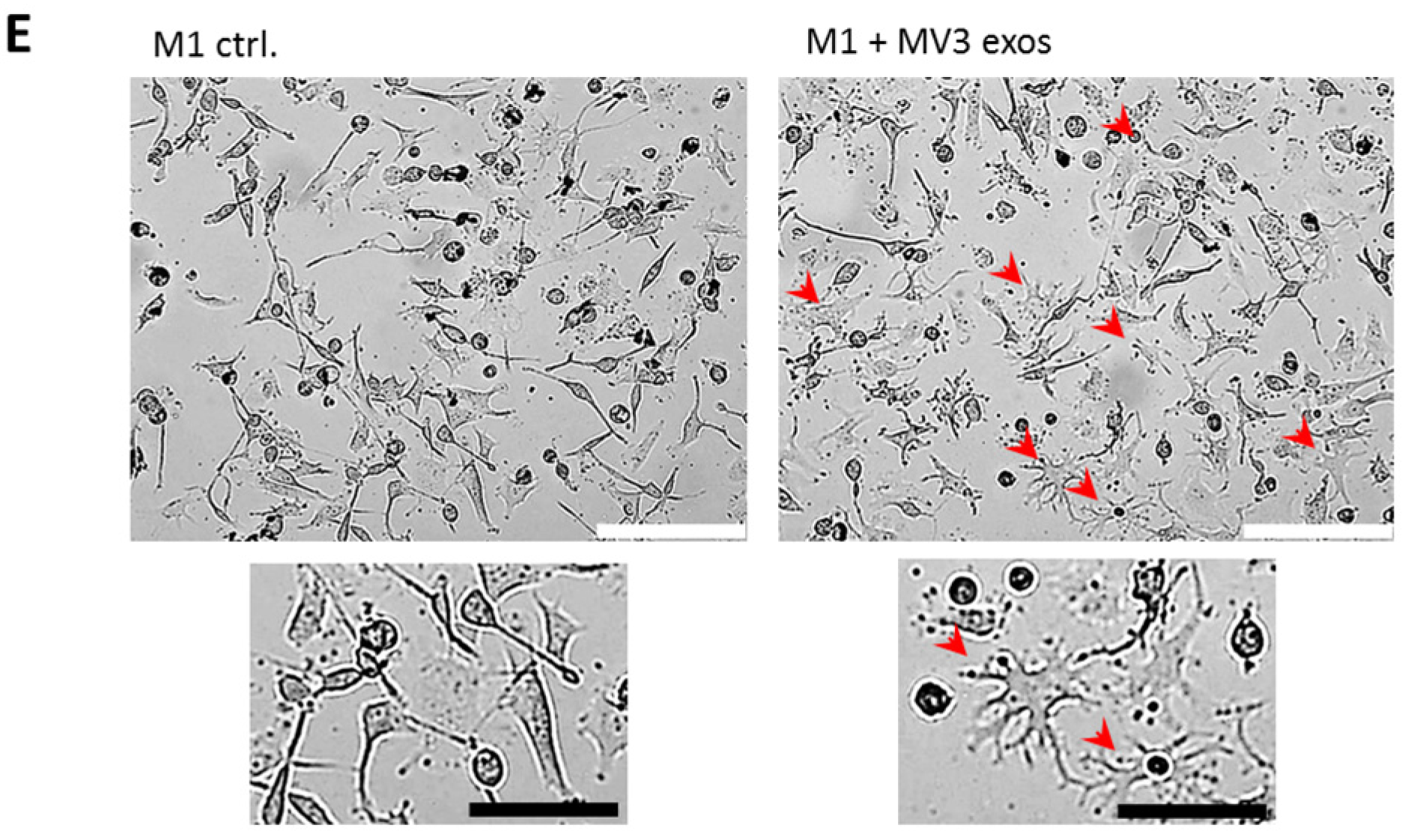
2.2. Melanoma Exosomes Induce a Proinflammatory and Proangiogenic Phenotype Macrophages
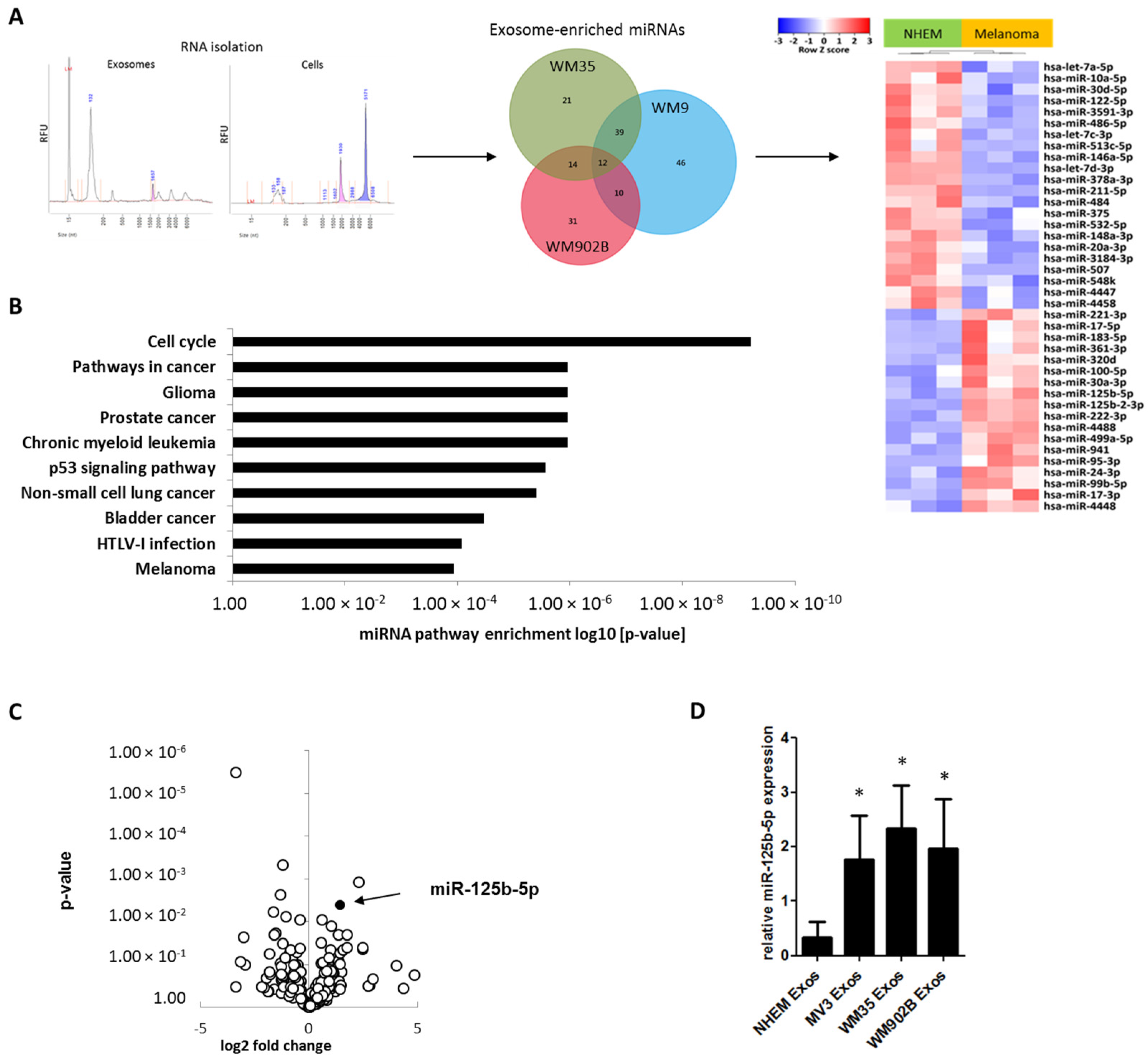
2.3. miR-125b-5p is Enriched in Melanoma-Derived Exosomes
2.4. miR-125b-5p is Delivered to M1 Macrophages by Melanoma Exosomes
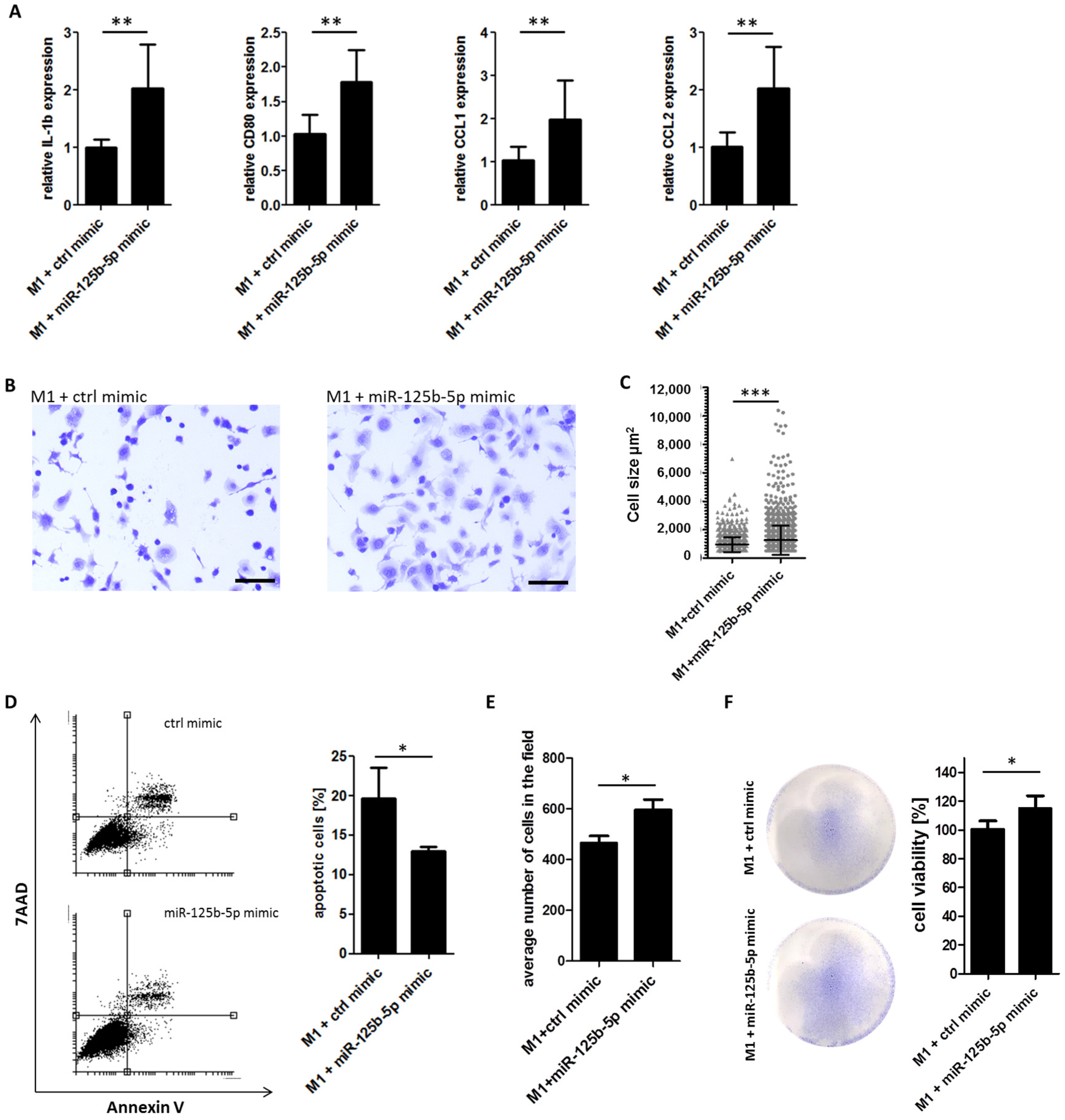
2.5. Overexpression of miR-125b-5p Partially Mimics Exosome-Induced Switch of TAM Phenotype and Promotes Survival of Macrophages
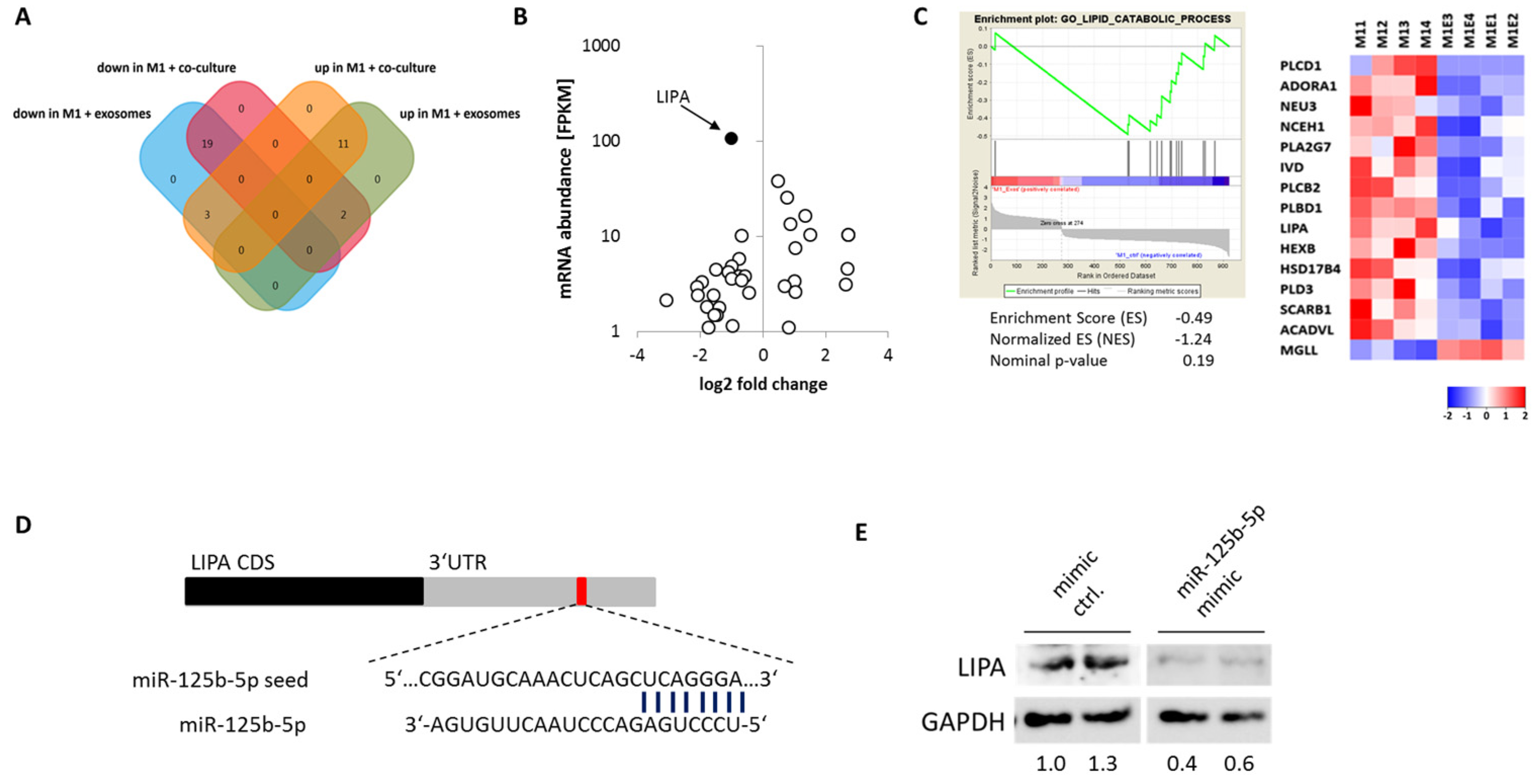
2.6. miR-125b-5p Targets Lysosomal Acid Lipase A (LIPA)
3. Discussion
4. Materials and Methods
4.1. Cell Cultures
4.2. Exosome Isolation, Analysis, and Staining
4.3. THP-1 Macrophage Polarization
4.4. Transcriptome Analysis
4.5. Small RNA-Seq Protocol
4.6. miRNA Detection by Quantitative Real-Time PCR
4.7. Transcriptional Analysis by qRT-PCR
- CCL1 for GCTTCACCAGGCTCATCAAA
- CCL1 rev TCAGGGGAATCTCTTGCTCC
- CCL2 for CTCTCGCCTCCAGCATGAAA
- CCL2 rev CTTGAAGATCACAGCTTCTTTGG
- CD80 for GGGAAATGTCGCCTCTCTGA
- CD80 rev TGCTCACGTAGAAGACCCTC
- GAPDH for ACCACAGTCCATGCCATCAC
- GAPDH rev TCCACCACCCTGTTGCTGTA
- IL1b for TGATGGCTTATTACAGTGGCAATG
- IL1b rev GTAGTGGTGGTGGGAGATTCG
- IL6 for GACAGCCACTCACCTCTTCAGA
- IL6 rev GTGCCTCTTTGCTGCTTTCAC
- IL8 for GCTAAAGAACTTCGATGTCAGTGC
- IL8 rev CTCAGCCCTCTTCAAAAACTTCTC
- TNFA for CTTCTGCCTGCTGCACTTTG
- TNFA rev GGCCAGAGGGCTGATTAGAGA
- VEGFA for CACCATGCAGATTATGCGGA
- VEGFA rev GAGGCTCCAGGGCATTAGAC
4.8. Transfections
4.9. Immunoblot Analyses
4.10. Apoptosis Assay
4.11. Viability Staining Assay
4.12. miRNA Pathway Analysis
4.13. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mrnas and micrornas is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Maia, J.; Caja, S.; Strano Moraes, M.C.; Couto, N.; Costa-Silva, B. Exosome-based cell-cell communication in the tumor microenvironment. Front. Cell Dev. Biol. 2018, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Deng, C.X. Current progresses of exosomes as cancer diagnostic and prognostic biomarkers. Int. J. Biol. Sci. 2019, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bland, C.L.; Byrne-Hoffman, C.N.; Fernandez, A.; Rellick, S.L.; Deng, W.; Klinke, D.J., 2nd. Exosomes derived from b16f0 melanoma cells alter the transcriptome of cytotoxic t cells that impacts mitochondrial respiration. FEBS J. 2018, 285, 1033–1050. [Google Scholar] [CrossRef]
- Bardi, G.T.; Smith, M.A.; Hood, J.L. Melanoma exosomes promote mixed m1 and m2 macrophage polarization. Cytokine 2018, 105, 63–72. [Google Scholar] [CrossRef]
- Peinado, H.; Alec kovic, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; Garcia-Santos, G.; Ghajar, C.M.; et al. Corrigendum: Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through met. Nat. Med. 2016, 22, 1502. [Google Scholar] [CrossRef]
- Plebanek, M.P.; Angeloni, N.L.; Vinokour, E.; Li, J.; Henkin, A.; Martinez-Marin, D.; Filleur, S.; Bhowmick, R.; Henkin, J.; Miller, S.D.; et al. Pre-metastatic cancer exosomes induce immune surveillance by patrolling monocytes at the metastatic niche. Nat. Commun. 2017, 8, 1319. [Google Scholar] [CrossRef]
- Ning, Y.; Shen, K.; Wu, Q.; Sun, X.; Bai, Y.; Xie, Y.; Pan, J.; Qi, C. Tumor exosomes block dendritic cells maturation to decrease the t cell immune response. Immunol. Lett. 2018, 199, 36–43. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Bronte, V. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 2012, 12, 253–268. [Google Scholar] [CrossRef]
- Pieniazek, M.; Matkowski, R.; Donizy, P. Macrophages in skin melanoma-the key element in melanomagenesis. Oncol. Lett. 2018, 15, 5399–5404. [Google Scholar] [CrossRef] [PubMed]
- Falleni, M.; Savi, F.; Tosi, D.; Agape, E.; Cerri, A.; Moneghini, L.; Bulfamante, G.P. M1 and m2 macrophages’ clinicopathological significance in cutaneous melanoma. Melanoma Res. 2017, 27, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Cassetta, L.; Pollard, J.W. Targeting macrophages: Therapeutic approaches in cancer. Nat. Rev. Drug Discov. 2018, 17, 887–904. [Google Scholar] [CrossRef] [PubMed]
- Poh, A.R.; Ernst, M. Targeting macrophages in cancer: From bench to bedside. Front. Oncol. 2018, 8, 49. [Google Scholar] [CrossRef]
- Torisu, H.; Ono, M.; Kiryu, H.; Furue, M.; Ohmoto, Y.; Nakayama, J.; Nishioka, Y.; Sone, S.; Kuwano, M. Macrophage infiltration correlates with tumor stage and angiogenesis in human malignant melanoma: Possible involvement of tnfalpha and il-1alpha. Int. J. Cancer 2000, 85, 182–188. [Google Scholar] [CrossRef]
- Salmi, S.; Siiskonen, H.; Sironen, R.; Tyynela-Korhonen, K.; Hirschovits-Gerz, B.; Valkonen, M.; Auvinen, P.; Pasonen-Seppanen, S. The number and localization of cd68+ and cd163+ macrophages in different stages of cutaneous melanoma. Melanoma Res. 2019, 29, 237–247. [Google Scholar] [CrossRef]
- Yang, M.; McKay, D.; Pollard, J.W.; Lewis, C.E. Diverse functions of macrophages in different tumor microenvironments. Cancer Res. 2018, 78, 5492–5503. [Google Scholar] [CrossRef]
- Baltimore, D.; Boldin, M.P.; O’Connell, R.M.; Rao, D.S.; Taganov, K.D. Micrornas: New regulators of immune cell development and function. Nat. Immunol. 2008, 9, 839–845. [Google Scholar] [CrossRef]
- Ying, X.; Wu, Q.; Wu, X.; Zhu, Q.; Wang, X.; Jiang, L.; Chen, X.; Wang, X. Epithelial ovarian cancer-secreted exosomal mir-222-3p induces polarization of tumor-associated macrophages. Oncotarget 2016, 7, 43076–43087. [Google Scholar] [CrossRef]
- Chen, X.; Ying, X.; Wang, X.; Wu, X.; Zhu, Q.; Wang, X. Exosomes derived from hypoxic epithelial ovarian cancer deliver microrna-940 to induce macrophage m2 polarization. Oncol. Rep. 2017, 38, 522–528. [Google Scholar] [CrossRef]
- Bartel, D.P. Micrornas: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Ambros, V. The functions of animal micrornas. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- Karp, X.; Ambros, V. Developmental biology. Encountering micrornas in cell fate signaling. Science 2005, 310, 1288–1289. [Google Scholar] [CrossRef]
- Jordan, S.D.; Kruger, M.; Willmes, D.M.; Redemann, N.; Wunderlich, F.T.; Bronneke, H.S.; Merkwirth, C.; Kashkar, H.; Olkkonen, V.M.; Bottger, T.; et al. Obesity-induced overexpression of mirna-143 inhibits insulin-stimulated akt activation and impairs glucose metabolism. Nat. Cell Biol. 2011, 13, 434–446. [Google Scholar] [CrossRef]
- Cheng, A.M.; Byrom, M.W.; Shelton, J.; Ford, L.P. Antisense inhibition of human mirnas and indications for an involvement of mirna in cell growth and apoptosis. Nucleic Acids Res. 2005, 33, 1290–1297. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. Microrna expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Ding, X.; Du, H.; Yoder, M.C.; Yan, C. Critical role of the mtor pathway in development and function of myeloid-derived suppressor cells in lal-/- mice. Am. J. Pathol. 2014, 184, 397–408. [Google Scholar] [CrossRef]
- Ding, X.; Wu, L.; Yan, C.; Du, H. Establishment of lal-/- myeloid lineage cell line that resembles myeloid-derived suppressive cells. PLoS ONE 2015, 10, e0121001. [Google Scholar] [CrossRef]
- Yan, C.; Lian, X.; Li, Y.; Dai, Y.; White, A.; Qin, Y.; Li, H.; Hume, D.A.; Du, H. Macrophage-specific expression of human lysosomal acid lipase corrects inflammation and pathogenic phenotypes in lal-/- mice. Am. J. Pathol. 2006, 169, 916–926. [Google Scholar] [CrossRef]
- Leopold, C.; Duta-Mare, M.; Sachdev, V.; Goeritzer, M.; Maresch, L.K.; Kolb, D.; Reicher, H.; Wagner, B.; Stojakovic, T.; Ruelicke, T.; et al. Hepatocyte-specific lysosomal acid lipase deficiency protects mice from diet-induced obesity but promotes hepatic inflammation. Biochim. et Biophys. Acta. Mol. Cell Biol. Lipids 2019, 1864, 500–511. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. Microrna genes are transcribed by rna polymerase ii. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Linton, S.S.; Abraham, T.; Liao, J.; Clawson, G.A.; Butler, P.J.; Fox, T.; Kester, M.; Matters, G.L. Tumor-promoting effects of pancreatic cancer cell exosomes on thp-1-derived macrophages. PLoS ONE 2018, 13, e0206759. [Google Scholar] [CrossRef]
- Huber, V.; Vallacchi, V.; Fleming, V.; Hu, X.; Cova, A.; Dugo, M.; Shahaj, E.; Sulsenti, R.; Vergani, E.; Filipazzi, P.; et al. Tumor-derived micrornas induce myeloid suppressor cells and predict immunotherapy resistance in melanoma. J. Clin. Investig. 2018, 128, 5505–5516. [Google Scholar] [CrossRef]
- Glud, M.; Manfe, V.; Biskup, E.; Holst, L.; Dirksen, A.M.; Hastrup, N.; Nielsen, F.C.; Drzewiecki, K.T.; Gniadecki, R. Microrna mir-125b induces senescence in human melanoma cells. Melanoma Res. 2011, 21, 253–256. [Google Scholar] [CrossRef]
- Glud, M.; Rossing, M.; Hother, C.; Holst, L.; Hastrup, N.; Nielsen, F.C.; Gniadecki, R.; Drzewiecki, K.T. Downregulation of mir-125b in metastatic cutaneous malignant melanoma. Melanoma Res. 2010, 20, 479–484. [Google Scholar] [CrossRef]
- Wandler, A.; Riber-Hansen, R.; Hager, H.; Hamilton-Dutoit, S.J.; Schmidt, H.; Nielsen, B.S.; Stougaard, M.; Steiniche, T. Quantification of microrna-21 and microrna-125b in melanoma tissue. Melanoma Res. 2017, 27, 417–428. [Google Scholar] [CrossRef]
- Duroux-Richard, I.; Roubert, C.; Ammari, M.; Presumey, J.; Grun, J.R.; Haupl, T.; Grutzkau, A.; Lecellier, C.H.; Boitez, V.; Codogno, P.; et al. Mir-125b controls monocyte adaptation to inflammation through mitochondrial metabolism and dynamics. Blood 2016, 128, 3125–3136. [Google Scholar] [CrossRef]
- Chaudhuri, A.A.; So, A.Y.; Sinha, N.; Gibson, W.S.; Taganov, K.D.; O’Connell, R.M.; Baltimore, D. Microrna-125b potentiates macrophage activation. J. Immunol. 2011, 187, 5062–5068. [Google Scholar] [CrossRef]
- Lee, C.W.; Schoenherr, C.; Battmer, K.; Ganser, A.; Hilfiker-Kleiner, D.; David, S.; Eder, M.; Scherr, M. Mir-125b regulates chemotaxis and survival of bone marrow derived granulocytes in vitro and in vivo. PLoS ONE 2018, 13, e0204942. [Google Scholar] [CrossRef]
- Batool, A.; Wang, Y.Q.; Hao, X.X.; Chen, S.R.; Liu, Y.X. A mir-125b/csf1-cx3cl1/tumor-associated macrophage recruitment axis controls testicular germ cell tumor growth. Cell Death Dis. 2018, 9, 962. [Google Scholar] [CrossRef]
- Xu, Z.; Zhao, L.; Yang, X.; Ma, S.; Ge, Y.; Liu, Y.; Liu, S.; Shi, J.; Zheng, D. Mmu-mir-125b overexpression suppresses no production in activated macrophages by targeting eef2k and ccna2. BMC Cancer 2016, 16, 252. [Google Scholar] [CrossRef][Green Version]
- Zhu, Y.; Zhang, S.; Li, Z.; Wang, H.; Li, Z.; Hu, Y.; Chen, H.; Zhang, X.; Cui, L.; Zhang, J.; et al. Mir-125b-5p and mir-99a-5p downregulate human gammadelta t-cell activation and cytotoxicity. Cell. Mol. Immunol. 2019, 16, 112–125. [Google Scholar] [CrossRef]
- Sansom, D.M. Cd28, ctla-4 and their ligands: Who does what and to whom? Immunology 2000, 101, 169–177. [Google Scholar] [CrossRef]
- Zheng, Z.; Qu, J.Q.; Yi, H.M.; Ye, X.; Huang, W.; Xiao, T.; Li, J.Y.; Wang, Y.Y.; Feng, J.; Zhu, J.F.; et al. Mir-125b regulates proliferation and apoptosis of nasopharyngeal carcinoma by targeting a20/nf-kappab signaling pathway. Cell Death Dis. 2017, 8, e2855. [Google Scholar] [CrossRef]
- Wei, M.; Gan, L.; Liu, Z.; Kong, L.H.; Chang, J.R.; Chen, L.H.; Su, X.L. Mir125b-5p protects endothelial cells from apoptosis under oxidative stress. Biomed. Pharmacother. 2017, 95, 453–460. [Google Scholar] [CrossRef]
- Nomura, M.; Liu, J.; Rovira, I.I.; Gonzalez-Hurtado, E.; Lee, J.; Wolfgang, M.J.; Finkel, T. Fatty acid oxidation in macrophage polarization. Nat. Immunol. 2016, 17, 216–217. [Google Scholar] [CrossRef]
- Huang, S.C.; Everts, B.; Ivanova, Y.; O’Sullivan, D.; Nascimento, M.; Smith, A.M.; Beatty, W.; Love-Gregory, L.; Lam, W.Y.; O’Neill, C.M.; et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat. Immunol. 2014, 15, 846–855. [Google Scholar] [CrossRef]
- Tili, E.; Michaille, J.J.; Luo, Z.; Volinia, S.; Rassenti, L.Z.; Kipps, T.J.; Croce, C.M. The down-regulation of mir-125b in chronic lymphocytic leukemias leads to metabolic adaptation of cells to a transformed state. Blood 2012, 120, 2631–2638. [Google Scholar] [CrossRef]
- Qu, P.; Yan, C.; Blum, J.S.; Kapur, R.; Du, H. Myeloid-specific expression of human lysosomal acid lipase corrects malformation and malfunction of myeloid-derived suppressor cells in lal-/- mice. J. Immunol. 2011, 187, 3854–3866. [Google Scholar] [CrossRef]
- Zhao, T.; Ding, X.; Du, H.; Yan, C. Lung epithelial cell-specific expression of human lysosomal acid lipase ameliorates lung inflammation and tumor metastasis in lipa (-/-) mice. Am. J. Pathol. 2016, 186, 2183–2192. [Google Scholar] [CrossRef]
- Zhao, T.; Du, H.; Ding, X.; Walls, K.; Yan, C. Activation of mtor pathway in myeloid-derived suppressor cells stimulates cancer cell proliferation and metastasis in lal (-/-) mice. Oncogene 2015, 34, 1938–1948. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Siklenka, K.; Arora, S.K.; Ribeiro, P.; Kimmins, S.; Xia, J. Mirnet—dissecting mirna-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016, 44, W135–W141. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Xia, J. Mirnet-functional analysis and visual exploration of mirna-target interactions in a network context. Methods Mol. Biol. 2018, 1819, 215–233. [Google Scholar] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gerloff, D.; Lützkendorf, J.; Moritz, R.K.C.; Wersig, T.; Mäder, K.; Müller, L.P.; Sunderkötter, C. Melanoma-Derived Exosomal miR-125b-5p Educates Tumor Associated Macrophages (TAMs) by Targeting Lysosomal Acid Lipase A (LIPA). Cancers 2020, 12, 464. https://doi.org/10.3390/cancers12020464
Gerloff D, Lützkendorf J, Moritz RKC, Wersig T, Mäder K, Müller LP, Sunderkötter C. Melanoma-Derived Exosomal miR-125b-5p Educates Tumor Associated Macrophages (TAMs) by Targeting Lysosomal Acid Lipase A (LIPA). Cancers. 2020; 12(2):464. https://doi.org/10.3390/cancers12020464
Chicago/Turabian StyleGerloff, Dennis, Jana Lützkendorf, Rose K.C. Moritz, Tom Wersig, Karsten Mäder, Lutz P. Müller, and Cord Sunderkötter. 2020. "Melanoma-Derived Exosomal miR-125b-5p Educates Tumor Associated Macrophages (TAMs) by Targeting Lysosomal Acid Lipase A (LIPA)" Cancers 12, no. 2: 464. https://doi.org/10.3390/cancers12020464
APA StyleGerloff, D., Lützkendorf, J., Moritz, R. K. C., Wersig, T., Mäder, K., Müller, L. P., & Sunderkötter, C. (2020). Melanoma-Derived Exosomal miR-125b-5p Educates Tumor Associated Macrophages (TAMs) by Targeting Lysosomal Acid Lipase A (LIPA). Cancers, 12(2), 464. https://doi.org/10.3390/cancers12020464





