A Highly Sensitive Next-Generation Sequencing-Based Genotyping Platform for EGFR Mutations in Plasma from Non-Small Cell Lung Cancer Patients
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Sample Preparation
2.3. EGFR Mutation Detection Platforms
2.4. Sel-Cap Mutation Enrichment PCR
2.5. NGS Library Preparation
2.6. Monitoring EGFR T790M in Plasma for EGFR-TKI Treatment
2.7. Statistical Analyses
3. Results
3.1. Study Population
3.2. Sel-Cap Showed High Sensitivity for EGFR Mutations in Plasma
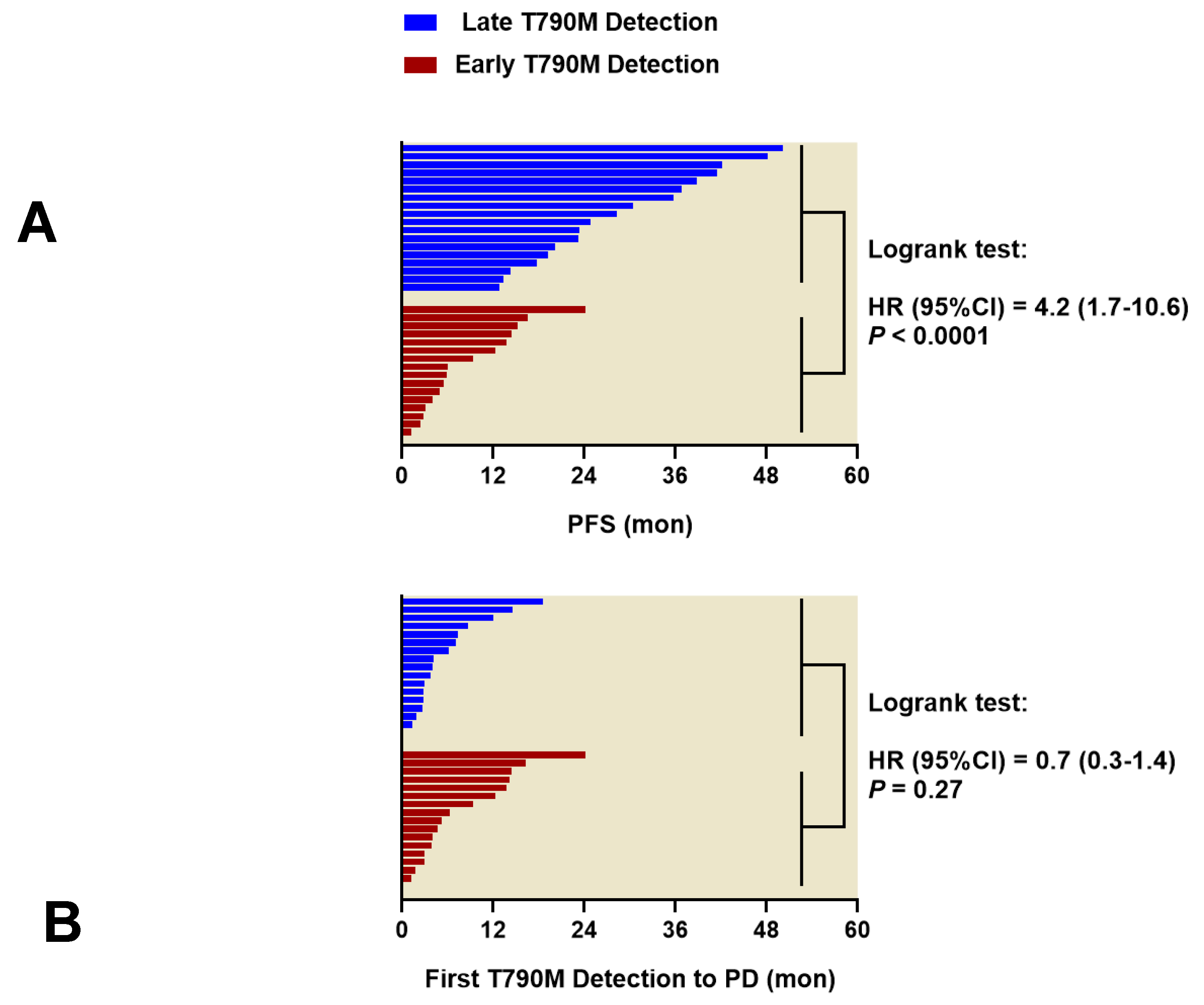
3.3. Timing of First T790M Detection in Plasma is Critical for PFS of First-Line EGFR-TKIs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hirsch, F.R.; Scagliotti, G.V.; Mulshine, J.L.; Kwon, R.; Curran, W.J., Jr.; Wu, Y.L.; Paz-Ares, L. Lung cancer: Current therapies and new targeted treatments. Lancet 2017, 389, 299–311. [Google Scholar] [CrossRef]
- Morgillo, F.; Della Corte, C.M.; Fasano, M.; Ciardiello, F. Mechanisms of resistance to EGFR-targeted drugs: Lung cancer. ESMO Open 2016, 1, e000060. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Wei, S.; Song, Y. T790M and acquired resistance of EGFR TKI: A literature review of clinical reports. J. Thorac. Dis. 2011, 3, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Diaz, L.A., Jr.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.M.; Tsui, D.W.Y. Circulating cell-free DNA for non-invasive cancer management. Cancer Genet. 2018, 228–229, 169–179. [Google Scholar] [CrossRef]
- Garcia, J.; Wozny, A.S.; Geiguer, F.; Delherme, A.; Barthelemy, D.; Merle, P.; Tissot, C.; Jones, F.S.; Johnson, C.; Xing, X.; et al. Profiling of circulating tumor DNA in plasma of non-small cell lung cancer patients, monitoring of epidermal growth factor receptor p.T790M mutated allelic fraction using beads, emulsion, amplification, and magnetics companion assay and evaluation in future application in mimicking circulating tumor cells. Cancer Med. 2019, 8, 3685–3697. [Google Scholar] [CrossRef]
- Xue, V.W.; Wong, C.S.C.; Cho, W.C.S. Early detection and monitoring of cancer in liquid biopsy: Advances and challenges. Expert Rev. Mol. Diagn. 2019, 19, 273–276. [Google Scholar] [CrossRef]
- Lee, B.; Lee, B.; Han, G.; Kwon, M.J.; Han, J.; Choi, Y.L. KRAS Mutation Detection in Non-small Cell Lung Cancer Using a Peptide Nucleic Acid-Mediated Polymerase Chain Reaction Clamping Method and Comparative Validation with Next-Generation Sequencing. Korean J. Pathol. 2014, 48, 100–107. [Google Scholar] [CrossRef]
- Xue, V.W.; Ng, S.S.M.; Leung, W.W.; Ma, B.B.Y.; Cho, W.C.S.; Au, T.C.C.; Yu, A.C.S.; Tsang, H.F.A.; Wong, S.C.C. The Effect of Centrifugal Force in Quantification of Colorectal Cancer-Related mRNA in Plasma Using Targeted Sequencing. Front. Genet. 2018, 9, 165. [Google Scholar] [CrossRef]
- Mouliere, F.; Robert, B.; Arnau Peyrotte, E.; Del Rio, M.; Ychou, M.; Molina, F.; Gongora, C.; Thierry, A.R. High fragmentation characterizes tumour-derived circulating DNA. PLoS ONE 2011, 6, e23418. [Google Scholar] [CrossRef]
- Kim, H.J.; Lee, K.Y.; Kim, Y.C.; Kim, K.S.; Lee, S.Y.; Jang, T.W.; Lee, M.K.; Shin, K.C.; Lee, G.H.; Lee, J.C.; et al. Detection and comparison of peptide nucleic acid-mediated real-time polymerase chain reaction clamping and direct gene sequencing for epidermal growth factor receptor mutations in patients with non-small cell lung cancer. Lung Cancer 2012, 75, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.O.; Shin, J.Y.; Kim, S.R.; Shin, K.S.; Kim, J.; Kim, M.Y.; Lee, M.R.; Kim, Y.; Kim, M.; Hong, S.H.; et al. Evaluation of Two EGFR Mutation Tests on Tumor and Plasma from Patients with Non-Small Cell Lung Cancer. Cancers 2020, 12, 785. [Google Scholar] [CrossRef] [PubMed]
- Traynor, A.M.; Weigel, T.L.; Oettel, K.R.; Yang, D.T.; Zhang, C.; Kim, K.; Salgia, R.; Iida, M.; Brand, T.M.; Hoang, T.; et al. Nuclear EGFR protein expression predicts poor survival in early stage non-small cell lung cancer. Lung Cancer 2013, 81, 138–141. [Google Scholar] [CrossRef]
- Douillard, J.Y.; Pirker, R.; O’Byrne, K.J.; Kerr, K.M.; Storkel, S.; von Heydebreck, A.; Grote, H.J.; Celik, I.; Shepherd, F.A. Relationship between EGFR expression, EGFR mutation status, and the efficacy of chemotherapy plus cetuximab in FLEX study patients with advanced non-small-cell lung cancer. J. Thorac. Oncol. 2014, 9, 717–724. [Google Scholar] [CrossRef]
- Li, C.; He, Q.; Liang, H.; Cheng, B.; Li, J.; Xiong, S.; Zhao, Y.; Guo, M.; Liu, Z.; He, J.; et al. Diagnostic Accuracy of Droplet Digital PCR and Amplification Refractory Mutation System PCR for Detecting EGFR Mutation in Cell-Free DNA of Lung Cancer: A Meta-Analysis. Front. Oncol. 2020, 10, 290. [Google Scholar] [CrossRef]
- Lee, J.Y.; Qing, X.; Xiumin, W.; Yali, B.; Chi, S.; Bak, S.H.; Lee, H.Y.; Sun, J.M.; Lee, S.H.; Ahn, J.S.; et al. Longitudinal monitoring of EGFR mutations in plasma predicts outcomes of NSCLC patients treated with EGFR TKIs: Korean Lung Cancer Consortium (KLCC-12-02). Oncotarget 2016, 7, 6984–6993. [Google Scholar] [CrossRef]
- Kebschull, J.M.; Zador, A.M. Sources of PCR-induced distortions in high-throughput sequencing data sets. Nucleic Acids Res. 2015, 43, e143. [Google Scholar] [CrossRef]
- Usui, K.; Yokoyama, T.; Naka, G.; Ishida, H.; Kishi, K.; Uemura, K.; Ohashi, Y.; Kunitoh, H. Plasma ctDNA monitoring during epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitor treatment in patients with EGFR-mutant non-small cell lung cancer (JP-CLEAR trial). Jpn. J. Clin. Oncol. 2019, 49, 554–558. [Google Scholar] [CrossRef]
- Ye, X.; Zhu, Z.Z.; Zhong, L.; Lu, Y.; Sun, Y.; Yin, X.; Yang, Z.; Zhu, G.; Ji, Q. High T790M detection rate in TKI-naive NSCLC with EGFR sensitive mutation: Truth or artifact? J. Thorac. Oncol. 2013, 8, 1118–1120. [Google Scholar] [CrossRef]
- Goldman, J.W.; Noor, Z.S.; Remon, J.; Besse, B.; Rosenfeld, N. Are liquid biopsies a surrogate for tissue EGFR testing? Ann. Oncol. 2018, 29, i38–i46. [Google Scholar] [CrossRef] [PubMed]
- Ichihara, E.; Hotta, K.; Kubo, T.; Higashionna, T.; Ninomiya, K.; Ohashi, K.; Tabata, M.; Maeda, Y.; Kiura, K. Clinical significance of repeat rebiopsy in detecting the EGFR T790M secondary mutation in patients with non-small cell lung cancer. Oncotarget 2018, 9, 29525–29531. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dal Maso, A.; Lorenzi, M.; Roca, E.; Pilotto, S.; Macerelli, M.; Polo, V.; Cecere, F.L.; Del Conte, A.; Nardo, G.; Buoro, V.; et al. Clinical Features and Progression Pattern of Acquired T790M-positive Compared with T790M-negative EGFR Mutant Non-small-cell Lung Cancer: Catching Tumor and Clinical Heterogeneity Over Time Through Liquid Biopsy. Clin. Lung Cancer 2020, 21, 1–14.e13. [Google Scholar] [CrossRef] [PubMed]
- Sueoka-Aragane, N.; Katakami, N.; Satouchi, M.; Yokota, S.; Aoe, K.; Iwanaga, K.; Otsuka, K.; Morita, S.; Kimura, S.; Negoro, S.; et al. Monitoring EGFR T790M with plasma DNA from lung cancer patients in a prospective observational study. Cancer Sci. 2016, 107, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Ye, X.; Zhang, M.Z.; Sun, Y.; Wang, J.Y.; Ni, J.; Zhang, H.P.; Zhang, L.; Luo, J.; Zhang, J.; et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci. Rep. 2016, 6, 20913. [Google Scholar] [CrossRef] [PubMed]
- Iwama, E.; Sakai, K.; Hidaka, N.; Inoue, K.; Fujii, A.; Nakagaki, N.; Ota, K.; Toyozawa, R.; Azuma, K.; Nakatomi, K.; et al. Longitudinal monitoring of somatic genetic alterations in circulating cell-free DNA during treatment with epidermal growth factor receptor-tyrosine kinase inhibitors. Cancer 2020, 126, 219–227. [Google Scholar] [CrossRef]
- Ouyang, W.; Yu, J.; Huang, Z.; Chen, G.; Liu, Y.; Liao, Z.; Zeng, W.; Zhang, J.; Xie, C. Risk factors of acquired T790M mutation in patients with epidermal growth factor receptor-mutated advanced non-small cell lung cancer. J. Cancer 2020, 11, 2060–2067. [Google Scholar] [CrossRef]
- Huang, Y.H.; Hsu, K.H.; Tseng, J.S.; Chen, K.C.; Hsu, C.H.; Su, K.Y.; Chen, J.J.W.; Chen, H.W.; Yu, S.L.; Yang, T.Y.; et al. The Association of Acquired T790M Mutation with Clinical Characteristics after Resistance to First-Line Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor in Lung Adenocarcinoma. Cancer Res. Treat. 2018, 50, 1294–1303. [Google Scholar] [CrossRef]
- Wang, X.; Wang, L.; Su, Y.; Yue, Z.; Xing, T.; Zhao, W.; Zhao, Q.; Duan, C.; Huang, C.; Zhang, D.; et al. Plasma cell-free DNA quantification is highly correlated to tumor burden in children with neuroblastoma. Cancer Med. 2018. [Google Scholar] [CrossRef]
- Valpione, S.; Gremel, G.; Mundra, P.; Middlehurst, P.; Galvani, E.; Girotti, M.R.; Lee, R.J.; Garner, G.; Dhomen, N.; Lorigan, P.C.; et al. Plasma total cell-free DNA (cfDNA) is a surrogate biomarker for tumour burden and a prognostic biomarker for survival in metastatic melanoma patients. Eur. J. Cancer 2018, 88, 1–9. [Google Scholar] [CrossRef]
- Bylicki, O.; Paleiron, N.; Assie, J.B.; Chouaid, C. Targeting the MET-Signaling Pathway in Non-Small-Cell Lung Cancer: Evidence to Date. OncoTargets Ther. 2020, 13, 5691–5706. [Google Scholar] [CrossRef] [PubMed]

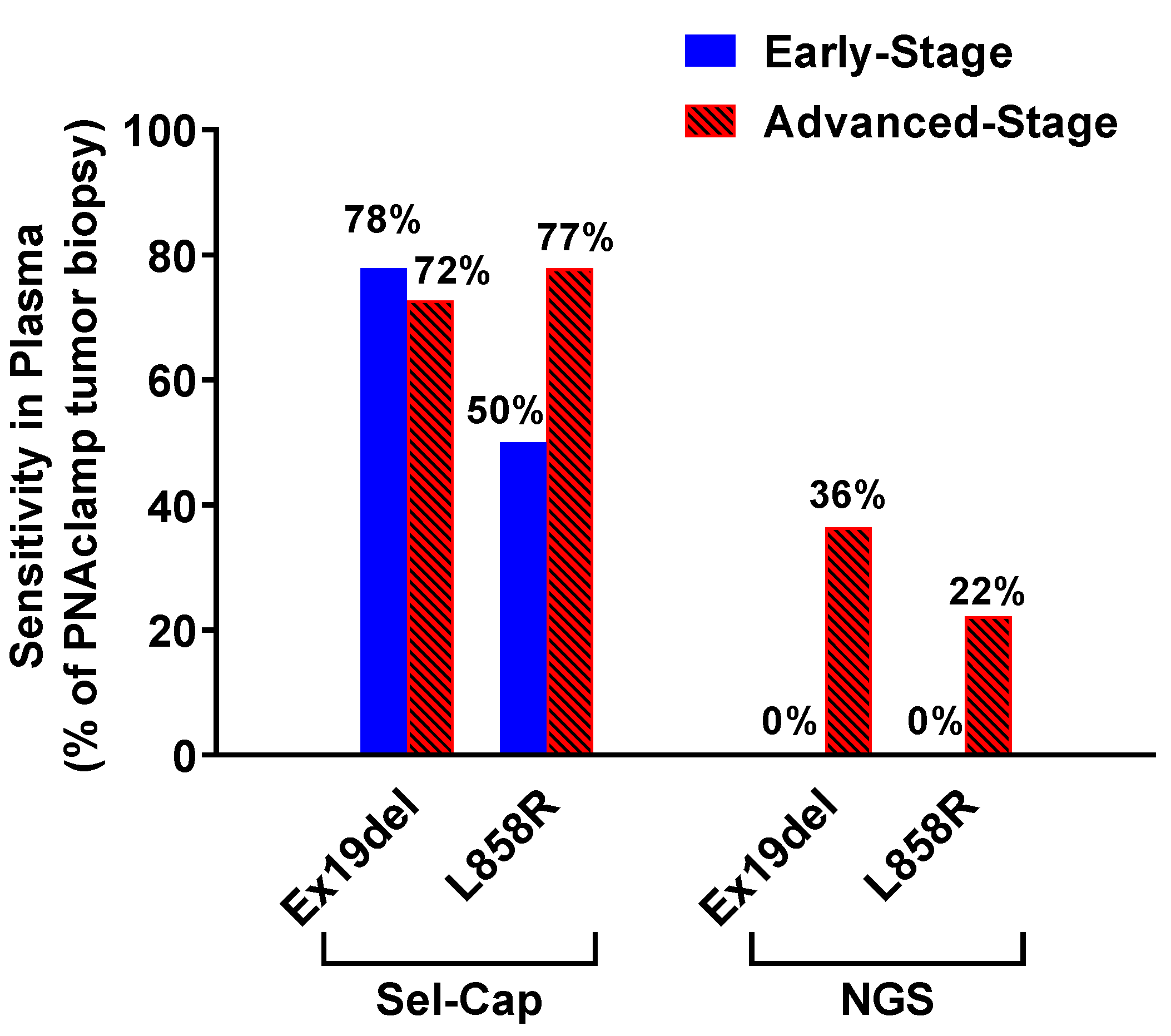
| Ex19del | L858R | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sel-Cap | NGS | Sel-Cap | NGS | ||||||||||
| Mutant | Wild | Total | Mutant | Wild | Total | Mutant | Wild | Total | Mutant | Wild | Total | ||
| PNAclamp | Mutant | 15 | 5 | 20 | 3 | 15 | 18 | 11 | 6 | 17 | 2 | 14 | 16 |
| Wild | 2 | 39 | 41 | 1 | 42 | 43 | 0 | 44 | 44 | 0 | 45 | 45 | |
| Total | 17 | 44 | 61 | 4 | 57 | 61 | 11 | 50 | 61 | 2 | 59 | 61 | |
| Sensitivity | (95% CI) | 75% | (53–89%) | 17% | (6–39%) | 65% | (41–83%) | 13% | (3–36%) | ||||
| Specificity | (95% CI) | 95% | (84–99%) | 98% | (88–100%) | 100% | (92–100%) | 100% | (92–100%) | ||||
| Accuracy | (95% CI) | 89% | (78–94%) | 74% | (62–83%) | 90% | (80–95%) | 77% | (65–86%) | ||||
| Kappa | (95% CI) | 0.73 | (0.54–0.92) | 0.19 | (−0.16–0.53) | 0.73 | (0.52–0.93) | 0.17 | (−0.21–0.55) | ||||
| Sel-Cap | Ex19del | L858R | T790M a | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mutant | Wild | Total | Mutant | Wild | Total | Mutant | Wild | Total | ||
| PNAclamp | Mutant | 50 | 19 | 69 | 24 | 12 | 36 | 7 | 14 | 21 |
| Wild | 1 | 115 | 116 | 3 | 146 | 149 | 1 | 63 | 64 | |
| Total | 51 | 134 | 185 | 27 | 158 | 185 | 8 | 77 | 85 | |
| Sensitivity | (95% CI) | 72% | (61–82%) | 67% | (50–80%) | 88% | (53–98%) | |||
| Specificity | (95% CI) | 99% | (95–100%) | 98% | (94–99%) | 82% | (72–89%) | |||
| Accuracy | (95% CI) | 89% | (84–93%) | 92% | (87–95%) | 82% | (73–89%) | |||
| Kappa | (95% CI) | 0.76 | (0.65–0.86) | 0.71 | (0.57–0.85) | 0.40 | (0.13–0.68) | |||
| No. | EGFR-TKI | Tumor | Plasma | |
|---|---|---|---|---|
| PNAclamp | NGS Cancer Panel | Sel-Cap | ||
| 1 | Erlotinib | Ex19del a | Ex19del | Ex19del, T790M d |
| 2 | Erlotinib | Ex19del a | Wild | T790M d |
| 3 | Erlotinib | L858R a | L858R, T790M d | Wild |
| 4 | Erlotinib | Ex19del a | Wild | T790M d |
| 5 | Erlotinib | Ex19del a | Ex19del d | Wild |
| 6 | Gefitinib | Ex19del, T790M a | Wild | Wild |
| 7 | Gefitinib | Ex19del a | Ex19del, T790M | Ex19del, T790M |
| 8 | Afatinib | Ex19del a | Wild | Wild |
| 9 | Afatinib | Ex19del a | Ex19del | Ex19del, T790M d |
| 10 | Gefitinib | L858R, T790M b | L858R, T790M d | L858R |
| 11 | Gefitinib | Ex19del, T790M b | Wild | Wild |
| 12 | Afatinib | Ex19del b | Ex19del | Ex19del, T790M d |
| 13 | Erlotinib | T790M c | Wild | Wild |
| 14 | Erlotinib | T790M c | Wild | Wild |
| 15 | Erlotinib | L858R, T790M c | L858R | L858R, T790M d |
| 16 | Erlotinib | Ex19del, T790M c | Ex19del | Ex19del |
| 17 | Erlotinib | Ex19del, T790M c | Wild | Ex19del, T790M d |
| 18 | Erlotinib | Ex19del, T790M c | Ex19del | Ex19del, T790M d |
| 19 | Gefitinib | Ex19del, T790M c | Wild | Ex19del, T790M d |
| 20 | Gefinitib | Ex19del, T790M c | Wild | Ex19del, T790M d |
| 21 | Gefinitib | L858R, T790M c | L858R, T790M | L858R, T790M |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, J.-Y.; Kim, J.-O.; Lee, M.-R.; Kim, S.R.; Beck, K.S.; Kang, J.H. A Highly Sensitive Next-Generation Sequencing-Based Genotyping Platform for EGFR Mutations in Plasma from Non-Small Cell Lung Cancer Patients. Cancers 2020, 12, 3579. https://doi.org/10.3390/cancers12123579
Shin J-Y, Kim J-O, Lee M-R, Kim SR, Beck KS, Kang JH. A Highly Sensitive Next-Generation Sequencing-Based Genotyping Platform for EGFR Mutations in Plasma from Non-Small Cell Lung Cancer Patients. Cancers. 2020; 12(12):3579. https://doi.org/10.3390/cancers12123579
Chicago/Turabian StyleShin, Jung-Young, Jeong-Oh Kim, Mi-Ran Lee, Seo Ree Kim, Kyongmin Sarah Beck, and Jin Hyoung Kang. 2020. "A Highly Sensitive Next-Generation Sequencing-Based Genotyping Platform for EGFR Mutations in Plasma from Non-Small Cell Lung Cancer Patients" Cancers 12, no. 12: 3579. https://doi.org/10.3390/cancers12123579
APA StyleShin, J.-Y., Kim, J.-O., Lee, M.-R., Kim, S. R., Beck, K. S., & Kang, J. H. (2020). A Highly Sensitive Next-Generation Sequencing-Based Genotyping Platform for EGFR Mutations in Plasma from Non-Small Cell Lung Cancer Patients. Cancers, 12(12), 3579. https://doi.org/10.3390/cancers12123579





