Molecular Genetics of Relapsed Diffuse Large B-Cell Lymphoma: Insight into Mechanisms of Therapy Resistance
Abstract
Simple Summary
Abstract
1. Introduction
2. Molecular Classifications of DLBCL
3. Immunochemotherapy Resistance in DLBCL
4. Clonal Evolution of Relapsed DLBCL
5. Genetic Alterations and Biological Pathways Selectively Enriched in R/R DLBCL
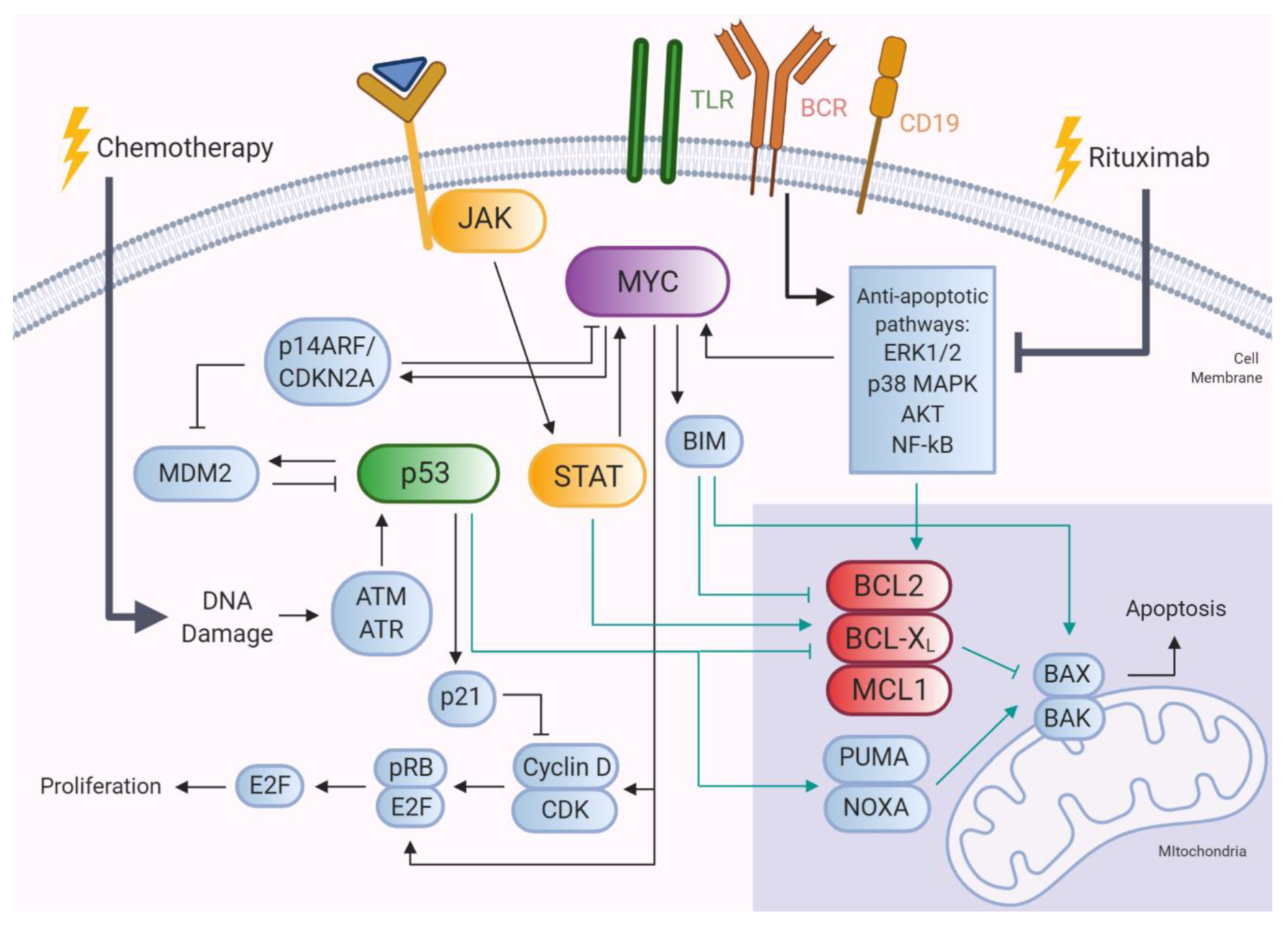
5.1. MYC, BCL2, and BCL6 Gene Alterations
5.1.1. BCL2
5.1.2. MYC
5.1.3. BCL6
5.2. TP53 Gene Alterations
5.3. Mutations Targeting JAK-STAT Signaling
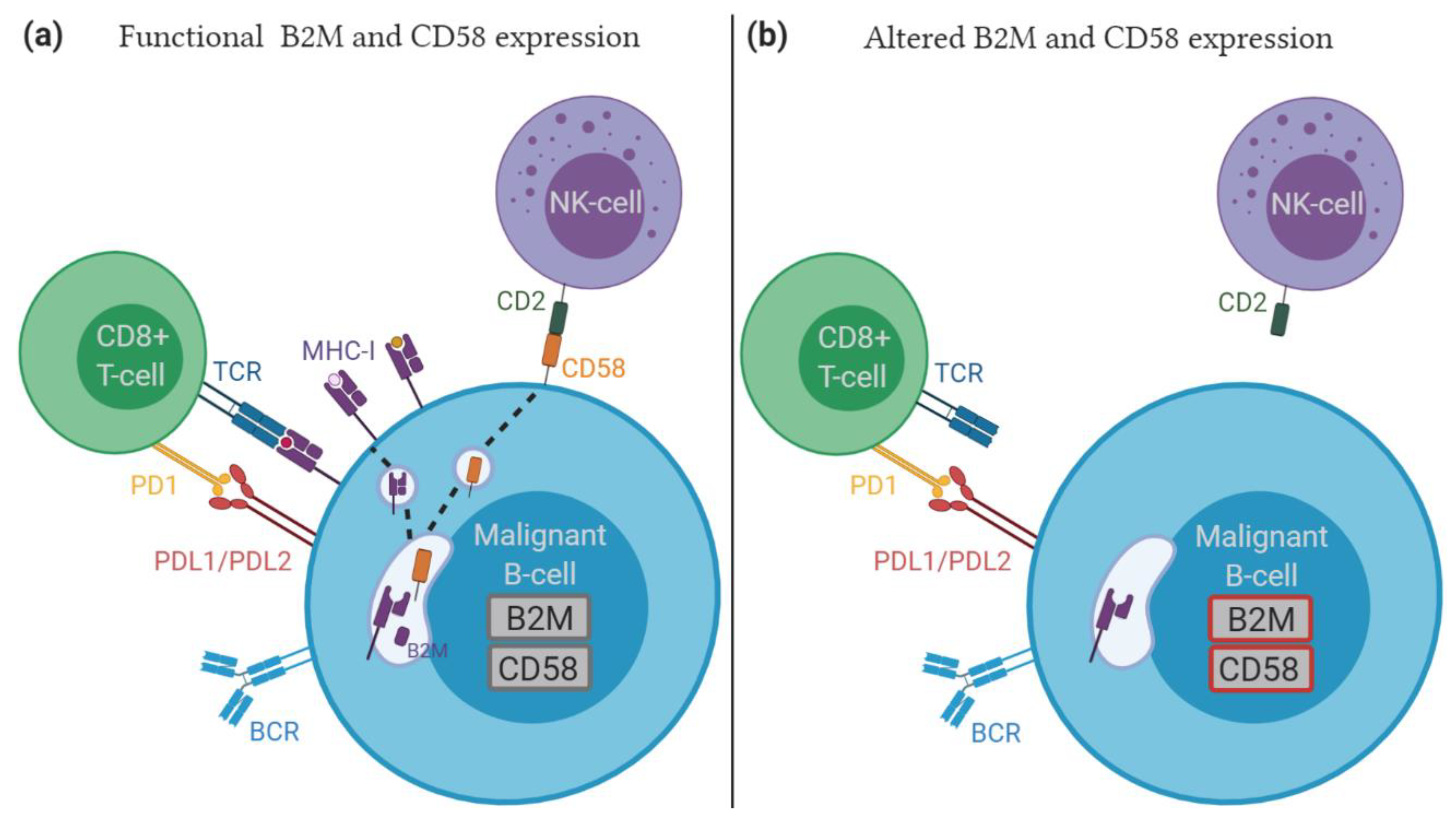
5.4. Role of Immune Escape in Relapsed/Refractory DLBCL
5.5. Gene Mutations Affecting Epigenetic Regulators in Relapsed/Refractory DLBCL
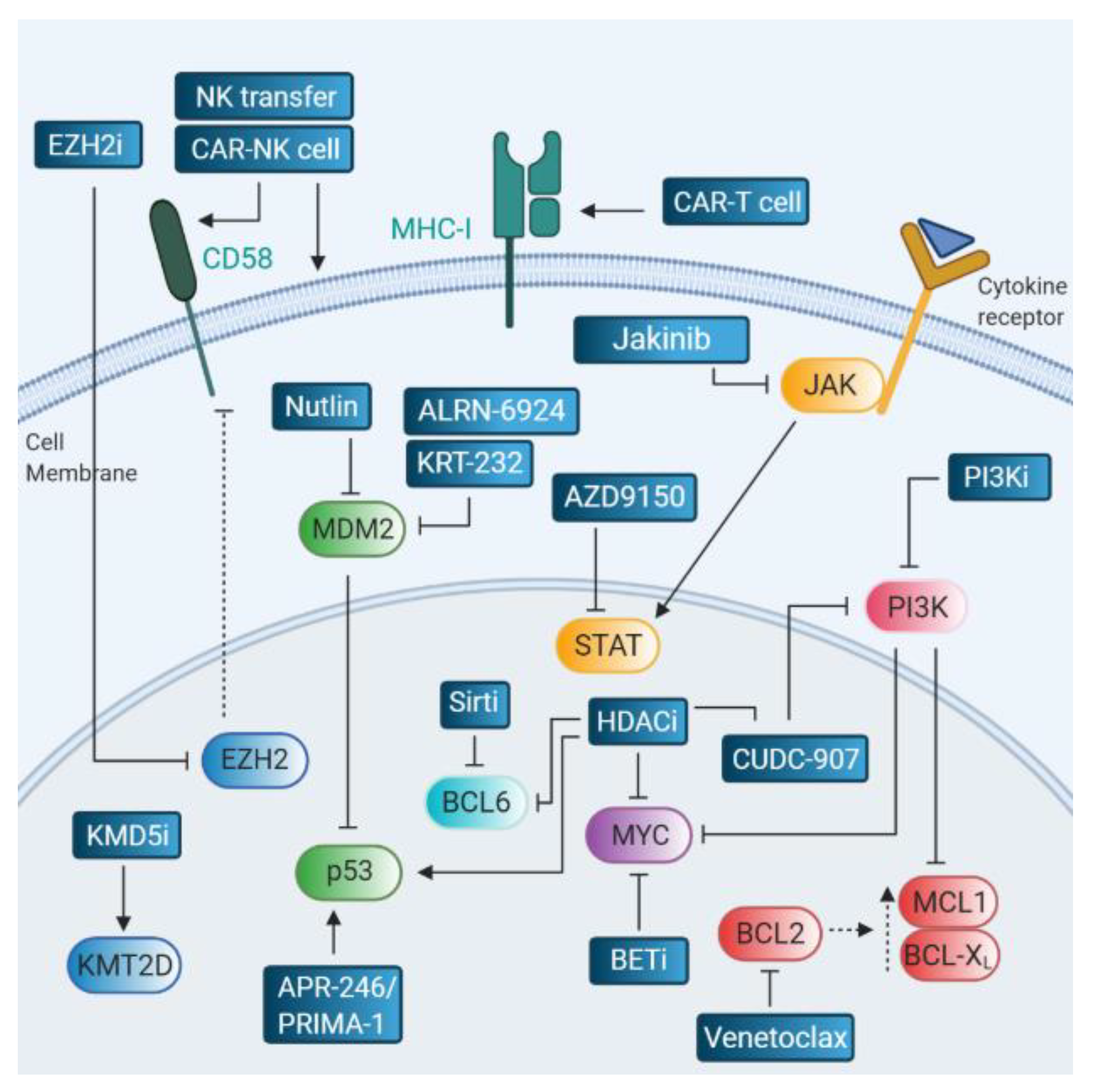
6. Therapies Targeting Relapse-Associated Drivers
6.1. BCL2 Inhibitors
6.2. MYC Inhibitors
6.3. Targeting the p53 Pathway
6.4. Targeting the JAK/STAT Pathway
6.5. Therapeutic Strategies in the Context of Immune Escape
6.6. Epigenetic Targeting
7. Conclusions and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Li, S.; Young, K.H.; Medeiros, L.J. Diffuse large B-cell lymphoma. Pathology 2018, 50, 74–87. [Google Scholar] [CrossRef]
- Martelli, M.; Ferreri, A.J.; Agostinelli, C.; Di Rocco, A.; Pfreundschuh, M.; Pileri, S.A. Diffuse large B-cell lymphoma. Crit. Rev. Oncol. Hematol. 2013, 87, 146–171. [Google Scholar] [CrossRef] [PubMed]
- Lossos, I.S.; Gascoyne, R.D. Transformation of follicular lymphoma. Best Pract. Res. Clin. Haematol. 2011, 24, 147–163. [Google Scholar] [CrossRef] [PubMed]
- Møller, M.B.; Pedersen, N.T.; Christensen, B.E. Diffuse large B-cell lymphoma: Clinical implications of extranodal versus nodal presentation—A population-based study of 1575 cases. Br. J. Haematol. 2004, 124, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Coiffier, B.; Thieblemont, C.; Van Den Neste, E.; Lepeu, G.; Plantier, I.; Castaigne, S.; Lefort, S.; Marit, G.; Macro, M.; Sebban, C.; et al. Long-term outcome of patients in the LNH-98.5 trial, the first randomized study comparing rituximab-CHOP to standard CHOP chemotherapy in DLBCL patients: A study by the Groupe d’Etudes des Lymphomes de l’Adulte. Blood 2010, 116, 2040–2045. [Google Scholar] [CrossRef] [PubMed]
- Fu, K.; Weisenburger, D.D.; Choi, W.W.; Perry, K.D.; Smith, L.M.; Shi, X.; Hans, C.P.; Greiner, T.C.; Bierman, P.J.; Bociek, R.G.; et al. Addition of rituximab to standard chemotherapy improves the survival of both the germinal center B-cell-like and non-germinal center B-cell-like subtypes of diffuse large B-cell lymphoma. J. Clin. Oncol. 2008, 26, 4587–4594. [Google Scholar] [CrossRef] [PubMed]
- Poeschel, V.; Held, G.; Ziepert, M.; Witzens-Harig, M.; Holte, H.; Thurner, L.; Borchmann, P.; Viardot, A.; Soekler, M.; Keller, U.; et al. Four versus six cycles of CHOP chemotherapy in combination with six applications of rituximab in patients with aggressive B-cell lymphoma with favourable prognosis (FLYER): A randomised, phase 3, non-inferiority trial. Lancet 2019, 394, 2271–2281. [Google Scholar] [CrossRef]
- Maurer, M.J.; Ghesquières, H.; Jais, J.P.; Witzig, T.E.; Haioun, C.; Thompson, C.A.; Delarue, R.; Micallef, I.N.; Peyrade, F.; Macon, W.R.; et al. Event-free survival at 24 months is a robust end point for disease-related outcome in diffuse large B-cell lymphoma treated with immunochemotherapy. J. Clin. Oncol. 2014, 32, 1066–1073. [Google Scholar] [CrossRef]
- Larouche, J.F.; Berger, F.; Chassagne-Clément, C.; Ffrench, M.; Callet-Bauchu, E.; Sebban, C.; Ghesquières, H.; Broussais-Guillaumot, F.; Salles, G.; Coiffier, B. Lymphoma recurrence 5 years or later following diffuse large B-cell lymphoma: Clinical characteristics and outcome. J. Clin. Oncol. 2010, 28, 2094–2100. [Google Scholar] [CrossRef]
- Stephens, D.M.; Li, H.; LeBlanc, M.L.; Puvvada, S.D.; Persky, D.; Friedberg, J.W.; Smith, S.M. Continued Risk of Relapse Independent of Treatment Modality in Limited-Stage Diffuse Large B-Cell Lymphoma: Final and Long-Term Analysis of Southwest Oncology Group Study S8736. J. Clin. Oncol. 2016, 34, 2997–3004. [Google Scholar] [CrossRef]
- Wang, Y.; Farooq, U.; Link, B.K.; Larson, M.C.; King, R.L.; Maurer, M.J.; Allmer, C.; Hefazi, M.; Thompson, C.A.; Micallef, I.N.; et al. Late Relapses in Patients With Diffuse Large B-Cell Lymphoma Treated with Immunochemotherapy. J. Clin. Oncol. 2019, 37, 1819–1827. [Google Scholar] [CrossRef] [PubMed]
- Bobillo, S.; Joffe, E.; Lavery, J.A.; Sermer, D.; Ghione, P.; Noy, A.; Caron, P.C.; Hamilton, A.M.; Hamlin, P.A.; Horwitz, S.M.; et al. Clinical Characteristics and Outcomes of Extranodal Stage I Diffuse Large B Cell Lymphoma in The Rituximab-Era. Blood 2020. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, A.A.; Eisen, M.B.; Davis, R.E.; Ma, C.; Lossos, I.S.; Rosenwald, A.; Boldrick, J.C.; Sabet, H.; Tran, T.; Yu, X.; et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000, 403, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Rosenwald, A.; Wright, G.; Chan, W.C.; Connors, J.M.; Campo, E.; Fisher, R.I.; Gascoyne, R.D.; Muller-Hermelink, H.K.; Smeland, E.B.; Giltnane, J.M.; et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N. Engl. J. Med. 2002, 346, 1937–1947. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.W.; Wright, G.W.; Williams, P.M.; Lih, C.J.; Walsh, W.; Jaffe, E.S.; Rosenwald, A.; Campo, E.; Chan, W.C.; Connors, J.M.; et al. Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood 2014, 123, 1214–1217. [Google Scholar] [CrossRef]
- Jais, J.P.; Molina, T.J.; Ruminy, P.; Gentien, D.; Reyes, C.; Scott, D.W.; Rimsza, L.M.; Wright, G.; Gascoyne, R.D.; Staudt, L.M.; et al. Reliable subtype classification of diffuse large B-cell lymphoma samples from GELA LNH2003 trials using the Lymph2Cx gene expression assay. Haematologica 2017, 102, e404–e406. [Google Scholar] [CrossRef]
- Davis, R.E.; Ngo, V.N.; Lenz, G.; Tolar, P.; Young, R.M.; Romesser, P.B.; Kohlhammer, H.; Lamy, L.; Zhao, H.; Yang, Y. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature 2010, 463, 88–92. [Google Scholar] [CrossRef]
- Sehn, L.H.; Gascoyne, R.D. Diffuse large B-cell lymphoma: Optimizing outcome in the context of clinical and biologic heterogeneity. Blood 2015, 125, 22–32. [Google Scholar] [CrossRef]
- Quintanilla-Martinez, L. The 2016 updated WHO classification of lymphoid neoplasias. Hematol. Oncol. 2017, 35 (Suppl. 1), 37–45. [Google Scholar] [CrossRef]
- Rosenthal, A.; Younes, A. High grade B-cell lymphoma with rearrangements of MYC and BCL2 and/or BCL6: Double hit and triple hit lymphomas and double expressing lymphoma. Blood Rev. 2017, 31, 37–42. [Google Scholar] [CrossRef]
- Sesques, P.; Johnson, N.A. Approach to the diagnosis and treatment of high-grade B-cell lymphomas with MYC and BCL2 and/or BCL6 rearrangements. Blood 2017, 129, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Sarkozy, C.; Traverse-Glehen, A.; Coiffier, B. Double-hit and double-protein-expression lymphomas: Aggressive and refractory lymphomas. Lancet Oncol. 2015, 16, e555–e567. [Google Scholar] [CrossRef]
- Ennishi, D.; Jiang, A.; Boyle, M.; Collinge, B.; Grande, B.M.; Ben-Neriah, S.; Rushton, C.; Tang, J.; Thomas, N.; Slack, G.W.; et al. Double-Hit Gene Expression Signature Defines a Distinct Subgroup of Germinal Center B-Cell-Like Diffuse Large B-Cell Lymphoma. J. Clin. Oncol. 2019, 37, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.; Wright, G.W.; Huang, D.W.; Johnson, C.A.; Phelan, J.D.; Wang, J.Q.; Roulland, S.; Kasbekar, M.; Young, R.M.; Shaffer, A.L.; et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2018, 378, 1396–1407. [Google Scholar] [CrossRef] [PubMed]
- Chapuy, B.; Stewart, C.; Dunford, A.J.; Kim, J.; Kamburov, A.; Redd, R.A.; Lawrence, M.S.; Roemer, M.G.M.; Li, A.J.; Ziepert, M.; et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med. 2018, 24, 679–690. [Google Scholar] [CrossRef]
- Wright, G.W.; Huang, D.W.; Phelan, J.D.; Coulibaly, Z.A.; Roulland, S.; Young, R.M.; Wang, J.Q.; Schmitz, R.; Morin, R.D.; Tang, J.; et al. A Probabilistic Classification Tool for Genetic Subtypes of Diffuse Large B Cell Lymphoma with Therapeutic Implications. Cancer Cell 2020, 37, 551–568.e514. [Google Scholar] [CrossRef]
- Lacy, S.E.; Barrans, S.L.; Beer, P.A.; Painter, D.; Smith, A.G.; Roman, E.; Cooke, S.L.; Ruiz, C.; Glover, P.; Van Hoppe, S.J.L.; et al. Targeted sequencing in DLBCL, molecular subtypes, and outcomes: A Haematological Malignancy Research Network report. Blood 2020, 135, 1759–1771. [Google Scholar] [CrossRef]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug resistance in cancer: An overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef]
- Scheijen, B. Molecular mechanisms contributing to glucocorticoid resistance in lymphoid malignancies. Cancer Drug Resist. 2019, 2, 647–664. [Google Scholar] [CrossRef]
- Maloney, D.G.; Smith, B.; Rose, A. Rituximab: Mechanism of action and resistance. Semin Oncol. 2002, 29, 2–9. [Google Scholar] [CrossRef]
- Bonavida, B. ‘Rituximab-induced inhibition of antiapoptotic cell survival pathways: Implications in chemo/immunoresistance, rituximab unresponsiveness, prognostic and novel therapeutic interventions’. Oncogene 2007, 26, 3629–3636. [Google Scholar] [CrossRef] [PubMed]
- Pescovitz, M.D. Rituximab, an anti-cd20 monoclonal antibody: History and mechanism of action. Am. J. Transpl. 2006, 6, 859–866. [Google Scholar] [CrossRef] [PubMed]
- Jazirehi, A.R.; Bonavida, B. Cellular and molecular signal transduction pathways modulated by rituximab (rituxan, anti-CD20 mAb) in non-Hodgkin’s lymphoma: Implications in chemosensitization and therapeutic intervention. Oncogene 2005, 24, 2121–2143. [Google Scholar] [CrossRef] [PubMed]
- Rushton, C.K.; Arthur, S.E.; Alcaide, M.; Cheung, M.; Jiang, A.; Coyle, K.M.; Cleary, K.L.S.; Thomas, N.; Hilton, L.K.; Michaud, N.; et al. Genetic and evolutionary patterns of treatment resistance in relapsed B-cell lymphoma. Blood Adv. 2020, 4, 2886–2898. [Google Scholar] [CrossRef] [PubMed]
- Tomita, A. Genetic and Epigenetic Modulation of CD20 Expression in B-Cell Malignancies: Molecular Mechanisms and Significance to Rituximab Resistance. J. Clin. Exp. Hematop. 2016, 56, 89–99. [Google Scholar] [CrossRef]
- Thomsen, E.A.; Rovsing, A.B.; Anderson, M.V.; Due, H.; Huang, J.; Luo, Y.; Dybkaer, K.; Mikkelsen, J.G. Identification of BLNK and BTK as mediators of rituximab-induced programmed cell death by CRISPR screens in GCB-subtype diffuse large B-cell lymphoma. Mol. Oncol. 2020, 14, 1978–1997. [Google Scholar] [CrossRef]
- Kuiper, R.P.; Waanders, E.; van der Velden, V.H.; van Reijmersdal, S.V.; Venkatachalam, R.; Scheijen, B.; Sonneveld, E.; van Dongen, J.J.; Veerman, A.J.; van Leeuwen, F.N.; et al. IKZF1 deletions predict relapse in uniformly treated pediatric precursor B-ALL. Leukemia 2010, 24, 1258–1264. [Google Scholar] [CrossRef]
- Mullighan, C.G.; Su, X.; Zhang, J.; Radtke, I.; Phillips, L.A.; Miller, C.B.; Ma, J.; Liu, W.; Cheng, C.; Schulman, B.A.; et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N. Engl. J. Med. 2009, 360, 470–480. [Google Scholar] [CrossRef]
- Landau, D.A.; Tausch, E.; Taylor-Weiner, A.N.; Stewart, C.; Reiter, J.G.; Bahlo, J.; Kluth, S.; Bozic, I.; Lawrence, M.; Bottcher, S.; et al. Mutations driving CLL and their evolution in progression and relapse. Nature 2015, 526, 525–530. [Google Scholar] [CrossRef]
- Rossi, D.; Cerri, M.; Deambrogi, C.; Sozzi, E.; Cresta, S.; Rasi, S.; De Paoli, L.; Spina, V.; Gattei, V.; Capello, D.; et al. The prognostic value of TP53 mutations in chronic lymphocytic leukemia is independent of Del17p13: Implications for overall survival and chemorefractoriness. Clin. Cancer Res. 2009, 15, 995–1004. [Google Scholar] [CrossRef]
- Rossi, D.; Khiabanian, H.; Spina, V.; Ciardullo, C.; Bruscaggin, A.; Fama, R.; Rasi, S.; Monti, S.; Deambrogi, C.; De Paoli, L.; et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood 2014, 123, 2139–2147. [Google Scholar] [CrossRef] [PubMed]
- Campo, E.; Cymbalista, F.; Ghia, P.; Jäger, U.; Pospisilova, S.; Rosenquist, R.; Schuh, A.; Stilgenbauer, S. TP53 aberrations in chronic lymphocytic leukemia: An overview of the clinical implications of improved diagnostics. Haematologica 2018, 103, 1956–1968. [Google Scholar] [CrossRef] [PubMed]
- Aitken, M.J.L.; Lee, H.J.; Post, S.M. Emerging treatment options for patients with p53-pathway-deficient CLL. Ther. Adv. Hematol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Melchardt, T.; Hufnagl, C.; Weinstock, D.M.; Kopp, N.; Neureiter, D.; Trankenschuh, W.; Hackl, H.; Weiss, L.; Rinnerthaler, G.; Hartmann, T.N.; et al. Clonal evolution in relapsed and refractory diffuse large B-cell lymphoma is characterized by high dynamics of subclones. Oncotarget 2016, 7, 51494–51502. [Google Scholar] [CrossRef]
- Juskevicius, D.; Lorber, T.; Gsponer, J.; Perrina, V.; Ruiz, C.; Stenner-Liewen, F.; Dirnhofer, S.; Tzankov, A. Distinct genetic evolution patterns of relapsing diffuse large B-cell lymphoma revealed by genome-wide copy number aberration and targeted sequencing analysis. Leukemia 2016, 30, 2385–2395. [Google Scholar] [CrossRef]
- Jiang, Y.; Redmond, D.; Nie, K.; Eng, K.W.; Clozel, T.; Martin, P.; Tan, L.H.; Melnick, A.M.; Tam, W.; Elemento, O. Deep sequencing reveals clonal evolution patterns and mutation events associated with relapse in B-cell lymphomas. Genome Biol. 2014, 15, 432. [Google Scholar] [CrossRef]
- Juskevicius, D.; Dirnhofer, S.; Tzankov, A. Genetic background and evolution of relapses in aggressive B-cell lymphomas. Haematologica 2017, 102, 1139–1149. [Google Scholar] [CrossRef]
- Isaev, K.; Ennishi, D.; Hilton, L.; Skinnider, B.; Mungall, K.L.; Mungall, A.J.; Bakhtiari, M.; Tremblay-LeMay, R.; Silva, A.; Ben-Neriah, S.; et al. Molecular Attributes Underlying Central Nervous System and Systemic Relapse in Diffuse Large B-cell Lymphoma. Haematologica 2020. [Google Scholar] [CrossRef]
- Waanders, E.; Gu, Z.; Dobson, S.M.; Antić, Ž.; Crawford, J.C.; Ma, X.; Edmonson, M.N.; Payne-Turner, D.; van der Vorst, M.; Jongmans, M.C.J.; et al. Mutational landscape and patterns of clonal evolution in relapsed pediatric acute lymphoblastic leukemia. Blood Cancer Discov. 2020, 1, 96–111. [Google Scholar] [CrossRef]
- Yu, S.L.; Zhang, H.; Ho, B.C.; Yu, C.H.; Chang, C.C.; Hsu, Y.C.; Ni, Y.L.; Lin, K.H.; Jou, S.T.; Lu, M.Y.; et al. FPGS relapse-specific mutations in relapsed childhood acute lymphoblastic leukemia. Sci. Rep. 2020, 10, 12074. [Google Scholar] [CrossRef]
- Morin, R.D.; Assouline, S.; Alcaide, M.; Mohajeri, A.; Johnston, R.L.; Chong, L.; Grewal, J.; Yu, S.; Fornika, D.; Bushell, K.; et al. Genetic Landscapes of Relapsed and Refractory Diffuse Large B-Cell Lymphomas. Clin. Cancer Res. 2016, 22, 2290–2300. [Google Scholar] [CrossRef] [PubMed]
- Greenawalt, D.M.; Liang, W.S.; Saif, S.; Johnson, J.; Todorov, P.; Dulak, A.; Enriquez, D.; Halperin, R.; Ahmed, A.; Saveliev, V.; et al. Comparative analysis of primary versus relapse/refractory DLBCL identifies shifts in mutation spectrum. Oncotarget 2017, 8, 99237–99244. [Google Scholar] [CrossRef] [PubMed]
- Nijland, M.; Seitz, A.; Terpstra, M.; van Imhoff, G.W.; Kluin, P.M.; van Meerten, T.; Atayar, C.; van Kempen, L.C.; Diepstra, A.; Kok, K.; et al. Mutational Evolution in Relapsed Diffuse Large B-Cell Lymphoma. Cancers (Basel) 2018, 10, 459. [Google Scholar] [CrossRef] [PubMed]
- Mareschal, S.; Dubois, S.; Viailly, P.J.; Bertrand, P.; Bohers, E.; Maingonnat, C.; Jais, J.P.; Tesson, B.; Ruminy, P.; Peyrouze, P.; et al. Whole exome sequencing of relapsed/refractory patients expands the repertoire of somatic mutations in diffuse large B-cell lymphoma. Genes Chromosomes Cancer 2016, 55, 251–267. [Google Scholar] [CrossRef]
- Park, H.Y.; Lee, S.B.; Yoo, H.Y.; Kim, S.J.; Kim, W.S.; Kim, J.I.; Ko, Y.H. Whole-exome and transcriptome sequencing of refractory diffuse large B-cell lymphoma. Oncotarget 2016, 7, 86433–86445. [Google Scholar] [CrossRef]
- Peroja, P.; Pedersen, M.; Mantere, T.; Norgaard, P.; Peltonen, J.; Haapasaari, K.M.; Bohm, J.; Jantunen, E.; Turpeenniemi-Hujanen, T.; Rapakko, K.; et al. Mutation of TP53, translocation analysis and immunohistochemical expression of MYC, BCL-2 and BCL-6 in patients with DLBCL treated with R-CHOP. Sci. Rep. 2018, 8, 14814. [Google Scholar] [CrossRef]
- Johnson, N.A.; Slack, G.W.; Savage, K.J.; Connors, J.M.; Ben-Neriah, S.; Rogic, S.; Scott, D.W.; Tan, K.L.; Steidl, C.; Sehn, L.H.; et al. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J. Clin. Oncol. 2012, 30, 3452–3459. [Google Scholar] [CrossRef]
- Green, T.M.; Young, K.H.; Visco, C.; Xu-Monette, Z.Y.; Orazi, A.; Go, R.S.; Nielsen, O.; Gadeberg, O.V.; Mourits-Andersen, T.; Frederiksen, M.; et al. Immunohistochemical double-hit score is a strong predictor of outcome in patients with diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J. Clin. Oncol. 2012, 30, 3460–3467. [Google Scholar] [CrossRef]
- Miura, K.; Takahashi, H.; Nakagawa, M.; Izu, A.; Sugitani, M.; Kurita, D.; Sakagami, M.; Ohtake, S.; Uchino, Y.; Hojo, A.; et al. Clinical significance of co-expression of MYC and BCL2 protein in aggressive B-cell lymphomas treated with a second line immunochemotherapy. Leuk Lymphoma 2016, 57, 1335–1341. [Google Scholar] [CrossRef]
- Herrera, A.F.; Mei, M.; Low, L.; Kim, H.T.; Griffin, G.K.; Song, J.Y.; Merryman, R.W.; Bedell, V.; Pak, C.; Sun, H.; et al. Relapsed or Refractory Double-Expressor and Double-Hit Lymphomas Have Inferior Progression-Free Survival After Autologous Stem-Cell Transplantation. J. Clin. Oncol. 2017, 35, 24–31. [Google Scholar] [CrossRef]
- Cuccuini, W.; Briere, J.; Mounier, N.; Voelker, H.U.; Rosenwald, A.; Sundstrom, C.; Cogliatti, S.; Hirchaud, E.; Ysebaert, L.; Bron, D.; et al. MYC+ diffuse large B-cell lymphoma is not salvaged by classical R-ICE or R-DHAP followed by BEAM plus autologous stem cell transplantation. Blood 2012, 119, 4619–4624. [Google Scholar] [CrossRef] [PubMed]
- Vaux, D.L.; Cory, S.; Adams, J.M. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature 1988, 335, 440–442. [Google Scholar] [CrossRef] [PubMed]
- Adams, C.M.; Clark-Garvey, S.; Porcu, P.; Eischen, C.M. Targeting the Bcl-2 Family in B Cell Lymphoma. Front. Oncol. 2018, 8, 636. [Google Scholar] [CrossRef] [PubMed]
- Visco, C.; Tzankov, A.; Xu-Monette, Z.Y.; Miranda, R.N.; Tai, Y.C.; Li, Y.; Liu, W.M.; d’Amore, E.S.; Li, Y.; Montes-Moreno, S.; et al. Patients with diffuse large B-cell lymphoma of germinal center origin with BCL2 translocations have poor outcome, irrespective of MYC status: A report from an International DLBCL rituximab-CHOP Consortium Program Study. Haematologica 2013, 98, 255–263. [Google Scholar] [CrossRef]
- Li, L.; Li, Y.; Que, X.; Gao, X.; Gao, Q.; Yu, M.; Ma, K.; Xi, Y.; Wang, T. Prognostic significances of overexpression MYC and/or BCL2 in R-CHOP-treated diffuse large B-cell lymphoma: A Systematic review and meta-analysis. Sci. Rep. 2018, 8, 6267. [Google Scholar] [CrossRef]
- Tsujimoto, Y.; Finger, L.R.; Yunis, J.; Nowell, P.C.; Croce, C.M. Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science 1984, 226, 1097–1099. [Google Scholar] [CrossRef]
- Barrans, S.L.; Evans, P.A.; O’Connor, S.J.; Kendall, S.J.; Owen, R.G.; Haynes, A.P.; Morgan, G.J.; Jack, A.S. The t(14;18) is associated with germinal center-derived diffuse large B-cell lymphoma and is a strong predictor of outcome. Clin. Cancer Res. 2003, 9, 2133–2139. [Google Scholar]
- Jost, P.J.; Ruland, J. Aberrant NF-kappaB signaling in lymphoma: Mechanisms, consequences, and therapeutic implications. Blood 2007, 109, 2700–2707. [Google Scholar] [CrossRef]
- García-Aranda, M.; Pérez-Ruiz, E.; Redondo, M. Bcl-2 Inhibition to Overcome Resistance to Chemo- and Immunotherapy. Int. J. Mol. Sci. 2018, 19, 3950. [Google Scholar] [CrossRef]
- Khan, N.; Kahl, B. Targeting BCL-2 in Hematologic Malignancies. Target Oncol. 2018, 13, 257–267. [Google Scholar] [CrossRef]
- Stolz, C.; Hess, G.; Hahnel, P.S.; Grabellus, F.; Hoffarth, S.; Schmid, K.W.; Schuler, M. Targeting Bcl-2 family proteins modulates the sensitivity of B-cell lymphoma to rituximab-induced apoptosis. Blood 2008, 112, 3312–3321. [Google Scholar] [CrossRef] [PubMed]
- Jazirehi, A.R.; Vega, M.I.; Bonavida, B. Development of rituximab-resistant lymphoma clones with altered cell signaling and cross-resistance to chemotherapy. Cancer Res. 2007, 67, 1270–1281. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, S.S.; Grau, M.; Mavis, C.; Hailfinger, S.; Wolf, A.; Madle, H.; Deeb, G.; Dörken, B.; Thome, M.; Lenz, P.; et al. MCL1 is deregulated in subgroups of diffuse large B-cell lymphoma. Leukemia 2013, 27, 1381–1390. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V. MYC on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef]
- Chisholm, K.M.; Bangs, C.D.; Bacchi, C.E.; Molina-Kirsch, H.; Cherry, A.; Natkunam, Y. Expression profiles of MYC protein and MYC gene rearrangement in lymphomas. Am. J. Surg. Pathol. 2015, 39, 294–303. [Google Scholar] [CrossRef]
- Karube, K.; Campo, E. MYC alterations in diffuse large B-cell lymphomas. Semin. Hematol. 2015, 52, 97–106. [Google Scholar] [CrossRef]
- Xu-Monette, Z.Y.; Deng, Q.; Manyam, G.C.; Tzankov, A.; Li, L.; Xia, Y.; Wang, X.X.; Zou, D.; Visco, C.; Dybkær, K.; et al. Clinical and Biologic Significance of MYC Genetic Mutations in De Novo Diffuse Large B-cell Lymphoma. Clin. Cancer Res. 2016, 22, 3593–3605. [Google Scholar] [CrossRef]
- Savage, K.J.; Johnson, N.A.; Ben-Neriah, S.; Connors, J.M.; Sehn, L.H.; Farinha, P.; Horsman, D.E.; Gascoyne, R.D. MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood 2009, 114, 3533–3537. [Google Scholar] [CrossRef]
- Epperla, N.; Maddocks, K.J.; Salhab, M.; Chavez, J.C.; Reddy, N.; Karmali, R.; Umyarova, E.; Bachanova, V.; Costa, C.; Glenn, M.; et al. C-MYC-positive relapsed and refractory, diffuse large B-cell lymphoma: Impact of additional “hits” and outcomes with subsequent therapy. Cancer 2017, 123, 4411–4418. [Google Scholar] [CrossRef]
- Hemann, M.T.; Bric, A.; Teruya-Feldstein, J.; Herbst, A.; Nilsson, J.A.; Cordon-Cardo, C.; Cleveland, J.L.; Tansey, W.P.; Lowe, S.W. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature 2005, 436, 807–811. [Google Scholar] [CrossRef]
- McMahon, S.B. MYC and the control of apoptosis. Cold Spring Harb. Perspect. Med. 2014, 4, a014407. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Yu, T.T.; Young, K.H. Cross-talk between Myc and p53 in B-cell lymphomas. Chronic Dis. Transl. Med. 2019, 5, 139–154. [Google Scholar] [CrossRef] [PubMed]
- Sewastianik, T.; Prochorec-Sobieszek, M.; Chapuy, B.; Juszczyński, P. MYC deregulation in lymphoid tumors: Molecular mechanisms, clinical consequences and therapeutic implications. Biochim. Biophys. Acta 2014, 1846, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Eischen, C.M.; Weber, J.D.; Roussel, M.F.; Sherr, C.J.; Cleveland, J.L. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999, 13, 2658–2669. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, A.L.; Yu, X.; He, Y.; Boldrick, J.; Chan, E.P.; Staudt, L.M. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity 2000, 13, 199–212. [Google Scholar] [CrossRef]
- Pasqualucci, L.; Bereshchenko, O.; Niu, H.; Klein, U.; Basso, K.; Guglielmino, R.; Cattoretti, G.; Dalla-Favera, R. Molecular pathogenesis of non-Hodgkin’s lymphoma: The role of Bcl-6. Leuk Lymphoma 2003, 44 (Suppl. 3), S5–S12. [Google Scholar] [CrossRef]
- Hatzi, K.; Melnick, A. Breaking bad in the germinal center: How deregulation of BCL6 contributes to lymphomagenesis. Trends Mol. Med. 2014, 20, 343–352. [Google Scholar] [CrossRef]
- Li, S.; Wang, Z.; Lin, L.; Wu, Z.; Yu, Q.; Gao, F.; Zhang, J.; Xu, Y. BCL6 Rearrangement Indicates Poor Prognosis in Diffuse Large B-cell Lymphoma Patients: A Meta-analysis of Cohort Studies. J. Cancer 2019, 10, 530–538. [Google Scholar] [CrossRef]
- Gebauer, N.; Bernard, V.; Gebauer, W.; Thorns, C.; Feller, A.C.; Merz, H. TP53 mutations are frequent events in double-hit B-cell lymphomas with MYC and BCL2 but not MYC and BCL6 translocations. Leuk Lymphoma 2015, 56, 179–185. [Google Scholar] [CrossRef]
- Phan, R.T.; Dalla-Favera, R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature 2004, 432, 635–639. [Google Scholar] [CrossRef]
- Margalit, O.; Amram, H.; Amariglio, N.; Simon, A.J.; Shaklai, S.; Granot, G.; Minsky, N.; Shimoni, A.; Harmelin, A.; Givol, D.; et al. BCL6 is regulated by p53 through a response element frequently disrupted in B-cell non-Hodgkin lymphoma. Blood 2006, 107, 1599–1607. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Lane, D.; Levine, A.J. Surfing the p53 network. Nature 2000, 408, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.J. P53 and The Immune Response: 40 Years of Exploration-A Plan for the Future. Int. J. Mol. Sci. 2020, 21, 541. [Google Scholar] [CrossRef] [PubMed]
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational landscape and significance across 12 major cancer types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.A.; Vousden, K.H. Mutant p53 in cancer: New functions and therapeutic opportunities. Cancer Cell 2014, 25, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Stiewe, T.; Haran, T.E. How mutations shape p53 interactions with the genome to promote tumorigenesis and drug resistance. Drug Resist. Updat. 2018, 38, 27–43. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Zhu, T.; Zhang, P.; Xiao, M.; Yi, S.; Yang, Y.; Li, Q.; Ling, S.; Wang, Y.; Gao, L.; et al. Mutations or copy number losses of CD58 and TP53 genes in diffuse large B cell lymphoma are independent unfavorable prognostic factors. Oncotarget 2016, 7, 83294–83307. [Google Scholar] [CrossRef]
- Zainuddin, N.; Berglund, M.; Wanders, A.; Ren, Z.P.; Amini, R.M.; Lindell, M.; Kanduri, M.; Roos, G.; Rosenquist, R.; Enblad, G. TP53 mutations predict for poor survival in de novo diffuse large B-cell lymphoma of germinal center subtype. Leuk. Res. 2009, 33, 60–66. [Google Scholar] [CrossRef]
- Xu-Monette, Z.Y.; Wu, L.; Visco, C.; Tai, Y.C.; Tzankov, A.; Liu, W.M.; Montes-Moreno, S.; Dybkaer, K.; Chiu, A.; Orazi, A.; et al. Mutational profile and prognostic significance of TP53 in diffuse large B-cell lymphoma patients treated with R-CHOP: Report from an International DLBCL Rituximab-CHOP Consortium Program Study. Blood 2012, 120, 3986–3996. [Google Scholar] [CrossRef]
- Stefancikova, L.; Moulis, M.; Fabian, P.; Vasova, I.; Zedek, F.; Ravcukova, B.; Muzik, J.; Kuglik, P.; Vranova, V.; Falkova, I.; et al. Prognostic impact of p53 aberrations for R-CHOP-treated patients with diffuse large B-cell lymphoma. Int. J. Oncol. 2011, 39, 1413–1420. [Google Scholar] [CrossRef]
- Karube, K.; Enjuanes, A.; Dlouhy, I.; Jares, P.; Martin-Garcia, D.; Nadeu, F.; Ordóñez, G.R.; Rovira, J.; Clot, G.; Royo, C.; et al. Integrating genomic alterations in diffuse large B-cell lymphoma identifies new relevant pathways and potential therapeutic targets. Leukemia 2018, 32, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Broseus, J.; Chen, G.; Hergalant, S.; Ramstein, G.; Mounier, N.; Gueant, J.L.; Feugier, P.; Gisselbrecht, C.; Thieblemont, C.; Houlgatte, R. Relapsed diffuse large B-cell lymphoma present different genomic profiles between early and late relapses. Oncotarget 2016, 7, 83987–84002. [Google Scholar] [CrossRef] [PubMed]
- Jardin, F.; Jais, J.P.; Molina, T.J.; Parmentier, F.; Picquenot, J.M.; Ruminy, P.; Tilly, H.; Bastard, C.; Salles, G.A.; Feugier, P.; et al. Diffuse large B-cell lymphomas with CDKN2A deletion have a distinct gene expression signature and a poor prognosis under R-CHOP treatment: A GELA study. Blood 2010, 116, 1092–1104. [Google Scholar] [CrossRef] [PubMed]
- Hientz, K.; Mohr, A.; Bhakta-Guha, D.; Efferth, T. The role of p53 in cancer drug resistance and targeted chemotherapy. Oncotarget 2017, 8, 8921–8946. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.; Tan, B.X.; Lane, D. How the Other Half Lives: What p53 Does When It Is Not Being a Transcription Factor. Int. J. Mol. Sci. 2019, 21, 13. [Google Scholar] [CrossRef]
- Blagih, J.; Buck, M.D.; Vousden, K.H. p53, cancer and the immune response. J. Cell Sci. 2020, 133, jcs237453. [Google Scholar] [CrossRef]
- Rawlings, J.S.; Rosler, K.M.; Harrison, D.A. The JAK/STAT signaling pathway. J. Cell Sci. 2004, 117, 1281–1283. [Google Scholar] [CrossRef]
- Villarino, A.V.; Kanno, Y.; O’Shea, J.J. Mechanisms and consequences of Jak-STAT signaling in the immune system. Nat. Immunol. 2017, 18, 374–384. [Google Scholar] [CrossRef]
- Morris, R.; Kershaw, N.J.; Babon, J.J. The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. 2018, 27, 1984–2009. [Google Scholar] [CrossRef]
- Turkson, J. STAT proteins as novel targets for cancer drug discovery. Expert Opin. Ther. Targets 2004, 8, 409–422. [Google Scholar] [CrossRef]
- Saint-Germain, E.; Mignacca, L.; Huot, G.; Acevedo, M.; Moineau-Vallée, K.; Calabrese, V.; Bourdeau, V.; Rowell, M.C.; Ilangumaran, S.; Lessard, F.; et al. Phosphorylation of SOCS1 Inhibits the SOCS1-p53 Tumor Suppressor Axis. Cancer Res. 2019, 79, 3306–3319. [Google Scholar] [CrossRef]
- Beaurivage, C.; Champagne, A.; Tobelaim, W.S.; Pomerleau, V.; Menendez, A.; Saucier, C. SOCS1 in cancer: An oncogene and a tumor suppressor. Cytokine 2016, 82, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Schif, B.; Lennerz, J.K.; Kohler, C.W.; Bentink, S.; Kreuz, M.; Melzner, I.; Ritz, O.; Trümper, L.; Loeffler, M.; Spang, R.; et al. SOCS1 mutation subtypes predict divergent outcomes in diffuse large B-Cell lymphoma (DLBCL) patients. Oncotarget 2013, 4, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Mellert, K.; Martin, M.; Lennerz, J.K.; Lüdeke, M.; Staiger, A.M.; Kreuz, M.; Löffler, M.; Schmitz, N.; Trümper, L.; Feller, A.C.; et al. The impact of SOCS1 mutations in diffuse large B-cell lymphoma. Br. J. Haematol. 2019, 187, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Lam, L.T.; Wright, G.; Davis, R.E.; Lenz, G.; Farinha, P.; Dang, L.; Chan, J.W.; Rosenwald, A.; Gascoyne, R.D.; Staudt, L.M. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-{kappa}B pathways in subtypes of diffuse large B-cell lymphoma. Blood 2008, 111, 3701–3713. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Zhu, F.; Zhang, M.; Li, Y.; Drennan, A.C.; Kimpara, S.; Rumball, I.; Selzer, C.; Cameron, H.; Kellicut, A.; et al. Gene regulation and suppression of type I interferon signaling by STAT3 in diffuse large B cell lymphoma. Proc. Natl. Acad. Sci. USA 2018, 115, E498–e505. [Google Scholar] [CrossRef] [PubMed]
- de Charette, M.; Houot, R. Hide or defend, the two strategies of lymphoma immune evasion: Potential implications for immunotherapy. Haematologica 2018, 103, 1256–1268. [Google Scholar] [CrossRef] [PubMed]
- Menter, T.; Tzankov, A. Mechanisms of Immune Evasion and Immune Modulation by Lymphoma Cells. Front. Oncol. 2018, 8, 54. [Google Scholar] [CrossRef] [PubMed]
- Garrido, F.; Aptsiauri, N.; Doorduijn, E.M.; Garcia Lora, A.M.; van Hall, T. The urgent need to recover MHC class I in cancers for effective immunotherapy. Curr. Opin. Immunol. 2016, 39, 44–51. [Google Scholar] [CrossRef]
- Williams, D.; Barber, B.; Flavell, R.; Allen, H. Role of beta 2-microglobulin in the intracellular transport and surface expression of murine class I histocompatibility molecules. J. Immunol. 1989, 142, 2796–2806. [Google Scholar]
- Ennishi, D.; Takata, K.; Béguelin, W.; Duns, G.; Mottok, A.; Farinha, P.; Bashashati, A.; Saberi, S.; Boyle, M.; Meissner, B.; et al. Molecular and Genetic Characterization of MHC Deficiency Identifies EZH2 as Therapeutic Target for Enhancing Immune Recognition. Cancer Discov. 2019, 9, 546–563. [Google Scholar] [CrossRef] [PubMed]
- Zaretsky, J.M.; Garcia-Diaz, A.; Shin, D.S.; Escuin-Ordinas, H.; Hugo, W.; Hu-Lieskovan, S.; Torrejon, D.Y.; Abril-Rodriguez, G.; Sandoval, S.; Barthly, L.; et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N. Engl. J. Med. 2016, 375, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Sade-Feldman, M.; Jiao, Y.J.; Chen, J.H.; Rooney, M.S.; Barzily-Rokni, M.; Eliane, J.P.; Bjorgaard, S.L.; Hammond, M.R.; Vitzthum, H.; Blackmon, S.M.; et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat. Commun. 2017, 8, 1136. [Google Scholar] [CrossRef] [PubMed]
- Middha, S.; Yaeger, R.; Shia, J.; Stadler, Z.K.; King, S.; Guercio, S.; Paroder, V.; Bates, D.D.; Rana, S.; Diaz Jr, L.A. Majority of B2M-mutant and-deficient colorectal carcinomas achieve clinical benefit from immune checkpoint inhibitor therapy and are microsatellite instability-high. JCO Precis. Oncol. 2019, 3, 1–14. [Google Scholar] [CrossRef]
- Kiyasu, J.; Miyoshi, H.; Hirata, A.; Arakawa, F.; Ichikawa, A.; Niino, D.; Sugita, Y.; Yufu, Y.; Choi, I.; Abe, Y.; et al. Expression of programmed cell death ligand 1 is associated with poor overall survival in patients with diffuse large B-cell lymphoma. Blood 2015, 126, 2193–2201. [Google Scholar] [CrossRef]
- Fang, X.; Xiu, B.; Yang, Z.; Qiu, W.; Zhang, L.; Zhang, S.; Wu, Y.; Zhu, X.; Chen, X.; Xie, S.; et al. The expression and clinical relevance of PD-1, PD-L1, and TP63 in patients with diffuse large B-cell lymphoma. Medicine (Baltimore) 2017, 96, e6398. [Google Scholar] [CrossRef]
- Godfrey, J.; Tumuluru, S.; Bao, R.; Leukam, M.; Venkataraman, G.; Phillip, J.; Fitzpatrick, C.; McElherne, J.; MacNabb, B.W.; Orlowski, R.; et al. PD-L1 gene alterations identify a subset of diffuse large B-cell lymphoma harboring a T-cell-inflamed phenotype. Blood 2019, 133, 2279–2290. [Google Scholar] [CrossRef]
- Challa-Malladi, M.; Lieu, Y.K.; Califano, O.; Holmes, A.B.; Bhagat, G.; Murty, V.V.; Dominguez-Sola, D.; Pasqualucci, L.; Dalla-Favera, R. Combined genetic inactivation of beta2-Microglobulin and CD58 reveals frequent escape from immune recognition in diffuse large B cell lymphoma. Cancer Cell 2011, 20, 728–740. [Google Scholar] [CrossRef]
- Sanchez-Madrid, F.; Krensky, A.M.; Ware, C.F.; Robbins, E.; Strominger, J.L.; Burakoff, S.J.; Springer, T.A. Three distinct antigens associated with human T-lymphocyte-mediated cytolysis: LFA-1, LFA-2, and LFA-3. Proc. Natl. Acad. Sci. USA 1982, 79, 7489–7493. [Google Scholar] [CrossRef]
- Krensky, A.M.; Sanchez-Madrid, F.; Robbins, E.; Nagy, J.A.; Springer, T.A.; Burakoff, S.J. The functional significance, distribution, and structure of LFA-1, LFA-2, and LFA-3: Cell surface antigens associated with CTL-target interactions. J. Immunol. 1983, 131, 611–616. [Google Scholar]
- Selvaraj, P.; Plunkett, M.L.; Dustin, M.; Sanders, M.E.; Shaw, S.; Springer, T.A. The T lymphocyte glycoprotein CD2 binds the cell surface ligand LFA-3. Nature 1987, 326, 400–403. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, Y.; Nishikori, M.; Arima, H.; Izumi, K.; Kitawaki, T.; Hishizawa, M.; Takaori-Kondo, A. EZH2 inhibitors restore epigenetically silenced CD58 expression in B-cell lymphomas. Mol. Immunol. 2020, 119, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Medeiros, L.J.; Li, Y.; Li, J.; Young, K.H. Genetic alterations and their clinical implications in DLBCL. Nat. Rev. Clin. Oncol. 2019, 16, 634–652. [Google Scholar] [CrossRef] [PubMed]
- Klemm, S.L.; Shipony, Z.; Greenleaf, W.J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet 2019, 20, 207–220. [Google Scholar] [CrossRef]
- Hamamoto, R.; Saloura, V.; Nakamura, Y. Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat. Rev. Cancer 2015, 15, 110–124. [Google Scholar] [CrossRef]
- Mullighan, C.G.; Zhang, J.; Kasper, L.H.; Lerach, S.; Payne-Turner, D.; Phillips, L.A.; Heatley, S.L.; Holmfeldt, L.; Collins-Underwood, J.R.; Ma, J.; et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature 2011, 471, 235–239. [Google Scholar] [CrossRef]
- Froimchuk, E.; Jang, Y.; Ge, K. Histone H3 lysine 4 methyltransferase KMT2D. Gene 2017, 627, 337–342. [Google Scholar] [CrossRef]
- Lee, J.; Kim, D.H.; Lee, S.; Yang, Q.H.; Lee, D.K.; Lee, S.K.; Roeder, R.G.; Lee, J.W. A tumor suppressive coactivator complex of p53 containing ASC-2 and histone H3-lysine-4 methyltransferase MLL3 or its paralogue MLL4. Proc. Natl. Acad. Sci. USA 2009, 106, 8513–8518. [Google Scholar] [CrossRef]
- Béguelin, W.; Teater, M.; Meydan, C.; Hoehn, K.B.; Phillip, J.M.; Soshnev, A.A.; Venturutti, L.; Rivas, M.A.; Calvo-Fernández, M.T.; Gutierrez, J.; et al. Mutant EZH2 Induces a Pre-malignant Lymphoma Niche by Reprogramming the Immune Response. Cancer Cell 2020, 37, 655–673.e611. [Google Scholar] [CrossRef]
- Pasqualucci, L.; Dominguez-Sola, D.; Chiarenza, A.; Fabbri, G.; Grunn, A.; Trifonov, V.; Kasper, L.H.; Lerach, S.; Tang, H.; Ma, J.; et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature 2011, 471, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Barta, S.K. Diffuse large B-cell lymphoma: 2019 update on diagnosis, risk stratification, and treatment. Am. J. Hematol. 2019, 94, 604–616. [Google Scholar] [CrossRef] [PubMed]
- Mondello, P.; Nowakowski, G.S. Treatment of Aggressive B Cell Lymphomas: Updates in 2019. Curr. Hematol. Malig. Rep. 2020, 15, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Kater, A.P.; Seymour, J.F.; Hillmen, P.; Eichhorst, B.; Langerak, A.W.; Owen, C.; Verdugo, M.; Wu, J.; Punnoose, E.A.; Jiang, Y.; et al. Fixed Duration of Venetoclax-Rituximab in Relapsed/Refractory Chronic Lymphocytic Leukemia Eradicates Minimal Residual Disease and Prolongs Survival: Post-Treatment Follow-Up of the MURANO Phase III Study. J. Clin. Oncol. 2019, 37, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Guerra, V.A.; DiNardo, C.; Konopleva, M. Venetoclax-based therapies for acute myeloid leukemia. Best Pract. Res. Clin. Haematol. 2019, 32, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.W.; Davids, M.S.; Pagel, J.M.; Kahl, B.S.; Puvvada, S.D.; Gerecitano, J.F.; Kipps, T.J.; Anderson, M.A.; Brown, J.R.; Gressick, L.; et al. Targeting BCL2 with Venetoclax in Relapsed Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2016, 374, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Zelenetz, A.D.; Salles, G.; Mason, K.D.; Casulo, C.; Le Gouill, S.; Sehn, L.H.; Tilly, H.; Cartron, G.; Chamuleau, M.E.D.; Goy, A.; et al. Venetoclax plus R- or G-CHOP in non-Hodgkin lymphoma: Results from the CAVALLI phase 1b trial. Blood 2019, 133, 1964–1976. [Google Scholar] [CrossRef]
- Morschhauser, F.; Feugier, P.; Flinn, I.W.; Gasiorowski, R.; Greil, R.; Illes, A.; Johnson, N.A.; Larouche, J.F.; Lugtenburg, P.J.; Patti, C.; et al. A phase II study of venetoclax plus R-CHOP as first-line treatment for patients with diffuse large B-cell lymphoma. Blood 2020. [Google Scholar] [CrossRef]
- Bose, P.; Gandhi, V.; Konopleva, M. Pathways and mechanisms of venetoclax resistance. Leuk Lymphoma 2017, 58, 1–17. [Google Scholar] [CrossRef]
- Liu, Y.; Mondello, P.; Erazo, T.; Tannan, N.B.; Asgari, Z.; de Stanchina, E.; Nanjangud, G.; Seshan, V.E.; Wang, S.; Wendel, H.G.; et al. NOXA genetic amplification or pharmacologic induction primes lymphoma cells to BCL2 inhibitor-induced cell death. Proc. Natl. Acad. Sci. USA 2018, 115, 12034–12039. [Google Scholar] [CrossRef]
- Li, L.; Pongtornpipat, P.; Tiutan, T.; Kendrick, S.L.; Park, S.; Persky, D.O.; Rimsza, L.M.; Puvvada, S.D.; Schatz, J.H. Synergistic induction of apoptosis in high-risk DLBCL by BCL2 inhibition with ABT-199 combined with pharmacologic loss of MCL1. Leukemia 2015, 29, 1702–1712. [Google Scholar] [CrossRef]
- Choudhary, G.S.; Al-Harbi, S.; Mazumder, S.; Hill, B.T.; Smith, M.R.; Bodo, J.; Hsi, E.D.; Almasan, A. MCL-1 and BCL-xL-dependent resistance to the BCL-2 inhibitor ABT-199 can be overcome by preventing PI3K/AKT/mTOR activation in lymphoid malignancies. Cell Death Dis. 2015, 6, e1593. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Atoyan, R.; Borek, M.A.; Dellarocca, S.; Rhyasen, G.; Fattaey, A.; Tuck, D.P. The combination of venetoclax and CUDC-907 exhibits synergistic activity in venetoclax-refractory DLBCL. Blood 2016, 128, 4184. [Google Scholar] [CrossRef]
- Mondello, P.; Derenzini, E.; Asgari, Z.; Philip, J.; Brea, E.J.; Seshan, V.; Hendrickson, R.C.; de Stanchina, E.; Scheinberg, D.A.; Younes, A. Dual inhibition of histone deacetylases and phosphoinositide 3-kinase enhances therapeutic activity against B cell lymphoma. Oncotarget 2017, 8, 14017–14028. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, W.; Gupta, S.K.; Han, W.; Kundson, R.A.; Nelson, S.; Knutson, D.; Greipp, P.T.; Elsawa, S.F.; Sotomayor, E.M.; Gupta, M. Targeting MYC activity in double-hit lymphoma with MYC and BCL2 and/or BCL6 rearrangements with epigenetic bromodomain inhibitors. J. Hematol. Oncol. 2019, 12, 73. [Google Scholar] [CrossRef]
- Mottok, A.; Gascoyne, R.D. Bromodomain inhibition in diffuse large B-cell lymphoma—Giving MYC a brake. Clin. Cancer Res. 2015, 21, 4–6. [Google Scholar] [CrossRef]
- Delmore, J.E.; Issa, G.C.; Lemieux, M.E.; Rahl, P.B.; Shi, J.; Jacobs, H.M.; Kastritis, E.; Gilpatrick, T.; Paranal, R.M.; Qi, J. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011, 146, 904–917. [Google Scholar] [CrossRef]
- Kumar, A.; Marqués, M.; Carrera, A.C. Phosphoinositide 3-kinase activation in late G1 is required for c-Myc stabilization and S phase entry. Mol. Cell Biol. 2006, 26, 9116–9125. [Google Scholar] [CrossRef]
- Fjorden, K.; Ekberg, S.; Kuric, N.; Ekstroem Smedby, K.; Lagerlöf, I.; Larsen, T.S.; Jørgensen, J.M.; Brown, P.d.N.; Jerkeman, M. Idelalisib in Patients with Relapsed/Refractory Diffuse Large B-Cell Lymphoma: First Results from the Nordic Lymphoma Group NLG-LBC-07 (ILIAD) Phase II Trial. Blood 2020, 136, 33. [Google Scholar] [CrossRef]
- Oki, Y.; Kelly, K.R.; Flinn, I.; Patel, M.R.; Gharavi, R.; Ma, A.; Parker, J.; Hafeez, A.; Tuck, D.; Younes, A. CUDC-907 in relapsed/refractory diffuse large B-cell lymphoma, including patients with MYC-alterations: Results from an expanded phase I trial. Haematologica 2017, 102, 1923–1930. [Google Scholar] [CrossRef]
- Jung, H.-S.; Kim, N.H.; Wang, J.; Son, M.K.; Kim, B.-K.; Jeon, B.; Choi, Y.; Myung, J.; Kim, D.-H. Combination of BR101801 and venetoclax demonstrates synergistic activity in DLBCL cell lines harboring double hit and double expressor alterations. Blood 2017, 130, 4114. [Google Scholar]
- Bykov, V.J.N.; Eriksson, S.E.; Bianchi, J.; Wiman, K.G. Targeting mutant p53 for efficient cancer therapy. Nat. Rev. Cancer 2018, 18, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.J. Targeting therapies for the p53 protein in cancer treatments. Annu. Rev. Cancer Biol. 2019, 3, 21–34. [Google Scholar] [CrossRef]
- Duffy, M.J.; Synnott, N.C.; Crown, J. Mutant p53 as a target for cancer treatment. Eur. J. Cancer 2017, 83, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tavana, O.; Gu, W. p53 modifications: Exquisite decorations of the powerful guardian. J. Mol. Cell Biol. 2019, 11, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Drakos, E.; Singh, R.R.; Rassidakis, G.Z.; Schlette, E.; Li, J.; Claret, F.X.; Ford, R.J., Jr.; Vega, F.; Medeiros, L.J. Activation of the p53 pathway by the MDM2 inhibitor nutlin-3a overcomes BCL2 overexpression in a preclinical model of diffuse large B-cell lymphoma associated with t(14;18)(q32;q21). Leukemia 2011, 25, 856–867. [Google Scholar] [CrossRef] [PubMed]
- Tisato, V.; Voltan, R.; Gonelli, A.; Secchiero, P.; Zauli, G. MDM2/X inhibitors under clinical evaluation: Perspectives for the management of hematological malignancies and pediatric cancer. J. Hematol. Oncol. 2017, 10, 133. [Google Scholar] [CrossRef]
- Ng, S.Y.; Yoshida, N.; Christie, A.L.; Ghandi, M.; Dharia, N.V.; Dempster, J.; Murakami, M.; Shigemori, K.; Morrow, S.N.; Van Scoyk, A.; et al. Targetable vulnerabilities in T- and NK-cell lymphomas identified through preclinical models. Nat. Commun. 2018, 9, 2024. [Google Scholar] [CrossRef]
- Cheok, C.F.; Lane, D.P. Exploiting the p53 Pathway for Therapy. Cold Spring Harb. Perspect. Med. 2017, 7. [Google Scholar] [CrossRef]
- Gadina, M.; Johnson, C.; Schwartz, D.; Bonelli, M.; Hasni, S.; Kanno, Y.; Changelian, P.; Laurence, A.; O’Shea, J.J. Translational and clinical advances in JAK-STAT biology: The present and future of jakinibs. J. Leukoc. Biol. 2018, 104, 499–514. [Google Scholar] [CrossRef]
- Drennan, A.C.; Rui, L. HiJAKing the epigenome in leukemia and lymphoma. Leuk Lymphoma 2017, 58, 2540–2547. [Google Scholar] [CrossRef]
- Phillips, T.J.; Forero-Torres, A.; Sher, T.; Diefenbach, C.S.; Johnston, P.; Talpaz, M.; Pulini, J.; Zhou, L.; Scherle, P.; Chen, X.; et al. Phase 1 study of the PI3Kδ inhibitor INCB040093 ± JAK1 inhibitor itacitinib in relapsed/refractory B-cell lymphoma. Blood 2018, 132, 293–306. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, J.J.; Schwartz, D.M.; Villarino, A.V.; Gadina, M.; McInnes, I.B.; Laurence, A. The JAK-STAT pathway: Impact on human disease and therapeutic intervention. Annu. Rev. Med. 2015, 66, 311–328. [Google Scholar] [CrossRef] [PubMed]
- Heppler, L.N.; Frank, D.A. Targeting Oncogenic Transcription Factors: Therapeutic Implications of Endogenous STAT Inhibitors. Trends Cancer 2017, 3, 816–827. [Google Scholar] [CrossRef] [PubMed]
- Del Campo, A.; Carretero, J.; Muñoz, J.; Zinchenko, S.; Ruiz-Cabello, F.; Gonzalez-Aseguinolaza, G.; Garrido, F.; Aptsiauri, N. Adenovirus expressing β2-microglobulin recovers HLA class I expression and antitumor immunity by increasing T-cell recognition. Cancer Gene Ther. 2014, 21, 317–332. [Google Scholar] [CrossRef]
- Neelapu, S.S.; Locke, F.L.; Bartlett, N.L.; Lekakis, L.J.; Miklos, D.B.; Jacobson, C.A.; Braunschweig, I.; Oluwole, O.O.; Siddiqi, T.; Lin, Y.; et al. Axicabtagene Ciloleucel CAR T-Cell Therapy in Refractory Large B-Cell Lymphoma. N. Engl. J. Med. 2017, 377, 2531–2544. [Google Scholar] [CrossRef]
- Schuster, S.J.; Bishop, M.R.; Tam, C.S.; Waller, E.K.; Borchmann, P.; McGuirk, J.P.; Jäger, U.; Jaglowski, S.; Andreadis, C.; Westin, J.R.; et al. Tisagenlecleucel in Adult Relapsed or Refractory Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2019, 380, 45–56. [Google Scholar] [CrossRef]
- Gonzalez-Rodriguez, A.P.; Villa-Álvarez, M.; Sordo-Bahamonde, C.; Lorenzo-Herrero, S.; Gonzalez, S. NK Cells in the Treatment of Hematological Malignancies. J. Clin. Med. 2019, 8, 1557. [Google Scholar] [CrossRef]
- Ansell, S.M.; Minnema, M.C.; Johnson, P.; Timmerman, J.M.; Armand, P.; Shipp, M.A.; Rodig, S.J.; Ligon, A.H.; Roemer, M.G.M.; Reddy, N.; et al. Nivolumab for Relapsed/Refractory Diffuse Large B-Cell Lymphoma in Patients Ineligible for or Having Failed Autologous Transplantation: A Single-Arm, Phase II Study. J. Clin. Oncol. 2019, 37, 481–489. [Google Scholar] [CrossRef]
- Herrera, A.F.; Goy, A.; Mehta, A.; Ramchandren, R.; Pagel, J.M.; Svoboda, J.; Guan, S.; Hill, J.S.; Kwei, K.; Liu, E.A.; et al. Safety and activity of ibrutinib in combination with durvalumab in patients with relapsed or refractory follicular lymphoma or diffuse large B-cell lymphoma. Am. J. Hematol. 2020, 95, 18–27. [Google Scholar] [CrossRef]
- Ribeiro, M.L.; Reyes-Garau, D.; Armengol, M.; Fernández-Serrano, M.; Roué, G. Recent advances in the targeting of epigenetic regulators in B-cell non-Hodgkin lymphoma. Front. Genet. 2019, 10, 986. [Google Scholar] [CrossRef]
- Duan, H.; Heckman, C.A.; Boxer, L.M. Histone deacetylase inhibitors down-regulate bcl-2 expression and induce apoptosis in t(14;18) lymphomas. Mol. Cell Biol. 2005, 25, 1608–1619. [Google Scholar] [CrossRef] [PubMed]
- Kurland, J.F.; Tansey, W.P. Myc-mediated transcriptional repression by recruitment of histone deacetylase. Cancer Res. 2008, 68, 3624–3629. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, X.; Fiskus, W.; Lin, J.; Lwin, T.; Rao, R.; Zhang, Y.; Chan, J.C.; Fu, K.; Marquez, V.E.; et al. Coordinated silencing of MYC-mediated miR-29 by HDAC3 and EZH2 as a therapeutic target of histone modification in aggressive B-Cell lymphomas. Cancer Cell 2012, 22, 506–523. [Google Scholar] [CrossRef] [PubMed]
- Bereshchenko, O.R.; Gu, W.; Dalla-Favera, R. Acetylation inactivates the transcriptional repressor BCL6. Nat. Genet. 2002, 32, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Crump, M.; Coiffier, B.; Jacobsen, E.D.; Sun, L.; Ricker, J.L.; Xie, H.; Frankel, S.R.; Randolph, S.S.; Cheson, B.D. Phase II trial of oral vorinostat (suberoylanilide hydroxamic acid) in relapsed diffuse large-B-cell lymphoma. Ann. Oncol. 2008, 19, 964–969. [Google Scholar] [CrossRef] [PubMed]
- Barnes, J.A.; Redd, R.; Fisher, D.C.; Hochberg, E.P.; Takvorian, T.; Neuberg, D.; Jacobsen, E.; Abramson, J.S. Panobinostat in combination with rituximab in heavily pretreated diffuse large B-cell lymphoma: Results of a phase II study. Hematol. Oncol. 2018, 36, 633–637. [Google Scholar] [CrossRef] [PubMed]
- Ribrag, V.; Kim, W.S.; Bouabdallah, R.; Lim, S.T.; Coiffier, B.; Illes, A.; Lemieux, B.; Dyer, M.J.S.; Offner, F.; Felloussi, Z.; et al. Safety and efficacy of abexinostat, a pan-histone deacetylase inhibitor, in non-Hodgkin lymphoma and chronic lymphocytic leukemia: Results of a phase II study. Haematologica 2017, 102, 903–909. [Google Scholar] [CrossRef] [PubMed]
- Amengual, J.E.; Clark-Garvey, S.; Kalac, M.; Scotto, L.; Marchi, E.; Neylon, E.; Johannet, P.; Wei, Y.; Zain, J.; O’Connor, O.A. Sirtuin and pan-class I/II deacetylase (DAC) inhibition is synergistic in preclinical models and clinical studies of lymphoma. Blood 2013, 122, 2104–2113. [Google Scholar] [CrossRef]
- Dai, C.; Gu, W. p53 post-translational modification: Deregulated in tumorigenesis. Trends Mol. Med. 2010, 16, 528–536. [Google Scholar] [CrossRef]
- Lue, J.K.; Amengual, J.E. Emerging EZH2 inhibitors and their application in lymphoma. Current Hematol. Malig. Rep. 2018, 13, 369–382. [Google Scholar] [CrossRef]
- McCabe, M.T.; Ott, H.M.; Ganji, G.; Korenchuk, S.; Thompson, C.; Van Aller, G.S.; Liu, Y.; Graves, A.P.; Della Pietra, A., 3rd; Diaz, E.; et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 2012, 492, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Béguelin, W.; Popovic, R.; Teater, M.; Jiang, Y.; Bunting, K.L.; Rosen, M.; Shen, H.; Yang, S.N.; Wang, L.; Ezponda, T.; et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 2013, 23, 677–692. [Google Scholar] [CrossRef] [PubMed]
- Plch, J.; Hrabeta, J.; Eckschlager, T. KDM5 demethylases and their role in cancer cell chemoresistance. Int. J. Cancer 2019, 144, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Heward, J.; Koniali, L.; D’Avola, A.; Close, K.; Yeomans, A.; Johnson, P.; Okosun, J.; Neve, R.; Gribben, J.; Oppermann, U. S844 KDM5 inhibition offers a novel therapeutic strategy for the treatment of kmt2d mutant lymphomas. HemaSphere 2019, 3, 376–377. [Google Scholar] [CrossRef]
| Study | Cohort Description | Cohort Size | Method | Genes Presenting R/R Enriched Variants in Paired Diagnosis-Relapse Analyses 1 | Genes Presenting R/R Enriched Variants in Comparison with Independent Primary Cohorts 2 |
|---|---|---|---|---|---|
| Jiang et al. 2014 [46] | Paired D-R samples | N = 7 (4/7 tLY) | WES | BCL2, EP300, B2M, CD58 | |
| Morin et al. 2016 [51] | Paired D-R/R samples | N = 12 (9/12 tLY) | Targeted panel | STAT6, EZH2, FOXO1, SOCS1, KMT2D, CD79B, NFKBIE | |
| R/R samples (taken after at least one cycle of immuno-chemotherapy) | N = 25 | WES Targeted panel | R/R samples compared with independent primary cohort: KMT2C, MPEG1, NFKBIZ, CCND3, STAT6, TP53, MYC, FOXO1 | ||
| Juskevicius et al. 2016 [45] | Paired D-R samples (relapse following complete remission) | N = 20 | Targeted panel | KMT2D, MEF2B, TET2, PRDM1, PTEN, EBF1 | |
| Non-relapsing samples (taken at diagnosis ≥4 years relapse-free) | N = 20 | Targeted panel | Diagnosis samples of relapsed patients compared with non-relapsing samples: KMT2D, BCL2, PTEN, PRDM1, MCL1, CARD11 | ||
| Melchardt et al. 2016 [44] | Paired D-R/R samples | N = 24 | Targeted panel | TP53, RB1, EZH2 | Diagnosis samples of R/R patients compared with independent primary cohort: NOTCH1, SMARCA4, PIM1, KMT2D R/R samples compared with independent primary cohort: TP53, BCL2, MYC, RB1, ATM, EZH2 |
| Park et al. 2016 [55] | Diagnosis samples of responsive (CR maintained > 1 year interval) vs. refractory patients (<1 year interval) | N = 7 responsive N = 6 refractory | WES | TP53, MYD88, B2M, PRDM15, FNBP4, AHR, CEP128, BRE, SORCS3, WDFY3, CXXC4 | |
| Mareschal et al. 2016 [54] | Diagnosis samples of R/R patients (≤1 year interval) | N = 14 | WES | ABC: MYD88, TBL1XR1, IRF4, CD58, PCDH17, HIST1H1B, HIST1H1C, HIST1H1D GCB: BCL2, DUSP2, NFKBIA, BTG2, MEF2B | |
| Greenawalt et al. 2017 [52] | Paired D-R/R samples | N = 8 | WES | CREBBP, BCL2 | |
| R/R samples (after 1–8 cycles of R-CHOP) | N = 47 | R/R samples compared with independent primary cohort: CREBBP, BCL2, TP53, B2M, MYC, BTK | |||
| Nijland et al. 2018 [53] | Paired D-R/R samples (patients that received 6–8 cycles of R-CHOP) | N = 6 | WES | SOCS1, PIM1, MYC, BCL2, BIRC3, BTG2, IRF4, SGK1, B2M, CALR, HLA-DR, HLA-B | Diagnosis and relapsed samples of R/R cohort compared with independent primary cohort: SOCS1, PIM1, MYC, HLA-DR, HLA-B |
| Rushton et al. 2020 [34] | Paired D-R/R samples (tissue biopsies/ctDNA) | N = 57 | Targeted panel | MS4A1, KMT2D, CD79B, TBL1XR1, ZFP36L1, CARD11, BTG2, MYC, SOCS1, PIM1, TNFAIP3, MYD88, HIST1H1E, NFKBIE, TNFRSF14, BCL2, IRF4, SGK1, GNA13, B2M, FBXO11, TP53, CD58, EP300 | |
| R/R ctDNA | N = 135 | R/R samples compared with independent primary cohort: KMT2D, TP53, CREBBP, FOXO1, NFKBIE, MS4A1 | |||
| Isaev et al. 2020 [48] | Paired D-CNS relapse samples | N = 5 | WES | PIM1, ETV6 | |
| Diagnosis samples of systemic and CNS relapsed patients (<1 year interval) vs. non-relapsing patients (≥5 years relapse free) | N = 62 systemic relapse N = 72 CNS relapse N = 89 Non-relapsing | Targeted panel WES | Diagnosis samples of CNS relapse compared with non-relapsing samples: MYD88, CD79B, PIM1 Diagnosis samples of refractory disease or systemic relapse compared with non-relapsing samples: TP53, MYD88, BCL2, HIST1H1E, HIST1H1C, FOXO1, BTG1, CIITA, CD58, ZFP36L1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berendsen, M.R.; Stevens, W.B.C.; van den Brand, M.; van Krieken, J.H.; Scheijen, B. Molecular Genetics of Relapsed Diffuse Large B-Cell Lymphoma: Insight into Mechanisms of Therapy Resistance. Cancers 2020, 12, 3553. https://doi.org/10.3390/cancers12123553
Berendsen MR, Stevens WBC, van den Brand M, van Krieken JH, Scheijen B. Molecular Genetics of Relapsed Diffuse Large B-Cell Lymphoma: Insight into Mechanisms of Therapy Resistance. Cancers. 2020; 12(12):3553. https://doi.org/10.3390/cancers12123553
Chicago/Turabian StyleBerendsen, Madeleine R., Wendy B. C. Stevens, Michiel van den Brand, J. Han van Krieken, and Blanca Scheijen. 2020. "Molecular Genetics of Relapsed Diffuse Large B-Cell Lymphoma: Insight into Mechanisms of Therapy Resistance" Cancers 12, no. 12: 3553. https://doi.org/10.3390/cancers12123553
APA StyleBerendsen, M. R., Stevens, W. B. C., van den Brand, M., van Krieken, J. H., & Scheijen, B. (2020). Molecular Genetics of Relapsed Diffuse Large B-Cell Lymphoma: Insight into Mechanisms of Therapy Resistance. Cancers, 12(12), 3553. https://doi.org/10.3390/cancers12123553






