The CHEK2 Variant C.349A>G Is Associated with Prostate Cancer Risk and Carriers Share a Common Ancestor
Simple Summary
Abstract
1. Introduction
2. Results
2.1. Frequency of the CHEK2 Variant c.349A>G
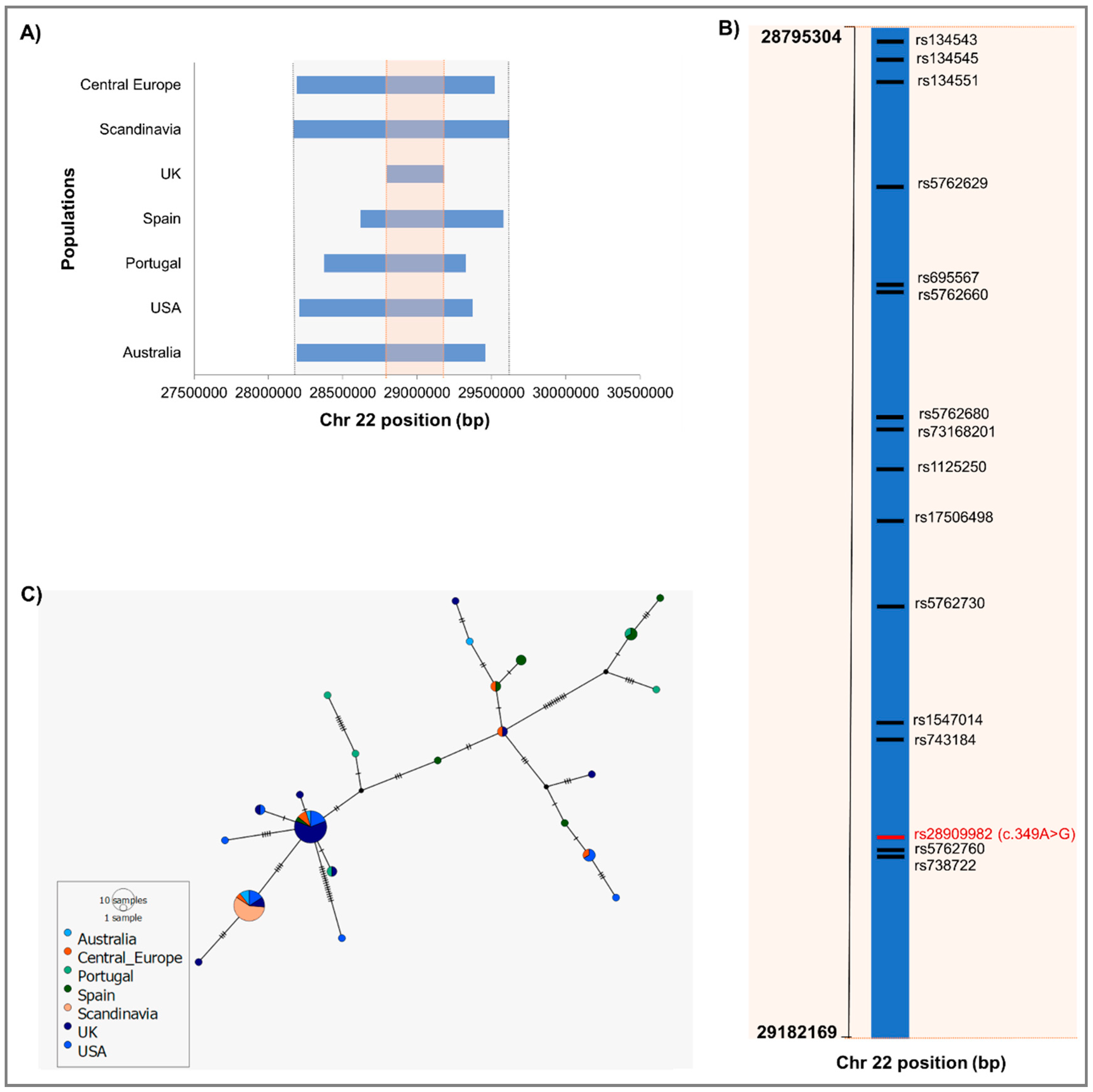
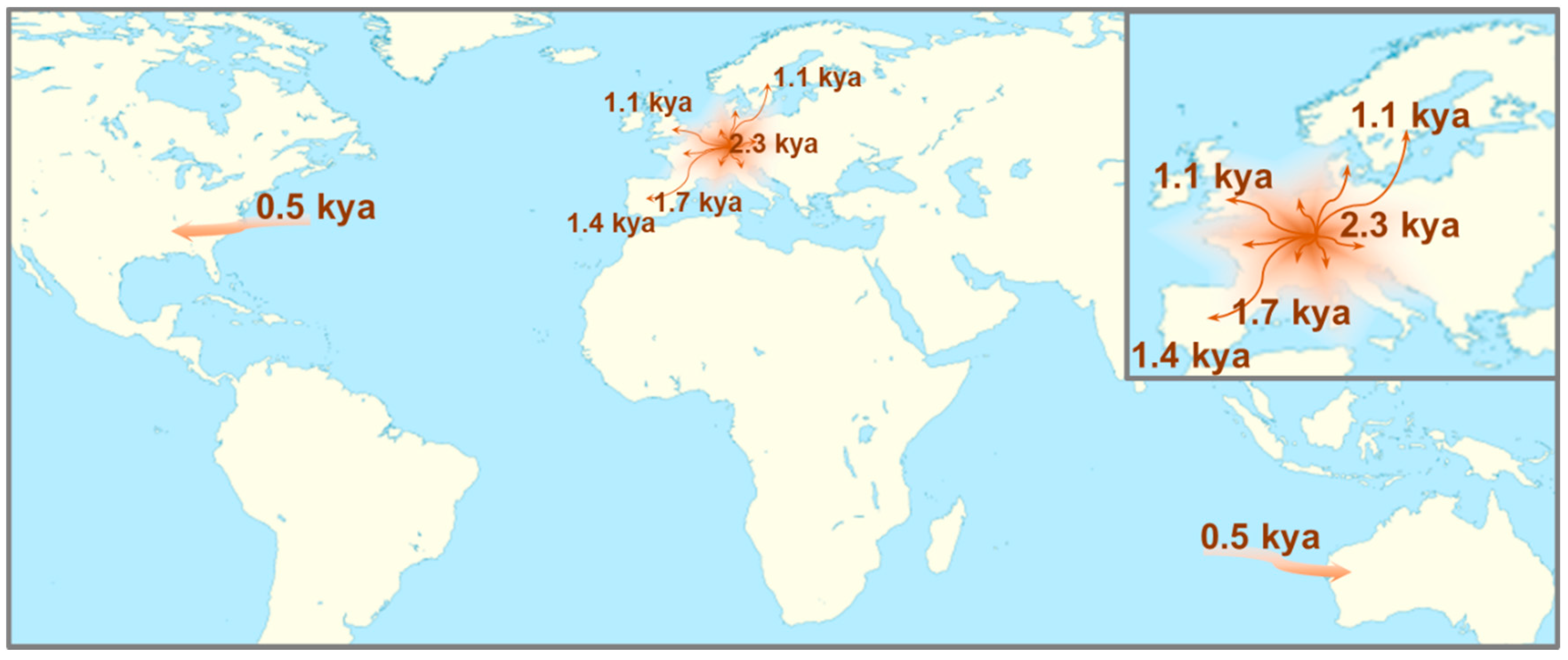
2.2. Identification of IBD Haplotype and Phylogeographic Analysis
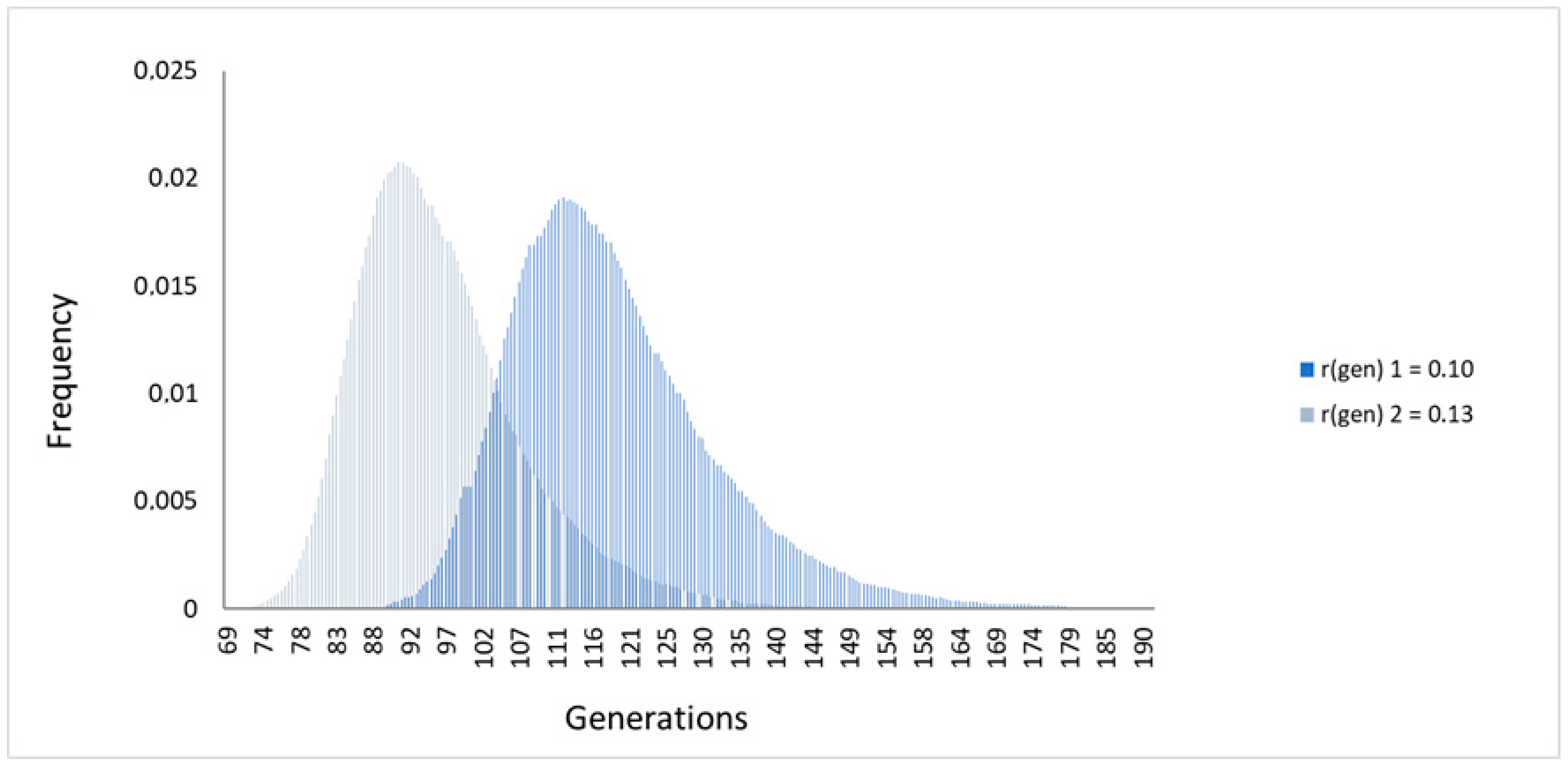
2.3. Age Estimation of the CHEK2 Variant c.349A>G
2.4. Haplotype Analysis Using Microsatellites
3. Discussion
4. Materials and Methods
4.1. Portuguese Early-Onset/Familial PrCa Sample Collection
4.2. Genotyping of the CHEK2 Variant c.349A>G
4.3. Statistical Analysis
4.4. Practical Sample Collection
4.5. OncoArray Genotyping and Quality Control
4.6. Identity-By-Descent Analysis and Phylogeographic Haplotype Reconstruction
4.7. Age Estimation of the CHEK2 Variant c.349A>G
4.8. Microsatellite Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Bashir, M.N. Epidemiology of prostate cancer. Asian Pac. J. Cancer Prev. 2015, 16, 5137–5141. [Google Scholar] [CrossRef] [PubMed]
- Al Olama, A.A.; Kote-Jarai, Z.; Schumacher, F.R.; Wiklund, F.; Berndt, S.I.; Benlloch, S.; Giles, G.G.; Severi, G.; Neal, D.E.; Hamdy, F.C.; et al. A meta-analysis of genome-wide association studies to identify prostate cancer susceptibility loci associated with aggressive and non-aggressive disease. Hum. Mol. Genet. 2013, 22, 408–415. [Google Scholar] [CrossRef]
- Schumacher, F.R.; Al Olama, A.A.; Berndt, S.I.; Benlloch, S.; Ahmed, M.; Saunders, E.J.; Dadaev, T.; Leongamornlert, D.; Anokian, E.; Cieza-Borrella, C.; et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet. 2018, 50, 928–936. [Google Scholar] [CrossRef] [PubMed]
- Al Olama, A.A.; Kote-Jarai, Z.; Giles, G.G.; Guy, M.; Morrison, J.; Severi, G.; Leongamornlert, D.A.; Tymrakiewicz, M.; Jhavar, S.; Saunders, E.; et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat. Genet. 2009, 41, 1058–1060. [Google Scholar] [CrossRef] [PubMed]
- Eeles, R.A.; Kote-Jarai, Z.; Al Olama, A.A.; Giles, G.G.; Guy, M.; Severi, G.; Muir, K.; Hopper, J.L.; Henderson, B.E.; Haiman, C.A.; et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat. Genet. 2009, 41, 1116–1121. [Google Scholar] [CrossRef]
- Eeles, R.; Goh, C.; Castro, E.; Bancroft, E.; Guy, M.; Al Olama, A.A.; Easton, D.; Kote-Jarai, Z. The genetic epidemiology of prostate cancer and its clinical implications. Nat. Rev. Urol. 2014, 11, 18–31. [Google Scholar] [CrossRef]
- Al Olama, A.A.; Kote-Jarai, Z.; Berndt, S.I.; Conti, D.V.; Schumacher, F.; Han, Y.; Benlloch, S.; Hazelett, D.J.; Wang, Z.; Saunders, E.; et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat. Genet. 2014, 46, 1103–1109. [Google Scholar] [CrossRef]
- Brechka, H.; Bhanvadia, R.R.; VanOpstall, C.; Griend, D.J.V. HOXB13 mutations and binding partners in prostate development and cancer: Function, clinical significance, and future directions. Genes Dis. 2017, 4, 75–87. [Google Scholar] [CrossRef]
- Ewing, C.M.; Ray, A.M.; Lange, E.M.; Zuhlke, K.A.; Robbins, C.M.; Tembe, W.D.; Wiley, K.E.; Isaacs, S.D.; Johng, D.; Wang, Y.; et al. Germline mutations in HOXB13 and prostate-cancer risk. N. Engl. J. Med. 2012, 366, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Kote-Jarai, Z.; Mikropoulos, C.; Leongamornlert, D.A.; Dadaev, T.; Tymrakiewicz, M.; Saunders, E.J.; Jones, M.; Jugurnauth-Little, S.; Govindasami, K.; Guy, M.; et al. Prevalence of the HOXB13 G84E germline mutation in British men and correlation with prostate cancer risk, tumour characteristics and clinical outcomes. Ann. Oncol. 2015, 26, 756–761. [Google Scholar] [CrossRef]
- Breyer, J.P.; Avritt, T.G.; McReynolds, K.M.; Dupont, W.D.; Smith, J.R. Confirmation of the HOXB13 G84E germline mutation in familial prostate cancer. Cancer Epidemiol. Biomark. Prev. 2012, 21, 1348–1353. [Google Scholar] [CrossRef]
- Castro, E.; Eeles, R. The role of BRCA1 and BRCA2 in prostate cancer. Asian J. Androl. 2012, 14, 409–414. [Google Scholar] [CrossRef]
- Maia, S.; Cardoso, M.; Paulo, P.; Pinheiro, M.; Pinto, P.; Santos, C.; Pinto, C.; Peixoto, A.; Henrique, R.; Teixeira, M.R. The role of germline mutations in the BRCA1/2 and mismatch repair genes in men ascertained for early-onset and/or familial prostate cancer. Fam. Cancer 2016, 15, 111–121. [Google Scholar] [CrossRef]
- Petrovics, G.; Price, D.K.; Lou, H.; Chen, Y.; Garland, L.; Bass, S.; Jones, K.; Kohaar, I.; Ali, A.; Ravindranath, L.; et al. Increased frequency of germline BRCA2 mutations associates with prostate cancer metastasis in a racially diverse patient population. Prostate Cancer Prostatic Dis. 2019, 22, 406–410. [Google Scholar] [CrossRef]
- Grindedal, E.M.; Møller, P.; Eeles, R.; Stormorken, A.T.; Bowitz-Lothe, I.M.; Landrø, S.M.; Clark, N.; Kvåle, R.; Shanley, S.; Mæhle, L. Germline mutations in mismatch repair genes associated with prostate cancer. Cancer Epidemiol. Biomark. Prev. 2009, 18, 2460–2467. [Google Scholar] [CrossRef]
- Guedes, L.B.; Antonarakis, E.S.; Schweizer, M.T.; Mirkheshti, N.; Almutairi, F.; Park, J.C.; Glavaris, S.; Hicks, J.; Eisenberger, M.A.; De Marzo, A.M.; et al. MSH2 loss in primary prostate cancer. Clin. Cancer Res. 2017, 23, 6863–6874. [Google Scholar] [CrossRef]
- Seppälä, E.H.; Ikonen, T.; Mononen, N.; Autio, V.; Rökman, A.; Matikainen, M.P.; Tammela, T.L.J.; Schleutker, J. CHEK2 variants associate with hereditary prostate cancer. Br. J. Cancer 2003, 89, 1966–1970. [Google Scholar] [CrossRef]
- Cybulski, C.; Huzarski, T.; Górski, B.; Masojć, B.; Mierzejewski, M.; Dȩbniak, T.; Gliniewicz, B.; Matyjasik, J.; Złowocka, E.; Kurzawski, G.; et al. A novel founder CHEK2 mutation is associated with increased prostate cancer risk. Cancer Res. 2004, 64, 2677–2679. [Google Scholar] [CrossRef]
- Ertych, N.; Stolz, A.; Valerius, O.; Braus, G.H.; Bastians, O. CHK2-BRCA1 tumor-suppressor axis restrains oncogenic Aurora-A kinase to ensure proper mitotic microtubule assembly. Proc. Natl. Acad. Sci. USA 2016, 113, 1817–1822. [Google Scholar] [CrossRef]
- Wang, Y.; Dai, B.; Ye, D. CHEK2 mutation and risk of prostate cancer: A systematic review and meta-analysis. Int. J. Clin. Exp. Med. 2015, 8, 15708–15715. [Google Scholar]
- Southey, M.C.C.; Goldgar, D.E.E.; Winqvist, R.; Pylkäs, K.; Couch, F.; Tischkowitz, M.; Foulkes, W.D.D.; Dennis, J.; Michailidou, K.; van Rensburg, E.J.J.; et al. PALB2, CHEK2 and ATM rare variants and cancer risk: Data from COGS. J. Med. Genet. 2016, 53, 800–811. [Google Scholar] [CrossRef]
- Xiang, H.P.; Geng, X.P.; Ge, W.W.; Li, H. Meta-analysis of CHEK2 1100delC variant and colorectal cancer susceptibility. Eur. J. Cancer 2011, 47, 2546–2551. [Google Scholar] [CrossRef] [PubMed]
- Aldubayan, S.H.; Pyle, L.C.; Gamulin, M.; Kulis, T.; Moore, N.D.; Taylor-Weiner, A.; Hamid, A.A.; Reardon, B.; Wubbenhorst, B.; Godse, R.; et al. Association of inherited pathogenic variants in checkpoint kinase 2 (CHEK2) with susceptibility to testicular germ cell tumors. JAMA Oncol. 2019, 5, 514–522. [Google Scholar] [CrossRef]
- Shaag, A.; Walsh, T.; Renbaum, P.; Kirchhoff, T.; Nafa, K.; Shiovitz, S.; Mandell, J.B.; Welcsh, P.; Lee, M.K.; Ellis, N.; et al. Functional and genomic approaches reveal an ancient CHEK2 allele associated with breast cancer in the Ashkenazi Jewish population. Hum. Mol. Genet. 2005, 14, 555–563. [Google Scholar] [CrossRef]
- Paulo, P.; Maia, S.; Pinto, C.; Pinto, P.; Monteiro, A.; Peixoto, A.; Teixeira, M.R. Targeted next generation sequencing identifies functionally deleterious germline mutations in novel genes in early-onset/familial prostate cancer. PLoS Genet. 2018, 14, e1007355. [Google Scholar] [CrossRef] [PubMed]
- Maia, S.; Cardoso, M.; Pinto, P.; Pinheiro, M.; Santos, C.; Peixoto, A.; Bento, M.J.; Oliveira, J.; Henrique, R.; Jerónimo, C.; et al. Identification of two novel HOXB13 germline mutations in Portuguese prostate cancer patients. PLoS ONE 2015, 10, e0132728. [Google Scholar] [CrossRef] [PubMed]
- Eccles, D. Worldmap Wdb Combined. Available online: http://user.interface.org.nz/~gringer/hacking/wdb2svg.txt (accessed on 6 January 2020).
- Easton, D. CHEK2*1100delC and susceptibility to breast cancer: A collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies. Am. J. Hum. Genet. 2004, 74, 1175–1182. [Google Scholar] [CrossRef]
- Peixoto, A.; Santos, C.; Pinto, P.; Pinheiro, M.; Rocha, P.; Pinto, C.; Bizarro, S.; Veiga, I.; Principe, A.S.; Maia, S.; et al. The role of targeted BRCA1/BRCA2 mutation analysis in hereditary breast/ovarian cancer families of Portuguese ancestry. Clin. Genet. 2015, 88, 41–48. [Google Scholar] [CrossRef]
- Rebbeck, T.R.; Friebel, T.M.; Friedman, E.; Hamann, U.; Huo, D.; Kwong, A.; Olah, E.; Olopade, O.I.; Solano, A.R.; Teo, S.H.; et al. Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Hum. Mutat. 2018, 39, 593–620. [Google Scholar] [CrossRef]
- Weischer, M.; Bojesen, S.E.; Ellervik, C.; Tybjærg-Hansen, A.; Nordestgaard, B.G. CHEK2*1100delC genotyping for clinical assessment of breast cancer risk: Meta-analyses of 26,000 patient cases and 27,000 controls. J. Clin. Oncol. 2008, 26, 542–548. [Google Scholar] [CrossRef]
- Apostolou, P.; Fostira, F. Hereditary breast cancer: The Era of new susceptibility genes. Biomed. Res. Int. 2013, 2013, 11. [Google Scholar] [CrossRef] [PubMed]
- Schutte, M.; Seal, S.; Barfoot, R.; Meijers-Heijboer, H.; Wasielewski, M.; Evans, D.G.; Eccles, D.; Meijers, C.; Lohman, F.; Klijn, J.; et al. Variants in CHEK2 other than 1100delC do not make a major contribution to breast cancer susceptibility. Am. J. Hum. Genet. 2003, 72, 1023–1028. [Google Scholar] [CrossRef]
- Li, J.; Williams, B.L.; Haire, L.F.; Goldberg, M.; Wilker, E.; Durocher, D.; Yaffe, M.B.; Jackson, S.P.; Smerdon, S.J. Structural and functional versatility of the FHA domain in DNA-damage signaling by the tumor suppressor kinase Chk2. Mol. Cell 2002, 9, 1045–1054. [Google Scholar] [CrossRef]
- Sodha, N.; Mantoni, T.S.; Tavtigian, S.V.; Eeles, R.; Garrett, M.D. Rare germ line CHEK2 variants identified in breast cancer families encode proteins that show impaired activation. Cancer Res. 2006, 66, 8966–8970. [Google Scholar] [CrossRef]
- Wu, X.; Dong, X.; Liu, W.; Chen, J. Characterization of CHEK2 mutations in prostate cancer. Hum. Mutat. 2006, 27, 742–747. [Google Scholar] [CrossRef]
- Angèle, S.; Falconer, A.; Edwards, S.M.; Dörk, T.; Bremer, M.; Moullan, N.; Chapot, B.; Muir, K.; Houlston, R.; Norman, A.R.; et al. ATM polymorphisms as risk factors for prostate cancer development. Br. J. Cancer 2004, 91, 783–787. [Google Scholar] [CrossRef] [PubMed]
- Leongamornlert, D.; Mahmud, N.; Tymrakiewicz, M.; Saunders, E.; Dadaev, T.; Castro, E.; Goh, C.; Govindasami, K.; Guy, M.; O’Brien, L.; et al. Germline BRCA1 mutations increase prostate cancer risk. Br. J. Cancer 2012, 106, 1697–1701. [Google Scholar] [CrossRef]
- Kote-Jarai, Z.; Leongamornlert, D.; Saunders, E.; Tymrakiewicz, M.; Castro, E.; Mahmud, N.; Guy, M.; Edwards, S.; O’Brien, L.; Sawyer, E.; et al. BRCA2 is a moderate penetrance gene contributing to young-onset prostate cancer: Implications for genetic testing in prostate cancer patients. Br. J. Cancer 2011, 105, 1230–1234. [Google Scholar] [CrossRef]
- Karlsson, R.; Aly, M.; Clements, M.; Zheng, L.; Adolfsson, J.; Xu, J.; Grönberg, H.; Wiklund, F. A population-based assessment of germline HOXB13 G84E mutation and prostate cancer risk. Eur. Urol. 2014, 65, 169–176. [Google Scholar] [CrossRef]
- Leslie, S.; Winney, B.; Hellenthal, G.; Davison, D.; Boumertit, A.; Day, T.; Hutnik, K.; Royrvik, E.C.; Cunliffe, B.; Lawson, D.J.; et al. The fine-scale genetic structure of the British population. Nature 2015, 519, 309–314. [Google Scholar] [CrossRef]
- Meijers-Heijboer, H.; Van den Ouweland, A.; Klijn, J.; Wasielewski, M.; De Shoo, A.; Oldenburg, R.; Hollestelle, A.; Houben, M.; Crepin, E.; Van Veghel-Plandsoen, M.; et al. Low-penetrance susceptibility to breast cancer due to CHEK2*1100delC in noncarriers of BRCA1 or BRCA2 mutations: The CHEK2-breast cancer consortium. Nat. Genet. 2002, 31, 55–59. [Google Scholar] [CrossRef]
- Vahteristo, P.; Bartkova, J.; Eerola, H.; Syrjäkoski, K.; Ojala, S.; Kilpivaara, O.; Tamminen, A.; Kononen, J.; Aittomäki, K.; Heikkilä, P.; et al. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am. J. Hum. Genet. 2002, 71, 432–438. [Google Scholar] [CrossRef]
- Heather, P. Empires and Barbarians: Migration, Development, and the Birth of Europe; Pan Macmillan: London, UK, 2010; pp. 1–734. [Google Scholar]
- Bryc, K.; Durand, E.Y.; Macpherson, J.M.; Reich, D.; Mountain, J.L. The genetic ancestry of african americans, latinos, and european Americans across the United States. Am. J. Hum. Genet. 2015, 96, 37–53. [Google Scholar] [CrossRef]
- McEvoy, B.P.; Lind, J.M.; Wang, E.T.; Moyzis, R.K.; Visscher, P.M.; Van Holst Pellekaan, S.M.; Wilton, A.N. Whole-genome genetic diversity in a sample of Australians with deep aboriginal ancestry. Am. J. Hum. Genet. 2010, 87, 297–305. [Google Scholar] [CrossRef]
- Greenwood, C.M.T.; Sun, S.; Veenstra, J.; Hamel, N.; Niell, B.; Gruber, S.; Foulkes, W.D. How old is this mutation?—A study of three Ashkenazi Jewish founder mutations. BMC Genet. 2010, 11, 39. [Google Scholar] [CrossRef] [PubMed]
- Cybulski, C.; Górski, B.; Huzarski, T.; Masojć, B.; Mierzejewski, M.; Dȩbniak, T.; Teodorczyk, U.; Byrski, T.; Gronwald, J.; Matyjasik, J.; et al. CHEK2 is a multiorgan cancer susceptibility gene. Am. J. Hum. Genet. 2004, 75, 1131–1135. [Google Scholar] [CrossRef] [PubMed]
- Pinto, P.; Paulo, P.; Santos, C.; Rocha, P.; Pinto, C.; Veiga, I.; Pinheiro, M.; Peixoto, A.; Teixeira, M.R. Implementation of next-generation sequencing for molecular diagnosis of hereditary breast and ovarian cancer highlights its genetic heterogeneity. Breast Cancer Res. Treat. 2016, 159, 245–256. [Google Scholar] [CrossRef]
- Roy, R.; Chun, J.; Powell, S.N. BRCA1 and BRCA2: Different roles in a common pathway of genome protection. Nat. Rev. Cancer 2012, 12, 68–78. [Google Scholar] [CrossRef]
- Susswein, L.R.; Marshall, M.L.; Nusbaum, R.; Vogel Postula, K.J.; Weissman, S.M.; Yackowski, L.; Vaccari, E.M.; Bissonnette, J.; Booker, J.K.; Cremona, M.L.; et al. Pathogenic and likely pathogenic variant prevalence among the first 10,000 patients referred for next-generation cancer panel testing. Genet. Med. 2016, 18, 823–832. [Google Scholar] [CrossRef]
- Pinto, P.; Peixoto, A.; Santos, C.; Rocha, P.; Pinto, C.; Pinheiro, M.; Leça, L.; Martins, A.T.; Ferreira, V.; Bartosch, C.; et al. Analysis of founder mutations in rare tumors associated with hereditary breast/ovarian cancer reveals a novel association of BRCA2 mutations with ampulla of vater carcinomas. PLoS ONE 2016, 11, e0161438. [Google Scholar] [CrossRef]
- Amos, C.I.; Dennis, J.; Wang, Z.; Byun, J.; Schumacher, F.R.; Gayther, S.A.; Casey, G.; Hunter, D.J.; Sellers, T.A.; Gruber, S.B.; et al. The oncoarray consortium: A network for understanding the genetic architecture of common cancers. Cancer Epidemiol. Biomark. Prev. 2017, 26, 126–135. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Browning, S.R.; Browning, B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007, 81, 1084–1097. [Google Scholar] [CrossRef]
- Browning, B.L.; Browning, S.R. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 2013, 194, 459–471. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Reeve, J.P.; Rannala, B. DMLE+: Bayesian linkage disequilibrium gene mapping. Bioinformatics 2002, 18, 894–895. [Google Scholar] [CrossRef]
- Genin, E.; Tullio-Pelet, A.; Begeot, F.; Lyonnet, S.; Abel, L. Estimating the age of rare disease mutations: The example of Triple-A syndrome. J. Med. Genet. 2004, 41, 445–449. [Google Scholar] [CrossRef]
- Pin, E.; Pastrello, C.; Tricarico, R.; Papi, L.; Quaia, M.; Fornasarig, M.; Carnevali, I.; Oliani, C.; Fornasin, A.; Agostini, M.; et al. MUTYH c.933+3A>C, associated with a severely impaired gene expression, is the first Italian founder mutation in MUTYH-Associated Polyposis. Int. J. Cancer 2013, 132, 1060–1069. [Google Scholar] [CrossRef]
- Nachman, M.W.; Crowell, S.L. Estimate of the mutation rate per nucleotide in humans. Genetics 2000, 156, 297–304. [Google Scholar]
- Consortium, T.G.P.; Auton, A.; Abecasis, G.R.; Altshuler, D.M.; Durbin, R.M.; Abecasis, G.R.; Bentley, D.R.; Chakravarti, A.; Clark, A.G.; Donnelly, P.; et al. A global reference for human genetic variation. Nature 2015, 526, 68. [Google Scholar] [CrossRef]
- Williams, J.E.; Zaremba, E.; Jackson, B.; Nikuni, T.; Griffin, A. Dynamical instability of a condensate induced by a rotating thermal gas. Phys. Rev. Lett. 2002, 88, 704011–704014. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
| Microsatellite Markers | ||||||
|---|---|---|---|---|---|---|
| Family | D22S310 | D22S689 | CHEK2 | D22S275 (Intragenic) | D22S1150 | D22S280 |
| 1 * | 181 | 294 | _ | 159 | 216 | 211 |
| 2 * | 183 | 294 | _ | 159 | 220 | 211 |
| 3 * | 181 | 294 | _ | 159 | 216 | 205/211 |
| 4 * | 179 | 294 | _ | 159 | 216/220 | 205 |
| 5 * | 183/187 | 290/294 | _ | 159/163 | 218/220 | 205/211 |
| 6 | 185/189 | 294 | _ | 159/161 | 216/220 | 205 |
| 7 | 187 | 294 | _ | 159 | 216 | 211 |
| 8 | 185 | 294 | _ | 159 | 216 | 211 |
| 9 | 177/185 | 294/298 | _ | 159/161 | 220 | 209 |
| 10 | 179/187 | 294 | _ | 159/167 | 218/220 | 205/209 |
| 11 | 177/189 | 294 | _ | 159 | 216 | 205 |
| 12 | 185 | 294 | _ | 159 | 216 | 205/209 |
| 13 | 185 | 294 | _ | 159 | 220 | 209 |
| 14 | 177/181 | 294/302 | _ | 159 | 216/226 | 205/213 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brandão, A.; Paulo, P.; Maia, S.; Pinheiro, M.; Peixoto, A.; Cardoso, M.; Silva, M.P.; Santos, C.; Eeles, R.A.; Kote-Jarai, Z.; et al. The CHEK2 Variant C.349A>G Is Associated with Prostate Cancer Risk and Carriers Share a Common Ancestor. Cancers 2020, 12, 3254. https://doi.org/10.3390/cancers12113254
Brandão A, Paulo P, Maia S, Pinheiro M, Peixoto A, Cardoso M, Silva MP, Santos C, Eeles RA, Kote-Jarai Z, et al. The CHEK2 Variant C.349A>G Is Associated with Prostate Cancer Risk and Carriers Share a Common Ancestor. Cancers. 2020; 12(11):3254. https://doi.org/10.3390/cancers12113254
Chicago/Turabian StyleBrandão, Andreia, Paula Paulo, Sofia Maia, Manuela Pinheiro, Ana Peixoto, Marta Cardoso, Maria P. Silva, Catarina Santos, Rosalind A. Eeles, Zsofia Kote-Jarai, and et al. 2020. "The CHEK2 Variant C.349A>G Is Associated with Prostate Cancer Risk and Carriers Share a Common Ancestor" Cancers 12, no. 11: 3254. https://doi.org/10.3390/cancers12113254
APA StyleBrandão, A., Paulo, P., Maia, S., Pinheiro, M., Peixoto, A., Cardoso, M., Silva, M. P., Santos, C., Eeles, R. A., Kote-Jarai, Z., Muir, K., UKGPCS Collaborators, Schleutker, J., Wang, Y., Pashayan, N., Batra, J., APCB BioResource, Grönberg, H., Neal, D. E., ... Teixeira, M. R. (2020). The CHEK2 Variant C.349A>G Is Associated with Prostate Cancer Risk and Carriers Share a Common Ancestor. Cancers, 12(11), 3254. https://doi.org/10.3390/cancers12113254









