DNA Repair Expression Profiling to Identify High-Risk Cytogenetically Normal Acute Myeloid Leukemia and Define New Therapeutic Targets
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
2.1. Linking Expression of DNA Repair Genes and AML Patient Overall Survival
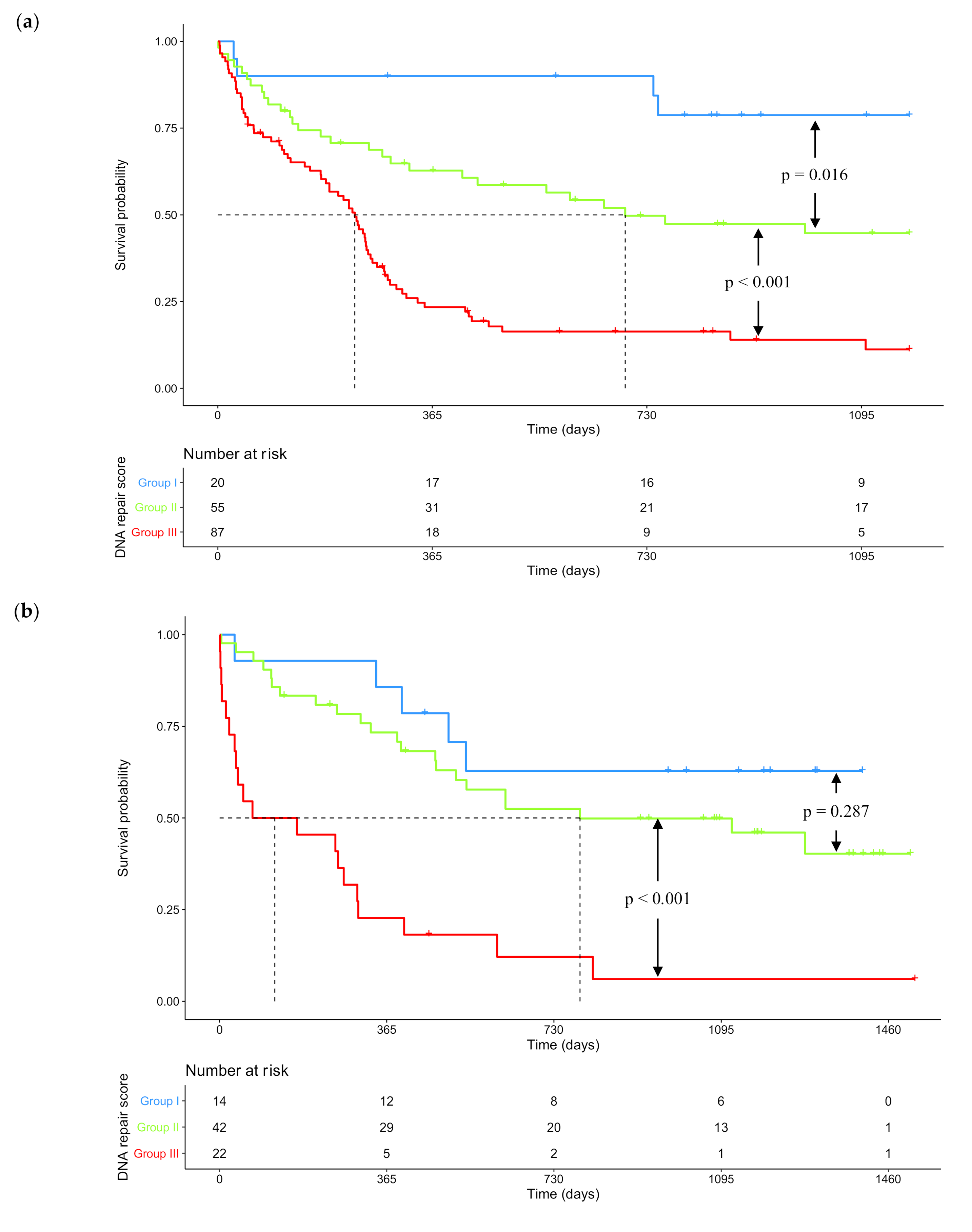
2.2. GEP-Based DNA Repair Score for Predicting CN-AML Patients’ Survival
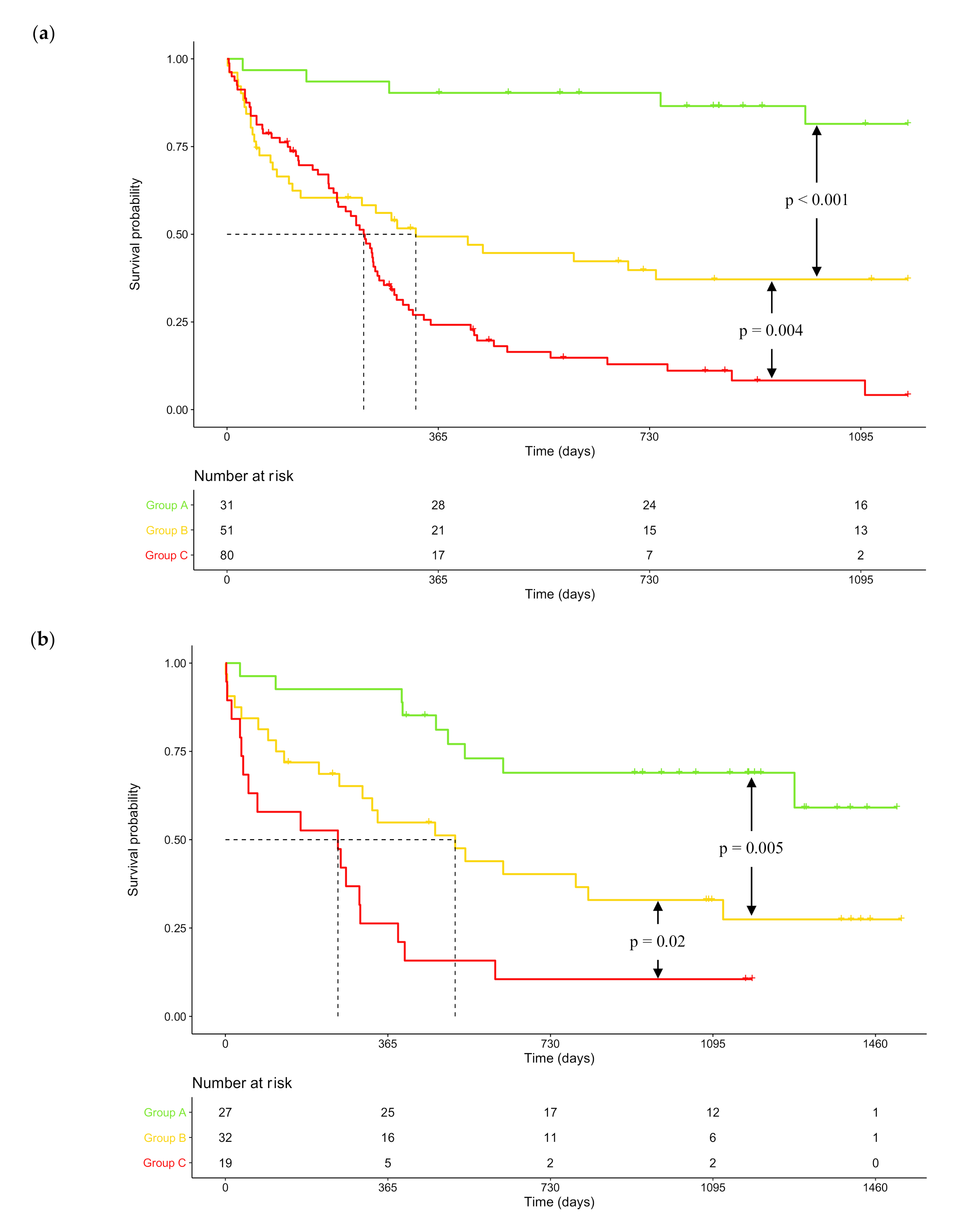
2.3. DNA Repair Score and NPM1 / FLT3 Mutational Status Combination as Prognosis Factors in CN-AML
3. Discussion
4. Materials and Methods
4.1. Patients and Gene Expression Data
4.2. Selection of Prognostic Genes
4.3. Building DNA Repair Gene Expression-Based Risk Score
4.4. Validation of the DNA Repair Score on Validation Cohort
4.5. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Grimwade, D.; Hills, R.K.; Moorman, A.V.; Walker, H.; Chatters, S.; Goldstone, A.H.; Wheatley, K.; Harrison, C.J.; Burnett, A.K.; on behalf of the National Cancer Research Institute Adult Leukaemia Working Group. Refinement of cytogenetic classification in acute myeloid leukemia: Determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 2010, 116, 354–365. [Google Scholar] [CrossRef]
- Port, M.; Böttcher, M.; Thol, F.; Ganser, A.; Schlenk, R.; Wasem, J.; Neumann, A.; Pouryamout, L. Prognostic significance of FLT3 internal tandem duplication, nucleophosmin 1, and CEBPA gene mutations for acute myeloid leukemia patients with normal karyotype and younger than 60 years: A systematic review and meta-analysis. Ann. Hematol. 2014, 93, 1279–1286. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef] [PubMed]
- An the Cancer Genome Atlas Research Network Genomic and Epigenomic Landscapes of Adult De Novo Acute Myeloid Leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [CrossRef] [PubMed]
- Ibáñez, M.; Carbonell-Caballero, J.; Such, E.; García-Alonso, L.; Liquori, A.; López-Pavía, M.; Llop, M.; Alonso, C.; Barragán, E.; Gómez-Seguí, I.; et al. The modular network structure of the mutational landscape of Acute Myeloid Leukemia. PLoS ONE 2018, 13, e0202926. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, V.; Tiacci, E.; Holmes, A.B.; Kohlmann, A.; Martelli, M.P.; Kern, W.; Spanhol-Rosseto, A.; Klein, H.-U.; Dugas, M.; Schindela, S.; et al. Whole-exome sequencing identifies somatic mutations of BCOR in acute myeloid leukemia with normal karyotype. Blood 2011, 118, 6153–6163. [Google Scholar] [CrossRef] [PubMed]
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational landscape and significance across 12 major cancer types. Nat. Cell Biol. 2013, 502, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Kayser, S.; Döhner, K.; Krauter, J.; Köhne, C.-H.; Horst, H.A.; Held, G.; Von Lilienfeld-Toal, M.; Wilhelm, S.; Kündgen, A.; Götze, K.; et al. The impact of therapy-related acute myeloid leukemia (AML) on outcome in 2853 adult patients with newly diagnosed AML. Blood 2011, 117, 2137–2145. [Google Scholar] [CrossRef]
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar] [CrossRef]
- Liu, D.; Wu, D.; Li, H.; Dong, M. The effect of XPD/ERCC2 Lys751Gln polymorphism on acute leukemia risk: A systematic review and meta-analysis. Gene 2014, 538, 209–216. [Google Scholar] [CrossRef]
- Alcalay, M.; Meani, N.; Gelmetti, V.; Fantozzi, A.; Fagioli, M.; Orleth, A.; Riganelli, D.; Sebastiani, C.; Cappelli, E.; Casciari, C.; et al. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J. Clin. Investig. 2003, 112, 1751–1761. [Google Scholar] [CrossRef] [PubMed]
- Krejci, O.; Wunderlich, M.; Geiger, H.; Chou, F.-S.; Schleimer, D.; Jansen, M.; Andreassen, P.R.; Mulloy, J.C. p53 signaling in response to increased DNA damage sensitizes AML1-ETO cells to stress-induced death. Blood 2008, 111, 2190–2199. [Google Scholar] [CrossRef] [PubMed]
- Yeung, P.L.; Denissova, N.G.; Nasello, C.; Hakhverdyan, Z.; Chen, J.D.; Brenneman, M.A. Promyelocytic leukemia nuclear bodies support a late step in DNA double-strand break repair by homologous recombination. J. Cell. Biochem. 2011, 113, 1787–1799. [Google Scholar] [CrossRef] [PubMed]
- Van Der Kouwe, E.; Staber, P.B. RUNX1-ETO: Attacking the Epigenome for Genomic Instable Leukemia. Int. J. Mol. Sci. 2019, 20, 350. [Google Scholar] [CrossRef]
- Alter, B.P. Fanconi anemia and the development of leukemia. Best Pract. Res. Clin. Haematol. 2014, 27, 214–221. [Google Scholar] [CrossRef]
- Quinn, E.; Nichols, K.E. Cancer predisposition syndromes associated with myeloid malignancy. Semin. Hematol. 2017, 54, 115–122. [Google Scholar] [CrossRef]
- Schoch, C.; Kern, W.; Kohlmann, A.; Hiddemann, W.; Schnittger, S.; Haferlach, T. Acute myeloid leukemia with a complex aberrant karyotype is a distinct biological entity characterized by genomic imbalances and a specific gene expression profile. Genes Chromosom. Cancer 2005, 43, 227–238. [Google Scholar] [CrossRef]
- Cavelier, C.; Didier, C.; Prade, N.; Mas, V.M.-D.; Manenti, S.; Recher, C.; Demur, C.; Ducommun, B. Constitutive Activation of the DNA Damage Signaling Pathway in Acute Myeloid Leukemia with Complex Karyotype: Potential Importance for Checkpoint Targeting Therapy. Cancer Res. 2009, 69, 8652–8661. [Google Scholar] [CrossRef]
- Wang, P.; Ma, D.; Wang, J.; Fang, Q.; Gao, R.; Wu, W.; Cao, L.; Hu, X.; Zhao, J.; Li, Y. INPP4B-mediated DNA repair pathway confers resistance to chemotherapy in acute myeloid leukemia. Tumor Biol. 2016, 37, 12513–12523. [Google Scholar] [CrossRef]
- Sallmyr, A.; Fan, J.; Datta, K.; Kim, K.-T.; Grosu, D.; Shapiro, P.; Small, D.; Rassool, F. Internal tandem duplication of FLT3 (FLT3/ITD) induces increased ROS production, DNA damage, and misrepair: Implications for poor prognosis in AML. Blood 2008, 111, 3173–3182. [Google Scholar] [CrossRef]
- Seedhouse, C.; Hunter, H.M.; Lloyd-Lewis, B.; Massip, A.-M.; Pallis, M.; I Carter, G.; Grundy, M.; Shang, S.; Russell, N.H. DNA repair contributes to the drug-resistant phenotype of primary acute myeloid leukaemia cells with FLT3 internal tandem duplications and is reversed by the FLT3 inhibitor PKC412. Leukemia 2006, 20, 2130–2136. [Google Scholar] [CrossRef] [PubMed]
- Aalbers, A.M.; Calado, R.T.; Young, N.S.; Zwaan, C.M.; Wu, C.; Kajigaya, S.; Coenen, E.A.; Baruchel, A.; Geleijns, K.; De Haas, V.; et al. Telomere length and telomerase complex mutations in pediatric acute myeloid leukemia. Leukemia 2013, 27, 1786–1789. [Google Scholar] [CrossRef] [PubMed]
- Bagrintseva, K.; Geisenhof, S.; Kern, R.; Eichenlaub, S.; Reindl, C.; Ellwart, J.W.; Hiddemann, W.; Spiekermann, K. FLT3-ITD-TKD dual mutants associated with AML confer resistance to FLT3 PTK inhibitors and cytotoxic agents by overexpression of Bcl-x(L). Blood 2005, 105, 3679–3685. [Google Scholar] [CrossRef]
- Alpermann, T.; Schnittger, S.; Eder, C.; Dicker, F.; Meggendorfer, M.; Kern, W.; Schmid, C.; Aul, C.; Staib, P.; Wendtner, C.-M.; et al. Molecular subtypes of NPM1 mutations have different clinical profiles, specific patterns of accompanying molecular mutations and varying outcomes in intermediate risk acute myeloid leukemia. Haematologica 2015, 101, e55–e58. [Google Scholar] [CrossRef] [PubMed]
- Okuda, M.; Horn, H.F.; Tarapore, P.; Tokuyama, Y.; Smulian, A.; Chan, P.-K.; Knudsen, E.S.; A Hofmann, I.; Snyder, J.D.; E Bove, K.; et al. Nucleophosmin/B23 Is a Target of CDK2/Cyclin E in Centrosome Duplication. Cell 2000, 103, 127–140. [Google Scholar] [CrossRef]
- Koike, A.; Nishikawa, H.; Wu, W.; Okada, Y.; Venkitaraman, A.R.; Ohta, T. Recruitment of Phosphorylated NPM1 to Sites of DNA Damage through RNF8-Dependent Ubiquitin Conjugates. Cancer Res. 2010, 70, 6746–6756. [Google Scholar] [CrossRef]
- Lirussi, L.; Antoniali, G.; Vascotto, C.; D’Ambrosio, C.; Poletto, M.; Romanello, M.; Marasco, D.; Leone, M.; Quadrifoglio, F.; Bhakat, K.K.; et al. Nucleolar accumulation of APE1 depends on charged lysine residues that undergo acetylation upon genotoxic stress and modulate its BER activity in cells. Mol. Biol. Cell 2012, 23, 4079–4096. [Google Scholar] [CrossRef]
- Colombo, E.; Marine, J.-C.; Danovi, D.; Falini, B.; Pelicci, P.G. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat. Cell Biol. 2002, 4, 529–533. [Google Scholar] [CrossRef]
- Vascotto, C.; Lirussi, L.; Poletto, M.; Tiribelli, M.; Damiani, D.; Fabbro, D.; Damante, G.; Demple, B.; Colombo, E.; Tell, G. Functional regulation of the apurinic/apyrimidinic endonuclease 1 by nucleophosmin: Impact on tumor biology. Oncogene 2013, 33, 2876–2887. [Google Scholar] [CrossRef]
- Meyers, R.M.; Bryan, J.G.; McFarland, J.M.; Weir, B.A.; Sizemore, A.E.; Xu, H.; Dharia, N.V.; Montgomery, P.G.; Cowley, G.S.; Pantel, S.; et al. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat. Genet. 2017, 49, 1779–1784. [Google Scholar] [CrossRef]
- Dempster, J.M.; Rossen, J.; Kazachkova, M.; Pan, J.; Kugener, G.; Root, D.E.; Tsherniak, A. Extracting Biological Insights from the Project Achilles Genome-Scale CRISPR Screens in Cancer Cell Lines. BioRxiv 2019. [Google Scholar] [CrossRef]
- Herviou, L.; Kassambara, A.; Boireau, S.; Robert, N.; Requirand, G.; Müller-Tidow, C.; Vincent, L.; Seckinger, A.; Goldschmidt, H.; Cartron, G.; et al. PRC2 targeting is a therapeutic strategy for EZ score defined high-risk multiple myeloma patients and overcome resistance to IMiDs. Clin. Epigenet. 2018, 10, 121. [Google Scholar] [CrossRef] [PubMed]
- Bret, C.; Klein, B.; Cartron, G.; Schved, J.; Constantinou, A.; Pasero, P.; Moreaux, J. DNA repair in diffuse large B-cell lymphoma: A molecular portrait. Br. J. Haematol. 2014, 169, 296–299. [Google Scholar] [CrossRef] [PubMed]
- Moreaux, J.; Reme, T.; Leonard, W.; Veyrune, J.-L.; Requirand, G.; Goldschmidt, H.; Hose, D.; Klein, B. Gene expression-based prediction of myeloma cell sensitivity to histone deacetylase inhibitors. Br. J. Cancer 2013, 109, 676–685. [Google Scholar] [CrossRef]
- De Boussac, H.; Bruyer, A.; Jourdan, M.; Maes, A.; Robert, N.; Gourzones, C.; Vincent, L.; Seckinger, A.; Cartron, G.; Hose, D.; et al. Kinome expression profiling to target new therapeutic avenues in multiple myeloma. Haematologica 2019, 105, 784–795. [Google Scholar] [CrossRef]
- Wouters, B.; Löwenberg, B.; Erpelinck-Verschueren, C.A.J.; Van Putten, W.; Valk, P.; Delwel, H. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood 2009, 113, 3088–3091. [Google Scholar] [CrossRef]
- Taskesen, E.; Bullinger, L.; Corbacioglu, A.; Sanders, M.A.; Erpelinck, C.A.J.; Wouters, B.J.; van der Poel-van de Luytgaarde, S.C.; Damm, F.; Krauter, J.; Ganser, A.; et al. Prognostic impact, concurrent genetic mutations, and gene expression features of AML with CEBPA mutations in a cohort of 1182 cytogenetically normal AML patients: Further evidence for CEBPA double mutant AML as a distinctive disease entity. Blood 2011, 117, 2469–2475. [Google Scholar] [CrossRef]
- Taskesen, E.; Babaei, S.; Reinders, M.M.; De Ridder, J. Integration of gene expression and DNA-methylation profiles improves molecular subtype classification in acute myeloid leukemia. BMC Bioinform. 2015, 16, S5. [Google Scholar] [CrossRef]
- Hartlerode, A.J.; Scully, R. Mechanisms of double-strand break repair in somatic mammalian cells. Biochem. J. 2009, 423, 157–168. [Google Scholar] [CrossRef]
- Wright, W.D.; Shah, S.S.; Heyer, W.-D. Homologous recombination and the repair of DNA double-strand breaks. J. Biol. Chem. 2018, 293, 10524–10535. [Google Scholar] [CrossRef]
- D’Amours, D.; Jackson, S.P. The MRE11 complex: At the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 2002, 3, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H. ATM Activation by DNA Double-Strand Breaks Through the Mre11-Rad50-Nbs1 Complex. Science 2005, 308, 551–554. [Google Scholar] [CrossRef] [PubMed]
- Esposito, M.T.; So, C.W.E. DNA damage accumulation and repair defects in acute myeloid leukemia: Implications for pathogenesis, disease progression, and chemotherapy resistance. Chromosom. 2014, 123, 545–561. [Google Scholar] [CrossRef] [PubMed]
- Uringa, E.-J.; Youds, J.L.; Lisaingo, K.; Lansdorp, P.M.; Boulton, S.J. RTEL1: An essential helicase for telomere maintenance and the regulation of homologous recombination. Nucleic Acids Res. 2010, 39, 1647–1655. [Google Scholar] [CrossRef]
- Dong, S.; Han, J.; Chen, H.; Liu, T.; Huen, M.S.Y.; Yang, Y.; Guo, C.; Huang, J. The Human SRCAP Chromatin Remodeling Complex Promotes DNA-End Resection. Curr. Biol. 2014, 24, 2097–2110. [Google Scholar] [CrossRef]
- Spivak, G. Nucleotide excision repair in humans. DNA Repair 2015, 36, 13–18. [Google Scholar] [CrossRef]
- Hashimoto, S.; Anai, H.; Hanada, K. Mechanisms of interstrand DNA crosslink repair and human disorders. Genes Environ. 2016, 38, 9. [Google Scholar] [CrossRef]
- Vonarx, E.J.; Tabone, E.K.; Osmond, M.J.; Anderson, H.J.; Kunz, B.A. Arabidopsis homologue of human transcription factor IIH/nucleotide excision repair factor p44 can function in transcription and DNA repair and interacts with AtXPD. Plant J. 2006, 46, 512–521. [Google Scholar] [CrossRef]
- London, R.E. The structural basis of XRCC1-mediated DNA repair. DNA Repair 2015, 30, 90–103. [Google Scholar] [CrossRef]
- Pietrzak, J.; Płoszaj, T.; Pułaski, Ł.; Robaszkiewicz, A. EP300-HDAC1-SWI/SNF functional unit defines transcription of some DNA repair enzymes during differentiation of human macrophages. Biochim. Biophys. Acta (BBA) Bioenerg. 2019, 1862, 198–208. [Google Scholar] [CrossRef]
- Seedhouse, C.; Faulkner, R.; Ashraf, N.; Das-Gupta, E.; Russell, N. Polymorphisms in genes involved in homologous recombination repair interact to increase the risk of developing acute myeloid leukemia. Clin. Cancer Res. 2004, 10, 2675–2680. [Google Scholar] [CrossRef] [PubMed]
- Jawad, M.; Seedhouse, C.; Russell, N.; Plumb, M. Polymorphisms in human homeobox HLX1 and DNA repair RAD51 genes increase the risk of therapy-related acute myeloid leukemia. Blood 2006, 108, 3916–3918. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, C.; Liu, Y.; Hu, Z.; Zhou, Y. Genetic polymorphisms of RAD51 and XRCC3 and acute myeloid leukemia risk: A meta-analysis. Leuk. Lymphoma 2013, 55, 1309–1319. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Long, Z.; Dai, Z. 135G/C polymorphism in the RAD51 gene and acute myeloid leukemia risk: A meta-analysis. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef]
- Allan, J.M. Genetic variation in XPD predicts treatment outcome and risk of acute myeloid leukemia following chemotherapy. Blood 2004, 104, 3872–3877. [Google Scholar] [CrossRef]
- Pratcorona, M.; Brunet, S.; Nomdedéu, J.F.; Ribera, J.; Tormo, M.; Duarte, R.; Escoda, L.; Guàrdia, R.; De Llano, M.P.Q.; Salamero, O.; et al. Favorable outcome of patients with acute myeloid leukemia harboring a low-allelic burden FLT3-ITD mutation and concomitant NPM1 mutation: Relevance to post-remission therapy. Blood 2013, 121, 2734–2738. [Google Scholar] [CrossRef]
- Lichtman, M.A. A historical perspective on the development of the cytarabine (7days) and daunorubicin (3days) treatment regimen for acute myelogenous leukemia: 2013 the 40th anniversary of 7+3. Blood Cells Mol. Dis. 2013, 50, 119–130. [Google Scholar] [CrossRef]
- Murphy, T.; Yee, K.W.L. Cytarabine and daunorubicin for the treatment of acute myeloid leukemia. Expert Opin. Pharmacother. 2017, 18, 1765–1780. [Google Scholar] [CrossRef]
- Bret, C.; Klein, B.; Moreaux, J. Nucleotide excision DNA repair pathway as a therapeutic target in patients with high-risk diffuse large B cell lymphoma. Cell Cycle 2013, 12, 1811–1812. [Google Scholar] [CrossRef][Green Version]
- Zhou, J.; Deng, Q.; Zhang, Y.; Tan, P.; Zhang, W.; Cui, H. CSN6 controls the proliferation and metastasis of glioblastoma by CHIP-mediated degradation of EGFR. Oncogene 2016, 36, 1134–1144. [Google Scholar] [CrossRef]
- Gao, S.; Fang, L.; Phan, L.M.; Qdaisat, A.; Yeung, S.-C.J.; Lee, M.-H. COP9 signalosome subunit 6 (CSN6) regulates E6AP/UBE3A in cervical cancer. Oncotarget 2015, 6, 28026–28041. [Google Scholar] [CrossRef] [PubMed]
- Wen, D.; Liao, T.; Ma, B.; Qu, N.; Shi, R.-L.; Lu, Z.-W.; Wang, Y.-L.; Wei, W.-J.; Ji, Q.-H. Downregulation of CSN6 attenuates papillary thyroid carcinoma progression by reducing Wnt/β-catenin signaling and sensitizes cancer cells to FH535 therapy. Cancer Med. 2018, 7, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, M.; Allen, C.; Nickoloff, J.A.; Hromas, R. Synthetic lethality: Exploiting the addiction of cancer to DNA repair. Blood 2011, 117, 6074–6082. [Google Scholar] [CrossRef] [PubMed]
- Curtin, N.J. Inhibiting the DNA damage response as a therapeutic manoeuvre in cancer. Br. J. Pharmacol. 2013, 169, 1745–1765. [Google Scholar] [CrossRef]
- Kohl, V.; Flach, J.; Naumann, N.; Brendel, S.; Kleiner, H.; Weiss, C.; Seifarth, W.; Nowak, D.; Hofmann, W.-K.; Fabarius, A.; et al. Antileukemic Efficacy in Vitro of Talazoparib and APE1 Inhibitor III Combined with Decitabine in Myeloid Malignancies. Cancers 2019, 11, 1493. [Google Scholar] [CrossRef]
- Nguyen, G.H.; Dexheimer, T.S.; Rosenthal, A.S.; Chu, W.K.; Singh, D.K.; Mosedale, G.; Bachrati, C.Z.; Schultz, L.; Sakurai, M.; Savitsky, P.; et al. A small molecule inhibitor of the BLM helicase modulates chromosome stability in human cells. Chem. Biol. 2013, 20, 55–62. [Google Scholar] [CrossRef]
- Aggarwal, M.; Banerjee, T.; A Sommers, J.; Brosh, J.R.M. Targeting an Achilles’ heel of cancer with a WRN helicase inhibitor. Cell Cycle. 2013, 12, 3329–3335. [Google Scholar] [CrossRef]
- Toma, M.; Sullivan-Reed, K.; Sliwinski, T.; Skorski, T.; Toma Reed, S. RAD52 as a Potential Target for Synthetic Lethality-Based Anticancer Therapies. Cancers 2019, 11, 1561. [Google Scholar] [CrossRef]
- Petroni, M.; Sardina, F.; Infante, P.; Bartolazzi, A.; Locatelli, E.; Fabretti, F.; Di Giulio, S.; Capalbo, C.; Cardinali, B.; Coppa, A.; et al. MRE11 inhibition highlights a replication stress-dependent vulnerability of MYCN-driven tumors. Cell Death Dis. 2018, 9, 895. [Google Scholar] [CrossRef]
- Metzeler, K.H.; Hummel, M.; Bloomfield, C.D.; Spiekermann, K.; Braess, J.; Sauerland, M.-C.; Heinecke, A.; Radmacher, M.; Marcucci, G.; Whitman, S.P.; et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood 2008, 112, 4193–4201. [Google Scholar] [CrossRef]
- Huber, W.; Von Heydebreck, A.; Sültmann, H.; Poustka, A.; Vingron, M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 2002, 18, S96–S104. [Google Scholar] [CrossRef] [PubMed]
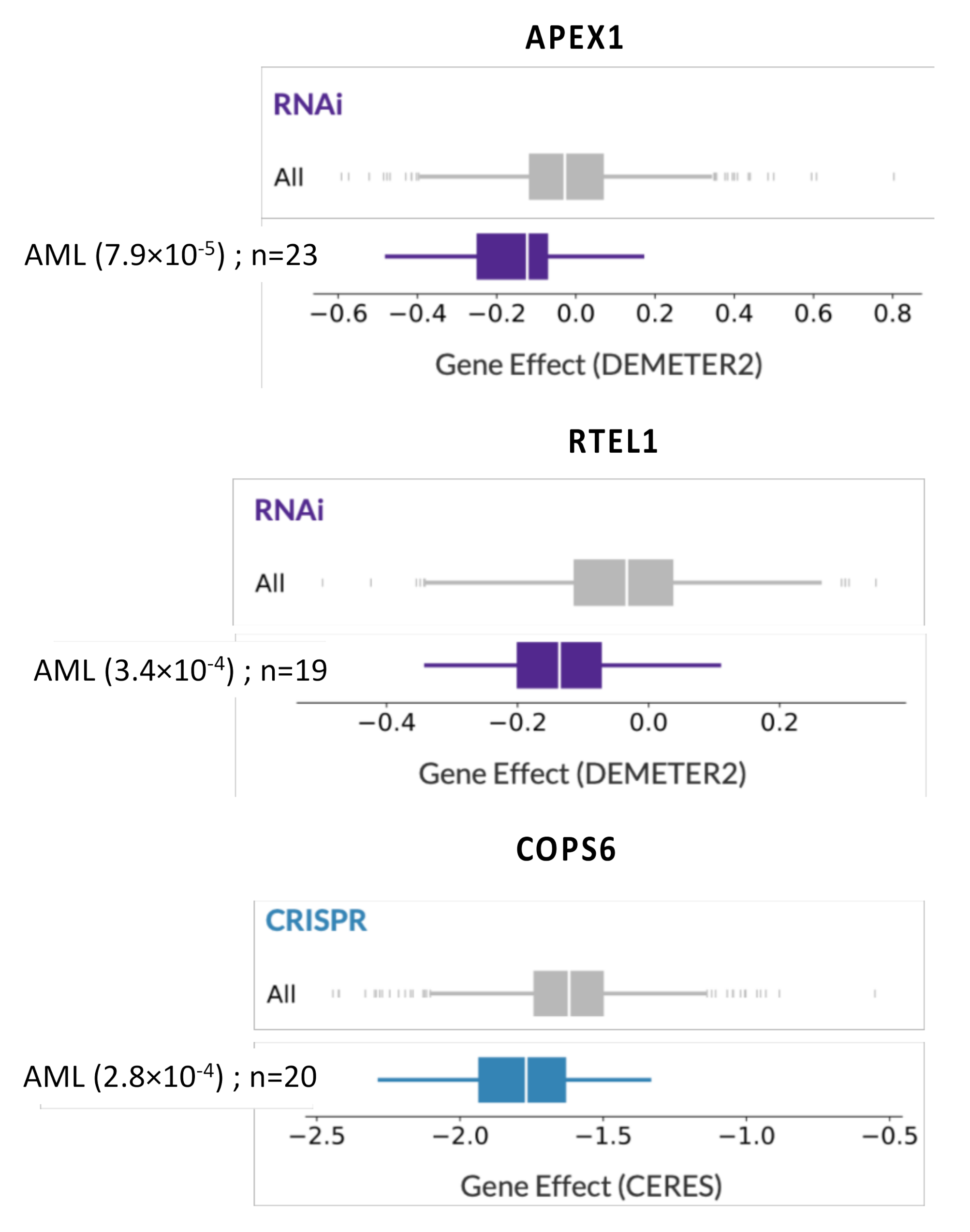
| DNA Repair Pathway | Probe Set | Gene Symbol | Benjamini–Hochberg Corrected p-Value | Hazard Ratio | Prognosis |
|---|---|---|---|---|---|
| Base Excision Repair pathway (BER) | 210027_s_at 209731_at 202330_s_at 203655_at | APEX1 NTHL1 UNG XRCC1 | 0.02 0.0016 0.0095 0.022 | 1.6 1.9 2 1.6 | Bad Bad Bad Bad |
| Fanconi pathway (FANC) | 209902_at 214727_at 203719_at 203678_at 221206_at 219317_at | ATR BRCA2 ERCC1 FAN1 PMS2 /// PMS2CL POLI | 0.0048 0.0049 0.0037 0.0028 0.024 0.0016 | 1.8 0.58 1.9 1.8 1.8 1.9 | Bad Good Bad Bad Bad Bad |
| Homologous Recombination Repair pathway (HRR) | 214727_at 205395_s_at 205647_at 206092_x_at 212275_s_at 207598_x_at | BRCA2 MRE11A RAD52 RTEL1 SRCAP XRCC2 | 0.0049 0.015 0.044 0.00047 0.014 0.007 | 0.58 1.8 1.9 2.5 0.6 1.7 | Good Bad Bad Bad Good Bad |
| Mismatch Repair pathway (MMR) | 205887_x_at 221206_at 1053_at | MSH3 PMS2 /// PMS2CL RFC2 | 0.000043 0.024 0.023 | 2.8 1.8 1.6 | Bad Bad Bad |
| Nucleotide Excision Repair pathway (NER) | 201405_s_at 213579_s_at 203719_at 205162_at 223758_s_at 201046_s_at 205672_at 203655_at | COPS6 EP300 ERCC1 ERCC8 GTF2H2 RAD23A XPA XRCC1 | 0.011 0.019 0.0037 0.04 0.033 0.0067 0.0035 0.022 | 1.7 0.59 1.9 1.5 1.5 0.53 1.8 1.6 | Bad Good Bad Bad Bad Good Bad Bad |
| DNA Repair Pathway Score | Univariate Cox Analysis | Multivariate Cox Analysis | ||
|---|---|---|---|---|
| HR | p-Value | HR | p-Value | |
| BER score | 1.97 | 1.44 × 10−3 | 0.93 | NS |
| FANC score | 2.32 | 2.98 × 10−5 | 1.30 | NS |
| HRR score | 3.23 | 2.16 × 10−7 | 2.36 | 5.89 × 10−4 |
| MMR score | 2.80 | 1.59 × 10−4 | 1.58 | NS |
| NER score | 3.83 | 2.90 × 10−4 | 2.54 | 1.66 × 10−2 |
| Scores | Univariate Cox Analysis | Multivariate Cox Analysis | ||
|---|---|---|---|---|
| HR | p-Value | HR | p-Value | |
| DNA repair score | 2.76 | 1.49 × 10−8 | 2.66 | 5.1 × 10−8 |
| NPM1 mutation/FLT3-ITD classification | 1.81 | 1.18 × 10−4 | 1.76 | 6.2 × 10−4 |
| Classification According to DNA Repair Score | ||||
|---|---|---|---|---|
| Group I 0 point | Group II 1 point | Group III 2 points | ||
| NPM1 and FLT3 mutational status | NPM1+ and FLT3-ITD- 0 point | 0 | 1 | 2 |
| NPM1+ and FLT3-ITD+ or NPM1- and FLT3-ITD- 1 point | 1 | 2 | 3 | |
| NPM1- and FLT3-ITD+ 2 points | 2 | 3 | 4 | |
| Target | Drug | Cancer | Phase | Intervention | Identifier |
|---|---|---|---|---|---|
| Base Excision Repair (BER) Pathway | |||||
| APEX1 | TRC-102 | Solid tumors & lymphomas | I/II | TRC-102 + temozolomide | NCT01851369 |
| PARP1/2 | Niraparib | Pancreatic ADK | II | Niraparib alone | NCT03601923 |
| Olaparib | Lymphomas (B/T/Hodgkin) | I | Olaparib + high-dose chemotherapy + ASCT | NCT03259503 | |
| Olaparib | AML or MDS with IDH1/2 mutation | II | Olaparib alone | NCT03953898 | |
| Talazoparib | R/R AML CD33+ | I/II | Talazoparib + GO | NCT04207190 | |
| Talazoparib | AML | I/II | Talazoparib + Decitabine | NCT02878785 | |
| Veliparib | Myeloproliferative disorders | II | Carboplatin + Topotecan +/− Veliparib | NCT03289910 | |
| Homologous Recombination Repair (HRR) Pathway | |||||
| ATM | AZD1390 | Glioblastoma | I | AZD1390 + radiotherapy | NCT03423628 |
| CHEK-1/2 | Prexasertib | R/R medulloblastoma | I | Prexasertib + Gemcitabine or Cyclophosphamide | NCT04023669 |
| RAD51 | CYT-0851 | Solid tumors & B-cell lymphomas | I/II | CYT-0851 alone | NCT03997968 |
| Fanconi (FANC) Pathway | |||||
| ATM | (see above) | ||||
| ATR | Ceralasertib | R/R non-Hodgkin’s lymphoma | I | Ceralasertib + Acalabrutinib | NCT03527147 |
| M6620 | Solid tumors | II | MS6620 alone | NCT03718091 | |
| RAD51 | (see above) | ||||
| Nucleotide Excision Repair (NER) pathway | |||||
| CDK7 | LY3405105 | Solid tumors | I | LY3405105 alone | NCT03770494 |
| SY5609 | Solid tumors | I | SY5609 +/− Fulvestrant | NCT04247126 | |
| CT7001 | Solid tumors | I/II | CT7001 +/− Fulvestrant | NCT03363893 | |
| Others | |||||
| WEE1 | Adavosertib | SQCLC | II | Adavosertib + Paclitaxel + Carboplatin | NCT02513563 |
| PARP1/2 + ATR or WEE1 | Olaparib Ceralasertib Adavosertib | Metastatic triple negative breast cancer | II | Olaparib alone or Olaparib + Ceralasertib or Olaparib + Adavosertib | NCT03330847 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gabellier, L.; Bret, C.; Bossis, G.; Cartron, G.; Moreaux, J. DNA Repair Expression Profiling to Identify High-Risk Cytogenetically Normal Acute Myeloid Leukemia and Define New Therapeutic Targets. Cancers 2020, 12, 2874. https://doi.org/10.3390/cancers12102874
Gabellier L, Bret C, Bossis G, Cartron G, Moreaux J. DNA Repair Expression Profiling to Identify High-Risk Cytogenetically Normal Acute Myeloid Leukemia and Define New Therapeutic Targets. Cancers. 2020; 12(10):2874. https://doi.org/10.3390/cancers12102874
Chicago/Turabian StyleGabellier, Ludovic, Caroline Bret, Guillaume Bossis, Guillaume Cartron, and Jérôme Moreaux. 2020. "DNA Repair Expression Profiling to Identify High-Risk Cytogenetically Normal Acute Myeloid Leukemia and Define New Therapeutic Targets" Cancers 12, no. 10: 2874. https://doi.org/10.3390/cancers12102874
APA StyleGabellier, L., Bret, C., Bossis, G., Cartron, G., & Moreaux, J. (2020). DNA Repair Expression Profiling to Identify High-Risk Cytogenetically Normal Acute Myeloid Leukemia and Define New Therapeutic Targets. Cancers, 12(10), 2874. https://doi.org/10.3390/cancers12102874






