Three-Dimensional Culture Systems in Gastric Cancer Research
Simple Summary
Abstract
1. Introduction
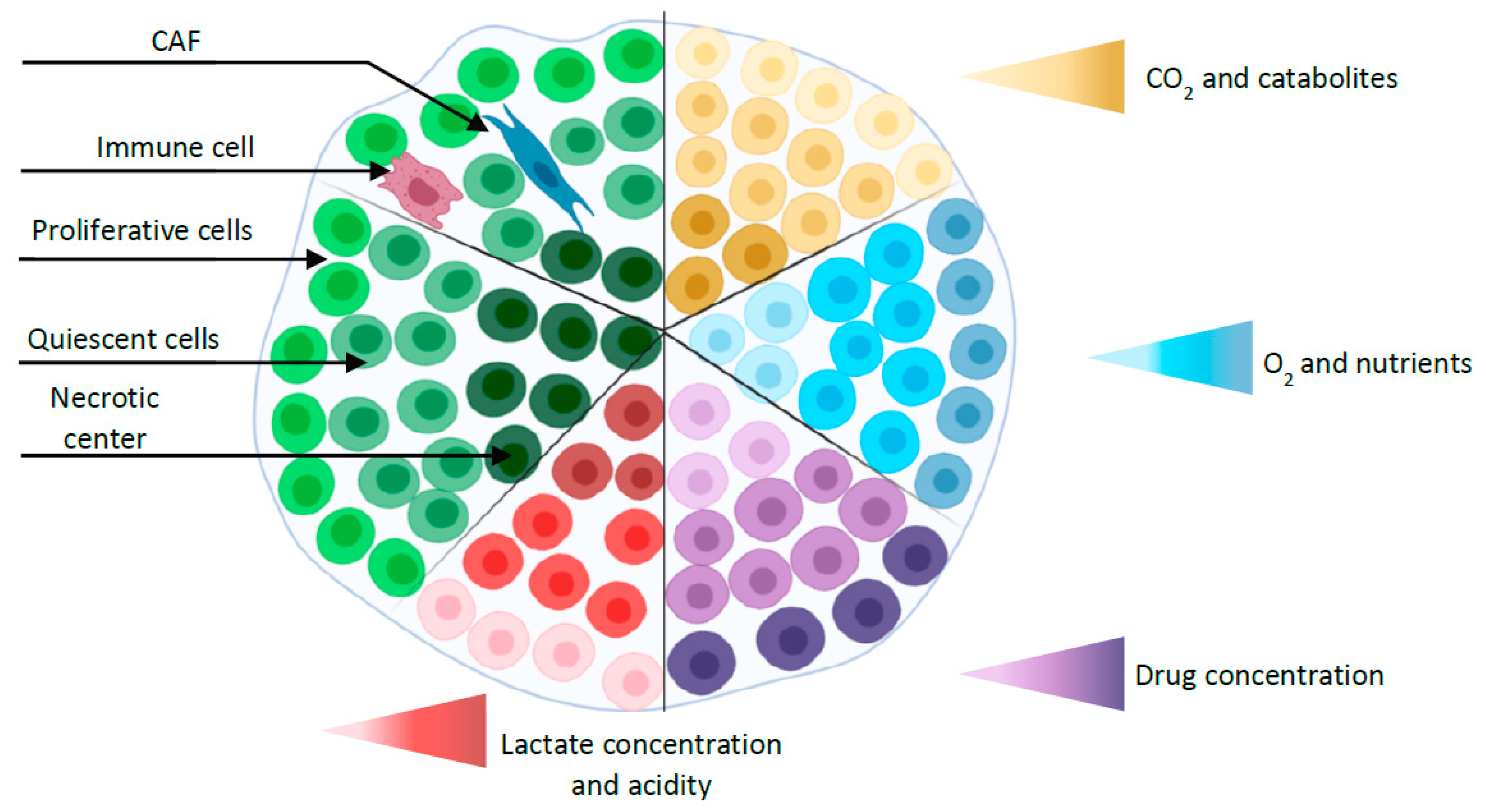
2. Three D Multicellular Tumor Spheroid Model
2.1. MCTS Production Methodologies
2.2. Applications of MCTS in Gastric Cancer
2.2.1. Gene Expression Profiling
2.2.2. Gastric Cancer Stem Cells: Biomarkers Identification
2.2.3. Drug Discovery
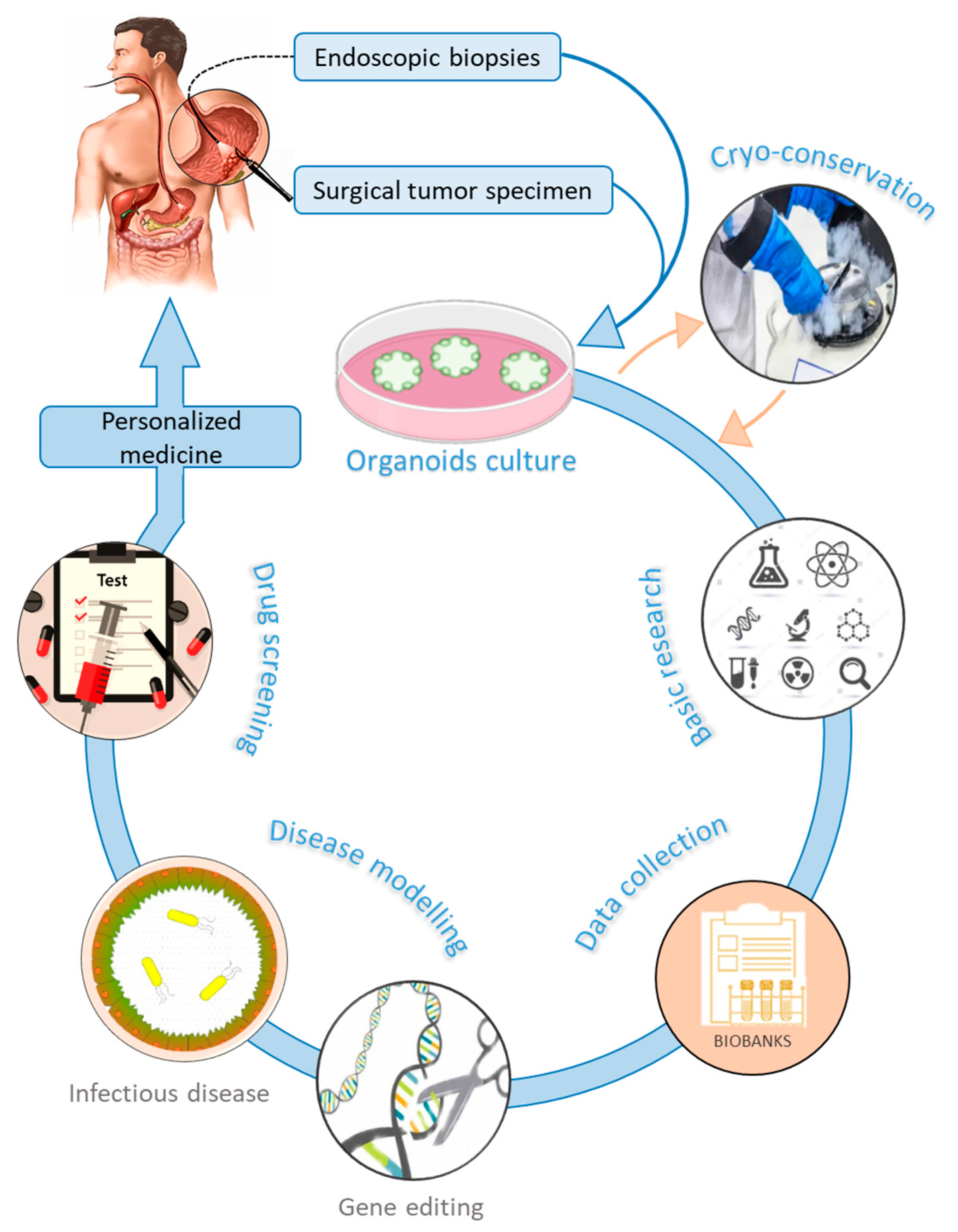
3. Gastric Organoids
3.1. Applications of Organoids in Gastric Cancer
3.1.1. Helicobacter Pylori Infection
3.1.2. Gastric Cancer Tumorigenesis
3.1.3. Drug Sensitivity and Personalized Medicine
4. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- McColl, K.E.L.; Going, J.J. Aetiology and classification of adenocarcinoma of the gastrooesophageal junction/cardia. Gut 2010, 59, 282–284. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Soerjomataram, I.; Ferlay, J.; Forman, D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut 2015, 64, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Lauren, P. The Two Histological Main Types of Gastric Carcinoma: Diffuse and So-Called Intestinal-Type Carcinoma. an Attempt At a Histo-Clinical Classification. Acta Pathol. Microbiol. Scand. 1965, 64, 31–49. [Google Scholar] [CrossRef] [PubMed]
- Venerito, M.; Vasapolli, R.; Rokkas, T.; Malfertheiner, P. Gastric cancer: Epidemiology, prevention, and therapy. Helicobacter 2018, 23, e12518. [Google Scholar] [CrossRef]
- Hu, B.; Hajj, N.E.; Sittler, S.; Lammert, N.; Barnes, R.; Meloni-Ehrig, A. Gastric cancer: Classification, histology and application of molecular pathology. J. Gastrointest. Oncol. 2012, 3, 251–261. [Google Scholar]
- Bass, A.J.; Thorsson, V.; Shmulevich, I.; Reynolds, S.M.; Miller, M.; Bernard, B.; Hinoue, T.; Laird, P.W.; Curtis, C.; Shen, H.; et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar]
- Kim, J.; Bowlby, R.; Mungall, A.J.; Robertson, A.G.; Odze, R.D.; Cherniack, A.D.; Shih, J.; Pedamallu, C.S.; Cibulskis, C.; Dunford, A.; et al. Integrated genomic characterization of oesophageal carcinoma. Nature 2017, 541, 169–174. [Google Scholar]
- Hohenberger, P.; Gretschel, S. Gastric cancer. Lancet 2003, 362, 305–315. [Google Scholar] [CrossRef]
- Cunningham, D.; Allum, W.H.; Stenning, S.P.; Thompson, J.N.; Van De Velde, C.J.H.; Nicolson, M.; Scarffe, J.H.; Lofts, F.J.; Falk, S.J.; Iveson, T.J.; et al. Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. N. Engl. J. Med. 2006, 355, 11–20. [Google Scholar] [CrossRef]
- Al-Batran, S.E.; Hofheinz, R.D.; Pauligk, C.; Kopp, H.G.; Haag, G.M.; Luley, K.B.; Meiler, J.; Homann, N.; Lorenzen, S.; Schmalenberg, H.; et al. Histopathological regression after neoadjuvant docetaxel, oxaliplatin, fluorouracil, and leucovorin versus epirubicin, cisplatin, and fluorouracil or capecitabine in patients with resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4-AIO): Results from the phase 2 part of a multicentre, open-label, randomised phase 2/3 trial. Lancet Oncol. 2016, 17, 1697–1708. [Google Scholar]
- Vanhoefer, U.; Rougier, P.; Wilke, H.; Ducreux, M.P.; Lacave, A.J.; Van Cutsem, E.; Planker, M.; Dos Santos, J.G.; Piedbois, P.; Paillot, B.; et al. Final Results of a Randomized Phase III Trial of Sequential High-Dose Methotrexate, Fluorouracil, and Doxorubicin Versus Etoposide, Leucovorin, and Fluorouracil Versus Infusional Fluorouracil and Cisplatin in Advanced Gastric Cancer: A Trial of the European Organization for Research and Treatment of Cancer Gastrointestinal Tract Cancer Cooperative Group. J. Clin. Oncol. 2000, 18, 2648–2657. [Google Scholar] [PubMed]
- Bang, Y.J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 2010, 376, 687–697. [Google Scholar] [CrossRef]
- Fuchs, C.S.; Tomasek, J.; Yong, C.J.; Dumitru, F.; Passalacqua, R.; Goswami, C.; Safran, H.; Dos Santos, L.V.; Aprile, G.; Ferry, D.R.; et al. Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): An international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet 2014, 383, 31–39. [Google Scholar] [CrossRef]
- Kim, S.T.; Cristescu, R.; Bass, A.J.; Kim, K.M.; Odegaard, J.I.; Kim, K.; Liu, X.Q.; Sher, X.; Jung, H.; Lee, M.; et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat. Med. 2018, 24, 1449–1458. [Google Scholar] [CrossRef]
- Thrift, A.P.; El-Serag, H.B. Burden of Gastric Cancer. Clin. Gastroenterol. Hepatol. 2020, 18, 534–542. [Google Scholar] [CrossRef]
- Gillet, J.-P.; Varma, S.; Gottesman, M.M. the Clinical relevance of Cancer Cell Lines. J. Natl. Cancer Inst. 2013, 105, 452–458. [Google Scholar] [CrossRef]
- Sensi, F.; D’Angelo, E.; D’Aronco, S.; Molinaro, R.; Agostini, M. Preclinical three-dimensional colorectal cancer model: The next generation of in vitro drug efficacy evaluation. J. Cell. Physiol. 2018, 234, 181–191. [Google Scholar] [CrossRef]
- Anton, D.; Burckel, H.; Josset, E.; Noel, G. Three-dimensional cell culture: A breakthrough in vivo. Int. J. Mol. Sci. 2015, 16, 5517–5527. [Google Scholar] [CrossRef]
- Fayad, W.; Brnjic, S.; Berglind, D.; Blixt, S.; Shoshan, M.C.; Berndtsson, M.; Olofsson, M.H.; Linder, S. Restriction of cisplatin induction of acute apoptosis to a subpopulation of cells in a three-dimensional carcinoma culture model. Int. J. Cancer 2009, 125, 2450–2455. [Google Scholar] [CrossRef]
- D’angelo, E.; Natarajan, D.; Sensi, F.; Ajayi, O.; Fassan, M.; Mammano, E.; Pilati, P.; Pavan, P.; Bresolin, S.; Preziosi, M.; et al. Patient-derived scaffolds of colorectal cancer metastases as an organotypic 3D model of the liver metastatic microenvironment. Cancers 2020, 12, 364. [Google Scholar] [CrossRef] [PubMed]
- Katt, M.E.; Placone, A.L.; Wong, A.D.; Xu, Z.S.; Searson, P.C. In vitro tumor models: Advantages, disadvantages, variables, and selecting the right platform. Front. Bioeng. Biotechnol. 2016, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, R.M.; Sordat, B.; Bamat, J.; Gabbert, H.; Bourrat, B.; Mueller-Klieser, W. Oxygenation and Differentiation in Multicellular Spheroids of Human Colon Carcinoma. Cancer Res. 1986, 46, 5320–5329. [Google Scholar] [PubMed]
- Drost, J.; Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 2018, 18, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Zanoni, M.; Cortesi, M.; Zamagni, A.; Arienti, C.; Pignatta, S.; Tesei, A. Modeling neoplastic disease with spheroids and organoids. J. Hematol. Oncol. 2020, 13, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, L.; Quintana, L.G.; Lopes, D.C.F.; Vidal, A.F.; Pereira, A.L.; D’Araujo Pinto, L.C.; De Jesus Viana Pinheiro, J.; Khayat, A.S.; Goulart, L.R.; Burbano, R.; et al. APC gene is modulated by hsa-miR-135b-5p in both diffuse and intestinal gastric cancer subtypes. BMC Cancer 2018, 18, 1055. [Google Scholar] [CrossRef] [PubMed]
- Sha, H.; Zou, Z.; Xin, K.; Bian, X.; Cai, X.; Lu, W.; Chen, J.; Chen, G.; Huang, L.; Blair, A.M.; et al. Tumor-penetrating peptide fused EGFR single-domain antibody enhances cancer drug penetration into 3D multicellular spheroids and facilitates effective gastric cancer therapy. J. Control. Release 2014, 200, 188–200. [Google Scholar] [CrossRef]
- Bekaii-Saab, T.; El-Rayes, B. Identifying and targeting cancer stem cells in the treatment of gastric cancer. Cancer 2017, 123, 1303–1312. [Google Scholar] [CrossRef]
- Sethi, N.S.; Kikuchi, O.; Duronio, G.N.; Stachler, M.D.; McFarland, J.M.; Ferrer-Luna, R.; Zhang, Y.; Bao, C.; Bronson, R.; Patil, D.; et al. Early TP53 alterations engage environmental exposures to promote gastric premalignancy in an integrative mouse model. Nat. Genet. 2020, 52, 219–230. [Google Scholar] [CrossRef]
- McCracken, K.W.; Catá, E.M.; Crawford, C.M.; Sinagoga, K.L.; Schumacher, M.; Rockich, B.E.; Tsai, Y.-H.; Mayhew, C.N.; Spence, J.R.; Zavros, Y.; et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 2014, 516, 400–404. [Google Scholar] [CrossRef]
- Friedrich, J.; Seidel, C.; Ebner, R.; Kunz-Schughart, L.A. Spheroid-based drug screen: Considerations and practical approach. Nat. Protoc. 2009, 4, 309–324. [Google Scholar] [CrossRef]
- Liao, C.P.; Adisetiyo, H.; Liang, M.; Roy-Burman, P. Cancer-associated fibroblasts enhance the gland-forming capability of prostate cancer stem cells. Cancer Res. 2010, 70, 7294–7303. [Google Scholar] [CrossRef] [PubMed]
- Togo, S.; Polanska, U.; Horimoto, Y.; Orimo, A. Carcinoma-Associated Fibroblasts Are a Promising Therapeutic Target. Cancers 2013, 5, 149–169. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.Y.; Kang, W.; Han, J.Y.; Min, S.; Kang, J.; Lee, A.; Kwon, J.Y.; Lee, C.; Park, H. An integrative approach to precision cancer medicine using patient-derived xenografts. Mol. Cells 2016, 39, 77–86. [Google Scholar] [PubMed]
- Jung, J.; Seol, H.S.; Chang, S. The generation and application of patient-derived xenograft model for cancer research. Cancer Res. Treat. 2018, 50, 1–10. [Google Scholar] [CrossRef]
- Byrne, A.T.; Alférez, D.G.; Amant, F.; Annibali, D.; Arribas, J.; Biankin, A.V.; Bruna, A.; Budinská, E.; Caldas, C.; Chang, D.K.; et al. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat. Rev. Cancer 2017, 17, 254–268. [Google Scholar] [CrossRef]
- Lin, M.; Gao, M.; Cavnar, M.J.; Kim, J. Utilizing gastric cancer organoids to assess tumor biology and personalize medicine. World J. Gastrointest. Oncol. 2019, 11, 509–517. [Google Scholar] [CrossRef]
- Aberle, M.R.; Burkhart, R.A.; Tiriac, H.; Olde Damink, S.W.M.; Dejong, C.H.C.; Tuveson, D.A.; van Dam, R.M. Patient-derived organoid models help define personalized management of gastrointestinal cancer. Br. J. Surg. 2018, 105, e48–e60. [Google Scholar] [CrossRef]
- Lau, H.C.H.; Kranenburg, O.; Xiao, H.; Yu, J. Organoid models of gastrointestinal cancers in basic and translational research. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 203–222. [Google Scholar] [CrossRef]
- Van De Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; Van Houdt, W.; Van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef]
- Schlaermann, P.; Toelle, B.; Berger, H.; Schmidt, S.C.; Glanemann, M.; Ordemann, J.; Bartfeld, S.; Mollenkopf, H.J.; Meyer, T.F. A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut 2016, 65, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Breslin, S.; O’Driscoll, L. Three-dimensional cell culture: The missing link in drug discovery. Drug Discov. Today 2013, 18, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Cancer Tissue Engineering—New Perspectives in Understand Google Scholar. Available online: https://scholar.google.com/scholar?hl=en&as_sdt=0%2C5&q=Cancer+tissue+engineering—New+perspectives+in+understanding+the+biology+of+solid+tumours—A+critical+review&btnG= (accessed on 11 September 2020).
- Fischbach, C.; Chen, R.; Matsumoto, T.; Schmelzle, T.; Brugge, J.S.; Polverini, P.J.; Mooney, D.J. Engineering tumors with 3D scaffolds. Nat. Methods 2007, 4, 855–860. [Google Scholar] [CrossRef]
- De-Souza, A.S.C.; Costa-Casagrande, T.A. Animal models for colorectal cancer. ABCD. Arq. Bras. Cir. Dig. 2018, 31. [Google Scholar] [CrossRef] [PubMed]
- Inch, W.; Credie, J.; Sutherland, R. Growth of nodular carcinomas in rodents compared with multi-cell spheroids in tissue culture. Subj. Strain Bibliogr. 1970, 34, 271–282. [Google Scholar]
- Hickman, J.A.; Graeser, R.; de Hoogt, R.; Vidic, S.; Brito, C.; Gutekunst, M.; van der Kuip, H. Imi Predect consortium Three-dimensional models of cancer for pharmacology and cancer cell biology: Capturing tumor complexity in vitro/ex vivo. Biotechnol. J. 2014, 9, 1115–1128. [Google Scholar] [CrossRef] [PubMed]
- Hirschhaeuser, F.; Menne, H.; Dittfeld, C.; West, J.; Mueller-Klieser, W.; Kunz-Schughart, L.A. Multicellular tumor spheroids: An underestimated tool is catching up again. J. Biotechnol. 2010, 148, 3–15. [Google Scholar] [CrossRef]
- Mehta, G.; Hsiao, A.Y.; Ingram, M.; Luker, G.D.; Takayama, S. Opportunities and challenges for use of tumor spheroids as models to test drug delivery and efficacy. J. Control. Release 2012, 164, 192–204. [Google Scholar] [CrossRef]
- Ghosh, S.; Spagnoli, G.C.; Martin, I.; Ploegert, S.; Demougin, P.; Heberer, M.; Reschner, A. Three-dimensional culture of melanoma cells profoundly affects gene expression profile: A high density oligonucleotide array study. J. Cell. Physiol. 2005, 204, 522–531. [Google Scholar] [CrossRef]
- Krueger, S.; Kalinski, T.; Wolf, H.; Kellner, U.; Roessner, A. Interactions between human colon carcinoma cells, fibroblasts and monocytic cells in coculture Regulation of cathepsin B expression and invasiveness. Cancer Lett. 2005, 223, 313–322. [Google Scholar] [CrossRef]
- Lamichhane, S.P.; Arya, N.; Kohler, E.; Xiang, S.; Christensen, J.; Shastri, V.P. Recapitulating epithelial tumor microenvironment in vitro using three dimensional tri-culture of human epithelial, endothelial, and mesenchymal cells. BMC Cancer 2016, 16, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sebrell, T.A.; Hashimi, M.; Sidar, B.; Wilkinson, R.A.; Kirpotina, L.; Quinn, M.T.; Malkoç, Z.; Taylor, P.J.; Wilking, J.N.; Bimczok, D. A Novel Gastric Spheroid Co-culture Model Reveals Chemokine-Dependent Recruitment of Human Dendritic Cells to the Gastric Epithelium. Cell. Mol. Gastroenterol. Hepatol. 2019, 8, 157–171.e3. [Google Scholar] [CrossRef]
- Ravi, M.; Paramesh, V.; Kaviya, S.R.; Anuradha, E.; Paul Solomon, F.D. 3D cell culture systems: Advantages and applications. J. Cell. Physiol. 2015, 230, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Place, E.S.; George, J.H.; Williams, C.K.; Stevens, M.M. Synthetic polymer scaffolds for tissue engineering. Chem. Soc. Rev. 2009, 38, 1139–1151. [Google Scholar] [CrossRef] [PubMed]
- Estrada, M.F.; Rebelo, S.P.; Davies, E.J.; Pinto, M.T.; Pereira, H.; Santo, V.E.; Smalley, M.J.; Barry, S.T.; Gualda, E.J.; Alves, P.M.; et al. Modelling the tumour microenvironment in long-term microencapsulated 3D co-cultures recapitulates phenotypic features of disease progression. Biomaterials 2016, 78, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Yuhas, J.M.; Li, A.P.; Martinez, A.O.; Ladman, A.J. A Simplified Method for Production and Growth of Multicellular Tumor Spheroids. Cancer Res. 1977, 37, 3639–3643. [Google Scholar]
- Yamada, K.M.; Cukierman, E. Modeling Tissue Morphogenesis and Cancer in 3D. Cell 2007, 130, 601–610. [Google Scholar] [CrossRef]
- Lin, R.-Z.; Chang, H.-Y. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol. J. 2008, 3, 1172–1184. [Google Scholar] [CrossRef]
- Jørgensen, A.; Young, J.; Nielsen, J.E.; Joensen, U.N.; Toft, B.G.; Rajpert-De Meyts, E.; Loveland, K.L. Hanging drop cultures of human testis and testis cancer samples: A model used to investigate activin treatment effects in a preserved niche. Br. J. Cancer 2014, 110, 2604–2614. [Google Scholar] [CrossRef]
- Kurosawa, H. Methods for inducing embryoid body formation: In vitro differentiation system of embryonic stem cells. J. Biosci. Bioeng. 2007, 103, 389–398. [Google Scholar] [CrossRef]
- Fennema, E.; Rivron, N.; Rouwkema, J.; van Blitterswijk, C.; De Boer, J. Spheroid culture as a tool for creating 3D complex tissues. Trends Biotechnol. 2013, 31, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, X.; Sha, H.; Zhang, L.; Bian, X.; Han, X.; Liu, B. STRAIL-iRGD is a promising therapeutic agent for gastric cancer treatment. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mayer, B.; Klement, G.; Kaneko, M.; Man, S.; Jothy, S.; Rak, J.; Kerbel, R.S. Multicellular Gastric Cancer Spheroids Recapitulate Growth Pattern and Differentiation Phenotype of Human Gastric Carcinomas. Gastroenterology 2001, 121, 839–852. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Pan, J.; Geng, Q.; Wang, G. Lnc RNA MALAT 1 increases the stemness of gastric cancer cells via enhancing SOX 2 mRNA stability. FEBS Open Bio 2019, 9, 1212–1222. [Google Scholar] [CrossRef]
- Song, H.; Shi, L.; Xu, Y.; Xu, T.; Fan, R.; Cao, M.; Xu, W.; Song, J. BRD4 promotes the stemness of gastric cancer cells via attenuating miR-216a-3p-mediated inhibition of Wnt/β-catenin signaling. Eur. J. Pharmacol. 2019, 852, 189–197. [Google Scholar] [CrossRef]
- Liu, J.; Ma, L.; Wang, Z.; Wang, L.; Liu, C.; Chen, R.; Zhang, J. MicroRNA Expression Profile of Gastric Cancer Stem Cells in the MKN-45 Cancer Cell Line. Acta Biochim. Biophys. Sin. 2014, 46, 92–99. [Google Scholar] [CrossRef]
- Kelm, J.M.; Timmins, N.E.; Brown, C.J.; Fussenegger, M.; Nielsen, L.K. Method for generation of homogeneous multicellular tumor spheroids applicable to a wide variety of cell types. Biotechnol. Bioeng. 2003, 83, 173–180. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, Z.; Fan, Y.; Zheng, Q. Expression of CD133 in SW620 colorectal cancer cells is modulated by the microenvironment. Oncol. Lett. 2012, 4, 75–79. [Google Scholar] [CrossRef]
- Al-Husaini, K.; Elkamel, E.; Han, X.; Chen, P. Therapeutic potential of a cell penetrating peptide (CPP, NP1) mediated siRNA delivery: Evidence in 3D spheroids of colon cancer cells. Can. J. Chem. Eng. 2020, 98, 1240–1254. [Google Scholar] [CrossRef]
- McLean, M.H.; El-Omar, E.M. Genetics of gastric cancer. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 664–674. [Google Scholar] [CrossRef]
- Baker, B.M.; Chen, C.S. Deconstructing the third dimension-how 3D culture microenvironments alter cellular cues. J. Cell Sci. 2012, 125, 3015–3024. [Google Scholar] [CrossRef] [PubMed]
- Ishimoto, T.; Nagano, O.; Yae, T.; Tamada, M.; Motohara, T.; Oshima, H.; Oshima, M.; Ikeda, T.; Asaba, R.; Yagi, H.; et al. CD44 Variant Regulates Redox Status in Cancer Cells by Stabilizing the xCT Subunit of System xc- and Thereby Promotes Tumor Growth. Cancer Cell 2011, 19, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Oue, N.; Mukai, S.; Imai, T.; Pham, T.T.B.; Oshima, T.; Sentani, K.; Sakamoto, N.; Yoshida, K.; Yasui, W. Induction of KIFC1 expression in gastric cancer spheroids. Oncol. Rep. 2016, 36, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Oue, N.; Sentani, K.; Sakamoto, N.; Uraoka, N.; Yasui, W. Molecular carcinogenesis of gastric cancer: Lauren classification, mucin phenotype expression, and cancer stem cells. Int. J. Clin. Oncol. 2019, 24, 771–778. [Google Scholar] [CrossRef] [PubMed]
- Imai, T.; Oue, N.; Nishioka, M.; Mukai, S.; Oshima, T.; Sakamoto, N.; Sentani, K.; Matsusaki, K.; Yoshida, K.; Yasui, W. Overexpression of KIF11 in Gastric Cancer with Intestinal Mucin Phenotype. Pathobiology 2017, 84, 16–24. [Google Scholar] [CrossRef]
- Kobayashi, G.; Sentani, K.; Hattori, T.; Yamamoto, Y.; Imai, T.; Sakamoto, N.; Kuraoka, K.; Oue, N.; Sasaki, N.; Taniyama, K.; et al. Clinicopathological significance of claspin overexpression and its association with spheroid formation in gastric cancer. Hum. Pathol. 2019, 84, 8–17. [Google Scholar] [CrossRef]
- Lee, J.W.; Sung, J.S.; Park, Y.S.; Chung, S.; Kim, Y.H. Isolation of spheroid-forming single cells from gastric cancer cell lines: Enrichment of cancer stem-like cells. Biotechniques 2018, 65, 197–203. [Google Scholar] [CrossRef]
- Lee, J.W.; Sung, J.S.; Park, Y.S.; Chung, S.; Kim, Y.H. Identification of different gene expressions between diffuse- and intestinal-type spheroid-forming gastric cancer cells. Gastric Cancer 2019, 22, 967–979. [Google Scholar] [CrossRef]
- Birgersdotter, A.; Sandberg, R.; Ernberg, I. Gene expression perturbation in vitro A growing case for three-dimensional (3D) culture systems. Semin. Cancer Biol. 2005, 15, 405–412. [Google Scholar] [CrossRef]
- Branco da Cunha, C.; Klumpers, D.D.; Koshy, S.T.; Weaver, J.C.; Chaudhuri, O.; Seruca, R.; Carneiro, F.; Granja, P.L.; Mooney, D.J. CD44 alternative splicing in gastric cancer cells is regulated by culture dimensionality and matrix stiffness. Biomaterials 2016, 98, 152–162. [Google Scholar] [CrossRef]
- Da Cunha, C.B.; Oliveira, C.; Wen, X.; Gomes, B.; Sousa, S.; Suriano, G.; Grellier, M.; Huntsman, D.G.; Carneiro, F.; Granja, P.L.; et al. De novo expression of CD44 variants in sporadic and hereditary gastric cancer. Lab. Investig. 2010, 90, 1604–1614. [Google Scholar] [CrossRef] [PubMed]
- Farnie, G.; Clarke, R.B. Mammary stem cells and breast cancer Role of notch signalling. Stem Cell Rev. 2007, 3, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Dubrovska, A.; Kim, S.; Salamone, R.J.; Walker, J.R.; Maira, S.M.; García-Echeverría, C.; Schultz, P.G.; Reddy, V.A. The role of PTEN/Akt/PI3K signaling in the maintenance and viability of prostate cancer stem-like cell populations. Proc. Natl. Acad. Sci. USA 2009, 106, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Alman, B.A. Side population cells in human cancers. Cancer Lett. 2008, 268, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Clarke, M.F.; Dick, J.E.; Dirks, P.B.; Eaves, C.J.; Jamieson, C.H.M.; Jones, D.L.; Visvader, J.; Weissman, I.L.; Wahl, G.M. Cancer stem cells Perspectives on current status and future directions: AACR workshop on cancer stem cells. Cancer Res. 2006, 66, 9339–9344. [Google Scholar] [CrossRef]
- Zhang, X.; Hua, R.; Wang, X.; Huang, M.; Gan, L.; Wu, Z.; Zhang, J.; Wang, H.; Cheng, Y.; Li, J.; et al. Identification of stem-like cells and clinical significance of candidate stem cell markers in gastric cancer. Oncotarget 2016, 7, 9815–9831. [Google Scholar] [CrossRef] [PubMed]
- Rocco, A.; Liguori, E.; Pirozzi, G.; Tirino, V.; Compare, D.; Franco, R.; Tatangelo, F.; Palaia, R.; D’Armiento, F.P.; Pollastrone, G.; et al. CD133 and CD44 Cell surface markers do not identify cancer stem cells in primary human gastric tumors. J. Cell. Physiol. 2012, 227, 2686–2693. [Google Scholar] [CrossRef]
- Takaishi, S.; Okumura, T.; Tu, S.; Wang, S.S.W.; Shibata, W.; Vigneshwaran, R.; Gordon, S.A.K.; Shimada, Y.; Wang, T.C. Identification of Gastric Cancer Stem Cells Using the Cell Surface Marker CD44. Stem Cells 2009, 27, 1006–1020. [Google Scholar] [CrossRef]
- Han, M.E.; Jeon, T.Y.; Hwang, S.H.; Lee, Y.S.; Kim, H.J.; Shim, H.E.; Yoon, S.; Baek, S.Y.; Kim, B.S.; Kang, C.D.; et al. Cancer spheres from gastric cancer patients provide an ideal model system for cancer stem cell research. Cell. Mol. Life Sci. 2011, 68, 3589–3605. [Google Scholar] [CrossRef]
- Liu, J.; Ma, L.; Xu, J.; Liu, C.; Zhang, J.; Liu, J.; Chen, R.; Zhou, Y. Spheroid body-forming cells in the human gastric cancer cell line MKN-45 possess cancer stem cell properties. Int. J. Oncol. 2013, 42, 453–459. [Google Scholar] [CrossRef]
- Liu, J.; Zhou, Y.; Ma, L.; Xu, J.; Liu, C.; Zhang, J.; Liu, J.; Chen, R.; Qian, H. Co-expression of CD44 and ABCG2 in spheroid body-forming cells of gastric cancer cell line MKN45. Hepatogastroenterology 2013, 60, 975–980. [Google Scholar] [PubMed]
- Jiang, J.; Zhang, Y.; Chuai, S.; Wang, Z.; Zheng, D.; Xu, F.; Zhang, Y.; Li, C.; Liang, Y.; Chen, Z. Trastuzumab (herceptin) targets gastric cancer stem cells characterized by CD90 phenotype. Oncogene 2012, 31, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Zhang, Y.; Wang, S.; Zhou, J.; Xu, S. Sox2 enhances the tumorigenicity and chemoresistance of cancer stem-like cells derived from gastric cancer. J. Biomed. Res. 2012, 26, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Yue, W.; Wei, B.; Wang, N.; Li, T.; Guan, L.; Shi, S.; Zeng, Q.; Pei, X.; Chen, L. Sonic hedgehog pathway is essential for maintenance of cancer stem-like cells in human gastric cancer. PLoS ONE 2011, 6, e17687. [Google Scholar] [CrossRef] [PubMed]
- Ohkuma, M.; Haraguchi, N.; Ishii, H.; Mimori, K.; Tanaka, F.; Kim, H.M.; Shimomura, M.; Hirose, H.; Yanaga, K.; Mori, M. Absence of CD71 transferrin receptor characterizes human gastric adenosquamous carcinoma stem cells. Ann. Surg. Oncol. 2012, 19, 1357–1364. [Google Scholar] [CrossRef]
- Yoon, C.; Cho, S.J.; Aksoy, B.A.; Park, D.J.; Schultz, N.; Ryeom, S.W.; Yoon, S.S. Chemotherapy resistance in diffuse-type gastric adenocarcinoma is mediated by RhoA activation in cancer stem-like cells. Clin. Cancer Res. 2016, 22, 971–983. [Google Scholar] [CrossRef]
- Atashzar, M.R.; Baharlou, R.; Karami, J.; Abdollahi, H.; Rezaei, R.; Pourramezan, F.; Zoljalali Moghaddam, S.H. Cancer stem cells: A review from origin to therapeutic implications. J. Cell. Physiol. 2020, 235, 790–803. [Google Scholar] [CrossRef]
- Costa, E.C.; Moreira, A.F.; de Melo-Diogo, D.; Gaspar, V.M.; Carvalho, M.P.; Correia, I.J. 3D tumor spheroids: An overview on the tools and techniques used for their analysis. Biotechnol. Adv. 2016, 34, 1427–1441. [Google Scholar] [CrossRef]
- Ocana, A.; Pandiella, A.; Siu, L.L.; Tannock, I.F. Preclinical development of molecular-targeted agents for cancer. Nat. Rev. Clin. Oncol. 2011, 8, 200–209. [Google Scholar] [CrossRef]
- Longati, P.; Jia, X.; Eimer, J.; Wagman, A.; Witt, M.R.; Rehnmark, S.; Verbeke, C.; Toftgård, R.; Löhr, M.; Heuchel, R.L. 3D pancreatic carcinoma spheroids induce a matrix-rich, chemoresistant phenotype offering a better model for drug testing. BMC Cancer 2013, 13, 1–13. [Google Scholar] [CrossRef]
- Courtois, S.; Durán, R.V.; Giraud, J.; Sifré, E.; Izotte, J.; Mégraud, F.; Lehours, P.; Varon, C.; Bessède, E. Metformin targets gastric cancer stem cells. Eur. J. Cancer 2017, 84, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Akrami, H.; Aminzadeh, S.; Fallahi, H. Inhibitory effect of ibuprofen on tumor survival and angiogenesis in gastric cancer cell. Tumor Biol. 2015, 36, 3237–3243. [Google Scholar] [CrossRef] [PubMed]
- Akrami, H.; Moradi, B.; Borzabadi Farahani, D.; Mehdizadeh, K. Ibuprofen reduces cell proliferation through inhibiting Wnt/β catenin signaling pathway in gastric cancer stem cells. Cell Biol. Int. 2018, 42, 949–958. [Google Scholar] [CrossRef]
- Nishikawa, S.; Konno, M.; Hamabe, A.; Hasegawa, S.; Kano, Y.; Ohta, K.; Fukusumi, T.; Sakai, D.; Kudo, T.; Haraguchi, N.; et al. Aldehyde dehydrogenasehigh gastric cancer stem cells are resistant to chemotherapy. Int. J. Oncol. 2013, 42, 1437–1442. [Google Scholar] [CrossRef] [PubMed]
- Koh, J.S.; Joo, M.K.; Park, J.J.; Yoo, H.S.; Choi, B.I.; Lee, B.J.; Chun, H.J.; Lee, S.W. Inhibition of STAT3 in gastric cancer: Role of pantoprazole as SHP-1 inducer. Cell Biosci. 2018, 8, 50. [Google Scholar] [CrossRef]
- Minchinton, A.I.; Tannock, I.F. Drug penetration in solid tumours. Nat. Rev. Cancer 2006, 6, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Sha, H.; Li, R.; Bian, X.; Liu, Q.; Xie, C.; Xin, X.; Kong, W.; Qian, X.; Jiang, X.; Hu, W.; et al. A tumor-penetrating recombinant protein anti-EGFR-iRGD enhance efficacy of paclitaxel in 3D multicellular spheroids and gastric cancer in vivo. Eur. J. Pharm. Sci. 2015, 77, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Ma, X.; Wang, H.; Xi, Y.; Qian, M.; Yang, W.; Luo, D.; Fan, L.; Xia, X.; Zhou, J.; et al. The novel fusion protein sTRAIL-TMTP1 exhibits a targeted inhibition of primary tumors and metastases. J. Mol. Med. 2014, 92, 165–175. [Google Scholar] [CrossRef]
- Sherman, H.; Gitschier, H.J.; Rossi, A.E. A novel three-dimensional immune oncology model for high-throughput testing of tumoricidal activity. Front. Immunol. 2018, 9, 857. [Google Scholar] [CrossRef]
- Zhou, S.; Zhu, M.; Meng, F.; Shao, J.; Xu, Q.; Wei, J.; Liu, B. Evaluation of PD-1 blockade using a multicellular tumor spheroid model. Am. J. Transl. Res. 2019, 11, 7471–7478. [Google Scholar]
- Salmaninejad, A.; Valilou, S.F.; Shabgah, A.G.; Aslani, S.; Alimardani, M.; Pasdar, A.; Sahebkar, A. PD-1/PD-L1 pathway: Basic biology and role in cancer immunotherapy. J. Cell. Physiol. 2019, 234, 16824–16837. [Google Scholar] [CrossRef] [PubMed]
- Langhans, S.A. Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front. Pharmacol. 2018, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Pattabiraman, D.R.; Weinberg, R.A. Tackling the cancer stem cells-what challenges do they pose? Nat. Rev. Drug Discov. 2014, 13, 497–512. [Google Scholar] [CrossRef] [PubMed]
- Das, V.; Fürst, T.; Gurská, S.; Džubák, P.; Hajdúch, M. Reproducibility of uniform spheroid formation in 384-well plates: The effect of medium evaporation. J. Biomol. Screen. 2016, 21, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Eiraku, M.; Sasai, Y. Self-formation of layered neural structures in three-dimensional culture of ES cells. Curr. Opin. Neurobiol. 2012, 22, 768–777. [Google Scholar] [CrossRef] [PubMed]
- Werner, K.; Weitz, J.; Stange, D.E. Organoids as Model Systems for Gastrointestinal Diseases: Tissue Engineering Meets Genetic Engineering. Curr. Pathobiol. Rep. 2016, 4, 1–9. [Google Scholar] [CrossRef]
- Neal, J.T.; Li, X.; Zhu, J.; Giangarra, V.; Grzeskowiak, C.L.; Ju, J.; Liu, I.H.; Chiou, S.H.; Salahudeen, A.A.; Smith, A.R.; et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell 2018, 175, 1972–1988.e16. [Google Scholar] [CrossRef]
- Sato, T.; Stange, D.E.; Ferrante, M.; Vries, R.G.J.; Van Es, J.H.; Van Den Brink, S.; Van Houdt, W.J.; Pronk, A.; Van Gorp, J.; Siersema, P.D.; et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 2011, 141, 1762–1772. [Google Scholar] [CrossRef]
- Barker, N.; Van Es, J.H.; Kuipers, J.; Kujala, P.; Van Den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007, 449, 1003–1007. [Google Scholar] [CrossRef]
- Sato, T.; Vries, R.G.; Snippert, H.J.; Van De Wetering, M.; Barker, N.; Stange, D.E.; Van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Boj, S.F.; Hwang, C.I.; Baker, L.A.; Chio, I.I.C.; Engle, D.D.; Corbo, V.; Jager, M.; Ponz-Sarvise, M.; Tiriac, H.; Spector, M.S.; et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 2015, 160, 324–338. [Google Scholar] [CrossRef]
- Huch, M.; Gehart, H.; Van Boxtel, R.; Hamer, K.; Blokzijl, F.; Verstegen, M.M.A.; Ellis, E.; Van Wenum, M.; Fuchs, S.A.; De Ligt, J.; et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 2015, 160, 299–312. [Google Scholar] [CrossRef]
- DeWard, A.D.; Cramer, J.; Lagasse, E. Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell Rep. 2014, 9, 701–711. [Google Scholar] [CrossRef]
- Chua, C.W.; Shibata, M.; Lei, M.; Toivanen, R.; Barlow, L.J.; Bergren, S.K.; Badani, K.K.; McKiernan, J.M.; Benson, M.C.; Hibshoosh, H.; et al. Single luminal epithelial progenitors can generate prostate organoids in culture. Nat. Cell Biol. 2014, 16, 951–961. [Google Scholar] [CrossRef]
- Wong, A.P.; Bear, C.E.; Chin, S.; Pasceri, P.; Thompson, T.O.; Huan, L.J.; Ratjen, F.; Ellis, J.; Rossant, J. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat. Biotechnol. 2012, 30, 876–882. [Google Scholar] [CrossRef]
- Sachs, N.; de Ligt, J.; Kopper, O.; Gogola, E.; Bounova, G.; Weeber, F.; Balgobind, A.V.; Wind, K.; Gracanin, A.; Begthel, H.; et al. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell 2018, 172, 373–386.e10. [Google Scholar] [CrossRef]
- Clevers, H. Modeling Development and Disease with Organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef] [PubMed]
- Barker, N.; Huch, M.; Kujala, P.; van de Wetering, M.; Snippert, H.J.; van Es, J.H.; Sato, T.; Stange, D.E.; Begthel, H.; van den Born, M.; et al. Lgr5+ve Stem Cells Drive Self-Renewal in the Stomach and Build Long-Lived Gastric Units In Vitro. Cell Stem Cell 2010, 6, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Stange, D.E.; Koo, B.K.; Huch, M.; Sibbel, G.; Basak, O.; Lyubimova, A.; Kujala, P.; Bartfeld, S.; Koster, J.; Geahlen, J.H.; et al. XDifferentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell 2013, 155, 357. [Google Scholar] [CrossRef] [PubMed]
- Seidlitz, T.; Merker, S.R.; Rothe, A.; Zakrzewski, F.; Von Neubeck, C.; Grützmann, K.; Sommer, U.; Schweitzer, C.; Schölch, S.; Uhlemann, H.; et al. Human gastric cancer modelling using organoids. Gut 2019, 68, 207–217. [Google Scholar] [CrossRef]
- Gao, D.; Vela, I.; Sboner, A.; Iaquinta, P.J.; Karthaus, W.R.; Gopalan, A.; Dowling, C.; Wanjala, J.N.; Undvall, E.A.; Arora, V.K.; et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014, 159, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Lin, M.; Rao, M.; Thompson, H.; Hirai, K.; Choi, M.; Georgakis, G.V.; Sasson, A.R.; Bucobo, J.C.; Tzimas, D.; et al. Development of Patient-Derived Gastric Cancer Organoids from Endoscopic Biopsies and Surgical Tissues. Ann. Surg. Oncol. 2018, 25, 2767–2775. [Google Scholar] [CrossRef] [PubMed]
- Hughes, C.S.; Postovit, L.M.; Lajoie, G.A. Matrigel: A complex protein mixture required for optimal growth of cell culture. Proteomics 2010, 10, 1886–1890. [Google Scholar] [CrossRef] [PubMed]
- Bartfeld, S.; Bayram, T.; Van De Wetering, M.; Huch, M.; Begthel, H.; Kujala, P.; Vries, R.; Peters, P.J.; Clevers, H. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 2015, 148, 126–136.e6. [Google Scholar] [CrossRef]
- Yan, H.H.N.; Siu, H.C.; Law, S.; Ho, S.L.; Yue, S.S.K.; Tsui, W.Y.; Chan, D.; Chan, A.S.; Ma, S.; Lam, K.O.; et al. A Comprehensive Human Gastric Cancer Organoid Biobank Captures Tumor Subtype Heterogeneity and Enables Therapeutic Screening. Cell Stem. Cell 2018, 23, 882–897.e11. [Google Scholar] [CrossRef]
- Nanki, K.; Toshimitsu, K.; Takano, A.; Fujii, M.; Shimokawa, M.; Ohta, Y.; Matano, M.; Seino, T.; Nishikori, S.; Ishikawa, K.; et al. Divergent Routes toward Wnt and R-spondin Niche Independency during Human Gastric Carcinogenesis. Cell 2018, 174, 856–869.e17. [Google Scholar] [CrossRef]
- Vlachogiannis, G.; Hedayat, S.; Vatsiou, A.; Jamin, Y.; Fernández-Mateos, J.; Khan, K.; Lampis, A.; Eason, K.; Huntingford, I.; Burke, R.; et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018, 359, 920–926. [Google Scholar] [CrossRef]
- Hooi, J.K.Y.; Lai, W.Y.; Ng, W.K.; Suen, M.M.Y.; Underwood, F.E.; Tanyingoh, D.; Malfertheiner, P.; Graham, D.Y.; Wong, V.W.S.; Wu, J.C.Y.; et al. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology 2017, 153, 420–429. [Google Scholar] [CrossRef]
- Salama, N.R.; Hartung, M.L.; Müller, A. Life in the human stomach: Persistence strategies of the bacterial pathogen Helicobacter pylori. Nat. Rev. Microbiol. 2013, 11, 385–399. [Google Scholar] [CrossRef]
- Camilo, V.; Sugiyama, T.; Touati, E. Pathogenesis of Helicobacter pylori infection. Helicobacter 2017, 22, e12405. [Google Scholar] [CrossRef]
- Odenbreit, S.; Püls, J.; Sedlmaier, B.; Gerland, E.; Fischer, W.; Haas, R. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 2000, 287, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Ogle, S.A.; Schumacher, M.A.; Orr-Asman, M.A.; Miller, M.L.; Lertkowit, N.; Varro, A.; Hollande, F.; Zavros, Y. Loss of Parietal Cell Expression of Sonic Hedgehog Induces Hypergastrinemia and Hyperproliferation of Surface Mucous Cells. Gastroenterology 2010, 138, 550–561. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, M.A.; Donnelly, J.M.; Engevik, A.C.; Xiao, C.; Yang, L.; Kenny, S.; Varro, A.; Hollande, F.; Samuelson, L.C.; Zavros, Y. Gastric sonic hedgehog acts as a macrophage chemoattractant during the immune response to Helicobacter pylori. Gastroenterology 2012, 142, 1150–1159.e6. [Google Scholar] [CrossRef]
- Schumacher, M.A.; Feng, R.; Aihara, E.; Engevik, A.C.; Montrose, M.H.; Ottemann, K.M.; Zavros, Y. Helicobacter pylori-induced sonic hedgehog expression is regulated by NFκB pathway activation: The use of a novel in vitro model to study epithelial response to infection. Helicobacter 2015, 20, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Wroblewski, L.E.; Piazuelo, M.B.; Chaturvedi, R.; Schumacher, M.; Aihara, E.; Feng, R.; Noto, J.M.; Delgado, A.; Israel, D.A.; Zavros, Y.; et al. Helicobacter pylori targets cancer-associated apical-junctional constituents in gastroids and gastric epithelial cells. Gut 2015, 64, 720–730. [Google Scholar] [CrossRef] [PubMed]
- Bertaux-Skeirik, N.; Feng, R.; Schumacher, M.A.; Li, J.; Mahe, M.M.; Engevik, A.C.; Javier, J.E.; Peek, R.M., Jr.; Ottemann, K.; Orian-Rousseau, V.; et al. CD44 Plays a Functional Role in Helicobacter pylori-induced Epithelial Cell Proliferation. PLoS Pathog. 2015, 11, e1004663. [Google Scholar] [CrossRef]
- Sigal, M.; Rothenberg, M.E.; Logan, C.Y.; Lee, J.Y.; Honaker, R.W.; Cooper, R.L.; Passarelli, B.; Camorlinga, M.; Bouley, D.M.; Alvarez, G.; et al. Helicobacter pylori activates and expands Lgr5+ stem cells through direct colonization of the gastric glands. Gastroenterology 2015, 148, 1392–1404.e21. [Google Scholar] [CrossRef]
- Holokai, L.; Chakrabarti, J.; Broda, T.; Chang, J.; Hawkins, J.A.; Sundaram, N.; Wroblewski, L.E.; Peek, R.M.; Wang, J.; Helmrath, M.; et al. Increased Programmed Death-Ligand 1 is an Early Epithelial Cell Response to Helicobacter pylori Infection. PLoS Pathog. 2019, 15, e1007468. [Google Scholar] [CrossRef]
- Hayakawa, Y.; Fox, J.G.; Gonda, T.; Worthley, D.L.; Muthupalani, S.; Wang, T.C. Mouse models of gastric cancer. Cancers 2013, 5, 92–130. [Google Scholar] [CrossRef]
- Matano, M.; Date, S.; Shimokawa, M.; Takano, A.; Fujii, M.; Ohta, Y.; Watanabe, T.; Kanai, T.; Sato, T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 2015, 21, 256–262. [Google Scholar] [CrossRef]
- Chen, J.; Lau, B.T.; Andor, N.; Grimes, S.M.; Handy, C.; Wood-Bouwens, C.; Ji, H.P. Single-cell transcriptome analysis identifies distinct cell types and niche signaling in a primary gastric organoid model. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liang, Q.; Zhang, L.; Gou, H.; Li, Z.; Chen, H.; Dong, Y.; Ji, J.; Yu, J. C8orf76 promotes gastric tumorigenicity and metastasis by directly inducing lncRNA DUSP5P1 and associates with patient outcomes. Clin. Cancer Res. 2019, 25, 3128–3140. [Google Scholar] [CrossRef]
- Roerink, S.F.; Sasaki, N.; Lee-Six, H.; Young, M.D.; Alexandrov, L.B.; Behjati, S.; Mitchell, T.J.; Grossmann, S.; Lightfoot, H.; Egan, D.A.; et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature 2018, 556, 437–462. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Shah, Y.M. In vitro organoid culture of primary mouse colon tumors. J. Vis. Exp. 2013, 17, e50210. [Google Scholar] [CrossRef] [PubMed]
- Steele, N.G.; Chakrabarti, J.; Wang, J.; Biesiada, J.; Holokai, L.; Chang, J.; Nowacki, L.M.; Hawkins, J.; Mahe, M.; Sundaram, N.; et al. An Organoid-Based Preclinical Model of Human Gastric Cancer. CMGH 2019, 7, 161–184. [Google Scholar] [CrossRef]
- Li, J.; Xu, H.; Zhang, L.; Song, L.; Feng, D.; Peng, X.; Wu, M.; Zou, Y.; Wang, B.; Zhan, L.; et al. Malignant ascites-derived organoid (MADO) cultures for gastric cancer in vitro modelling and drug screening. J. Cancer Res. Clin. Oncol. 2019, 145, 2637–2647. [Google Scholar] [CrossRef]
- Gambardella, V.; Tarazona, N.; Cejalvo, J.M.; Lombardi, P.; Huerta, M.; Roselló, S.; Fleitas, T.; Roda, D.; Cervantes, A. Personalized Medicine: Recent Progress in Cancer Therapy. Cancers 2020, 12, 1009. [Google Scholar] [CrossRef]
- OPPOSITE: Outcome Prediction Of Systemic Treatment in Esophagogastric Carcinoma Full Text View ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT03429816 (accessed on 16 May 2020).
| 3D Culture Models | Spheroids | Organoids |
|---|---|---|
| Origin | Cancer cell lines | Patient tumor |
| Protocol | Easy to use | Delicate |
| Architecture | Simplified | Closer to organ |
| Reproducibility | High | Medium-high |
| Cellularity | Defined cell type | Different cell types |
| Cancer subtype modeling | + | ++ |
| Biological material produced | Abundant | Abundant |
| Genetic manipulation | ++ | ++ |
| Co-culture | ++ | |
| High-throughput drug screening | ++ | + |
| Prediction of clinical drug response in vitro | + | ++ |
| Cost | Low | Medium |
| Biobanks | Unavailable | Available |
| Test System Main Features | 2D Cell Culture [42,43] | 3D Cell Culture [44] | PDX Animal Model [45] |
|---|---|---|---|
| Physiological relevance | Limited | Better than 2D standard culture | Most physiological environment |
| Model complexity | Limited | Complex | Very complex |
| Gene expression | Stable at early passages | Close to in vivo tumors | Close to in vivo tumors |
| Immune system | No but co-cultures of cancer cells and immune cells possible | No but co-cultures of cancer cells and immune cells possible | No |
| Efficient drug screening | Yes | Yes | No |
| Disease modeling | Naive | Feasible | Sophisticated methods |
| Data provider | Easily exploitable | Easily exploitable | Hardly exploitable |
| Controlled microenvironment | Yes | Yes | No |
| Reproducibility | Favorable | Not so favorable | Unsuited |
| Cost | Low | Some expensive materials and special equipment required | High |
| Ethical and regulatory issues | No | No | No |
| Spheroids Production Methods | Description | Advantages | Disadvantages | References |
|---|---|---|---|---|
| Scaffold-based models | 3D construction that provides an ECM capable of supporting cells | Simple Mimic in vivo microenvironment Cell-ECM interactions Long-term culture Directly visualize and analyze | Difficulty of cell recovery Scaffold consistency can be variable across lots Nonuniform control (composition, size) Co-culture delicate Not suitable for drug testing | [27,63] |
| Non-adherent surfaces | Prevent attachment to the support | Simple Available pre-coated plates Uniform spheroid size control Ease of cell recovery Long-term culture Co-culture feasible Useful for drug screening Directly visualize and analyze | Low throughput production Defined co-culture cellular ratio Requires transfer of spheroids for analysis | [64,65,66,67] |
| Suspension culture | Keeps the cells in suspension to avoid sedimentation | High throughput production Homogeneous media composition Long-term culture | High shear force Nonuniform control (composition, size) Not easily suitable for drug testing Requires special equipment Requires a centrifugation step | [23] * |
| Hanging drop technique | Gravity based spheroid formation | Simple Uniform spheroid size control Co-culture feasible | Small size of spheroids Low throughput production Tedious spheroid handling and transfer Difficulty of long-term culture Defined co-culture cellular ratio Not suitable for drug testing | [68,69,70] * |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alzeeb, G.; Metges, J.-P.; Corcos, L.; Le Jossic-Corcos, C. Three-Dimensional Culture Systems in Gastric Cancer Research. Cancers 2020, 12, 2800. https://doi.org/10.3390/cancers12102800
Alzeeb G, Metges J-P, Corcos L, Le Jossic-Corcos C. Three-Dimensional Culture Systems in Gastric Cancer Research. Cancers. 2020; 12(10):2800. https://doi.org/10.3390/cancers12102800
Chicago/Turabian StyleAlzeeb, George, Jean-Philippe Metges, Laurent Corcos, and Catherine Le Jossic-Corcos. 2020. "Three-Dimensional Culture Systems in Gastric Cancer Research" Cancers 12, no. 10: 2800. https://doi.org/10.3390/cancers12102800
APA StyleAlzeeb, G., Metges, J.-P., Corcos, L., & Le Jossic-Corcos, C. (2020). Three-Dimensional Culture Systems in Gastric Cancer Research. Cancers, 12(10), 2800. https://doi.org/10.3390/cancers12102800







