Identification of Genes Whose Expression Overlaps Age Boundaries and Correlates with Risk Groups in Paediatric and Adult Acute Myeloid Leukaemia
Simple Summary
Abstract
1. Introduction
2. Results
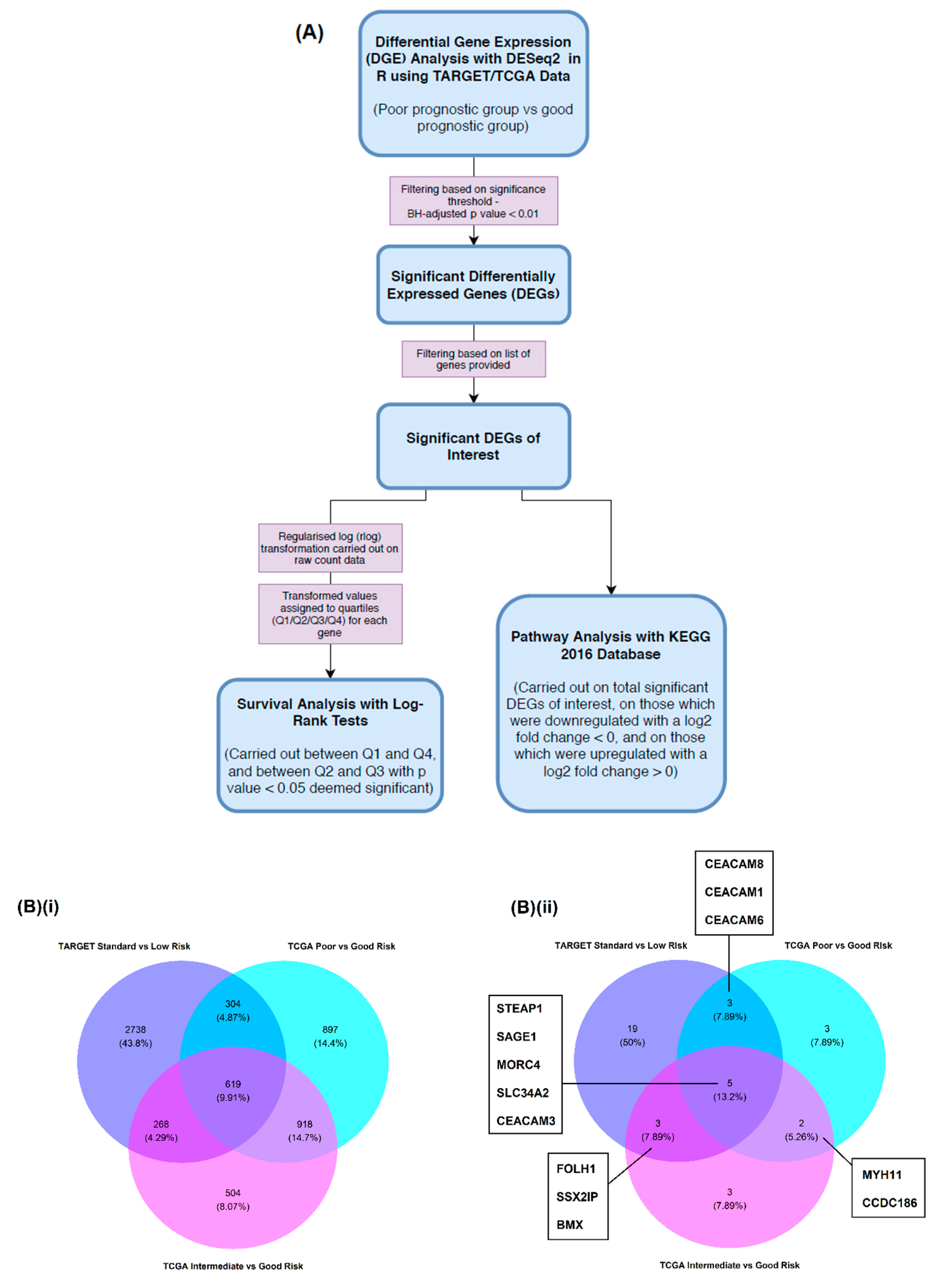
2.1. Differential Gene Expression (DGE) Analysis
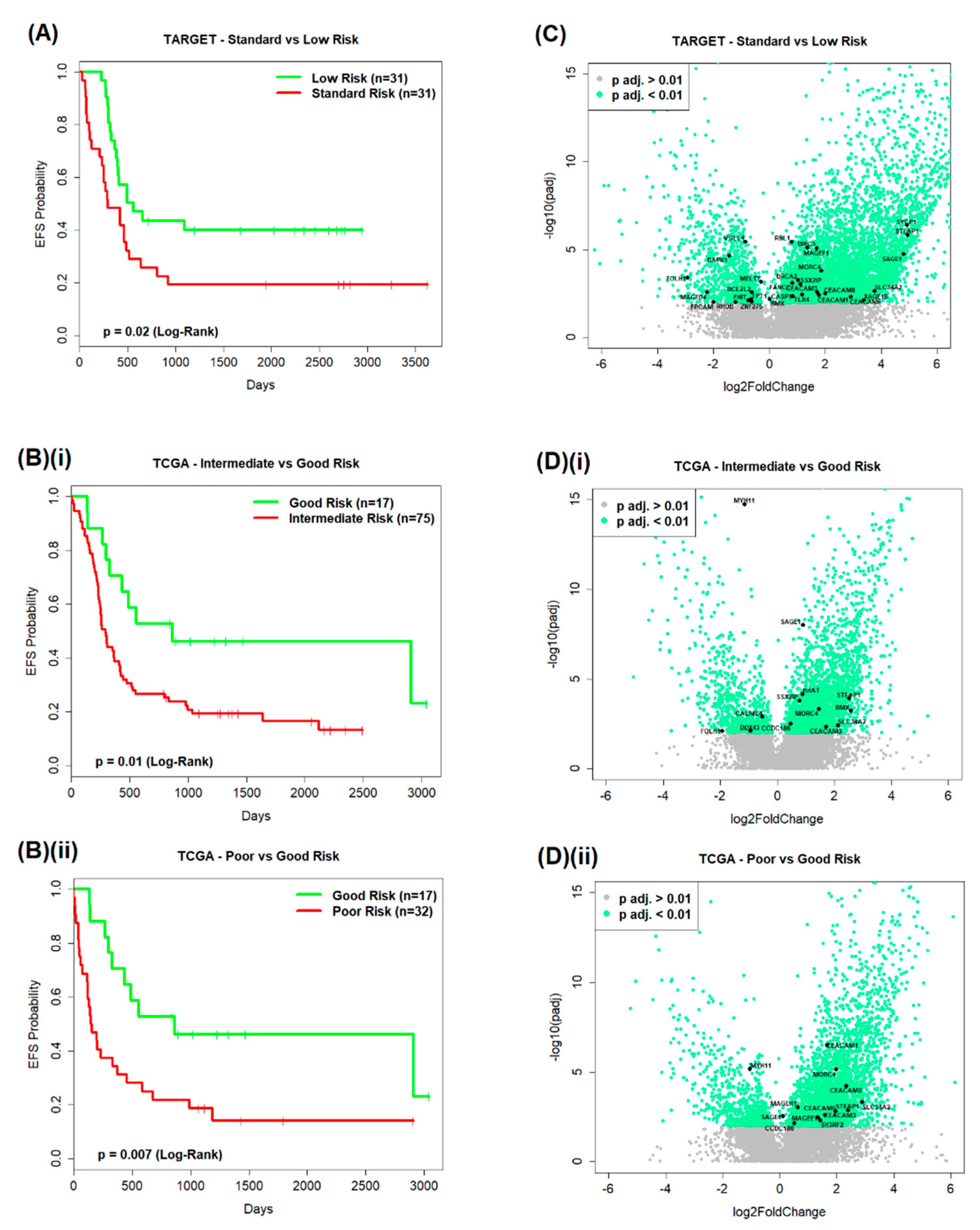
2.1.1. TARGET AML Data
2.1.2. TCGA AML Data
2.2. Survival Analysis
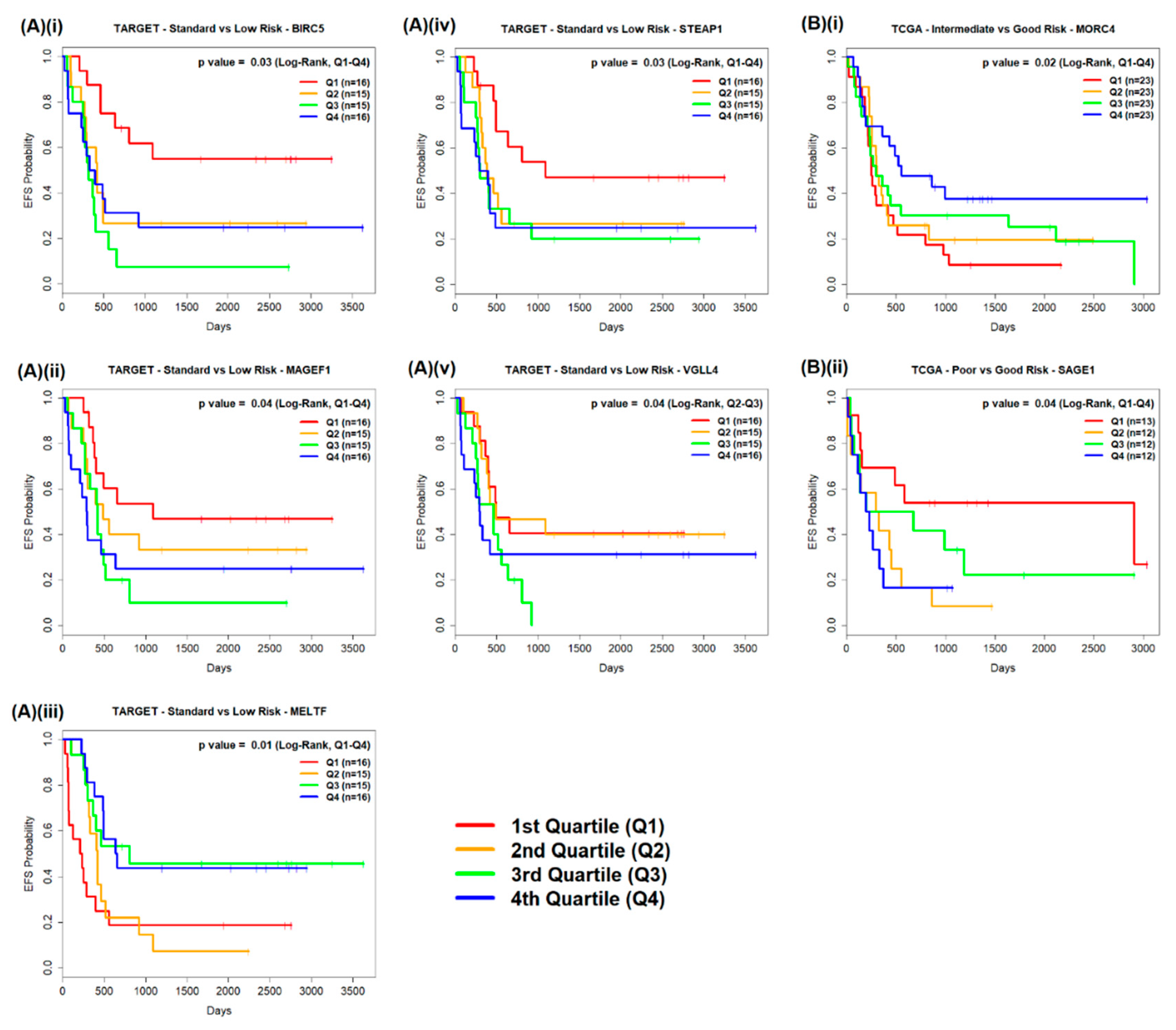
2.2.1. TARGET AML Data
2.2.2. TCGA AML Data
2.3. Pathway Analysis
3. Discussion
4. Materials and Methods
4.1. Identification of Antigens and Databases for Investigation
4.1.1. TARGET Dataset
4.1.2. TCGA Dataset
4.2. DGE Analysis
4.2.1. Prognostic Subgroup Formation
4.2.2. DGE Analysis with DESeq2
4.2.3. DESeq2 Workflow
4.3. Survival Analysis
4.4. Pathway Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AML | Acute myeloid leukaemia | ITD | Internal tandem duplication |
| BH | Benjamini–Hochberg | KEGG | Kyoto Encyclopaedia of Genes and Genomes |
| BIRC5 | Baculoviral Inhibitor Apoptosis Proteins (IAP) Repeat Containing 5 | MAGEF1 | Melanoma antigen family F1 |
| CAR | Chimeric antigen receptor | MELTF | Melanotransferrin |
| CBFβ | Core binding factor β | MLL | Mixed-lineage leukaemia |
| CCDC186 | Coiled coil domain containing 186 | MORC4 | MORC Family CW-Type Zinc Finger 4 |
| CEACAM | Carcinoembryonic antigen-related cell adhesion molecule | MYH11 | Myosin heavy chain 11 |
| DEG | Differentially expressed gene | NK | Natural killer |
| DGE | Differential gene expression | OS | Overall survival |
| DMNT3B | DNA methyltransferase 3B | PRAME | Preferentially Expressed Antigen in Melanoma |
| EFS | Event-free survival | RUNX1T1 | Runt-related transcription factor-1 Partner Transcriptional Co-Repressor 1 |
| ETO | Eight twenty-one | SAGE1 | Sarcoma antigen 1 |
| FOLH-1 | Folate Hydrolase 1 | SLC34A2 | Solute Carrier Family 34 Member 2 |
| GOI | Genes of interest | SOX | SRY-related HMG-box |
| HAGE | Helicase antigen | STEAP1 | Six-transmembrane epithelial antigen of prostate member 1 |
| HMG | High mobility group | TARGET | Therapeutically Applicable Research to Generate Effective Treatments |
| HNMT | Histamine N-Methyltransferase | TCGA | The Cancer Genome Atlas |
| HOX | Homeobox | TET | Ten-eleven translocation |
| IAP | Inhibitors of apoptosis | TRIM | Tripartite motif family |
| IL | Interleukin | VGLL4 | Vestigial-like 4 |
References
- Creutzig, U.; Kutny, M.A.; Barr, R.; Schlenk, R.F.; Ribeiro, R.C. Acute myelogenous leukemia in adolescents and young adults. Pediatr. Blood Cancer 2018, 65, e27089. [Google Scholar] [CrossRef]
- Creutzig, U.; van den Heuvel-Eibrink, M.M.; Gibson, B.; Dworzak, M.N.; Adachi, S.; de Bont, E.; Harbott, J.; Hasle, H.; Johnston, D.; Kinoshita, A.; et al. Diagnosis and management of acute myeloid leukemia in children and adolescents: Recommendations from an international expert panel. Blood 2012, 120, 3187–3205. [Google Scholar] [CrossRef]
- Elgarten, C.W.; Aplenc, R. Pediatric acute myeloid leukemia: Updates on biology, risk stratification, and therapy. Curr. Opin. Pediatr. 2020, 32, 57–66. [Google Scholar] [CrossRef]
- Paschka, P.; Marcucci, G.; Ruppert, A.S.; Mrozek, K.; Chen, H.; Kittles, R.A.; Vukosavljevic, T.; Perrotti, D.; Vardiman, J.W.; Carroll, A.J.; et al. Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): A Cancer and Leukemia Group B Study. J. Clin. Oncol. 2006, 24, 3904–3911. [Google Scholar] [CrossRef]
- Wei, H.; Wang, Y.; Zhou, C.; Lin, D.; Liu, B.; Liu, K.; Qiu, S.; Gong, B.; Li, Y.; Zhang, G.; et al. Distinct genetic alteration profiles of acute myeloid leukemia between Caucasian and Eastern Asian population. J. Hematol. Oncol. 2018, 11, 18. [Google Scholar] [CrossRef]
- Stirewalt, D.L.; Radich, J.P. The role of FLT3 in haematopoietic malignancies. Nat. Rev. Cancer 2003, 3, 650–665. [Google Scholar] [CrossRef] [PubMed]
- Mannelli, F.; Ponziani, V.; Bencini, S.; Bonetti, M.I.; Benelli, M.; Cutini, I.; Gianfaldoni, G.; Scappini, B.; Pancani, F.; Piccini, M.; et al. CEBPA-double-mutated acute myeloid leukemia displays a unique phenotypic profile: A reliable screening method and insight into biological features. Haematologica 2017, 102, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, H.; Kajihara, M.; Tomizawa, D.; Watanabe, T.; Saito, A.M.; Fujimoto, J.; Horibe, K.; Kodama, K.; Tokumasu, M.; Itoh, H.; et al. Prognostic implications of CEBPA mutations in pediatric acute myeloid leukemia: A report from the Japanese Pediatric Leukemia/Lymphoma Study Group. Blood Cancer J. 2014, 4, e226. [Google Scholar] [CrossRef]
- Suzuki, T.; Kiyoi, H.; Ozeki, K.; Tomita, A.; Yamaji, S.; Suzuki, R.; Kodera, Y.; Miyawaki, S.; Asou, N.; Kuriyama, K.; et al. Clinical characteristics and prognostic implications of NPM1 mutations in acute myeloid leukemia. Blood 2005, 106, 2854–2861. [Google Scholar] [CrossRef] [PubMed]
- Khan, G.N.; Orchard, K.; Guinn, B.A. Antigenic Targets for the Immunotherapy of Acute Myeloid Leukaemia. J. Clin. Med. 2019, 8, 134. [Google Scholar] [CrossRef] [PubMed]
- Bonifant, C.L.; Velasquez, M.P.; Gottschalk, S. Advances in immunotherapy for pediatric acute myeloid leukemia. Expert Opin. Biol. Ther. 2018, 18, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Bonifant, C.L.; Tasian, S.K. The future of cellular immunotherapy for childhood leukemia. Curr. Opin. Pediatr. 2020, 32, 13–25. [Google Scholar] [CrossRef]
- Tasian, S.K. Acute myeloid leukemia chimeric antigen receptor T-cell immunotherapy: How far up the road have we traveled? Ther. Adv. Hematol. 2018, 9, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Perna, F.; Berman, S.H.; Soni, R.K.; Mansilla-Soto, J.; Eyquem, J.; Hamieh, M.; Hendrickson, R.C.; Brennan, C.W.; Sadelain, M. Integrating Proteomics and Transcriptomics for Systematic Combinatorial Chimeric Antigen Receptor Therapy of AML. Cancer Cell 2017, 32, 506–519.e5. [Google Scholar] [CrossRef]
- Liao, D.; Wang, M.; Liao, Y.; Li, J.; Niu, T. A Review of Efficacy and Safety of Checkpoint Inhibitor for the Treatment of Acute Myeloid Leukemia. Front. Pharmacol. 2019, 10, 609. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Correa, B.; Bergua, J.M.; Campos, C.; Gayoso, I.; Arcos, M.J.; Banas, H.; Morgado, S.; Casado, J.G.; Solana, R.; Tarazona, R. Cytokine profiles in acute myeloid leukemia patients at diagnosis: Survival is inversely correlated with IL-6 and directly correlated with IL-10 levels. Cytokine 2013, 61, 885–891. [Google Scholar] [CrossRef]
- Szczepanski, M.J.; Szajnik, M.; Welsh, A.; Foon, K.A.; Whiteside, T.L.; Boyiadzis, M. Interleukin-15 enhances natural killer cell cytotoxicity in patients with acute myeloid leukemia by upregulating the activating NK cell receptors. Cancer Immunol. Immunother. 2010, 59, 73–79. [Google Scholar] [CrossRef]
- Liggins, A.P.; Guinn, B.A.; Hatton, C.S.; Pulford, K.; Banham, A.H. Serologic detection of diffuse large B-cell lymphoma-associated antigens. Int. J. Cancer 2004, 110, 563–569. [Google Scholar] [CrossRef]
- Liggins, A.P.; Cooper, C.D.; Lawrie, C.H.; Brown, P.J.; Collins, G.P.; Hatton, C.S.; Pulford, K.; Banham, A.H. MORC4, a novel member of the MORC family, is highly expressed in a subset of diffuse large B-cell lymphomas. Br. J. Haematol. 2007, 138, 479–486. [Google Scholar] [CrossRef]
- Noren, E.; Verma, D.; Soderkvist, P.; Weisselberg, T.; Soderman, J.; Lotfi, K.; Almer, S. Single Nucleotide Polymorphisms in MORC4, CD14, and TLR4 Are Related to Outcome of Allogeneic Stem Cell Transplantation. Ann. Transplant. 2016, 21, 56–67. [Google Scholar] [CrossRef]
- Ishihara, M.; Kageyama, S.; Miyahara, Y.; Ishikawa, T.; Ueda, S.; Soga, N.; Naota, H.; Mukai, K.; Harada, N.; Ikeda, H.; et al. MAGE-A4, NY-ESO-1 and SAGE expression rates and co-expression relationships in solid tumours. BMC Cancer 2020, 20, 606. [Google Scholar] [CrossRef] [PubMed]
- Liberante, F.G.; Pellagatti, A.; Boncheva, V.; Bowen, D.T.; Mills, K.I.; Boultwood, J.; Guinn, B.A. High and low, but not intermediate, PRAME expression levels are poor prognostic markers in myelodysplastic syndrome at disease presentation. Br. J. Haematol. 2013, 162, 282–285. [Google Scholar] [CrossRef] [PubMed]
- Tajonar, A.; Maehr, R.; Hu, G.; Sneddon, J.B.; Rivera-Feliciano, J.; Cohen, D.E.; Elledge, S.J.; Melton, D.A. Brief report: VGLL4 is a novel regulator of survival in human embryonic stem cells. Stem Cells 2013, 31, 2833–2841. [Google Scholar] [CrossRef]
- Deng, X.; Fang, L. VGLL4 is a transcriptional cofactor acting as a novel tumor suppressor via interacting with TEADs. Am. J. Cancer Res. 2018, 8, 932–943. [Google Scholar] [PubMed]
- Boullosa, L.F.; Savaliya, P.; Bonney, S.; Orchard, L.; Wickenden, H.; Lee, C.; Smits, E.; Banham, A.H.; Mills, K.I.; Orchard, K.; et al. Identification of survivin as a promising target for the immunotherapy of adult B-cell acute lymphoblastic leukemia. Oncotarget 2018, 9, 3853–3866. [Google Scholar] [CrossRef]
- Adams, S.P.; Sahota, S.S.; Mijovic, A.; Czepulkowski, B.; Padua, R.A.; Mufti, G.J.; Guinn, B.A. Frequent expression of HAGE in presentation chronic myeloid leukaemias. Leukemia 2002, 16, 2238–2242. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Chen, Q.; Yang, J.; Qian, J.; Deng, Z.Q.; Qian, W.; Chen, X.X.; Ma, J.C.; Xiong, D.S.; Ma, Y.J.; et al. DDX43 promoter is frequently hypomethylated and may predict a favorable outcome in acute myeloid leukemia. Leuk. Res. 2014, 38, 601–607. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Fatah, T.M.; McArdle, S.E.; Johnson, C.; Moseley, P.M.; Ball, G.R.; Pockley, A.G.; Ellis, I.O.; Rees, R.C.; Chan, S.Y. HAGE (DDX43) is a biomarker for poor prognosis and a predictor of chemotherapy response in breast cancer. Br. J. Cancer 2014, 110, 2450–2461. [Google Scholar] [CrossRef]
- Jordaens, S.; Cooksey, L.; Bonney, S.; Orchard, L.; Coutinho, M.; Van Tendeloo, V.; Mills, K.I.; Orchard, K.; Guinn, B.A. Serum profiling identifies ibrutinib as a treatment option for young adults with B-cell acute lymphoblastic leukaemia. Br. J. Haematol. 2020, 189, 500–512. [Google Scholar] [CrossRef]
- Coutre, S.E.; Byrd, J.C.; Hillmen, P.; Barrientos, J.C.; Barr, P.M.; Devereux, S.; Robak, T.; Kipps, T.J.; Schuh, A.; Moreno, C.; et al. Long-term safety of single-agent ibrutinib in patients with chronic lymphocytic leukemia in 3 pivotal studies. Blood Adv. 2019, 3, 1799–1807. [Google Scholar] [CrossRef]
- Nuttall, E.; Tung, J.; Trounce, E.; Johnston, R.; Chevassut, T. Real-world experience of ibrutinib therapy in relapsed chronic lymphocytic leukemia: Results of a single-center retrospective analysis. J. Blood Med. 2019, 10, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Shanafelt, T.D.; Wang, X.V.; Kay, N.E.; Hanson, C.A.; O’Brien, S.; Barrientos, J.; Jelinek, D.F.; Braggio, E.; Leis, J.F.; Zhang, C.C.; et al. Ibrutinib-Rituximab or Chemoimmunotherapy for Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2019, 381, 432–443. [Google Scholar] [CrossRef] [PubMed]
- Rushworth, S.A.; Murray, M.Y.; Zaitseva, L.; Bowles, K.M.; MacEwan, D.J. Identification of Bruton’s tyrosine kinase as a therapeutic target in acute myeloid leukemia. Blood 2014, 123, 1229–1238. [Google Scholar] [CrossRef] [PubMed]
- Langabeer, S.E.; Walker, H.; Gale, R.E.; Wheatley, K.; Burnett, A.K.; Goldstone, A.H.; Linch, D.C. Frequency of CBF beta/MYH11 fusion transcripts in patients entered into the U.K. MRC AML trials. The MRC Adult Leukaemia Working Party. Br. J. Haematol. 1997, 96, 736–739. [Google Scholar] [CrossRef]
- Singh, A.A.; Mandoli, A.; Prange, K.H.; Laakso, M.; Martens, J.H. AML associated oncofusion proteins PML-RARA, AML1-ETO and CBFB-MYH11 target RUNX/ETS-factor binding sites to modulate H3ac levels and drive leukemogenesis. Oncotarget 2017, 8, 12855–12865. [Google Scholar] [CrossRef]
- Guinn, B.A.; Bland, E.A.; Lodi, U.; Liggins, A.P.; Tobal, K.; Petters, S.; Wells, J.W.; Banham, A.H.; Mufti, G.J. Humoral detection of leukaemia-associated antigens in presentation acute myeloid leukaemia. Biochem. Biophys. Res. Commun. 2005, 335, 1293–1304. [Google Scholar] [CrossRef]
- Sood, R.; Kamikubo, Y.; Liu, P. Role of RUNX1 in hematological malignancies. Blood 2017, 129, 2070–2082. [Google Scholar] [CrossRef]
- Erickson, P.; Gao, J.; Chang, K.S.; Look, T.; Whisenant, E.; Raimondi, S.; Lasher, R.; Trujillo, J.; Rowley, J.; Drabkin, H. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood 1992, 80, 1825–1831. [Google Scholar] [CrossRef]
- Elagib, K.E.; Goldfarb, A.N. Oncogenic pathways of AML1-ETO in acute myeloid leukemia: Multifaceted manipulation of marrow maturation. Cancer Lett. 2007, 251, 179–186. [Google Scholar] [CrossRef]
- Cho, E.K.; Bang, S.M.; Ahn, J.Y.; Yoo, S.M.; Park, P.W.; Seo, Y.H.; Shin, D.B.; Lee, J.H. Prognostic value of AML 1/ETO fusion transcripts in patients with acute myelogenous leukemia. Korean J. Intern. Med. 2003, 18, 13–20. [Google Scholar] [CrossRef]
- Majeti, R.; Becker, M.W.; Tian, Q.; Lee, T.L.; Yan, X.; Liu, R.; Chiang, J.H.; Hood, L.; Clarke, M.F.; Weissman, I.L. Dysregulated gene expression networks in human acute myelogenous leukemia stem cells. Proc. Natl. Acad. Sci. USA 2009, 106, 3396–3401. [Google Scholar] [CrossRef]
- Guinn, B.; Greiner, J.; Schmitt, M.; Mills, K.I. Elevated expression of the leukemia-associated antigen SSX2IP predicts survival in acute myeloid leukemia patients who lack detectable cytogenetic rearrangements. Blood 2009, 113, 1203–1204. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Krivtsov, A.V.; Sinha, A.U.; North, T.E.; Goessling, W.; Feng, Z.; Zon, L.I.; Armstrong, S.A. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science 2010, 327, 1650–1653. [Google Scholar] [CrossRef] [PubMed]
- Scholl, C.; Bansal, D.; Dohner, K.; Eiwen, K.; Huntly, B.J.; Lee, B.H.; Rucker, F.G.; Schlenk, R.F.; Bullinger, L.; Dohner, H.; et al. The homeobox gene CDX2 is aberrantly expressed in most cases of acute myeloid leukemia and promotes leukemogenesis. J. Clin. Investig. 2007, 117, 1037–1048. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Mistri, T.K. Transcription factors in SOX family: Potent regulators for cancer initiation and development in the human body. Semin. Cancer Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Tosic, N.; Petrovic, I.; Grujicic, N.K.; Davidovic, S.; Virijevic, M.; Vukovic, N.S.; Pavlovic, S.; Stevanovic, M. Prognostic significance of SOX2, SOX3, SOX11, SOX14 and SOX18 gene expression in adult de novo acute myeloid leukemia. Leuk. Res. 2018, 67, 32–38. [Google Scholar] [CrossRef]
- Lamba, J.K.; Cao, X.; Raimondi, S.C.; Rafiee, R.; Downing, J.R.; Lei, S.; Gruber, T.; Ribeiro, R.C.; Rubnitz, J.E.; Pounds, S.B. Integrated epigenetic and genetic analysis identifies markers of prognostic significance in pediatric acute myeloid leukemia. Oncotarget 2018, 9, 26711–26723. [Google Scholar] [CrossRef]
- Boncheva-Henderson, V.B.; Linnebacher, M.; Tangney, M.; Mills, K.I.; O’Sullivan, G.; Guinn, B.A. Sero-recognition of novel tumour antigens by patients with immunologically ‘hot’ colon cancer. Submitted.
- Ge, G.; Zhou, C.; Ren, Y.; Tang, X.; Wang, K.; Zhang, W.; Niu, L.; Zhou, Y.; Yan, Y.; He, J. Enhanced SLC34A2 in breast cancer stem cell-like cells induces chemotherapeutic resistance to doxorubicin via SLC34A2-Bmi1-ABCC5 signaling. Tumor Biol. 2016, 37, 5049–5062. [Google Scholar] [CrossRef]
- Moreaux, J.; Kassambara, A.; Hose, D.; Klein, B. STEAP1 is overexpressed in cancers: A promising therapeutic target. Biochem. Biophys. Res. Commun. 2012, 429, 148–155. [Google Scholar] [CrossRef]
- Guinn, B.A.; Gilkes, A.F.; Woodward, E.; Westwood, N.B.; Mufti, G.J.; Linch, D.; Burnett, A.K.; Mills, K.I. Microarray analysis of tumour antigen expression in presentation acute myeloid leukaemia. Biochem. Biophys. Res. Commun. 2005, 333, 703–713. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network; Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, A.; Hoadley, K.; Triche, T.J., Jr.; Laird, P.W.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [CrossRef] [PubMed]
- Bolouri, H.; Farrar, J.E.; Triche, T., Jr.; Ries, R.E.; Lim, E.L.; Alonzo, T.A.; Ma, Y.; Moore, R.; Mungall, A.J.; Marra, M.A.; et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat. Med. 2018, 24, 103–112. [Google Scholar] [CrossRef]
- Portal, G.D. TCGA-LAML [Internet]. 2018. Available online: https://portal.gdc.cancer.gov/projects/TCGA-LAML (accessed on 11 January 2019).
- Therneau, T.M. Survival: A Package for Survival Analysis in S. R Package Version 2.43-3. 2018. Available online: https://cran.r-project.org/package=survival (accessed on 15 February 2019).
- Team, R.C. R: A Language and Environment for Statistical Computing; Foundation for Statistical Computing: Vienna, Austria, 2017; Available online: https://www.r-project.org/ (accessed on 28 January 2019).
- Soneson, C.; Delorenzi, M. A comparison of methods for differential expression analysis of RNA-seq data. BMC Bioinform. 2013, 14, 91. [Google Scholar] [CrossRef]
- Seyednasrollah, F.; Laiho, A.; Elo, L.L. Comparison of software packages for detecting differential expression in RNA-seq studies. Brief. Bioinform. 2015, 16, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Pagès, H.; Carlson, M.; Falcon, S.; Li, N. AnnotationDbi: Annotation Database Interface; R Package Version 1.40.0. 2017. Available online: http://bioconductor.org/packages/release/bioc/html/AnnotationDbi.html (accessed on 30 January 2019).
- Carlson, M. org.Hs.eg.db: Genome Wide Annotation for Human; R Package Version 3.5.0. 2017. Available online: https://bioconductor.org/packages/release/data/annotation/html/org.Hs.eg.db.html (accessed on 30 January 2019).
- Graffelman, J. Calibrate: Calibration of Scatterplot and Biplot Axes. R Package Version 1.7.2. 2013. Available online: https://cran.r-project.org/web/packages/calibrate/index.html (accessed on 30 January 2019).
- Chan, F.C. Survutils: Utility Functions for Survival Analysis. R Package Survutils Version 1.0.2. 2018. Available online: https://cran.r-project.org/web/packages/survutils/index.html (accessed on 1 February 2019).
- Jawaid, W. EnrichR: Provides an R Interface to “Enrichr”. R Package Version 1.0. 2017. Available online: https://cran.r-project.org/web/packages/enrichR/index.html (accessed on 4 February 2019).
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
| (A) | Gene Symbol | Log2 Fold Change | BH-Adjusted p Value | (B) | Gene Symbol | Log2 Fold Change | BH-Adjusted p Value | (C) | Gene Symbol | Log2 Fold Change | BH-Adjusted p Value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GOI | BCL2L2 | −0.63 | 2.61 × 10−3 | GOI | BMX | 2.57 | 5.74 × 10−4 | GOI | CCDC186 | 0.51 | 7.05 × 10−3 |
| BIRC5 | 1.35 | 7.48 × 10−6 | CALML4 | −0.53 | 1.25 × 10−3 | CEACAM1 | 1.67 | 3.10 × 10−7 | |||
| BMX | 0.01 | 6.36 × 10−3 | CCDC186 | 0.46 | 2.96 × 10−3 | CEACAM3 | 1.59 | 2.36 × 10−3 | |||
| BRCA2 | 1.02 | 5.12 × 10−2 | CEACAM3 | 1.71 | 4.33 × 10−3 | CEACAM6 | 1.96 | 1.57 × 10−3 | |||
| CASP1 | 0.84 | 4.31 × 10−3 | DDX43 | −0.95 | 7.69 × 10−3 | CEACAM8 | 2.33 | 5.66 × 10−5 | |||
| CEACAM1 | 1.76 | 3.89 × 10−3 | FOLH1 | −1.94 | 7.52 × 10−3 | MAGEE1 | 1.33 | 3.34 × 10−3 | |||
| CEACAM3 | 1.70 | 2.57 × 10−3 | HHAT | 0.87 | 6.75 × 10−5 | MAGEH1 | 0.62 | 8.86 × 10−4 | |||
| CEACAM6 | 2.91 | 4.80 × 10−3 | MORC4 | 1.45 | 4.40 × 10−4 | MORC4 | 1.98 | 7.20 × 10−6 | |||
| CEACAM8 | 1.99 | 3.13 × 10−3 | MYH11 | −1.15 | 1.74 × 10−15 | MYH11 | −1.05 | 6.70 × 10−6 | |||
| DAPK1 | −1.45 | 2.12 × 10−5 | SAGE1 | 0.89 | 9.64 × 10−9 | SAGE1 | 0.11 | 2.85 × 10−3 | |||
| EPCAM | −2.00 | 9.72 × 10−3 | SLC34A2 | 2.12 | 3.72 × 10−3 | SH3RF2 | 1.42 | 4.72 × 10−3 | |||
| FANCC | 0.81 | 7.45 × 10−4 | SSX2IP | 0.77 | 1.55 × 10−4 | SLC34A2 | 2.89 | 4.65 × 10−4 | |||
| FHIT | −0.76 | 7.58 × 10−3 | STEAP1 | 2.51 | 1.19 × 10−4 | STEAP1 | 2.38 | 1.29 × 10−3 | |||
| FOLH1 | −2.95 | 3.97 × 10−4 | Other Notable Genes | CD52 | −1.60 | 1.31 × 10−4 | Other Notable Genes | CD19 | −1.92 | 2.52 × 10−6 | |
| MAGED4 | −2.24 | 2.65 × 10−3 | DNMT3B | 1.95 | 2.17 × 10−11 | CD276 | 2.19 | 1.67 × 10−4 | |||
| MELTF | −0.30 | 6.72 × 10−4 | HMNT | 2.99 | 5.48 × 10−10 | CD7 | 1.97 | 7.27 × 10−6 | |||
| MORC4 | 1.86 | 1.54 × 10−4 | HOXA10 | 3.55 | 1.59 × 10−13 | CD70 | 2.32 | 1.80 × 10−4 | |||
| RBL1 | 0.80 | 3.51 × 10−6 | HOXA10-AS | 3.54 | 8.13 × 10−13 | CD81 | 1.33 | 1.67 × 10−5 | |||
| RHOB | −1.22 | 9.32 × 10−3 | HOXA11 | 2.70 | 1.10 × 10−12 | CD82 | 1.82 | 6.60 × 10−11 | |||
| SAGE1 | 4.80 | 1.78 × 10−5 | HOXA11-AS | 2.25 | 1.96 × 10−11 | DNMT3B | 2.62 | 3.94 × 10−14 | |||
| SLC34A2 | 3.75 | 2.20 × 10−3 | HOXA13 | 2.70 | 4.16 × 10−8 | HOXA1 | 1.33 | 8.85 × 10−4 | |||
| SSX2IP | 1.11 | 8.69 × 10−4 | HOXA2 | 3.34 | 6.09 × 10−15 | HOXA10 | 3.42 | 1.18 × 10−9 | |||
| STEAP1 | 4.95 | 1.37 × 10−6 | HOXA3 | 5.30 | 6.50 × 10−33 | HOXA10-AS | 3.58 | 7.04 × 10−11 | |||
| SYCP1 | 4.91 | 3.92 × 10−7 | HOXA4 | 4.48 | 1.50 × 10−21 | HOXA11 | 3.09 | 3.10 × 10−8 | |||
| TLR4 | 1.16 | 3.56 × 10−3 | HOXA5 | 4.58 | 7.43 × 10−22 | HOXA11-AS | 2.30 | 1.52 × 10−5 | |||
| TPT1 | −0.65 | 6.73 × 10−3 | HOXA6 | 5.83 | 1.04 × 10−34 | HOXA2 | 2.91 | 7.86 × 10−11 | |||
| VGLL4 | −0.86 | 3.43 × 10−6 | HOXA7 | 4.92 | 1.19 × 10−24 | HOXA3 | 5.08 | 3.57 × 10−26 | |||
| XAGE1B | 3.37 | 7.41 × 10−3 | HOXA9 | 4.19 | 6.04 × 10−18 | HOXA4 | 3.94 | 3.99 × 10−14 | |||
| ZNF275 | −0.64 | 9.11 × 10−3 | HOXA-AS2 | 5.39 | 6.77 × 10−35 | HOXA5 | 4.19 | 1.69 × 10−14 | |||
| Other Notable Genes | CD34 | −2.91 | 1.53 × 10−7 | HOXA-AS3 | 6.25 | 1.30 × 10−39 | HOXA6 | 6.17 | 7.87 × 10−32 | ||
| CD52 | −2.68 | 2.79 × 10−12 | HOXB1 | 1.86 | 5.60 × 10−3 | HOXA7 | 4.59 | 5.05 × 10−16 | |||
| CD99 | −1.51 | 2.48 × 10−8 | HOXB3 | 2.39 | 1.22 × 10−6 | HOXA9 | 4.21 | 1.67 × 10−14 | |||
| DNMT3B | 1.13 | 4.92 × 10−3 | HOXB4 | 1.83 | 3.63 × 10−4 | HOXA-AS2 | 5.07 | 9.73 × 10−26 | |||
| HOXA10 | 3.29 | 4.57 × 10−9 | HOXB5 | 4.66 | 5.13 × 10−22 | HOXA-AS3 | 6.31 | 3.19 × 10−33 | |||
| HOXA10-AS | 2.69 | 3.65 × 10−4 | HOXB6 | 4.60 | 1.48 × 10−21 | HOXB5 | 3.43 | 3.50 × 10−7 | |||
| HOXA7 | 3.90 | 2.05 × 10−7 | HOXB7 | 3.69 | 2.53 × 10−13 | HOXB6 | 3.50 | 6.34 × 10−4 | |||
| HOXA-AS3 | 6.41 | 3.79 × 10−19 | HOXB8 | 4.49 | 2.09 × 10−12 | HOXB-AS3 | 4.06 | 1.02 × 10−5 | |||
| HOXB7 | 3.35 | 1.13 × 10−10 | HOXB9 | 4.73 | 1.48 × 10−13 | HOXC4 | 1.74 | 2.15 × 10−3 | |||
| HOXB-AS3 | 1.22 | 2.20 × 10−12 | HOXB-AS3 | 5.12 | 3.85 × 10−27 | IL10 | 1.61 | 6.55 × 10−7 | |||
| IL11 | −2.55 | 6.73 × 10−4 | IL10 | 0.41 | 9.84 × 10−3 | IL11 | 1.40 | 1.30 × 10−4 | |||
| IL15 | 1.98 | 4.86 × 10−4 | IL15 | 2.28 | 1.15 × 10−7 | IL15 | 1.85 | 1.21 × 10−5 | |||
| IL19 | 3.18 | 1.45 × 10−4 | IL4 | −1.24 | 1.36 × 10−3 | IL7 | 4.23 | 3.14 × 10−21 | |||
| IL24 | 4.56 | 3.41 × 10−5 | IL7 | 2.93 | 5.27 × 10−7 | PRICKLE1 | 1.55 | 3.78 × 10−4 | |||
| IL3 | −2.92 | 7.12 × 10−3 | PRICKLE1 | 2.16 | 5.06 × 10−9 | PRICKLE4 | 1.16 | 3.71 × 10−3 | |||
| IL7 | 2.63 | 3.73 × 10−5 | PRICKLE2 | 2.32 | 1.56 × 10−5 | RUNX1 | −0.48 | 6.12 × 10−3 | |||
| RUNX1 | −0.98 | 7.78 × 10−4 | RUNX1T1 | −5.72 | 2.92 × 10−17 | RUNX1T1 | −6.70 | 1.28 × 10−22 | |||
| RUNX3 | 1.24 | 1.21 × 10−3 | RUNX3 | 1.98 | 6.50 × 10−12 | RUNX3 | 2.50 | 1.67 × 10−14 | |||
| SOX11 | 6.43 | 2.45 × 10−8 | SOX6 | 1.17 | 6.86 × 10−4 | SOX15 | −0.68 | 7.94 × 10−3 | |||
| SOX15 | −2.65 | 7.94 × 10−9 | TRIM10 | 2.47 | 2.77 × 10−7 | SOX18 | 2.17 | 2.01 × 10−4 | |||
| SOX18 | −2.21 | 4.93 × 10−4 | TRIM15 | 2.09 | 2.87 × 10−4 | SOX5 | 2.50 | 1.65 × 10−3 | |||
| SOX30 | 2.56 | 3.86 × 10−4 | TRIM24 | −0.54 | 3.49 × 10−3 | SOX6 | 1.66 | 1.61 × 10−5 | |||
| SOX5 | 2.26 | 6.16 × 10−5 | TRIM29 | 2.96 | 1.20 × 10−7 | TOX2 | 2.11 | 5.61 × 10−6 | |||
| SOX8 | −1.99 | 3.69 × 10−3 | TRIM47 | −2.08 | 7.03 × 10−10 | TRIM10 | 2.91 | 4.65 × 10−8 | |||
| TET1 | −1.22 | 1.08 × 10−3 | TRIM6 | −1.11 | 5.88 × 10−4 | TRIM24 | −0.76 | 1.34 × 10−6 | |||
| TRIM10 | 0.67 | 4.54 × 10−4 | TRIM68 | 0.42 | 5.52 × 10−3 | TRIM29 | 3.77 | 6.26 × 10−6 | |||
| TRIM24 | −1.16 | 2.33 × 10−5 | TRIM71 | −2.01 | 3.94 × 10−5 | TRIM36 | −1.50 | 4.51 × 10−3 | |||
| TRIM29 | 1.68 | 3.33 × 10−6 | TRIM8 | 0.88 | 8.27 × 10−5 | TRIM40 | 2.23 | 4.01 × 10−3 | |||
| TRIM31 | 3.20 | 3.33 × 10−3 | TRIM9 | 2.72 | 7.94 × 10−13 | TRIM72 | −1.25 | 3.32 × 10−3 | |||
| TRIM35 | 0.59 | 4.38 × 10−3 | TRIM8 | 1.06 | 1.55 × 10−4 | ||||||
| TRIM38 | 0.60 | 3.51 × 10−3 | TRIM9 | 2.68 | 2.00 × 10−9 | ||||||
| TRIM47 | −2.65 | 3.25 × 10−9 | WNT6 | 3.14 | 3.99 × 10−10 | ||||||
| TRIM59 | 0.95 | 1.64 × 10−3 | |||||||||
| TRIM61 | 2.30 | 5.56 × 10−3 | |||||||||
| TRIM69 | 0.67 | 9.44 × 10−3 | |||||||||
| TRIM7 | 2.46 | 3.84 × 10−6 | |||||||||
| TRIM71 | −3.09 | 4.81 × 10−7 | |||||||||
| TRIM8 | 0.75 | 2.92 × 10−3 | |||||||||
| TRIM9 | 2.49 | 2.82 × 10−6 |
| Patient Subgroup Comparison | Significant DEGs of Interest | n of Q1, Q4 | Q1–Q4 Log-Rank p Value | Q1 Median EFS Survival in Days (95% CL) | Q4 Median EFS Survival in Days (95% CL) | n of Q2, Q3 | Q2-Q3 Log-Rank p Value | Q2 Median EFS Survival in Days (95% CL) | Q3 Median EFS Survival in Days (95% CL) | |
|---|---|---|---|---|---|---|---|---|---|---|
| TARGET | Standard Risk vs. Low Risk (n = 31 vs. n = 31) | BIRC5 | 16, 16 | 0.0293 | NR (637–NA) | 362 (237–NA) | 15, 15 | 0.2169 | 419 (291–NA) | 321 (269–655) |
| MAGEF1 | 16, 16 | 0.0376 | 1093 (404–NA) | 292 (102–NA) | 15, 15 | 0.2338 | 488 (294–NA) | 419 (269–NA) | ||
| MELTF | 16, 16 | 0.0127 | 225 (71–NA) | 646 (488 – NA) | 15, 15 | 0.0849 | 419 (321–1093) | 809 (366–NA) | ||
| STEAP1 | 16, 16 | 0.0305 | 1093 (497–NA) | 344 (77–NA) | 15, 15 | 0.5139 | 383 (321–NA) | 299 (269–NA) | ||
| VGLL4 | 16, 16 | 0.2087 | 497 (395–NA) | 296 (109–NA) | 15, 15 | 0.0368 | 496 (383–NA) | 461 (269–NA) | ||
| TCGA | Intermediate Risk vs. Good Risk (n = 75 vs. n = 17) | MORC4 | 23, 23 | 0.0235 | 253 (219–517) | 554 (362–NA) | 23, 23 | 0.8741 | 310 (259–834) | 298 (234–2121) |
| Poor Risk vs. Good Risk (n = 32 vs. n = 17) | SAGE1 | 13, 12 | 0.0394 | 2910 (158–NA) | 216 (113–NA) | 12, 12 | 0.2502 | 314 (119–NA) | 438 (128–NA) |
| KEGG (2016) Term | Overlapping KEGG (2016) Genes | No. of Subgroup Comparisons | |
|---|---|---|---|
| (A) | Mineral absorption Homo sapiens_hsa04978 | SLC34A2, STEAP1 | 3 |
| Vitamin digestion and absorption_Homo sapiens_hsa04977 | FOLH1 | 2 | |
| (B) | Alanine, aspartate and glutamate metabolism_Homo sapiens_hsa00250 | FOLH1 | 2 |
| Vitamin digestion and absorption_Homo sapiences_hsa04977 | FOLH1 | 2 | |
| (C) | Mineral absorption_Homo sapiens_hsa04978 | SLC34A2, STEAP1 | 3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davis, L.; Mills, K.I.; Orchard, K.H.; Guinn, B.-A. Identification of Genes Whose Expression Overlaps Age Boundaries and Correlates with Risk Groups in Paediatric and Adult Acute Myeloid Leukaemia. Cancers 2020, 12, 2769. https://doi.org/10.3390/cancers12102769
Davis L, Mills KI, Orchard KH, Guinn B-A. Identification of Genes Whose Expression Overlaps Age Boundaries and Correlates with Risk Groups in Paediatric and Adult Acute Myeloid Leukaemia. Cancers. 2020; 12(10):2769. https://doi.org/10.3390/cancers12102769
Chicago/Turabian StyleDavis, Lindsay, Ken I. Mills, Kim H. Orchard, and Barbara-Ann Guinn. 2020. "Identification of Genes Whose Expression Overlaps Age Boundaries and Correlates with Risk Groups in Paediatric and Adult Acute Myeloid Leukaemia" Cancers 12, no. 10: 2769. https://doi.org/10.3390/cancers12102769
APA StyleDavis, L., Mills, K. I., Orchard, K. H., & Guinn, B.-A. (2020). Identification of Genes Whose Expression Overlaps Age Boundaries and Correlates with Risk Groups in Paediatric and Adult Acute Myeloid Leukaemia. Cancers, 12(10), 2769. https://doi.org/10.3390/cancers12102769







