Targeting Liver Cancer Stem Cells: An Alternative Therapeutic Approach for Liver Cancer
Abstract
Simple Summary
Abstract
1. Introduction
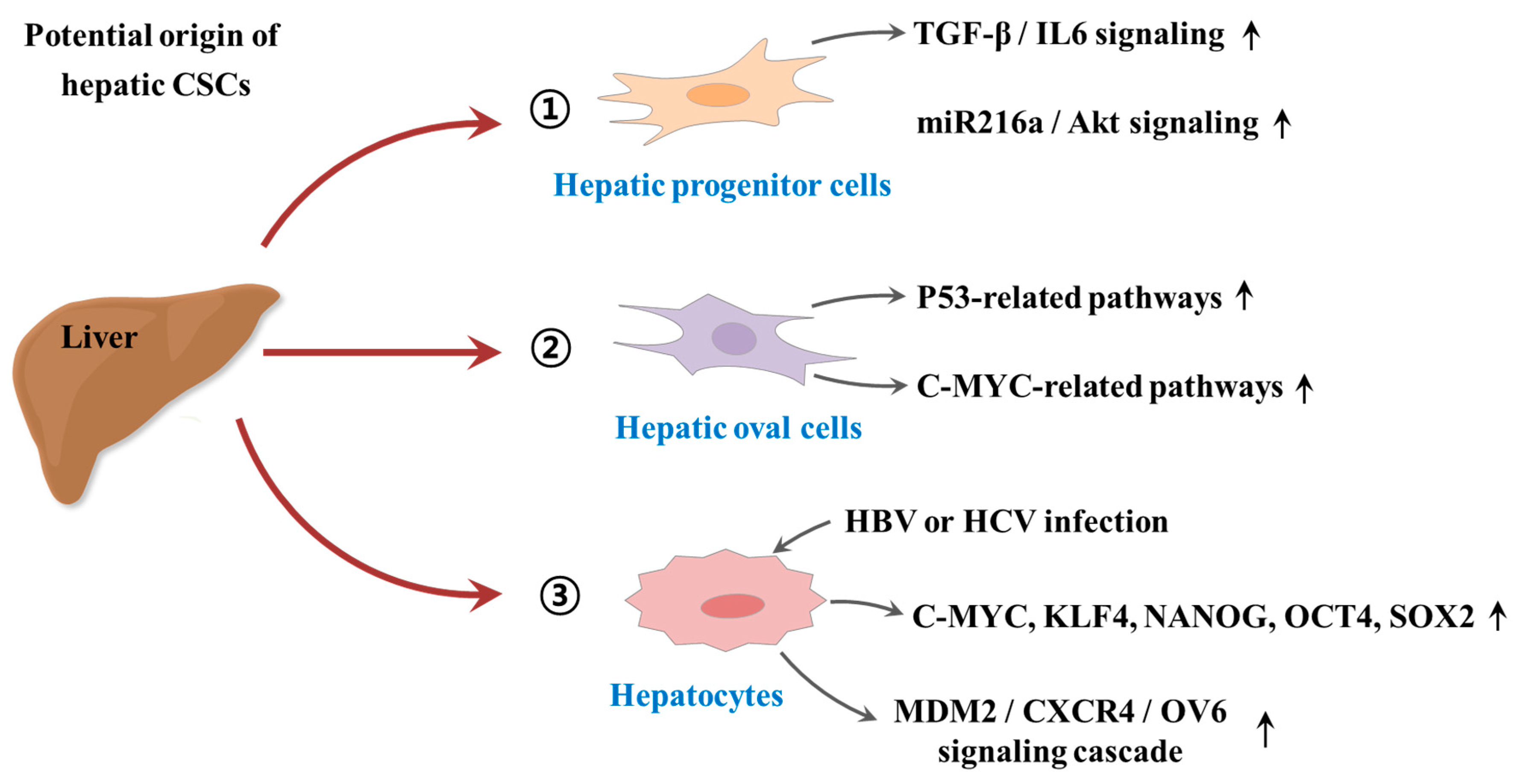
2. The Origin of Cancer Stem Cells
3. Cancer Stem Cells: Implications for Hepatocarcinogenesis
3.1. Identification of CSCs in Various Types of Tumors
3.2. CSCs as a Novel Therapeutic Target
3.3. The Hepatic CSC Microenvironment
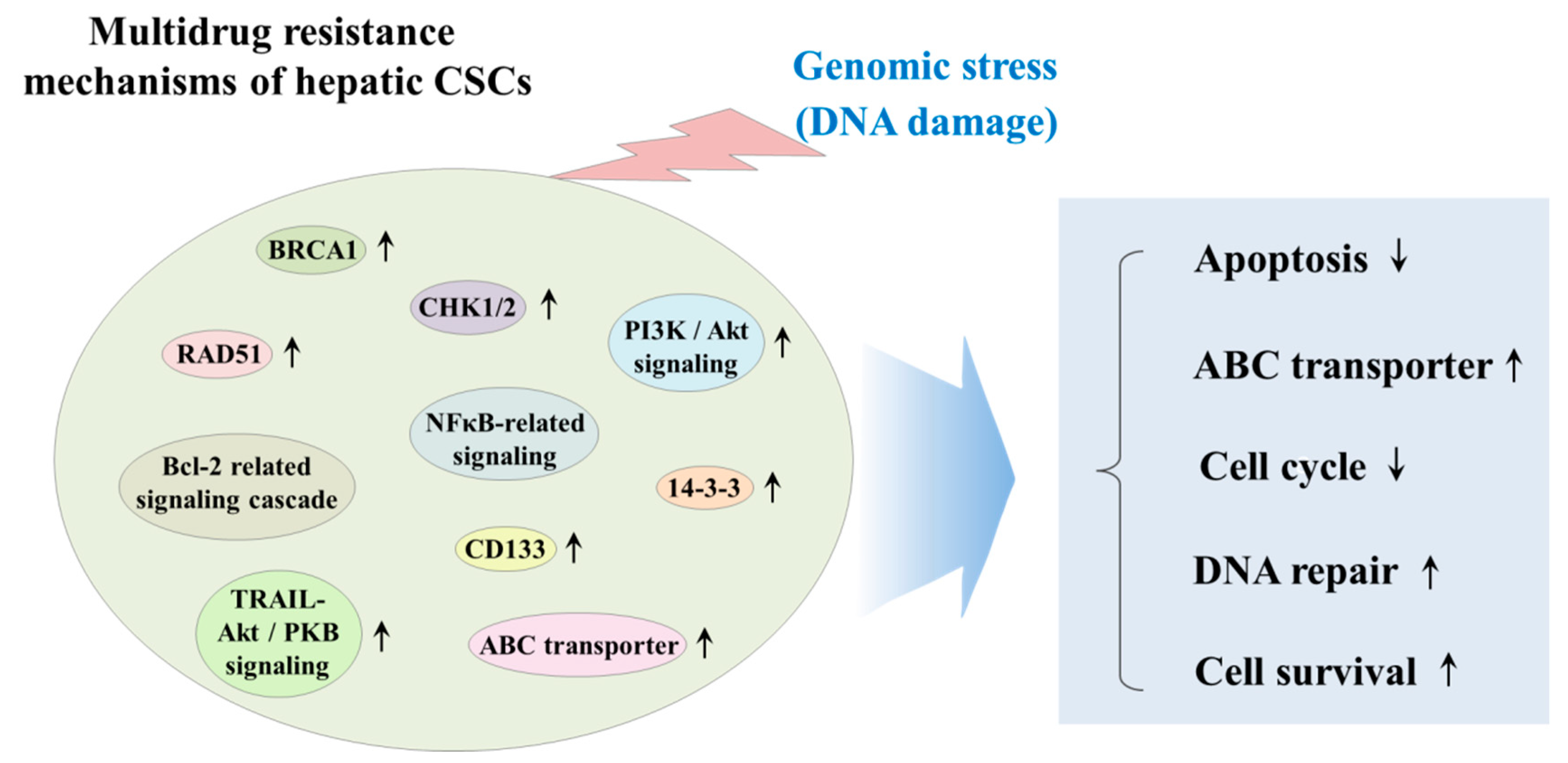
3.4. The Effect of Chemotherapy/Radiotherapy on Hepatic CSCs
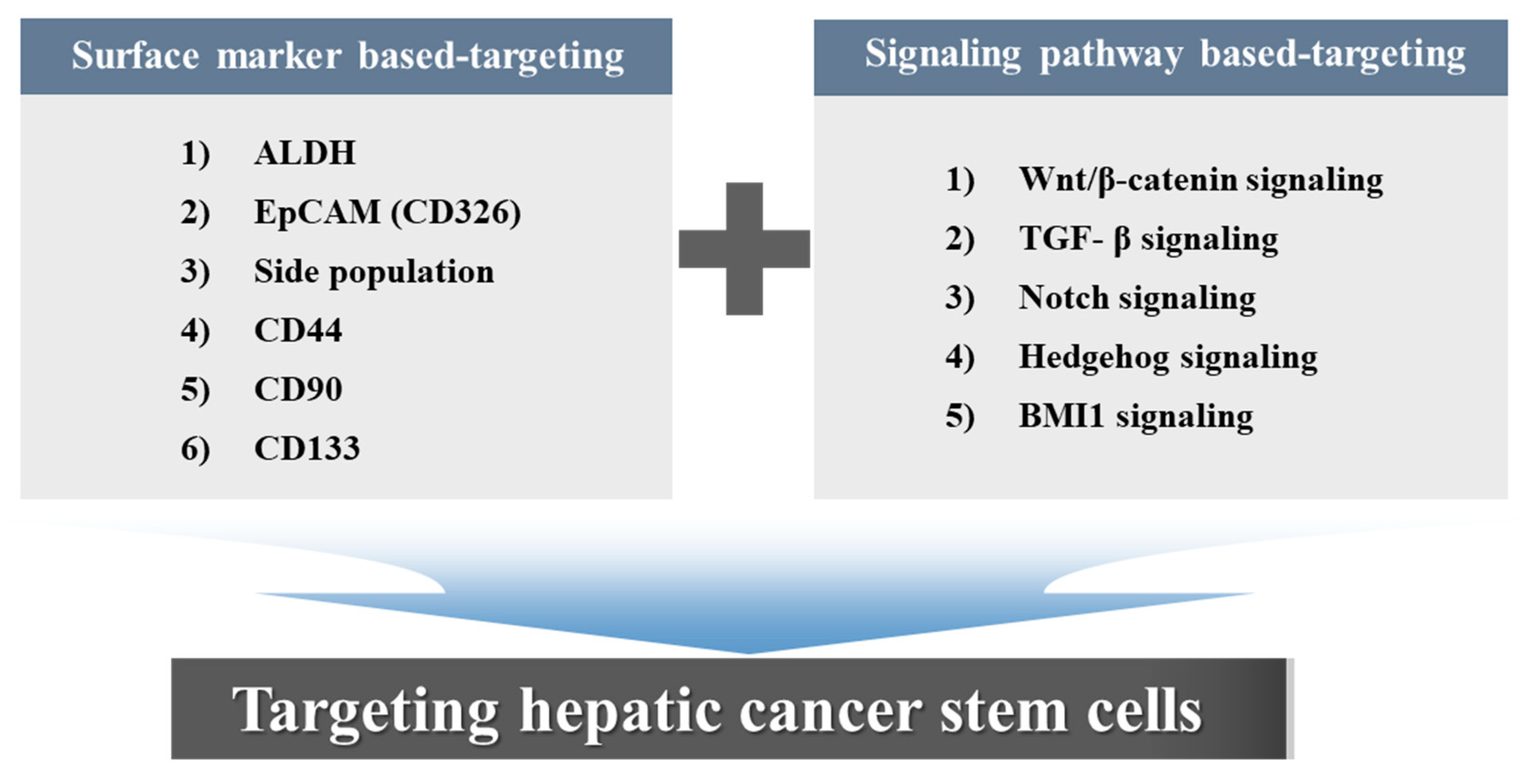
4. Surface Marker-Based Therapies
4.1. Aldehyde Dehydrogenase (ALDHs)
4.2. EpCAM
4.3. Side Population
4.4. CD44
4.5. CD90
4.6. CD133
5. Signaling Pathway-Based Therapies
5.1. Wnt/β-Catenin Signaling
5.2. Transforming Growth Factor (TGF)-β Signaling
5.3. Notch Signaling
5.4. Hedgehog Signaling
5.5. BMI1 Signaling
6. Potential Clinical Application of Liver Cancer Stem Cells
6.1. Aspects of Diagnosis and Prognosis
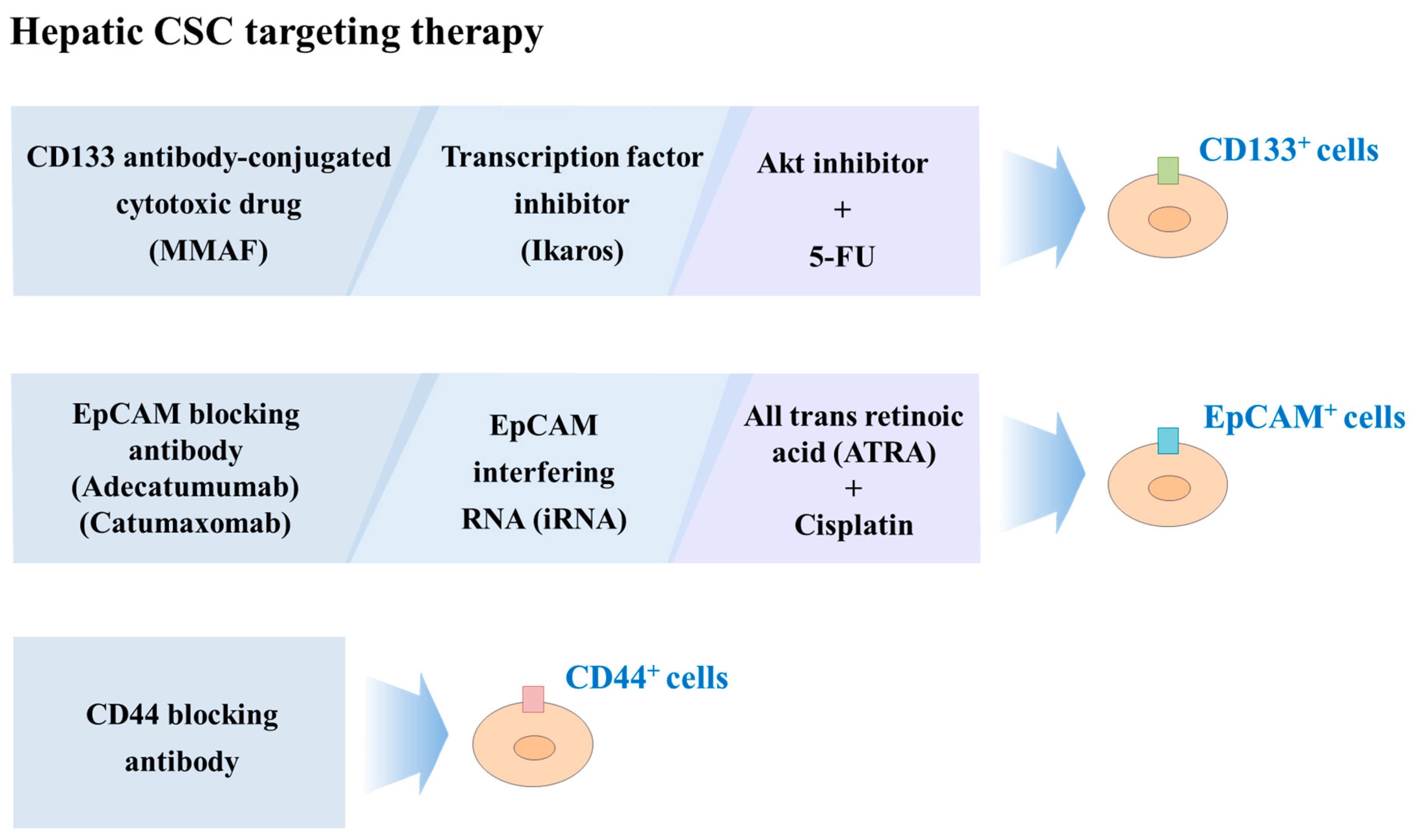
6.2. Liver CSCs-Targeted Therapy
7. Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Rawla, P.; Sunkara, T.; Muralidharan, P.; Raj, J.P. Update in global trends and aetiology of hepatocellular carcinoma. Contemp. Oncol. 2018, 22, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Wu, Y.; Zhou, J.; Zhao, J. Novel targets and small molecular interventions for liver cancer. Biomed. Res. Int. 2014, 2014, 148783. [Google Scholar] [CrossRef]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Hoyos, S.; Navas, M.C.; Restrepo, J.C.; Botero, R.C. Current controversies in cholangiocarcinoma. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 1461–1467. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Li, H.; Kaplan, D.E. Murine hepatoblast-derived liver tumors resembling human combined hepatocellular-cholangiocarcinoma with stem cell features. Cell Biosci. 2020, 10, 38. [Google Scholar] [CrossRef] [PubMed]
- Nio, K.; Yamashita, T.; Kaneko, S. The evolving concept of liver cancer stem cells. Mol. Cancer 2017, 16, 4. [Google Scholar] [CrossRef]
- Sun, J.H.; Luo, Q.; Liu, L.L.; Song, G.B. Liver cancer stem cell markers: Progression and therapeutic implications. World J. Gastroenterol. 2016, 22, 3547–3557. [Google Scholar] [CrossRef]
- Thorgeirsson, S.S. Stemness and reprogramming in liver cancer. Hepatology 2016, 63, 1068–1070. [Google Scholar] [CrossRef]
- Romano, M.; De Francesco, F.; Pirozzi, G.; Gringeri, E.; Boetto, R.; Di Domenico, M.; Zavan, B.; Ferraro, G.A.; Cillo, U. Expression of cancer stem cell biomarkers as a tool for a correct therapeutic approach to hepatocellular carcinoma. Oncoscience 2015, 2, 443–456. [Google Scholar] [CrossRef]
- Glumac, P.M.; LeBeau, A.M. The role of CD133 in cancer: A concise review. Clin. Transl. Med. 2018, 7, 18. [Google Scholar] [CrossRef]
- Akiba, J.; Nakashima, O.; Hattori, S.; Naito, Y.; Kusano, H.; Kondo, R.; Nakayama, M.; Tanikawa, K.; Todoroki, K.; Umeno, Y.; et al. The expression of arginase-1, keratin (K) 8 and K18 in combined hepatocellular-cholangiocarcinoma, subtypes with stem-cell features, intermediate-cell type. J. Clin. Pathol. 2016, 69, 846–851. [Google Scholar] [CrossRef] [PubMed]
- Thanee, M.; Loilome, W.; Techasen, A.; Sugihara, E.; Okazaki, S.; Abe, S.; Ueda, S.; Masuko, T.; Namwat, N.; Khuntikeo, N.; et al. CD44 variant-dependent redox status regulation in liver fluke-associated cholangiocarcinoma: A target for cholangiocarcinoma treatment. Cancer Sci. 2016, 107, 991–1000. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.J.; Chu, P.Y. Role of Cancer Stem Cells in Cholangiocarcinoma and Therapeutic Implications. Int. J. Mol. Sci. 2019, 20, 4154. [Google Scholar] [CrossRef] [PubMed]
- Bragazzi, M.C.; Ridola, L.; Safarikia, S.; Matteo, S.D.; Costantini, D.; Nevi, L.; Cardinale, V. New insights into cholangiocarcinoma: Multiple stems and related cell lineages of origin. Ann. Gastroenterol. 2018, 31, 42–55. [Google Scholar] [CrossRef] [PubMed]
- Rycaj, K.; Tang, D.G. Cell-of-Origin of Cancer versus Cancer Stem Cells: Assays and Interpretations. Cancer Res. 2015, 75, 4003–4011. [Google Scholar] [CrossRef] [PubMed]
- White, A.C.; Lowry, W.E. Refining the role for adult stem cells as cancer cells of origin. Trends Cell Biol. 2015, 25, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Zakrzewski, W.; Dobrzynski, M.; Szymonowicz, M.; Rybak, Z. Stem cells: Past, present, and future. Stem Cell Res. Ther. 2019, 10, 68. [Google Scholar] [CrossRef]
- Afify, S.M.; Seno, M. Conversion of Stem Cells to Cancer Stem Cells: Undercurrent of Cancer Initiation. Cancers 2019, 11, 345. [Google Scholar] [CrossRef]
- Chopra, M.; Bohlander, S.K. The cell of origin and the leukemia stem cell in acute myeloid leukemia. Genes Chromosomes Cancer 2019, 58, 850–858. [Google Scholar] [CrossRef]
- Wang, X.; Huang, S.; Chen, J.L. Understanding of leukemic stem cells and their clinical implications. Mol. Cancer 2017, 16, 2. [Google Scholar] [CrossRef]
- Prasetyanti, P.R.; Medema, J.P. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer 2017, 16, 41. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Luo, Y.; Dingli, D.; Traulsen, A. The invasion of de-differentiating cancer cells into hierarchical tissues. PLoS Comput. Biol. 2019, 15, e1007167. [Google Scholar] [CrossRef]
- Gong, L.; Yan, Q.; Zhang, Y.; Fang, X.; Liu, B.; Guan, X. Cancer cell reprogramming: A promising therapy converting malignancy to benignity. Cancer Commun. 2019, 39, 48. [Google Scholar] [CrossRef] [PubMed]
- Brazhnik, K.; Sun, S.; Alani, O.; Kinkhabwala, M.; Wolkoff, A.W.; Maslov, A.Y.; Dong, X.; Vijg, J. Single-cell analysis reveals different age-related somatic mutation profiles between stem and differentiated cells in human liver. Sci. Adv. 2020, 6, eaax2659. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Won, M.; Kim, J.H.; Jung, E.; Min, K.; Jangili, P.; Kim, J.S. Cancer stem cell-targeted bio-imaging and chemotherapeutic perspective. Chem. Soc. Rev. 2020. [Google Scholar] [CrossRef]
- Andrews, A.; Warner, K.; Rodriguez-Ramirez, C.; Pearson, A.T.; Nor, F.; Zhang, Z.; Kerk, S.; Kulkarni, A.; Helman, J.I.; Brenner, J.C.; et al. Ablation of Cancer Stem Cells by Therapeutic Inhibition of the MDM2-p53 Interaction in Mucoepidermoid Carcinoma. Clin. Cancer Res. 2019, 25, 1588–1600. [Google Scholar] [CrossRef]
- Bruce, W.R.; Van Der Gaag, H. A Quantitative Assay for the Number of Murine Lymphoma Cells Capable of Proliferation in Vivo. Nature 1963, 199, 79–80. [Google Scholar] [CrossRef]
- Karunanithi, S.; Levi, L.; DeVecchio, J.; Karagkounis, G.; Reizes, O.; Lathia, J.D.; Kalady, M.F.; Noy, N. RBP4-STRA6 Pathway Drives Cancer Stem Cell Maintenance and Mediates High-Fat Diet-Induced Colon Carcinogenesis. Stem Cell Rep. 2017, 9, 438–450. [Google Scholar] [CrossRef]
- Parada, L.F.; Dirks, P.B.; Wechsler-Reya, R.J. Brain Tumor Stem Cells Remain in Play. J. Clin. Oncol. 2017, 35, 2428–2431. [Google Scholar] [CrossRef] [PubMed]
- Phiboonchaiyanan, P.P.; Chanvorachote, P. Suppression of a cancer stem-like phenotype mediated by alpha-lipoic acid in human lung cancer cells through down-regulation of beta-catenin and Oct-4. Cell. Oncol. 2017, 40, 497–510. [Google Scholar] [CrossRef] [PubMed]
- Li, X.F.; Chen, C.; Xiang, D.M.; Qu, L.; Sun, W.; Lu, X.Y.; Zhou, T.F.; Chen, S.Z.; Ning, B.F.; Cheng, Z.; et al. Chronic Inflammation-elicited Liver Progenitor Cell Conversion to Liver Cancer Stem Cell with Clinical Significance. Hepatology 2017, 66, 1934–1951. [Google Scholar] [CrossRef]
- Chen, S.M.; Wang, B.Y.; Lee, C.H.; Lee, H.T.; Li, J.J.; Hong, G.C.; Hung, Y.C.; Chien, P.J.; Chang, C.Y.; Hsu, L.S.; et al. Hinokitiol up-regulates miR-494-3p to suppress BMI1 expression and inhibits self-renewal of breast cancer stem/progenitor cells. Oncotarget 2017, 8, 76057. [Google Scholar] [CrossRef]
- Qin, G.; Lian, J.; Yue, D.; Chen, X.; Nan, S.; Qi, Y.; Li, B.; Cui, G.; Li, X.; Zhao, S.; et al. Musashi1, a potential prognostic marker in esophageal squamous cell carcinoma. Oncol. Rep. 2017, 38, 1724–1732. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.W.; Song, Y.; Kim, S.H.; Kim, J.; Seo, H.R. Potential mechanisms of CD133 in cancer stem cells. Life Sci. 2017, 184, 25–29. [Google Scholar] [CrossRef]
- Durko, L.; Wlodarski, W.; Stasikowska-Kanicka, O.; Wagrowska-Danilewicz, M.; Danilewicz, M.; Hogendorf, P.; Strzelczyk, J.; Malecka-Panas, E. Expression and Clinical Significance of Cancer Stem Cell Markers CD24, CD44, and CD133 in Pancreatic Ductal Adenocarcinoma and Chronic Pancreatitis. Dis. Markers 2017, 2017, 3276806. [Google Scholar] [CrossRef]
- Deng, L.; Li, D.; Gu, W.; Liu, A.; Cheng, X. Formation of spherical cancer stem-like cell colonies with resistance to chemotherapy drugs in the human malignant fibrous histiocytoma NMFH-1 cell line. Oncol. Lett. 2015, 10, 3323–3331. [Google Scholar] [CrossRef][Green Version]
- De Angelis, M.L.; Francescangeli, F.; La Torre, F.; Zeuner, A. Stem Cell Plasticity and Dormancy in the Development of Cancer Therapy Resistance. Front. Oncol. 2019, 9, 626. [Google Scholar] [CrossRef]
- Arnold, C.R.; Mangesius, J.; Skvortsova, I.I.; Ganswindt, U. The Role of Cancer Stem Cells in Radiation Resistance. Front. Oncol. 2020, 10, 164. [Google Scholar] [CrossRef]
- Thomas, D.; Majeti, R. Biology and relevance of human acute myeloid leukemia stem cells. Blood 2017, 129, 1577–1585. [Google Scholar] [CrossRef] [PubMed]
- Izumi, D.; Ishimoto, T.; Miyake, K.; Eto, T.; Arima, K.; Kiyozumi, Y.; Uchihara, T.; Kurashige, J.; Iwatsuki, M.; Baba, Y.; et al. Colorectal Cancer Stem Cells Acquire Chemoresistance through the Upregulation of FBXW7 and the Consequent Degradation of c-Myc. Stem Cells 2017, 35, 2027–2036. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.Y.; Lin, C.H.; Chuang, J.Y.; Chang, W.C.; Hsu, T.I. MDM2 Degrades Deacetylated Nucleolin through Ubiquitination to Promote Glioma Stem-Like Cell Enrichment for Chemotherapeutic Resistance. Mol. Neurobiol. 2018, 55, 3211–3223. [Google Scholar] [CrossRef]
- Chaudhary, A.K.; Mondal, G.; Kumar, V.; Kattel, K.; Mahato, R.I. Chemosensitization and inhibition of pancreatic cancer stem cell proliferation by overexpression of microRNA-205. Cancer Lett. 2017, 402, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Redmer, T.; Walz, I.; Klinger, B.; Khouja, S.; Welte, Y.; Schafer, R.; Regenbrecht, C. The role of the cancer stem cell marker CD271 in DNA damage response and drug resistance of melanoma cells. Oncogenesis 2017, 6, e291. [Google Scholar] [CrossRef]
- Woo, Y.; Oh, J.; Kim, J.S. Suppression of Nrf2 Activity by Chestnut Leaf Extract Increases Chemosensitivity of Breast Cancer Stem Cells to Paclitaxel. Nutrients 2017, 9, 760. [Google Scholar] [CrossRef]
- Yan, Y.; Wang, Y.; Zhao, P.; Ma, W.; Hu, Z.; Zhang, K. BMI-1 Promotes Self-Renewal of Radio- and Temozolomide (TMZ)-Resistant Breast Cancer Cells. Reprod. Sci. 2017, 24, 1620–1629. [Google Scholar] [CrossRef]
- Yang, W.; Yu, H.; Shen, Y.; Liu, Y.; Yang, Z.; Sun, T. MiR-146b-5p overexpression attenuates stemness and radioresistance of glioma stem cells by targeting HuR/lincRNA-p21/beta-catenin pathway. Oncotarget 2016, 7, 41505–41526. [Google Scholar] [CrossRef]
- Tang, Y.; Kitisin, K.; Jogunoori, W.; Li, C.; Deng, C.X.; Mueller, S.C.; Ressom, H.W.; Rashid, A.; He, A.R.; Mendelson, J.S.; et al. Progenitor/stem cells give rise to liver cancer due to aberrant TGF-beta and IL-6 signaling. Proc. Natl. Acad. Sci. USA 2008, 105, 2445–2450. [Google Scholar] [CrossRef]
- Wu, K.; Ding, J.; Chen, C.; Sun, W.; Ning, B.F.; Wen, W.; Huang, L.; Han, T.; Yang, W.; Wang, C.; et al. Hepatic transforming growth factor beta gives rise to tumor-initiating cells and promotes liver cancer development. Hepatology 2012, 56, 2255–2267. [Google Scholar] [CrossRef]
- Xu, R.H.; Zheng, L.Y.; He, D.L.; Meng, J.; Xia, L.P.; Hao, X.B.; Zhang, Z.Z. Profiling of differentially expressed microRNAs (miRNAs) during differentiation of rat hepatic oval cells (HOCs) into hepatocellular carcinoma (HCC) cells. Clin. Transl. Oncol. 2015, 17, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Dumble, M.L.; Croager, E.J.; Yeoh, G.C.; Quail, E.A. Generation and characterization of p53 null transformed hepatic progenitor cells: Oval cells give rise to hepatocellular carcinoma. Carcinogenesis 2002, 23, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.H.; Gong, J.Q.; Zhang, W. Function of oval cells in hepatocellular carcinoma in rats. World J. Gastroenterol. 2004, 10, 2482–2487. [Google Scholar] [CrossRef]
- El-Serag, H.B. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 2012, 142, 1264–1273.e1. [Google Scholar] [CrossRef]
- Liu, Z.; Dai, X.; Wang, T.; Zhang, C.; Zhang, W.; Zhang, W.; Zhang, Q.; Wu, K.; Liu, F.; Liu, Y.; et al. Hepatitis B virus PreS1 facilitates hepatocellular carcinoma development by promoting appearance and self-renewal of liver cancer stem cells. Cancer Lett. 2017, 400, 149–160. [Google Scholar] [CrossRef]
- Wang, C.; Wang, M.D.; Cheng, P.; Huang, H.; Dong, W.; Zhang, W.W.; Li, P.P.; Lin, C.; Pan, Z.Y.; Wu, M.C.; et al. Hepatitis B virus X protein promotes the stem-like properties of OV6(+) cancer cells in hepatocellular carcinoma. Cell Death Dis. 2017, 8, e2560. [Google Scholar] [CrossRef]
- Li, H.; Zhang, L. Liver regeneration microenvironment of hepatocellular carcinoma for prevention and therapy. Oncotarget 2017, 8, 1805–1813. [Google Scholar] [CrossRef]
- Verhulst, S.; Best, J.; van Grunsven, L.A.; Dolle, L. Advances in hepatic stem/progenitor cell biology. EXCLI J. 2015, 14, 33–47. [Google Scholar] [CrossRef]
- Carpino, G.; Renzi, A.; Franchitto, A.; Cardinale, V.; Onori, P.; Reid, L.; Alvaro, D.; Gaudio, E. Stem/Progenitor Cell Niches Involved in Hepatic and Biliary Regeneration. Stem Cells Int. 2016, 2016, 3658013. [Google Scholar] [CrossRef]
- Wu, Y.; Bu, F.; Yu, H.; Li, W.; Huang, C.; Meng, X.; Zhang, L.; Ma, T.; Li, J. Methylation of Septin9 mediated by DNMT3a enhances hepatic stellate cells activation and liver fibrogenesis. Toxicol. Appl. Pharmacol. 2017, 315, 35–49. [Google Scholar] [CrossRef]
- Junttila, M.R.; de Sauvage, F.J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 2013, 501, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Zeisberg, M. Fibroblasts in cancer. Nat. Rev. Cancer 2006, 6, 392–401. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Bai, S.; Wang, R.; Xiong, S.; Li, Y.; Wang, X.; Chen, W.; Cheng, B. Cancer-associated fibroblasts endow stem-like qualities to liver cancer cells by modulating autophagy. Cancer Manag. Res. 2019, 11, 5737–5744. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, G.J.; Azuma, A.; Miura, Y.; Orimo, A. Activated Fibroblast Program Orchestrates Tumor Initiation and Progression; Molecular Mechanisms and the Associated Therapeutic Strategies. Int. J. Mol. Sci. 2019, 20, 2256. [Google Scholar] [CrossRef]
- Cuiffo, B.G.; Karnoub, A.E. Mesenchymal stem cells in tumor development: Emerging roles and concepts. Cell Adhes. Migr. 2012, 6, 220–230. [Google Scholar] [CrossRef]
- Nishimura, K.; Semba, S.; Aoyagi, K.; Sasaki, H.; Yokozaki, H. Mesenchymal stem cells provide an advantageous tumor microenvironment for the restoration of cancer stem cells. Pathobiology 2012, 79, 290–306. [Google Scholar] [CrossRef]
- Yan, L.; Xu, F.; Dai, C.L. Relationship between epithelial-to-mesenchymal transition and the inflammatory microenvironment of hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2018, 37, 203. [Google Scholar] [CrossRef]
- Mi, F.; Gong, L. Secretion of interleukin-6 by bone marrow mesenchymal stem cells promotes metastasis in hepatocellular carcinoma. Biosci. Rep. 2017, 37. [Google Scholar] [CrossRef]
- Cabarcas, S.M.; Mathews, L.A.; Farrar, W.L. The cancer stem cell niche--there goes the neighborhood? Int. J. Cancer 2011, 129, 2315–2327. [Google Scholar] [CrossRef]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 2018, 9, 7204–7218. [Google Scholar] [CrossRef]
- Dong, H.J.; Jang, G.B.; Lee, H.Y.; Park, S.R.; Kim, J.Y.; Nam, J.S.; Hong, I.S. The Wnt/beta-catenin signaling/Id2 cascade mediates the effects of hypoxia on the hierarchy of colorectal-cancer stem cells. Sci. Rep. 2016, 6, 22966. [Google Scholar] [CrossRef] [PubMed]
- Morrison, R.; Schleicher, S.M.; Sun, Y.; Niermann, K.J.; Kim, S.; Spratt, D.E.; Chung, C.H.; Lu, B. Targeting the mechanisms of resistance to chemotherapy and radiotherapy with the cancer stem cell hypothesis. J. Oncol. 2011, 2011, 941876. [Google Scholar] [CrossRef] [PubMed]
- Frosina, G. DNA repair in normal and cancer stem cells, with special reference to the central nervous system. Curr. Med. Chem. 2009, 16, 854–866. [Google Scholar] [CrossRef] [PubMed]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef]
- Lee, Y.K.; Hur, W.; Lee, S.W.; Hong, S.W.; Kim, S.W.; Choi, J.E.; Yoon, S.K. Knockdown of 14-3-3zeta enhances radiosensitivity and radio-induced apoptosis in CD133(+) liver cancer stem cells. Exp. Mol. Med. 2014, 46, e77. [Google Scholar] [CrossRef]
- Ma, S.; Lee, T.K.; Zheng, B.J.; Chan, K.W.; Guan, X.Y. CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene 2008, 27, 1749–1758. [Google Scholar] [CrossRef]
- Dean, M. ABC transporters, drug resistance, and cancer stem cells. J. Mammary Gland Biol. Neoplasia 2009, 14, 3–9. [Google Scholar] [CrossRef]
- Fung, S.W.; Cheung, P.F.; Yip, C.W.; Ng, L.W.; Cheung, T.T.; Chong, C.C.; Lee, C.; Lai, P.B.; Chan, A.W.; Tsao, G.S.; et al. The ATP-binding cassette transporter ABCF1 is a hepatic oncofetal protein that promotes chemoresistance, EMT and cancer stemness in hepatocellular carcinoma. Cancer Lett. 2019, 457, 98–109. [Google Scholar] [CrossRef]
- Nishimura, T.; Nakata, A.; Chen, X.; Nishi, K.; Meguro-Horike, M.; Sasaki, S.; Kita, K.; Horike, S.I.; Saitoh, K.; Kato, K.; et al. Cancer stem-like properties and gefitinib resistance are dependent on purine synthetic metabolism mediated by the mitochondrial enzyme MTHFD2. Oncogene 2019, 38, 2464–2481. [Google Scholar] [CrossRef]
- Kahraman, D.C.; Kahraman, T.; Cetin-Atalay, R. Targeting PI3K/Akt/mTOR Pathway Identifies Differential Expression and Functional Role of IL8 in Liver Cancer Stem Cell Enrichment. Mol. Cancer Ther. 2019, 18, 2146–2157. [Google Scholar] [CrossRef]
- Walczak, H.; Miller, R.E.; Ariail, K.; Gliniak, B.; Griffith, T.S.; Kubin, M.; Chin, W.; Jones, J.; Woodward, A.; Le, T.; et al. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat. Med. 1999, 5, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Tang, H.; Zhang, L.; Gong, L.; Wu, G.; Ni, J.; Tang, X. Suppression of miR-21-3p enhances TRAIL-mediated apoptosis in liver cancer stem cells by suppressing the PI3K/Akt/Bad cascade via regulating PTEN. Cancer Manag. Res. 2019, 11, 955–968. [Google Scholar] [CrossRef]
- Rausch, V.; Liu, L.; Kallifatidis, G.; Baumann, B.; Mattern, J.; Gladkich, J.; Wirth, T.; Schemmer, P.; Buchler, M.W.; Zoller, M.; et al. Synergistic activity of sorafenib and sulforaphane abolishes pancreatic cancer stem cell characteristics. Cancer Res. 2010, 70, 5004–5013. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Cao, X.; Xiao, Q.; Sheng, X.; Ren, K.; Quan, M.; Song, Z.; Li, D.; Zheng, Y.; Zeng, W.; et al. Synergistic inhibition of characteristics of liver cancer stem-like cells with a combination of sorafenib and 8-bromo-7-methoxychrysin in SMMC-7721 cell line. Oncol. Rep. 2016, 36, 1731–1738. [Google Scholar] [CrossRef] [PubMed]
- Tomita, H.; Tanaka, K.; Tanaka, T.; Hara, A. Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget 2016, 7, 11018–11032. [Google Scholar] [CrossRef] [PubMed]
- Koenders, S.T.A.; Wijaya, L.S.; Erkelens, M.N.; Bakker, A.T.; van der Noord, V.E.; van Rooden, E.J.; Burggraaff, L.; Putter, P.C.; Botter, E.; Wals, K.; et al. Development of a Retinal-Based Probe for the Profiling of Retinaldehyde Dehydrogenases in Cancer Cells. ACS Cent. Sci. 2019, 5, 1965–1974. [Google Scholar] [CrossRef]
- Rodriguez-Torres, M.; Allan, A.L. Aldehyde dehydrogenase as a marker and functional mediator of metastasis in solid tumors. Clin. Exp. Metastasis 2016, 33, 97–113. [Google Scholar] [CrossRef]
- Nguyen, P.H.; Giraud, J.; Chambonnier, L.; Dubus, P.; Wittkop, L.; Belleannee, G.; Collet, D.; Soubeyran, I.; Evrard, S.; Rousseau, B.; et al. Characterization of Biomarkers of Tumorigenic and Chemoresistant Cancer Stem Cells in Human Gastric Carcinoma. Clin. Cancer Res. 2017, 23, 1586–1597. [Google Scholar] [CrossRef]
- Qiu, Y.; Pu, T.; Guo, P.; Wei, B.; Zhang, Z.; Zhang, H.; Zhong, X.; Zheng, H.; Chen, L.; Bu, H.; et al. ALDH(+)/CD44(+) cells in breast cancer are associated with worse prognosis and poor clinical outcome. Exp. Mol. Pathol. 2016, 100, 145–150. [Google Scholar] [CrossRef]
- Ma, S.; Chan, K.W.; Lee, T.K.; Tang, K.H.; Wo, J.Y.; Zheng, B.J.; Guan, X.Y. Aldehyde dehydrogenase discriminates the CD133 liver cancer stem cell populations. Mol. Cancer Res. MCR 2008, 6, 1146–1153. [Google Scholar] [CrossRef]
- Wang, N.; Wang, S.; Li, M.Y.; Hu, B.G.; Liu, L.P.; Yang, S.L.; Yang, S.; Gong, Z.; Lai, P.B.S.; Chen, G.G. Cancer stem cells in hepatocellular carcinoma: An overview and promising therapeutic strategies. Ther. Adv. Med. Oncol. 2018, 10, 1758835918816287. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Chen, Y.W.; Hsu, H.S.; Tseng, L.M.; Huang, P.I.; Lu, K.H.; Chen, D.T.; Tai, L.K.; Yung, M.C.; Chang, S.C.; et al. Aldehyde dehydrogenase 1 is a putative marker for cancer stem cells in head and neck squamous cancer. Biochem. Biophys. Res. Commun. 2009, 385, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Yang, Y.; Yang, F.; Liu, S.; Zhu, Z.; Lei, Z.; Guo, J. Functions of EpCAM in physiological processes and diseases (Review). Int. J. Mol. Med. 2018, 42, 1771–1785. [Google Scholar] [CrossRef] [PubMed]
- Litvinov, S.V.; Velders, M.P.; Bakker, H.A.; Fleuren, G.J.; Warnaar, S.O. Ep-CAM: A human epithelial antigen is a homophilic cell-cell adhesion molecule. J. Cell Biol. 1994, 125, 437–446. [Google Scholar] [CrossRef]
- Litvinov, S.V.; Bakker, H.A.; Gourevitch, M.M.; Velders, M.P.; Warnaar, S.O. Evidence for a role of the epithelial glycoprotein 40 (Ep-CAM) in epithelial cell-cell adhesion. Cell Adhes. Commun. 1994, 2, 417–428. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, H.; Ding, J.; Liu, H.; Li, H.; Li, H.; Lu, M.; Miao, Y.; Li, L.; Zheng, J. Combination Therapy with EpCAM-CAR-NK-92 Cells and Regorafenib against Human Colorectal Cancer Models. J. Immunol. Res. 2018, 2018, 4263520. [Google Scholar] [CrossRef]
- Leng, Z.; Xia, Q.; Chen, J.; Li, Y.; Xu, J.; Zhao, E.; Zheng, H.; Ai, W.; Dong, J. Lgr5+CD44+EpCAM+ Strictly Defines Cancer Stem Cells in Human Colorectal Cancer. Cell. Physiol. Biochem. 2018, 46, 860–872. [Google Scholar] [CrossRef]
- Nio, K.; Yamashita, T.; Okada, H.; Kondo, M.; Hayashi, T.; Hara, Y.; Nomura, Y.; Zeng, S.S.; Yoshida, M.; Hayashi, T.; et al. Defeating EpCAM(+) liver cancer stem cells by targeting chromatin remodeling enzyme CHD4 in human hepatocellular carcinoma. J. Hepatol. 2015, 63, 1164–1172. [Google Scholar] [CrossRef]
- Dai, X.M.; Huang, T.; Yang, S.L.; Zheng, X.M.; Chen, G.G.; Zhang, T. Peritumoral EpCAM Is an Independent Prognostic Marker after Curative Resection of HBV-Related Hepatocellular Carcinoma. Dis. Markers 2017, 2017, 8495326. [Google Scholar] [CrossRef]
- Hao, S.; Chen, S.; Tu, C.; Huang, T. Anterior Approach to Improve the Prognosis in HCC Patients via Decreasing Dissemination of EpCAM+ Circulating Tumor Cells. J. Gastrointest. Surg. 2017, 21, 1112–1120. [Google Scholar] [CrossRef]
- Khosla, R.; Rastogi, A.; Ramakrishna, G.; Pamecha, V.; Mukhopadhyay, A.; Vasudevan, M.; Sarin, S.K.; Trehanpati, N. EpCAM+ Liver Cancer Stem-Like Cells Exhibiting Autocrine Wnt Signaling Potentially Originate in Cirrhotic Patients. Stem Cells Transl. Med. 2017, 6, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Ji, J.; Budhu, A.; Forgues, M.; Yang, W.; Wang, H.Y.; Jia, H.; Ye, Q.; Qin, L.X.; Wauthier, E.; et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology 2009, 136, 1012–1024. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.J.; Li, C.J.; Wu, M.Y.; Chu, P.Y. Overexpression of epithelial cell adhesion molecule as a predictor of poor outcome in patients with hepatocellular carcinoma. Exp. Ther. Med. 2018, 16, 4810–4816. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, A.; Suda, T.; Oda, C.; Kimura, A.; Hosaka, K.; Kimura, N.; Tominaga, K.; Hayashi, K.; Takamura, M.; Terai, S. EpCAM- and/or NCAM-Expressing Hepatocellular Carcinoma in Which Behavior of Hepatic Progenitor Cell Marker-Positive Cells Are Followed. Case Rep. Gastroenterol. 2019, 13, 118–124. [Google Scholar] [CrossRef]
- Wang, C.; Yang, W.; Yan, H.X.; Luo, T.; Zhang, J.; Tang, L.; Wu, F.Q.; Zhang, H.L.; Yu, L.X.; Zheng, L.Y.; et al. Hepatitis B virus X (HBx) induces tumorigenicity of hepatic progenitor cells in 3,5-diethoxycarbonyl-1,4-dihydrocollidine-treated HBx transgenic mice. Hepatology 2012, 55, 108–120. [Google Scholar] [CrossRef]
- Arzumanyan, A.; Friedman, T.; Ng, I.O.; Clayton, M.M.; Lian, Z.; Feitelson, M.A. Does the hepatitis B antigen HBx promote the appearance of liver cancer stem cells? Cancer Res. 2011, 71, 3701–3708. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Tang, J.; Deng, L.; Xie, Y.; Jiang, R.; Li, G.; Sun, B. LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget 2015, 6, 42813–42824. [Google Scholar] [CrossRef] [PubMed]
- Robey, R.W.; Pluchino, K.M.; Hall, M.D.; Fojo, A.T.; Bates, S.E.; Gottesman, M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 2018, 18, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, M.; Ota, M.; Okada, Y. Isolation of Cancer Stem Cells by Side Population Method. Methods Mol. Biol. 2018, 1692, 49–59. [Google Scholar] [CrossRef]
- Chiba, T.; Kita, K.; Zheng, Y.W.; Yokosuka, O.; Saisho, H.; Iwama, A.; Nakauchi, H.; Taniguchi, H. Side population purified from hepatocellular carcinoma cells harbors cancer stem cell-like properties. Hepatology 2006, 44, 240–251. [Google Scholar] [CrossRef]
- Hu, C.; Li, H.; Li, J.; Zhu, Z.; Yin, S.; Hao, X.; Yao, M.; Zheng, S.; Gu, J. Analysis of ABCG2 expression and side population identifies intrinsic drug efflux in the HCC cell line MHCC-97L and its modulation by Akt signaling. Carcinogenesis 2008, 29, 2289–2297. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Han, J.; Kim, J.B.; Yang, M.G.; Kim, Y.J.; Lim, H.J.; An, S.Y.; Kim, J.H. Interleukin-8 is related to poor chemotherapeutic response and tumourigenicity in hepatocellular carcinoma. Eur. J. Cancer 2014, 50, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Chiba, T.; Miyagi, S.; Saraya, A.; Aoki, R.; Seki, A.; Morita, Y.; Yonemitsu, Y.; Yokosuka, O.; Taniguchi, H.; Nakauchi, H.; et al. The polycomb gene product BMI1 contributes to the maintenance of tumor-initiating side population cells in hepatocellular carcinoma. Cancer Res. 2008, 68, 7742–7749. [Google Scholar] [CrossRef] [PubMed]
- Barat, S.; Chen, X.; Cuong Bui, K.; Bozko, P.; Gotze, J.; Christgen, M.; Krech, T.; Malek, N.P.; Plentz, R.R. Gamma-Secretase Inhibitor IX (GSI) Impairs Concomitant Activation of Notch and Wnt-Beta-Catenin Pathways in CD44+ Gastric Cancer Stem Cells. Stem Cells Transl. Med. 2017, 6, 819–829. [Google Scholar] [CrossRef]
- Keymoosi, H.; Gheytanchi, E.; Asgari, M.; Shariftabrizi, A.; Madjd, Z. ALDH1 in combination with CD44 as putative cancer stem cell markers are correlated with poor prognosis in urothelial carcinoma of the urinary bladder. Asian Pac. J. Cancer Prev. 2014, 15, 2013–2020. [Google Scholar] [CrossRef]
- Liu, H.; Wang, Y.J.; Bian, L.; Fang, Z.H.; Zhang, Q.Y.; Cheng, J.X. CD44+/CD24+ cervical cancer cells resist radiotherapy and exhibit properties of cancer stem cells. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 1745–1754. [Google Scholar]
- Nurwidya, F.; Takahashi, F.; Kato, M.; Baskoro, H.; Hidayat, M.; Wirawan, A.; Takahashi, K. CD44 silencing decreases the expression of stem cell-related factors induced by transforming growth factor beta1 and tumor necrosis factor alpha in lung cancer: Preliminary findings. Bosn. J. Basic Med. Sci. 2017, 17, 228. [Google Scholar] [CrossRef]
- Stotz, M.; Herzog, S.A.; Pichler, M.; Smolle, M.; Riedl, J.; Rossmann, C.; Bezan, A.; Stoger, H.; Renner, W.; Berghold, A.; et al. Cancer Stem Cell Gene Variants in CD44 Predict Outcome in Stage II and Stage III Colon Cancer Patients. Anticancer Res. 2017, 37, 2011–2018. [Google Scholar] [CrossRef]
- Lin, J.; Ding, D. The prognostic role of the cancer stem cell marker CD44 in ovarian cancer: A meta-analysis. Cancer Cell Int. 2017, 17, 8. [Google Scholar] [CrossRef]
- Denisov, E.V.; Skryabin, N.A.; Gerashchenko, T.S.; Tashireva, L.A.; Wilhelm, J.; Buldakov, M.A.; Sleptcov, A.A.; Lebedev, I.N.; Vtorushin, S.V.; Zavyalova, M.V.; et al. Clinically relevant morphological structures in breast cancer represent transcriptionally distinct tumor cell populations with varied degrees of epithelial-mesenchymal transition and CD44+CD24- stemness. Oncotarget 2017, 8, 61163. [Google Scholar] [CrossRef]
- Kalantari, E.; Asgari, M.; Nikpanah, S.; Salarieh, N.; Asadi Lari, M.H.; Madjd, Z. Co-Expression of Putative Cancer Stem Cell Markers CD44 and CD133 in Prostate Carcinomas. Pathol. Oncol. Res. 2017, 23, 793–802. [Google Scholar] [CrossRef]
- Seto, K.; Sakabe, T.; Itaba, N.; Azumi, J.; Oka, H.; Morimoto, M.; Umekita, Y.; Shiota, G. A Novel Small-molecule WNT Inhibitor, IC-2, Has the Potential to Suppress Liver Cancer Stem Cells. Anticancer Res. 2017, 37, 3569–3579. [Google Scholar] [PubMed]
- Zhao, Q.; Zhou, H.; Liu, Q.; Cao, Y.; Wang, G.; Hu, A.; Ruan, L.; Wang, S.; Bo, Q.; Chen, W.; et al. Prognostic value of the expression of cancer stem cell-related markers CD133 and CD44 in hepatocellular carcinoma: From patients to patient-derived tumor xenograft models. Oncotarget 2016, 7, 47431–47443. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Na, J.; Ryu, J.; Kim, H.J.; Nam, S.H.; Kang, M.; Jung, J.W.; Lee, M.S.; Song, H.E.; Choi, J.; et al. Interaction of tetraspan(in) TM4SF5 with CD44 promotes self-renewal and circulating capacities of hepatocarcinoma cells. Hepatology 2015, 61, 1978–1997. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Hao, X.; Yan, M.; Yao, M.; Ge, C.; Gu, J.; Li, J. Cancer stem/progenitor cells are highly enriched in CD133+CD44+ population in hepatocellular carcinoma. Int. J. Cancer 2010, 126, 2067–2078. [Google Scholar] [CrossRef]
- Fernando, J.; Malfettone, A.; Cepeda, E.B.; Vilarrasa-Blasi, R.; Bertran, E.; Raimondi, G.; Fabra, A.; Alvarez-Barrientos, A.; Fernandez-Salguero, P.; Fernandez-Rodriguez, C.M.; et al. A mesenchymal-like phenotype and expression of CD44 predict lack of apoptotic response to sorafenib in liver tumor cells. Int. J. Cancer 2015, 136, E161–E172. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.Y.; Wu, F.J.; Chang, C.L.; Li, Z.; Luo, C.W. NMU signaling promotes endometrial cancer cell progression by modulating adhesion signaling. Oncotarget 2016, 7, 10228–10242. [Google Scholar] [CrossRef]
- Cortes-Dericks, L.; Schmid, R.A. CD44 and its ligand hyaluronan as potential biomarkers in malignant pleural mesothelioma: Evidence and perspectives. Respir Res. 2017, 18, 58. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.W. Matrix Hyaluronan-CD44 Interaction Activates MicroRNA and LncRNA Signaling Associated with Chemoresistance, Invasion, and Tumor Progression. Front. Oncol. 2019, 9, 492. [Google Scholar] [CrossRef]
- Ouhtit, A.; Rizeq, B.; Saleh, H.A.; Rahman, M.M.; Zayed, H. Novel CD44-downstream signaling pathways mediating breast tumor invasion. Int. J. Biol. Sci. 2018, 14, 1782–1790. [Google Scholar] [CrossRef]
- Orian-Rousseau, V.; Sleeman, J. CD44 is a multidomain signaling platform that integrates extracellular matrix cues with growth factor and cytokine signals. Adv. Cancer Res. 2014, 123, 231–254. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.; Steinway, S.N.; Ding, W.; Rountree, C.B. Induction of tumor initiation is dependent on CD44s in c-Met(+) hepatocellular carcinoma. BMC Cancer 2015, 15, 161. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Lee, J.W. Self-renewal and circulating capacities of metastatic hepatocarcinoma cells required for collaboration between TM4SF5 and CD44. BMB Rep. 2015, 48, 127–128. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Tian, Y.; Yuan, X.; Wu, H.; Liu, Q.; Pestell, R.G.; Wu, K. The role of CD44 in epithelial-mesenchymal transition and cancer development. Onco Targets Ther. 2015, 8, 3783–3792. [Google Scholar] [CrossRef] [PubMed]
- Ronn, R.E.; Guibentif, C.; Saxena, S.; Woods, N.B. Reactive Oxygen Species Impair the Function of CD90+ Hematopoietic Progenitors Generated from Human Pluripotent Stem Cells. Stem Cells 2017, 35, 197–206. [Google Scholar] [CrossRef]
- Xia, W.; Lo, C.M.; Poon, R.Y.C.; Cheung, T.T.; Chan, A.C.Y.; Chen, L.; Yang, S.; Tsao, G.S.W.; Wang, X.Q. Smad inhibitor induces CSC differentiation for effective chemosensitization in cyclin D1- and TGF-beta/Smad-regulated liver cancer stem cell-like cells. Oncotarget 2017, 8, 38811–38824. [Google Scholar] [CrossRef]
- Chen, W.C.; Chang, Y.S.; Hsu, H.P.; Yen, M.C.; Huang, H.L.; Cho, C.Y.; Wang, C.Y.; Weng, T.Y.; Lai, P.T.; Chen, C.S.; et al. Therapeutics targeting CD90-integrin-AMPK-CD133 signal axis in liver cancer. Oncotarget 2015, 6, 42923–42937. [Google Scholar] [CrossRef]
- Ho, D.W.; Yang, Z.F.; Yi, K.; Lam, C.T.; Ng, M.N.; Yu, W.C.; Lau, J.; Wan, T.; Wang, X.; Yan, Z.; et al. Gene expression profiling of liver cancer stem cells by RNA-sequencing. PLoS ONE 2012, 7, e37159. [Google Scholar] [CrossRef]
- Fujita, T. Subpopulations of circulating cancer stem cell-like cells. Ann. Surg. 2014, 259, e53. [Google Scholar] [CrossRef]
- Yamashita, T.; Honda, M.; Nakamoto, Y.; Baba, M.; Nio, K.; Hara, Y.; Zeng, S.S.; Hayashi, T.; Kondo, M.; Takatori, H.; et al. Discrete nature of EpCAM+ and CD90+ cancer stem cells in human hepatocellular carcinoma. Hepatology 2013, 57, 1484–1497. [Google Scholar] [CrossRef] [PubMed]
- Barzegar Behrooz, A.; Syahir, A.; Ahmad, S. CD133: Beyond a cancer stem cell biomarker. J. Drug Target. 2019, 27, 257–269. [Google Scholar] [CrossRef]
- Suetsugu, A.; Nagaki, M.; Aoki, H.; Motohashi, T.; Kunisada, T.; Moriwaki, H. Characterization of CD133+ hepatocellular carcinoma cells as cancer stem/progenitor cells. Biochem. Biophys. Res. Commun. 2006, 351, 820–824. [Google Scholar] [CrossRef]
- Zhang, J.; Qi, Y.P.; Ma, N.; Lu, F.; Gong, W.F.; Chen, B.; Ma, L.; Zhong, J.H.; Xiang, B.D.; Li, L.Q. Overexpression of Epcam and CD133 Correlates with Poor Prognosis in Dual-phenotype Hepatocellular Carcinoma. J. Cancer 2020, 11, 3400–3406. [Google Scholar] [CrossRef] [PubMed]
- Lathia, J.D.; Liu, H. Overview of Cancer Stem Cells and Stemness for Community Oncologists. Target. Oncol. 2017, 12, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Chan, K.W.; Hu, L.; Lee, T.K.; Wo, J.Y.; Ng, I.O.; Zheng, B.J.; Guan, X.Y. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology 2007, 132, 2542–2556. [Google Scholar] [CrossRef]
- Cordero-Espinoza, L.; Huch, M. The balancing act of the liver: Tissue regeneration versus fibrosis. J. Clin. Investig. 2018, 128, 85–96. [Google Scholar] [CrossRef]
- Hur, W.; Ryu, J.Y.; Kim, H.U.; Hong, S.W.; Lee, E.B.; Lee, S.Y.; Yoon, S.K. Systems approach to characterize the metabolism of liver cancer stem cells expressing CD133. Sci. Rep. 2017, 7, 45557. [Google Scholar] [CrossRef]
- Ma, S.; Tang, K.H.; Chan, Y.P.; Lee, T.K.; Kwan, P.S.; Castilho, A.; Ng, I.; Man, K.; Wong, N.; To, K.F.; et al. miR-130b Promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell 2010, 7, 694–707. [Google Scholar] [CrossRef]
- Chen, Y.L.; Lin, P.Y.; Ming, Y.Z.; Huang, W.C.; Chen, R.F.; Chen, P.M.; Chu, P.Y. The effects of the location of cancer stem cell marker CD133 on the prognosis of hepatocellular carcinoma patients. BMC Cancer 2017, 17, 474. [Google Scholar] [CrossRef]
- Jang, J.W.; Song, Y.; Kim, S.H.; Kim, J.S.; Kim, K.M.; Choi, E.K.; Kim, J.; Seo, H.R. CD133 confers cancer stem-like cell properties by stabilizing EGFR-AKT signaling in hepatocellular carcinoma. Cancer Lett. 2017, 389, 1–10. [Google Scholar] [CrossRef]
- Tang, K.H.; Ma, S.; Lee, T.K.; Chan, Y.P.; Kwan, P.S.; Tong, C.M.; Ng, I.O.; Man, K.; To, K.F.; Lai, P.B.; et al. CD133(+) liver tumor-initiating cells promote tumor angiogenesis, growth, and self-renewal through neurotensin/interleukin-8/CXCL1 signaling. Hepatology 2012, 55, 807–820. [Google Scholar] [CrossRef]
- Xu, Z.; Robitaille, A.M.; Berndt, J.D.; Davidson, K.C.; Fischer, K.A.; Mathieu, J.; Potter, J.C.; Ruohola-Baker, H.; Moon, R.T. Wnt/beta-catenin signaling promotes self-renewal and inhibits the primed state transition in naive human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2016, 113, E6382–E6390. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.L.; Yang, Z.W.; Wang, J.; Zhang, X.L.; Chen, X.M.; Lu, F.M. Integrative analysis of aberrant Wnt signaling in hepatitis B virus-related hepatocellular carcinoma. World J. Gastroenterol. 2015, 21, 6317–6328. [Google Scholar] [CrossRef] [PubMed]
- Vilchez, V.; Turcios, L.; Marti, F.; Gedaly, R. Targeting Wnt/beta-catenin pathway in hepatocellular carcinoma treatment. World J. Gastroenterol. 2016, 22, 823–832. [Google Scholar] [CrossRef]
- Zhang, J.G.; Shi, Y.; Hong, D.F.; Song, M.; Huang, D.; Wang, C.Y.; Zhao, G. MiR-148b suppresses cell proliferation and invasion in hepatocellular carcinoma by targeting WNT1/beta-catenin pathway. Sci. Rep. 2015, 5, 8087. [Google Scholar] [CrossRef] [PubMed]
- Bell, D.; Ranganathan, S.; Tao, J.; Monga, S.P. Novel Advances in Understanding of Molecular Pathogenesis of Hepatoblastoma: A Wnt/beta-Catenin Perspective. Gene Expr. 2017, 17, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Tang, F.; Wang, Z.; Qi, G.; Liang, X.; Li, B.; Yuan, S.; Liu, J.; Yu, S.; He, S. Overexpression of CTNND1 in hepatocellular carcinoma promotes carcinous characters through activation of Wnt/beta-catenin signaling. J. Exp. Clin. Cancer Res. 2016, 35, 82. [Google Scholar] [CrossRef] [PubMed]
- Javanmard, D.; Najafi, M.; Babaei, M.R.; Karbalaie Niya, M.H.; Esghaei, M.; Panahi, M.; Safarnezhad Tameshkel, F.; Tavakoli, A.; Jazayeri, S.M.; Ghaffari, H.; et al. Investigation of CTNNB1 gene mutations and expression in hepatocellular carcinoma and cirrhosis in association with hepatitis B virus infection. Infect. Agent Cancer 2020, 15, 37. [Google Scholar] [CrossRef]
- Takigawa, Y.; Brown, A.M. Wnt signaling in liver cancer. Curr. Drug Targets 2008, 9, 1013–1024. [Google Scholar] [CrossRef]
- Jung, Y.S.; Park, J.I. Wnt signaling in cancer: Therapeutic targeting of Wnt signaling beyond beta-catenin and the destruction complex. Exp. Mol. Med. 2020, 52, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Lisby, A.; Ma, C.; Lo, N.; Ehmer, U.; Hayer, K.E.; Furth, E.E.; Viatour, P. Promotion of growth factor signaling as a critical function of beta-catenin during HCC progression. Nat. Commun. 2019, 10, 1909. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Wang, J.; Karagoz, E.; Liang, B.; Song, X.; Shang, R.; Evert, K.; Xu, M.; Che, L.; Evert, M.; et al. Axis inhibition protein 1 (Axin1) Deletion-Induced Hepatocarcinogenesis Requires Intact beta-Catenin but Not Notch Cascade in Mice. Hepatology 2019, 70, 2003–2017. [Google Scholar] [CrossRef]
- Huang, H.; He, X. Wnt/beta-catenin signaling: New (and old) players and new insights. Curr. Opin. Cell Biol. 2008, 20, 119–125. [Google Scholar] [CrossRef]
- Gan, X.Q.; Wang, J.Y.; Xi, Y.; Wu, Z.L.; Li, Y.P.; Li, L. Nuclear Dvl, c-Jun, beta-catenin, and TCF form a complex leading to stabilization of beta-catenin-TCF interaction. J. Cell Biol. 2008, 180, 1087–1100. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Budhu, A.; Forgues, M.; Wang, X.W. Activation of hepatic stem cell marker EpCAM by Wnt-beta-catenin signaling in hepatocellular carcinoma. Cancer Res. 2007, 67, 10831–10839. [Google Scholar] [CrossRef]
- Ji, J.; Yamashita, T.; Wang, X.W. Wnt/beta-catenin signaling activates microRNA-181 expression in hepatocellular carcinoma. Cell Biosci. 2011, 1, 4. [Google Scholar] [CrossRef]
- Song, Y.; Kim, J.S.; Choi, E.K.; Kim, J.; Kim, K.M.; Seo, H.R. TGF-beta-independent CTGF induction regulates cell adhesion mediated drug resistance by increasing collagen I in HCC. Oncotarget 2017, 8, 21650–21662. [Google Scholar] [CrossRef]
- Wang, X.H.; Chen, Z.G.; Xu, R.L.; Lv, C.Q.; Liu, J.; Du, B. TGF-beta1 signaling pathway serves a role in HepG2 cell regulation by affecting the protein expression of PCNA, gankyrin, p115, XIAP and survivin. Oncol. Lett. 2017, 13, 3239–3246. [Google Scholar] [CrossRef][Green Version]
- Lin, T.H.; Shao, Y.Y.; Chan, S.Y.; Huang, C.Y.; Hsu, C.H.; Cheng, A.L. High Serum Transforming Growth Factor-beta1 Levels Predict Outcome in Hepatocellular Carcinoma Patients Treated with Sorafenib. Clin. Cancer Res. 2015, 21, 3678–3684. [Google Scholar] [CrossRef]
- Dooley, S.; ten Dijke, P. TGF-beta in progression of liver disease. Cell Tissue Res. 2012, 347, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Iwamura, A.; Shimada, Y.J.; Wakai, K.; Tamakoshi, A.; Iso, H.; Group, J.S. Transforming Growth Factor-beta1 as a Predictor for the Development of Hepatocellular Carcinoma: A Nested Case-Controlled Study. EBioMedicine 2016, 12, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Teama, S.; Fawzy, A.; Teama, S.; Helal, A.; Drwish, A.D.; Elbaz, T.; Desouky, E. Increased Serum Endoglin and Transforming Growth Factor beta1 mRNA Expression and Risk of Hepatocellular Carcinoma in Cirrhotic Egyptian Patients. Asian Pac. J. Cancer Prev. 2016, 17, 2429–2434. [Google Scholar] [PubMed]
- Reichl, P.; Haider, C.; Grubinger, M.; Mikulits, W. TGF-beta in epithelial to mesenchymal transition and metastasis of liver carcinoma. Curr. Pharm. Des. 2012, 18, 4135–4147. [Google Scholar] [CrossRef]
- Baek, H.J.; Lim, S.C.; Kitisin, K.; Jogunoori, W.; Tang, Y.; Marshall, M.B.; Mishra, B.; Kim, T.H.; Cho, K.H.; Kim, S.S.; et al. Hepatocellular cancer arises from loss of transforming growth factor beta signaling adaptor protein embryonic liver fodrin through abnormal angiogenesis. Hepatology 2008, 48, 1128–1137. [Google Scholar] [CrossRef]
- van Caam, A.; Madej, W.; Garcia de Vinuesa, A.; Goumans, M.J.; Ten Dijke, P.; Blaney Davidson, E.; van der Kraan, P. TGFbeta1-induced SMAD2/3 and SMAD1/5 phosphorylation are both ALK5-kinase-dependent in primary chondrocytes and mediated by TAK1 kinase activity. Arthritis Res. Ther. 2017, 19, 112. [Google Scholar] [CrossRef]
- Thien, A.; Prentzell, M.T.; Holzwarth, B.; Klasener, K.; Kuper, I.; Boehlke, C.; Sonntag, A.G.; Ruf, S.; Maerz, L.; Nitschke, R.; et al. TSC1 activates TGF-beta-Smad2/3 signaling in growth arrest and epithelial-to-mesenchymal transition. Dev. Cell 2015, 32, 617–630. [Google Scholar] [CrossRef]
- Yan, X.; Liao, H.; Cheng, M.; Shi, X.; Lin, X.; Feng, X.H.; Chen, Y.G. Smad7 Protein Interacts with Receptor-regulated Smads (R-Smads) to Inhibit Transforming Growth Factor-beta (TGF-beta)/Smad Signaling. J. Biol. Chem. 2016, 291, 382–392. [Google Scholar] [CrossRef]
- Giannelli, G.; Fransvea, E.; Marinosci, F.; Bergamini, C.; Colucci, S.; Schiraldi, O.; Antonaci, S. Transforming growth factor-beta1 triggers hepatocellular carcinoma invasiveness via alpha3beta1 integrin. Am. J. Pathol. 2002, 161, 183–193. [Google Scholar] [CrossRef]
- Godoy, P.; Hengstler, J.G.; Ilkavets, I.; Meyer, C.; Bachmann, A.; Muller, A.; Tuschl, G.; Mueller, S.O.; Dooley, S. Extracellular matrix modulates sensitivity of hepatocytes to fibroblastoid dedifferentiation and transforming growth factor beta-induced apoptosis. Hepatology 2009, 49, 2031–2043. [Google Scholar] [CrossRef]
- Sjoqvist, M.; Andersson, E.R. Do as I say, Not(ch) as I do: Lateral control of cell fate. Dev. Biol. 2019, 447, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yu, L.; Zhu, L.Y.; He, H.; Ren, J.; Pan, J.; Xie, X.; Cai, C.; Lu, L.; Tian, H.; et al. Cytokines Induce Monkey Neural Stem Cell Differentiation through Notch Signaling. Biomed. Res. Int. 2020, 2020, 1308526. [Google Scholar] [CrossRef] [PubMed]
- Sueda, R.; Kageyama, R. Regulation of active and quiescent somatic stem cells by Notch signaling. Dev. Growth Differ. 2020, 62, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Hirano, K.I.; Suganami, A.; Tamura, Y.; Yagita, H.; Habu, S.; Kitagawa, M.; Sato, T.; Hozumi, K. Delta-like 1 and Delta-like 4 differently require their extracellular domains for triggering Notch signaling in mice. eLife 2020, 9, e50979. [Google Scholar] [CrossRef]
- Katoh, M.; Katoh, M. Precision medicine for human cancers with Notch signaling dysregulation (Review). Int. J. Mol. Med. 2020, 45, 279–297. [Google Scholar] [CrossRef]
- Aburjania, Z.; Jang, S.; Whitt, J.; Jaskula-Stzul, R.; Chen, H.; Rose, J.B. The Role of Notch3 in Cancer. Oncologist 2018, 23, 900–911. [Google Scholar] [CrossRef]
- Shimojo, H.; Isomura, A.; Ohtsuka, T.; Kori, H.; Miyachi, H.; Kageyama, R. Oscillatory control of Delta-like1 in cell interactions regulates dynamic gene expression and tissue morphogenesis. Genes Dev. 2016, 30, 102–116. [Google Scholar] [CrossRef]
- Hosseini-Alghaderi, S.; Baron, M. Notch3 in Development, Health and Disease. Biomolecules 2020, 10, 485. [Google Scholar] [CrossRef]
- Sweeney, M.; Foldes, G. It Takes Two: Endothelial-Perivascular Cell Cross-Talk in Vascular Development and Disease. Front. Cardiovasc. Med. 2018, 5, 154. [Google Scholar] [CrossRef]
- Jin, H.; Chen, L.; Wang, S.; Chao, D. Portulaca oleracea extract can inhibit nodule formation of colon cancer stem cells by regulating gene expression of the Notch signal transduction pathway. Tumor Biol. 2017, 39, 1010428317708699. [Google Scholar] [CrossRef]
- Dahan, P.; Martinez Gala, J.; Delmas, C.; Monferran, S.; Malric, L.; Zentkowski, D.; Lubrano, V.; Toulas, C.; Cohen-Jonathan Moyal, E.; Lemarie, A. Ionizing radiations sustain glioblastoma cell dedifferentiation to a stem-like phenotype through survivin: Possible involvement in radioresistance. Cell Death Dis. 2014, 5, e1543. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.Y.; Lee, Y.S.; Huang, J.J.; Chang, C.C.; Chang, J.M.; Chuang, S.H.; Kao, K.J.; Tsai, Y.J.; Tsai, P.Y.; Liu, C.W.; et al. Characterization of the biological activity of a potent small molecule Hec1 inhibitor TAI-1. J. Exp. Clin. Cancer Res. 2014, 33, 6. [Google Scholar] [CrossRef] [PubMed]
- Giovannini, C.; Bolondi, L.; Gramantieri, L. Targeting Notch3 in Hepatocellular Carcinoma: Molecular Mechanisms and Therapeutic Perspectives. Int. J. Mol. Sci. 2016, 18, 56. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Xia, Y.; Chen, K.; Zheng, Y.; Wang, J.; Lu, W.; Yin, Q.; Wang, F.; Zhou, Y.; Guo, C. Oncogenic role of the Notch pathway in primary liver cancer. Oncol. Lett. 2016, 12, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, A.; Alsinet, C.; Yanger, K.; Hoshida, Y.; Zong, Y.; Toffanin, S.; Rodriguez-Carunchio, L.; Sole, M.; Thung, S.; Stanger, B.Z.; et al. Notch signaling is activated in human hepatocellular carcinoma and induces tumor formation in mice. Gastroenterology 2012, 143, 1660–1669.e7. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, D.S.; Li, Q.J.; Sun, W.; Zhang, Y.; Dou, K.F. Downregulation of the Notch signaling pathway inhibits hepatocellular carcinoma cell invasion by inactivation of matrix metalloproteinase-2 and -9 and vascular endothelial growth factor. Oncol. Rep. 2012, 28, 874–882. [Google Scholar] [CrossRef]
- Mosaliganti, K.R.; Swinburne, I.A.; Chan, C.U.; Obholzer, N.D.; Green, A.A.; Tanksale, S.; Mahadevan, L.; Megason, S.G. Size control of the inner ear via hydraulic feedback. Elife 2019, 8, e39596. [Google Scholar] [CrossRef]
- Tickle, C.; Towers, M. Sonic Hedgehog Signaling in Limb Development. Front. Cell Dev. Biol. 2017, 5, 14. [Google Scholar] [CrossRef]
- Barbier, I.; Perez-Carrasco, R.; Schaerli, Y. Controlling spatiotemporal pattern formation in a concentration gradient with a synthetic toggle switch. Mol. Syst. Biol. 2020, 16, e9361. [Google Scholar] [CrossRef]
- Sasai, N.; Toriyama, M.; Kondo, T. Hedgehog Signal and Genetic Disorders. Front. Genet. 2019, 10, 1103. [Google Scholar] [CrossRef]
- Cortes, J.E.; Gutzmer, R.; Kieran, M.W.; Solomon, J.A. Hedgehog signaling inhibitors in solid and hematological cancers. Cancer Treat. Rev. 2019, 76, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Pietrobono, S.; Gagliardi, S.; Stecca, B. Non-canonical Hedgehog Signaling Pathway in Cancer: Activation of GLI Transcription Factors Beyond Smoothened. Front. Genet. 2019, 10, 556. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.H.; Siebold, C.; Rohatgi, R. Biochemical mechanisms of vertebrate hedgehog signaling. Development 2019, 146. [Google Scholar] [CrossRef] [PubMed]
- Della Corte, C.M.; Viscardi, G.; Papaccio, F.; Esposito, G.; Martini, G.; Ciardiello, D.; Martinelli, E.; Ciardiello, F.; Morgillo, F. Implication of the Hedgehog pathway in hepatocellular carcinoma. World J. Gastroenterol. 2017, 23, 4330–4340. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Miao, S.; Miao, M.; Li, R.; Cao, X.; Zhang, K.; Huang, G.; Fu, B. Hedgehog Signaling Activation in Hepatic Stellate Cells Promotes Angiogenesis and Vascular Mimicry in Hepatocellular Carcinoma. Cancer Investig. 2016, 34, 424–430. [Google Scholar] [CrossRef]
- Cai, H.; Li, H.; Li, J.; Li, X.; Li, Y.; Shi, Y.; Wang, D. Sonic hedgehog signaling pathway mediates development of hepatocellular carcinoma. Tumor Biol. 2016, 37, 16199–16205. [Google Scholar] [CrossRef]
- Chan, I.S.; Guy, C.D.; Chen, Y.; Lu, J.; Swiderska-Syn, M.; Michelotti, G.A.; Karaca, G.; Xie, G.; Kruger, L.; Syn, W.K.; et al. Paracrine Hedgehog signaling drives metabolic changes in hepatocellular carcinoma. Cancer Res. 2012, 72, 6344–6350. [Google Scholar] [CrossRef]
- Pereira Tde, A.; Witek, R.P.; Syn, W.K.; Choi, S.S.; Bradrick, S.; Karaca, G.F.; Agboola, K.M.; Jung, Y.; Omenetti, A.; Moylan, C.A.; et al. Viral factors induce Hedgehog pathway activation in humans with viral hepatitis, cirrhosis, and hepatocellular carcinoma. Lab. Investig. J. Tech. Methods Pathol. 2010, 90, 1690–1703. [Google Scholar] [CrossRef]
- Philips, G.M.; Chan, I.S.; Swiderska, M.; Schroder, V.T.; Guy, C.; Karaca, G.F.; Moylan, C.; Venkatraman, T.; Feuerlein, S.; Syn, W.K.; et al. Hedgehog signaling antagonist promotes regression of both liver fibrosis and hepatocellular carcinoma in a murine model of primary liver cancer. PLoS ONE 2011, 6, e23943. [Google Scholar] [CrossRef]
- Ye, Y.; Wei, Y.; Xu, Y.; Li, Y.; Wang, R.; Chen, J.; Zhou, Y.; Fu, Z.; Chen, Y.; Wang, X.; et al. Induced MiR-1249 expression by aberrant activation of Hedegehog signaling pathway in hepatocellular carcinoma. Exp. Cell Res. 2017, 355, 9–17. [Google Scholar] [CrossRef]
- Hanna, A.; Shevde, L.A. Hedgehog signaling: Modulation of cancer properies and tumor mircroenvironment. Mol. Cancer 2016, 15, 24. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Han, C.; Lu, L.; Magliato, S.; Wu, T. Hedgehog signaling pathway regulates autophagy in human hepatocellular carcinoma cells. Hepatology 2013, 58, 995–1010. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.J.; Chun, T. Structural heterogeneity of the mammalian polycomb repressor complex in immune regulation. Exp. Mol. Med. 2020, 52, 1004–1015. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Kitano, A.; Luu, V.; Dawson, B.; Hoegenauer, K.A.; Lee, B.H.; Nakada, D. Bmi1 Suppresses Adipogenesis in the Hematopoietic Stem Cell Niche. Stem Cell Rep. 2019, 13, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Lin, Y.; Mishra, A.; Shelly, C.; Gao, R.; Reeh, C.W.; Wang, P.Z.; Xi, R.; Liu, Y.; Wenzel, P.; et al. Bmi1 Maintains the Self-Renewal Property of Innate-like B Lymphocytes. J. Immunol. 2020, 204, 3262–3272. [Google Scholar] [CrossRef]
- Abdel-Khalek, S.; Saleh, L.M.; Abdel-Aziz, S.; Hyder, A.; Abdel-Ghaffar, H. Utility of BMI-1 and NANOG expression levels in survival prediction of pediatric acute lymphoblastic leukemia. Hematol. Transfus. Cell Ther. 2020. [Google Scholar] [CrossRef]
- Hsu, Y.C.; Luo, C.W.; Huang, W.L.; Wu, C.C.; Chou, C.L.; Chen, C.I.; Chang, S.J.; Chai, C.Y.; Wang, H.C.; Chen, T.Y.; et al. BMI1-KLF4 axis deficiency improves responses to neoadjuvant concurrent chemoradiotherapy in patients with rectal cancer. Radiother. Oncol. 2020, 149, 249–258. [Google Scholar] [CrossRef]
- Bedewy, A.M.L.; Elmaghraby, S.M.; Kandil, N.S. ABCB1 and BMI1 mRNA expression in patients with chronic myeloid leukemia: Impact on imatinib efficacy. Blood Res. 2019, 54, 57–62. [Google Scholar] [CrossRef]
- Bartucci, M.; Hussein, M.S.; Huselid, E.; Flaherty, K.; Patrizii, M.; Laddha, S.V.; Kui, C.; Bigos, R.A.; Gilleran, J.A.; El Ansary, M.M.S.; et al. Synthesis and Characterization of Novel BMI1 Inhibitors Targeting Cellular Self-Renewal in Hepatocellular Carcinoma. Target. Oncol. 2017, 12, 449–462. [Google Scholar] [CrossRef]
- Zhang, R.; Real, C.I.; Liu, C.; Baba, H.A.; Gerken, G.; Lu, M.; Broering, R. Hepatic expression of oncogenes Bmi1 and Dkk1 is up-regulated in hepatitis B virus surface antigen-transgenic mice and can be induced by treatment with HBV particles or lipopolysaccharides in vitro. Int. J. Cancer 2017, 141, 354–363. [Google Scholar] [CrossRef]
- Zhai, R.; Tang, F.; Gong, J.; Zhang, J.; Lei, B.; Li, B.; Wei, Y.; Liang, X.; Tang, B.; He, S. The relationship between the expression of USP22, BMI1, and EZH2 in hepatocellular carcinoma and their impacts on prognosis. Onco Targets Ther. 2016, 9, 6987–6998. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Srinivasan, M.; Bharali, D.J.; Sudha, T.; Khedr, M.; Guest, I.; Sell, S.; Glinsky, G.V.; Mousa, S.A. Downregulation of Bmi1 in breast cancer stem cells suppresses tumor growth and proliferation. Oncotarget 2017, 8, 38731–38742. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Wu, M.; Li, Y.; Chang, I.; Yuan, Q.; Ekimyan-Salvo, M.; Deng, P.; Yu, B.; Yu, Y.; Dong, J.; et al. Targeting BMI1+ Cancer Stem Cells Overcomes Chemoresistance and Inhibits Metastases in Squamous Cell Carcinoma. Cell Stem Cell 2017, 20, 621–634.e6. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Atsumi, N.; Nakamura, N.; Yanai, H.; Komai, Y.; Omachi, T.; Tanaka, K.; Ishigaki, K.; Saiga, K.; Ohsugi, H.; et al. Bmi1-positive cells in the lingual epithelium could serve as cancer stem cells in tongue cancer. Sci. Rep. 2016, 6, 39386. [Google Scholar] [CrossRef]
- Kim, J.H.; Yoon, S.Y.; Kim, C.N.; Joo, J.H.; Moon, S.K.; Choe, I.S.; Choe, Y.K.; Kim, J.W. The Bmi-1 oncoprotein is overexpressed in human colorectal cancer and correlates with the reduced p16INK4a/p14ARF proteins. Cancer Lett. 2004, 203, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, M.; Ikeda, H.; Itatsu, K.; Yamaguchi, J.; Sawada, S.; Minato, H.; Ohta, T.; Nakanuma, Y. The overexpression of polycomb group proteins Bmi1 and EZH2 is associated with the progression and aggressive biological behavior of hepatocellular carcinoma. Lab. Investig. 2008, 88, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Vrzalikova, K.; Skarda, J.; Ehrmann, J.; Murray, P.G.; Fridman, E.; Kopolovic, J.; Knizetova, P.; Hajduch, M.; Klein, J.; Kolek, V.; et al. Prognostic value of Bmi-1 oncoprotein expression in NSCLC patients: A tissue microarray study. J. Cancer Res. Clin. Oncol. 2008, 134, 1037–1042. [Google Scholar] [CrossRef]
- Park, I.K.; Morrison, S.J.; Clarke, M.F. Bmi1, stem cells, and senescence regulation. J. Clin. Investig. 2004, 113, 175–179. [Google Scholar] [CrossRef]
- Lobo, N.A.; Shimono, Y.; Qian, D.; Clarke, M.F. The biology of cancer stem cells. Annu. Rev. Cell Dev. Biol. 2007, 23, 675–699. [Google Scholar] [CrossRef]
- Ruan, Z.P.; Xu, R.; Lv, Y.; Tian, T.; Wang, W.J.; Guo, H.; Nan, K.J. Bmi1 knockdown inhibits hepatocarcinogenesis. Int. J. Oncol. 2013, 42, 261–268. [Google Scholar] [CrossRef]
- Xu, C.R.; Lee, S.; Ho, C.; Bommi, P.; Huang, S.A.; Cheung, S.T.; Dimri, G.P.; Chen, X. Bmi1 functions as an oncogene independent of Ink4A/Arf repression in hepatic carcinogenesis. Mol. Cancer Res. 2009, 7, 1937–1945. [Google Scholar] [CrossRef]
- Effendi, K.; Mori, T.; Komuta, M.; Masugi, Y.; Du, W.; Sakamoto, M. Bmi-1 gene is upregulated in early-stage hepatocellular carcinoma and correlates with ATP-binding cassette transporter B1 expression. Cancer Sci. 2010, 101, 666–672. [Google Scholar] [CrossRef] [PubMed]
- Chiba, T.; Seki, A.; Aoki, R.; Ichikawa, H.; Negishi, M.; Miyagi, S.; Oguro, H.; Saraya, A.; Kamiya, A.; Nakauchi, H.; et al. Bmi1 promotes hepatic stem cell expansion and tumorigenicity in both Ink4a/Arf-dependent and -independent manners in mice. Hepatology 2010, 52, 1111–1123. [Google Scholar] [CrossRef] [PubMed]
- Chiba, T.; Zheng, Y.W.; Kita, K.; Yokosuka, O.; Saisho, H.; Onodera, M.; Miyoshi, H.; Nakano, M.; Zen, Y.; Nakanuma, Y.; et al. Enhanced self-renewal capability in hepatic stem/progenitor cells drives cancer initiation. Gastroenterology 2007, 133, 937–950. [Google Scholar] [CrossRef] [PubMed]
- Franco, D.; Usatoff, V. Resection of hepatocellular carcinoma. Hepatogastroenterology 2001, 48, 33–36. [Google Scholar]
- Wakizaka, K.; Yokoo, H.; Kamiyama, T.; Kakisaka, T.; Ohira, M.; Tani, M.; Kato, K.; Fujii, Y.; Sugiyama, K.; Nagatsu, A.; et al. CD133 and epithelial cell adhesion molecule expressions in the cholangiocarcinoma component are prognostic factors for combined hepatocellular cholangiocarcinoma. Hepatol. Res. 2020, 50, 258–267. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, G.; Fu, X.; Xu, S.; Wang, T.; Zhang, Q.; Yang, Y. Aquaporin 3 maintains the stemness of CD133+ hepatocellular carcinoma cells by activating STAT3. Cell Death Dis. 2019, 10, 465. [Google Scholar] [CrossRef]
- Wang, R.; Li, Y.; Tsung, A.; Huang, H.; Du, Q.; Yang, M.; Deng, M.; Xiong, S.; Wang, X.; Zhang, L.; et al. iNOS promotes CD24(+)CD133(+) liver cancer stem cell phenotype through a TACE/ADAM17-dependent Notch signaling pathway. Proc. Natl. Acad. Sci. USA 2018, 115, E10127–E10136. [Google Scholar] [CrossRef]
- Song, Y.; Kim, S.; Lee, H.; No, J.H.; Ryu, H.C.; Kim, J.; Lim, J.W.; Kim, M.; Choi, I.; Seo, H.R. Chromenopyrimidinone Controls Stemness and Malignancy by suppressing CD133 Expression in Hepatocellular Carcinoma. Cancers 2020, 12, 1193. [Google Scholar] [CrossRef]
- Song, Y.J.; Zhang, S.S.; Guo, X.L.; Sun, K.; Han, Z.P.; Li, R.; Zhao, Q.D.; Deng, W.J.; Xie, X.Q.; Zhang, J.W.; et al. Autophagy contributes to the survival of CD133+ liver cancer stem cells in the hypoxic and nutrient-deprived tumor microenvironment. Cancer Lett. 2013, 339, 70–81. [Google Scholar] [CrossRef]
- Hagiwara, S.; Kudo, M.; Nagai, T.; Inoue, T.; Ueshima, K.; Nishida, N.; Watanabe, T.; Sakurai, T. Activation of JNK and high expression level of CD133 predict a poor response to sorafenib in hepatocellular carcinoma. Br. J. Cancer 2012, 106, 1997–2003. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.F.; Ngai, P.; Ho, D.W.; Yu, W.C.; Ng, M.N.; Lau, C.K.; Li, M.L.; Tam, K.H.; Lam, C.T.; Poon, R.T.; et al. Identification of local and circulating cancer stem cells in human liver cancer. Hepatology 2008, 47, 919–928. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.F.; Ho, D.W.; Ng, M.N.; Lau, C.K.; Yu, W.C.; Ngai, P.; Chu, P.W.; Lam, C.T.; Poon, R.T.; Fan, S.T. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell 2008, 13, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Thakolwiboon, S.; Zhu, J.; Liang, Q.; Welling, T.H.; Zhang, M.; Lubman, D.M. Heterogeneity of The CD90 Population in Different Stages of Hepatocarcinogenesis. J. Proteom. Bioinform. 2014, 7, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.W.; Chang, J.G.; Yeh, K.T.; Chen, R.M.; Tsai, J.J.; Hu, R.M. Overexpression of Thy1/CD90 in human hepatocellular carcinoma is associated with HBV infection and poor prognosis. Acta Histochem. 2011, 113, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Zou, Q.; Ge, R.; Shen, F.; Wang, Y. The critical role of CD133(+)CD44(+/high) tumor cells in hematogenous metastasis of liver cancers. Cell Res. 2012, 22, 259–272. [Google Scholar] [CrossRef]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef]
- Yang, M.H.; Chen, C.L.; Chau, G.Y.; Chiou, S.H.; Su, C.W.; Chou, T.Y.; Peng, W.L.; Wu, J.C. Comprehensive analysis of the independent effect of twist and snail in promoting metastasis of hepatocellular carcinoma. Hepatology 2009, 50, 1464–1474. [Google Scholar] [CrossRef]
- Mima, K.; Hayashi, H.; Imai, K.; Kuroki, H.; Nakagawa, S.; Okabe, H.; Chikamoto, A.; Watanabe, M.; Beppu, T.; Baba, H. High CD44s expression is associated with the EMT expression profile and intrahepatic dissemination of hepatocellular carcinoma after local ablation therapy. J. Hepatobiliary Pancreat. Sci. 2013, 20, 429–434. [Google Scholar] [CrossRef]
- Smith, L.M.; Nesterova, A.; Ryan, M.C.; Duniho, S.; Jonas, M.; Anderson, M.; Zabinski, R.F.; Sutherland, M.K.; Gerber, H.P.; Van Orden, K.L.; et al. CD133/prominin-1 is a potential therapeutic target for antibody-drug conjugates in hepatocellular and gastric cancers. Br. J. Cancer 2008, 99, 100–109. [Google Scholar] [CrossRef]
- Zhang, L.; Li, H.; Ge, C.; Li, M.; Zhao, F.Y.; Hou, H.L.; Zhu, M.X.; Tian, H.; Zhang, L.X.; Chen, T.Y.; et al. Inhibitory effects of transcription factor Ikaros on the expression of liver cancer stem cell marker CD133 in hepatocellular carcinoma. Oncotarget 2014, 5, 10621–10635. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Guan, D.X.; Shi, J.; Gao, H.; Li, J.J.; Zhao, J.S.; Qiu, L.; Liu, J.; Li, N.; Guo, W.X.; et al. All-trans retinoic acid potentiates the chemotherapeutic effect of cisplatin by inducing differentiation of tumor initiating cells in liver cancer. J. Hepatol. 2013, 59, 1255–1263. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Honda, M.; Nio, K.; Nakamoto, Y.; Yamashita, T.; Takamura, H.; Tani, T.; Zen, Y.; Kaneko, S. Oncostatin m renders epithelial cell adhesion molecule-positive liver cancer stem cells sensitive to 5-Fluorouracil by inducing hepatocytic differentiation. Cancer Res. 2010, 70, 4687–4697. [Google Scholar] [CrossRef] [PubMed]
- Magee, J.A.; Piskounova, E.; Morrison, S.J. Cancer stem cells: Impact, heterogeneity, and uncertainty. Cancer Cell 2012, 21, 283–296. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.-Y.; Hong, I.-S. Targeting Liver Cancer Stem Cells: An Alternative Therapeutic Approach for Liver Cancer. Cancers 2020, 12, 2746. https://doi.org/10.3390/cancers12102746
Lee H-Y, Hong I-S. Targeting Liver Cancer Stem Cells: An Alternative Therapeutic Approach for Liver Cancer. Cancers. 2020; 12(10):2746. https://doi.org/10.3390/cancers12102746
Chicago/Turabian StyleLee, Hwa-Yong, and In-Sun Hong. 2020. "Targeting Liver Cancer Stem Cells: An Alternative Therapeutic Approach for Liver Cancer" Cancers 12, no. 10: 2746. https://doi.org/10.3390/cancers12102746
APA StyleLee, H.-Y., & Hong, I.-S. (2020). Targeting Liver Cancer Stem Cells: An Alternative Therapeutic Approach for Liver Cancer. Cancers, 12(10), 2746. https://doi.org/10.3390/cancers12102746



