Germline Variants in Driver Genes of Breast Cancer and Their Association with Familial and Early-Onset Breast Cancer Risk in a Chilean Population
Abstract
1. Introduction
2. Results
2.1. Association Study between rs10497520, rs2242442, rs11168827, rs702688 and rs702689 with Familial Breast Cancer and Early-Onset Non-Familial Breast Cancer in Non-Carriers of BRCA1/2 Mutations
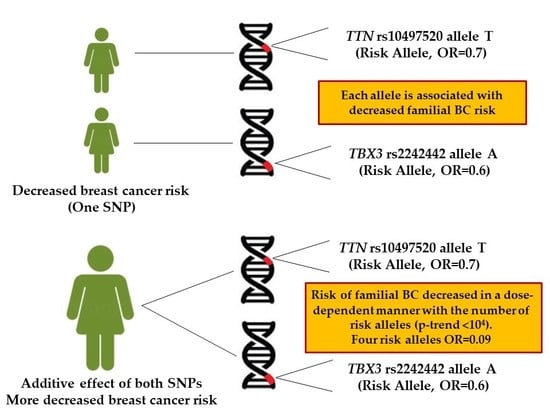
2.2. Combined Effect between TTN rs10497520-T and TBX3 rs2242442-G Alleles with Breast Cancer Risk
3. Discussion
4. Materials and Methods
4.1. Families
4.2. Control Population
4.3. Mutation Analysis
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Ethical Approval and Informed Consent
References
- Parkin, D.M.; Fernandez, L.M. Use of statistics to assess the global burden of breast cancer. Breast J. 2006, 12 (Suppl. 1), S70–S80. [Google Scholar] [CrossRef]
- Oldenburg, R.A.; Meijers-Heijboer, H.; Cornelisse, C.J.; Devilee, P. Genetic susceptibility for breast cancer: How many more genes to be found? Crit. Rev. Oncol. Hematol. 2007, 63, 125–149. [Google Scholar] [CrossRef]
- Jemal, A.; Siegel, R.; Ward, E.; Hao, Y.; Xu, J.; Thun, M.J. Cancer statistics, 2009. CA Cancer J. Clin. 2009, 59, 225–249. [Google Scholar] [CrossRef]
- Peralta, O. Cáncer de mama en Chile. Datos Epidemiológicos. Rev. Chil. Obstet. Ginecol. 2002, 67, 439–445. [Google Scholar] [CrossRef]
- Chile Government, Plan Nacional de Cáncer 2018–2028, by Department of Comprehensive Management of Cancer and Other Tumors, Division of Disease Prevention and Control, Undersecretary of Public Health, Ministry of Health. 2019. Available online: https://www.minsal.cl/wp-content/uploads/2019/01/2019.01.23_PLAN-NACIONAL-DE-CANCER_web.pdf (accessed on 20 January 2020).
- Prieto, M. Situación epidemiológica del cáncer de mama en Chile 1994-2003. Rev. Med. Clin. Condes 2006, 17, 142–148. [Google Scholar] [CrossRef][Green Version]
- Miki, Y.; Swensen, J.; Shattuck-Eidens, D.; Futreal, P.A.; Harshman, K.; Tavtigian, S.; Liu, Q.; Cochran, C.; Bennett, L.M.; Ding, W.; et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994, 266, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Wooster, R.; Neuhausen, S.L.; Mangion, J.; Quirk, Y.; Ford, D.; Collins, N.; Nguyen, K.; Seal, S.; Tran, T.; Averill, D.; et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science 1994, 265, 2088–2090. [Google Scholar] [CrossRef] [PubMed]
- Tavtigian, S.V.; Simard, J.; Rommens, J.; Couch, F.; Shattuck-Eidens, D.; Neuhausen, S.; Merajver, S.; Thorlacius, S.; Offit, K.; Stoppa-Lyonnet, D.; et al. The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds. Nat. Genet. 1996, 12, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, C.; Rahman, N. Genetic predisposition to breast cancer: Past, present, and future. Annu. Rev. Genomics Hum. Genet. 2008, 9, 321–345. [Google Scholar] [CrossRef]
- Couch, F.J.; Hart, S.N.; Sharma, P.; Toland, A.E.; Wang, X.; Miron, P.; Olson, J.E.; Godwin, A.K.; Pankratz, V.S.; Olswold, C.; et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J. Clin. Oncol. 2015, 33, 304–311. [Google Scholar] [CrossRef]
- Anglian Breast Cancer Study Group. Prevalence and penetrance of BRCA1 and BRCA2 mutations in a population-based series of breast cancer cases. Br. J. Cancer 2000, 83, 1301–1308. [Google Scholar] [CrossRef] [PubMed]
- Stratton, M.R.; Rahman, N. The emerging landscape of breast cancer susceptibility. Nat. Genet. 2008, 40, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Ford, D.; Easton, D.F.; Stratton, M.; Narod, S.; Goldgar, D.; Devilee, P.; Bishop, D.T.; Weber, B.; Lenoir, G.; Chang-Claude, J.; et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The breast cancer linkage consortium. Am. J. Hum. Genet. 1998, 62, 676–689. [Google Scholar] [CrossRef] [PubMed]
- Chandler, M.R.; Bilgili, E.P.; Merner, N.D. A review of whole-exome sequencing efforts toward hereditary breast cancer susceptibility gene discovery. Hum. Mutat. 2016, 37, 835–846. [Google Scholar] [CrossRef] [PubMed]
- Stratton, M.R.; Campbell, P.J.; Futreal, P.A. The cancer genome. Nature 2009, 458, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Stephens, P.J.; Tarpey, P.S.; Davies, H.; Van Loo, P.; Greenman, C.; Wedge, D.C.; Nik-Zainal, S.; Martin, S.; Varela, I.; Bignell, G.R.; et al. The landscape of cancer genes and mutational processes in breast cancer. Nature 2012, 486, 400–404. [Google Scholar] [CrossRef]
- Teschendorff, A.E.; Caldas, C. The breast cancer somatic ‘muta-ome’: Tackling the complexity. Breast Cancer Res. 2009, 11, 301. [Google Scholar] [CrossRef]
- Tomasetti, C.; Marchionni, L.; Nowak, M.A.; Parmigiani, G.; Vogelstein, B. Only three driver gene mutations are required for the development of lung and colorectal cancers. Proc. Natl. Acad. Sci. USA 2015, 112, 118–123. [Google Scholar] [CrossRef]
- Gohler, S.; Da Silva Filho, M.I.; Johansson, R.; Enquist-Olsson, K.; Henriksson, R.; Hemminki, K.; Lenner, P.; Försti, A. Functional germline variants in driver genes of breast cancer. Cancer Causes Control 2017, 28, 259–271. [Google Scholar] [CrossRef]
- Douglas, N.C.; Papaioannou, V.E. The T-box transcription factors TBX2 and TBX3 in mammary gland development and breast cancer. J. Mammary Gland Biol. Neoplasia 2013, 18, 143–147. [Google Scholar] [CrossRef]
- Washkowitz, A.J.; Gavrilov, S.; Begum, S.; Papaioannou, V.E. Diverse functional networks of Tbx3 in development and disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 2012, 4, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Banerji, S.; Cibulskis, K.; Rangel-Escareno, C.; Brown, K.K.; Carter, S.L.; Frederick, A.M.; Lawrence, M.S.; Sivachenko, A.Y.; Sougnez, C.; Zou, L.; et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 2012, 486, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Ellis, M.J.; Ding, L.; Shen, D.; Luo, J.; Suman, V.J.; Wallis, J.W.; Van Tine, B.A.; Hoog, J.; Goiffon, R.J.; Goldstein, T.C.; et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 2012, 486, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.P.; Roth, A.; Goya, R.; Oloumi, A.; Ha, G.; Zhao, Y.; Turashvili, G.; Ding, J.; Tse, K.; Haffari, G.; et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 2012, 486, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Stojanov, P.; Mermel, C.H.; Robinson, J.T.; Garraway, L.A.; Golub, T.R.; Meyerson, M.; Gabriel, S.B.; Lander, E.S.; Getz, G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014, 505, 495–501. [Google Scholar] [CrossRef]
- Marouf, C.; Göhler, S.; Filho, M.I.; Hajji, O.; Hemminki, K.; Nadifi, S.; Försti, A. Analysis of functional germline variants in APOBEC3 and driver genes on breast cancer risk in Moroccan study population. BMC Cancer 2016, 16, 165. [Google Scholar] [CrossRef]
- Granzier, H.L.; Labeit, S. Titin and its associated proteins: The third myofilament system of the sarcomere. Adv. Protein Chem. 2005, 71, 89–119. [Google Scholar] [CrossRef]
- Machado, C.; Andrew, D.J. D-Titin: A giant protein with dual roles in chromosomes and muscles. J. Cell Biol. 2000, 151, 639–652. [Google Scholar] [CrossRef]
- Rabello Ddo, A.; de Moura, C.A.; de Andrade, R.V.; Motoyama, A.B.; Silva, F.P. Altered expression of MLL methyltransferase family genes in breast cancer. Int. J. Oncol. 2013, 43, 653–660. [Google Scholar] [CrossRef]
- Gala, K.; Li, Q.; Sinha, A.; Razavi, P.; Dorso, M.; Sanchez-Vega, F.; Chung, Y.R.; Hendrickson, R.; Hsieh, J.J.; Berger, M.; et al. KMT2C mediates the estrogen dependence of breast cancer through regulation of ERalpha enhancer function. Oncogene 2018, 37, 4692–4710. [Google Scholar] [CrossRef]
- Tan, H.; Bao, J.; Zhou, X. Genome-wide mutational spectra analysis reveals significant cancer-specific heterogeneity. Sci. Rep. 2015, 5, 12566. [Google Scholar] [CrossRef] [PubMed]
- Skaaby, T.; Husemoen, L.L.; Thyssen, J.P.; Meldgaard, M.; Thuesen, B.H.; Pisinger, C.; Jørgensen, T.; Carlsen, K.; Johansen, J.D.; Menné, T.; et al. Filaggrin loss-of-function mutations and incident cancer: A population-based study. Br. J. Dermatol. 2014, 171, 1407–1414. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Coke, R. Ethnic origin and evolution of the Chilean population. Rev. Med. Chil. 1976, 104, 365–368. [Google Scholar] [PubMed]
- Valenzuela, C.Y.; Acuna, M.P.; Harb, Z. Sociogenetic gradient in the Chilean population. Rev. Med. Chile 1987, 115, 295–299. [Google Scholar]
- Valenzuela, C.Y.; Harb, Z. Socioeconomic assortative mating in Santiago, Chile: A demonstration using stochastic matrices of mother-child relationships applied to ABO blood groups. Soc. Biol. 1977, 24, 225–233. [Google Scholar] [CrossRef]
- Fuentes, M.; Pulgar, I.; Gallo, C.; Bortolini, M.C.; Canizales-Quinteros, S.; Bedoya, G.; González-José, R.; Ruiz-Linares, A.; Rothhammer, F. Gene geography of Chile: Regional distribution of American, European and African genetic contributions. Rev. Med. Chile 2014, 142, 281–289. [Google Scholar] [CrossRef]
- Gonzalez-Hormazabal, P.; Gutierrez-Enriquez, S.; Gaete, D.; Reyes, J.M.; Peralta, O.; Waugh, E.; Gomez, F.; Margarit, S.; Bravo, T.; Blanco, R. Spectrum of BRCA1/2 point mutations and genomic rearrangements in high-risk breast/ovarian cancer Chilean families. Breast Cancer Res. Treat. 2011, 126, 705–716. [Google Scholar] [CrossRef]
- Chomczynski, P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 1993, 15, 532–534, 536–537. [Google Scholar]
| Genotype or Allele | Controls (%) (n = 1078) | All BC Cases (n = 489) | Families with ≥2 BC and/or OC Cases (n = 311) | A Single case, Diagnosis at ≤50 Years of Age (n = 178) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BC Cases (%) | OR [95% CI] | p-Value a | BC Cases (%) | OR [95% CI] | p-Value a | BC Cases (%) | OR [95% CI] | p-Value a | ||
| rs10497520 (TTN) | ||||||||||
| C/C | 314 (29.1) | 182 (37.2) | 1.0 (Ref) | - | 122 (39.2) | 1.0 (ref) | - | 60 (33.7) | 1.0 Ref | - |
| C/T | 504 (46.8) | 226 (46.2) | 0.7 [0.6–0.9] | 0.04 | 139 (44.7) | 0.7 [0.5–0.9] | 0.02 | 87 (48.9) | 0.9 [0.6–1.2] | 0.58 |
| T/T | 260 (24.1) | 81 (16.6) | 0.5 [0.3–0.9] | <0.0001 | 50 (16.1) | 0.4 [0.3–07] | 0.0001 | 31 (17.4) | 0.6 [0.3–0.9] | 0.05 |
| C/T + T/T | 764 (70.9) | 307(62.8) | 0.6 [0.5–0.8] | 0.001 | 189 (60.8) | 0.6 [0.4–0.8] | 0.0009 | 118 (66.3) | 0.8 [0.5–1.1] | 0.21 |
| Allele C | 1132 (52.5) | 590 (60.3) | 1.0 (Ref) | - | 383 (61.6) | 1.0 (ref) | - | 207 (58.1) | 1.0 (Ref) | - |
| Allele T | 1024 (47.5) | 388 (39.7) | 0.7 [0.6–0.8] | <0.0001 | 239 (38.4) | 0.6 [0.5–0.8] | <0.0001 | 149 (41.9) | 0.7 [0.6–0.9] | 0.05 |
| rs2242442 (TBX3) | ||||||||||
| G/G | 674 (62.5) | 328 (67.1) | 1.0 (Ref) | - | 210 (67.5) | 1.0 (Ref) | - | 118 (66.3) | 1.0 (Ref) | - |
| G/A | 358 (33.2) | 146 (29.9) | 0.8 [0.6–1.0] | 0.14 | 90 (28.9) | 0.8 [0.6–1.0] | 0.14 | 56 (31.5) | 0.8 [0.6–1.2] | 0.54 |
| A/A | 46 (4.3) | 15 (3.0) | 0.6 [0.3–1.1] | 0.20 | 11 (3.5) | 0.7 [0.3–1.5] | 0.52 | 4 (2.2) | 0.4 [0.1–1.4] | 0.21 |
| G/A + A/A | 404 (37.5) | 161 (32.9) | 0.8 [0.6–1.0] | 0.08 | 101 (32.5) | 0.8 [0.6–1.0] | 0.10 | 60 (33.7) | 0.8 [0.6–1.1] | 0.35 |
| Allele G | 1706 (79.1) | 802 (82.0) | 1.0 (Ref) | - | 510 (82.0) | 1.0 (Ref) | - | 292 (82.0) | 1.0 (Ref) | - |
| Allele A | 450 (20.9) | 176 (18.0) | 0.8 [0.6–1.0] | 0.06 | 112 (18.0) | 0.8 [0.6–1.0] | 0.13 | 64 (18.0) | 0.8 [0.6–1.1] | 0.23 |
| rs11168827 (KMT2D) | ||||||||||
| G/G | 439 (40.7) | 198 (40.5) | 1.0 (ref) | - | 121 (38.9) | 1.0 (ref) | - | 77 (43.3) | 1.0 (ref) | - |
| G/C | 510 (47.3) | 239 (48.9) | 1.0 [0.8–1.3] | 0.77 | 157 (50.5) | 1.1 [0.8–1.4] | 0.45 | 82 (46.1) | 0.9 [0.6–1.3] | 0.66 |
| C/C | 129 (12.0) | 52 (10.6) | 0.8 [0.6–1.2] | 0.58 | 33 (10.6) | 0.9 [0.6–1.4] | 0.82 | 19 (10.7) | 0.8 [0.4–1.4] | 0.59 |
| G/C + C/C | 639 (59.3) | 291 (59.5) | 1.0 [0.8–1.2] | 0.95 | 190 (61.1) | 1.0 [0.8–1.3] | 0.59 | 101 (56.7) | 0.9 [0.6–1.2] | 0.56 |
| Allele G | 1388 (64.4) | 635 (64.9) | 1.0 (ref) | - | 399 (64.1) | 1.0 (ref) | - | 236 (66.3) | 1.0 (ref) | - |
| Allele C | 768 (35.6) | 343 (35.1) | 0.9 [0.8–1.1] | 0.79 | 223 (35.9) | 1.0 [0.8–1.2] | 0.95 | 120 (33.7) | 0.9 [0.7–1.1] | 0.52 |
| rs702688 (MAP3K1) | ||||||||||
| A/A | 345 (32.0) | 167 (34.2) | 1.0 (Ref) | - | 100 (32.3) | 1.0 (Ref) | - | 67 (37.6) | 1.0 (Ref) | - |
| A/G | 525 (48.7) | 236 (48.3) | 0.9 [0.7–1.1] | 0.58 | 150 (48.6) | 0.9 [0.7–1.3] | 0.94 | 85 (47.8) | 0.8 [0,.5–1.1] | 0.32 |
| G/G | 208 (19.3) | 86 (17.6) | 0.8 [0.6–1.1] | 0.34 | 60 (19.3) | 0.9 [0.6–1.4] | 1.0 | 26 (14.6) | 0.6 [0.3–1.0] | 0.08 |
| A/G + G/G | 733 (68.0) | 322 (65.8) | 0.9 [0.7–1.1] | 0.41 | 210 (67.8) | 0.9 [0.7–1.3] | 0.94 | 111 (62.4) | 0.7 [0.5–1.0] | 0.14 |
| Allele A | 1215 (56.4) | 570 (58.3) | 1,0 (Ref) | - | 350 (56.4) | 1.0 (Ref) | - | 219 (61.5) | 1.0 (Ref) | - |
| Allele G | 941 (43.6) | 408 (41.7) | 0.9 [0.7–1.0] | 0.33 | 270 (43.6) | 0.9 [0.8–1.1] | 0.99 | 137 (38.5) | 0.8 [0.6–1.0] | 0.07 |
| rs702689 (MAP3K1) | ||||||||||
| G/G | 274 (25.4) | 132 (27.0) | 1.0 (Ref) | - | 82 (26.4) | 1.0 (Ref) | - | 48 (27.0) | 1.0 (Ref) | - |
| G/A | 544 (50.5) | 226 (46.2) | 0.8 [0.6–1.1] | 0.28 | 133 (42.8) | 0.8 [0.5–1.1] | 0.22 | 92 (51.7) | 0.9 [0.6–1.4] | 0.84 |
| A/A | 260 (24.1) | 131 (26.8) | 1.0 [0.7–1.4] | 0.82 | 96 (30.9) | 1.2 [0.8–1.7] | 0.26 | 38 (21.3) | 0.8 [0.5–1.3] | 0.48 |
| G/A + A/A | 804 (74.6) | 357 (73.0) | 0.9 [0.7–1.1] | 0.53 | 229 (73.6) | 0.9 [0.7–1.2] | 0.76 | 130 (73.0) | 0.9 [0.6–1.3] | 0.64 |
| Allele G | 1092 (50.6) | 490 (50.1) | 1.0 (Ref) | - | 297 (47.7) | 1.0 (Ref) | - | 188 (52.8) | 1.0 (Ref) | - |
| Allele A | 1064 (49.4) | 488 (49.9) | 1.02 [0.8–1.1] | 0.81 | 325 (52.3) | 1.1 [0.9–1.3] | 0.21 | 168 (47.2) | 0.9 [0.7–1.1] | 0.48 |
| Genotype or Allele | Controls (%) (n = 1078) | Families with 2 BC and/or OC Cases (n = 166) | Families with ≥3 BC and/or OC Cases (n = 145) | ||||
|---|---|---|---|---|---|---|---|
| BC Cases (%) | OR [95% CI] | p Value a | BC Cases (%) | OR [95% CI] | p Value a | ||
| rs10497520 (TTN) | |||||||
| C/C | 314 (29.1) | 65 (39.2) | 1.0 (ref) | - | 57 (39.3) | 1.0 (ref) | - |
| C/T | 504 (46.8) | 77 (46.4) | 0.7 [0.5–1.0] | 0.11 | 62 (42.8) | 0.6 [0.4–0.9] | 0.05 |
| T/T | 260 (24.1) | 24 (14.5) | 0.4 [0.2–0.7] | 0.001 | 26 (17.9) | 0.5 [0.3–0.9] | 0.01 |
| C/T + T/T | 764 (70.9) | 101 (60.8) | 0.6 [0.4–0.8] | 0.01 | 88 (60.7) | 0.6 [0.4–0.9] | 0.01 |
| Allele C | 1132 (52.5) | 207 (62.3) | 1.0 (ref) | - | 176 (60.7) | 1.0 (ref) | - |
| Allele T | 1024 (47.5) | 125 (37.7) | 0.6 [0.5–0.8] | 0.001 | 114 (39.3) | 0.7 [0.5–0.9] | 0.01 |
| rs2242442 (TBX3) | |||||||
| G/G | 674 (62.5) | 105 (63.3) | 1.0 (Ref) | - | 105 (72.4) | 1.0 (Ref) | - |
| G/A | 358 (33.2) | 54 (32.5) | 0.9 [0.6–1.3] | 0.92 | 36 (24.8) | 0.6 [0.4–0.9] | 0.03 |
| A/A | 46 (4.3) | 7 (4.2) | 0.9 [0.4–2.2] | 1.00 | 4 (2.8) | 0.5 [0.8–1.5] | 0.38 |
| G/A + A/A | 404 (37.5) | 61 (36.7) | 0.9 [0.6–1.3] | 0.93 | 40 (27.6) | 0.6 [0.4–0.9] | 0.02 |
| Allele G | 1706 (79.1) | 264 (79.5) | 1.0 (Ref) | - | 246 (84.8) | 1.0 (Ref) | - |
| Allele A | 450 (20.9) | 68 (20.5) | 0.9 [0.7–1.2] | 0.92 | 44 (15.2) | 0.6 [0.4–0.9] | 0.02 |
| rs11168827 (KMT2D) | |||||||
| G/G | 439 (40.7) | 72 (43.4) | 1.0 (ref) | - | 49 (33.8) | 1.0 (ref) | - |
| G/C | 510 (47.3) | 74 (44.6) | 0.8 [0.6–1.3] | 0.53 | 83 (57.2) | 1.4 [1.0–2.1] | 0.05 |
| C/C | 129 (12.0) | 20 (12.0) | 0.9 [0.5–1.6] | 0.89 | 13 (9.0) | 0.9 [0.4–1.7] | 0.87 |
| G/C + C/C | 639 (59.3) | 94 (56.6) | 0.9 [0.6–1.2] | 0.55 | 96 (66.2) | 1.3 [0.9–1.9] | 0.12 |
| Allele G | 1388 (64.4) | 218 (65.7) | 1.0 (ref) | - | 181 (62.4) | 1.0 (ref) | - |
| Allele C | 768 (35.6) | 114 (34.3) | 0.9 [0.7–1.2] | 0.69 | 109 (37.6) | 1.0 [0.8–1.4] | 0.55 |
| rs702688 (MAP3K1) | |||||||
| A/A | 345 (32.0) | 51 (30.7) | 1.0 (Ref) | - | 49 (33.8) | 1.0 (Ref) | - |
| A/G | 525 (48.7) | 83 (50.0) | 1.0 [0.7–1.5] | 0.77 | 68 (46.9) | 0.9 [0.6–1.3] | 0.68 |
| G/G | 208 (19.3) | 32 (19.3) | 1.0 [0.6–1.6] | 0.90 | 28 (19.3) | 0.9 [0.5–1.5] | 0.90 |
| A/G + G/G | 733 (68.0) | 115 (69.3) | 1.0 [0.7–1.5] | 0.78 | 96 (66.2) | 0.9 [0.6–1.3] | 0.70 |
| Allele A | 1215 (56.4) | 185 (55.7) | 1.0 (Ref) | - | 166 (57.2) | 1.0 (Ref) | - |
| Allele G | 941 (43.6) | 147 (44.3) | 1.0 [0.8–1.2] | 0.87 | 124 (42.8) | 0.9 [0.7–1.2] | 0.82 |
| rs702689 (MAP3K1) | |||||||
| G/G | 274 (25.4) | 41 (24.7) | 1.0 (Ref) | - | 41 (28.3) | 1.0 (Ref) | - |
| G/A | 544 (50.5) | 71 (42.8) | 0.8 [0.5–1.3] | 0.52 | 62 (42.8) | 0.7 [0.5–1.1] | 0.22 |
| A/A | 260 (24.1) | 54 (32.5) | 1.3 [0.8–2.1] | 0.14 | 42 (29.0) | 1.0 [0.6–1.7] | 0.81 |
| G/A + A/A | 804 (74.6) | 125 (75.3) | 1.0 [0.7–1.5] | 0.92 | 104 (71.7) | 0.8 [0.5–1.2] | 0.47 |
| Allele G | 1092 (50.6) | 153 (46.1) | 1.0 (Ref) | - | 144 (49.7) | 1.0 (Ref) | - |
| Allele A | 1064 (49.4) | 179 (53.9) | 1.2 [0.9–1.5] | 0.13 | 146 (50.3) | 1.0 [0.8–1.3] | 0.79 |
| Number of Risk Alleles (a) | Controls (n = 1078) (%) | All BC Cases (n = 489) | Families with ≥2 BC and/or OC Cases (n = 3 11) | A Single Case, Diagnosis at ≤50 Years of Age (n = 178) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BC Cases (%) | OR [95% CI] | p Value (b) | BC Cases (%) | OR [95% CI] | p Value (b) | BC Cases (%) | OR [95% CI] | p Value (b) | ||
| 0 risk alleles | 200 (18.6) | 125 (25.6) | 1.0 (Ref) | - | 84 (27.0) | 1.0 (Ref) | - | 41 (23.0) | 1.0 (Ref) | - |
| 1 risk allele | 409 (37.9) | 196 (40.1) | 0.7 [0.5–1.0] | 0.07 | 127 (40.8) | 0.7 [0.5–1.0] | 0.07 | 69 (38.8) | 0.8 [0.5–1.2] | 0.3 |
| 2 risk alleles | 354 (32.8) | 139 (28.4) | 0.6 [0.5–0.8] | 0.002 | 79 (25.4) | 0.5 [0.4–0.7] | 0.0005 | 60 (33.7) | 0.8 [0.5–1.2] | 0.4 |
| 3 risk alleles | 103 (9.6) | 29 (5.9) | 0.4 [0.2–0.7] | 0.0007 | 21 (6.8) | 0.4 [0.2–0.8] | 0.006 | 8 (4.5) | 1.3 [0.5–3.1] | 0.4 |
| 4 risk alleles | 12 (1.1) | 0 (0) | 0.06 [0.003–1.0] | 0.004 | 0 (0) | 0.09 [0.005–1.6] | 0.02 | 0 (0) | 0.1 [0.01–3.3] | 0.2 |
| P-trend (c) | <10−4 | <10−4 | 0.02 | |||||||
| Global P (d) | 0.0003 | 0.0008 | 0.08 | |||||||
| Number of Risk Alleles (a) | Controls (n = 1078) (%) | Families with Two BC and/or OC Cases (n = 166) | Families with ≥3 BC and/or OC Cases (n = 145) | ||||
|---|---|---|---|---|---|---|---|
| BC Cases (%) | OR [95% CI] | p-Value (b) | BC Cases (%) | OR [95% CI] | p-Value (b) | ||
| 0 risk alleles | 200 (18.6) | 43 (25.9) | 1.0 (Ref) | - | 41 (28.3) | 1.0 (Ref) | - |
| 1 risk allele | 409 (37.9) | 67 (40.4) | 0.7 [0.4–1.1] | 0.1 | 60 (41.4) | 0.7 [0.5–1.1] | 0.2 |
| 2 risk alleles | 354 (32.8) | 45 (27.1) | 0.4 [0.2–0.7] | 0.002 | 34 (23.4) | 0.5 [0.3–0.9] | 0.02 |
| 3 risk alleles | 103 (9.6) | 11 (6.6) | 0.4 [0.2–0.9] | 0.05 | 10 (6.9) | 0.4 [0.2–1.0] | 0.05 |
| 4 risk alleles | 12 (1.1) | 0 (0.0) | 0.1 [0.01–3.3] | 0.2 | 0 (0.0) | 0.1 [0.01–3.1] | 0.2 |
| p-trend(c) | 0.0007 | 0.004 | |||||
| Global p (d) | 0.01 | 0.06 | |||||
| Inclusion Criteria | Families n (%) |
|---|---|
| Three or more family members with breast and/or ovarian cancer | 145 (29.7%) |
| Two family members with breast and/or ovarian cancer | 166 (33.9%) |
| Single affected individual with breast cancer, onset ≤35 years of age | 91 (18.6%) |
| Single affected individual with breast cancer, onset 36–50 years of age | 87 (17.8%) |
| TOTAL | 489 (100%) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandez-Moya, A.; Morales, S.; Arancibia, T.; Gonzalez-Hormazabal, P.; Tapia, J.C.; Godoy-Herrera, R.; Reyes, J.M.; Gomez, F.; Waugh, E.; Jara, L. Germline Variants in Driver Genes of Breast Cancer and Their Association with Familial and Early-Onset Breast Cancer Risk in a Chilean Population. Cancers 2020, 12, 249. https://doi.org/10.3390/cancers12010249
Fernandez-Moya A, Morales S, Arancibia T, Gonzalez-Hormazabal P, Tapia JC, Godoy-Herrera R, Reyes JM, Gomez F, Waugh E, Jara L. Germline Variants in Driver Genes of Breast Cancer and Their Association with Familial and Early-Onset Breast Cancer Risk in a Chilean Population. Cancers. 2020; 12(1):249. https://doi.org/10.3390/cancers12010249
Chicago/Turabian StyleFernandez-Moya, Alejandro, Sebastian Morales, Trinidad Arancibia, Patricio Gonzalez-Hormazabal, Julio C. Tapia, Raul Godoy-Herrera, Jose Miguel Reyes, Fernando Gomez, Enrique Waugh, and Lilian Jara. 2020. "Germline Variants in Driver Genes of Breast Cancer and Their Association with Familial and Early-Onset Breast Cancer Risk in a Chilean Population" Cancers 12, no. 1: 249. https://doi.org/10.3390/cancers12010249
APA StyleFernandez-Moya, A., Morales, S., Arancibia, T., Gonzalez-Hormazabal, P., Tapia, J. C., Godoy-Herrera, R., Reyes, J. M., Gomez, F., Waugh, E., & Jara, L. (2020). Germline Variants in Driver Genes of Breast Cancer and Their Association with Familial and Early-Onset Breast Cancer Risk in a Chilean Population. Cancers, 12(1), 249. https://doi.org/10.3390/cancers12010249